Abstract
The D2 dopamine receptor mediates neuropsychiatric symptoms and is a target of pharmacotherapy. Inter-individual variation of D2 receptor density is thought to influence disease risk and pharmacological response. Numerous molecular imaging studies have tested whether common genetic variants influence D2 receptor binding potential (BP) in humans, but demonstration of robust effects has been limited by small sample sizes. We performed a systematic search of published human in vivo molecular imaging studies to estimate effect sizes of common genetic variants on striatal D2 receptor BP. We identified 21 studies examining 19 variants in 11 genes. The most commonly studied variant was a single-nucleotide polymorphism in ANKK1 (rs1800497, Glu713Lys, also called ‘Taq1A'). Fixed- and random-effects meta-analyses of this variant (5 studies, 194 subjects total) revealed that striatal BP was significantly and robustly lower among carriers of the minor allele (Lys713) relative to major allele homozygotes. The weighted standardized mean difference was −0.57 under the fixed-effect model (95% confidence interval=(−0.87, −0.27), P=0.0002). The normal relationship between rs1800497 and BP was not apparent among subjects with neuropsychiatric diseases. Significant associations with baseline striatal D2 receptor BP have been reported for four DRD2 variants (rs1079597, rs1076560, rs6277 and rs1799732) and a PER2 repeat polymorphism, but none have yet been tested in more than two independent samples. Our findings resolve apparent discrepancies in the literature and establish that rs1800497 robustly influences striatal D2 receptor availability. This genetic variant is likely to contribute to important individual differences in human striatal function, neuropsychiatric disease risk and pharmacological response.
Introduction
The dopamine D2 receptor is a G-protein-coupled receptor that is highly expressed in the striatum. D2 receptors mediate an array of fundamental brain functions, including reward behavior, regulation of movement, learning and memory, and attention. The D2 receptor is also important clinically as a target of pharmacotherapy for psychosis, parkinsonism, brain injuries and restless legs syndrome.1 These diverse roles attest to the clinical and neurobiological importance of the D2 receptor.
Substantial inter-individual variation in D2 receptor expression has been observed in human postmortem and in vivo imaging studies, with typical coefficients of variation in the range of 20–60%.2, 3, 4, 5, 6, 7 Individual differences in D2 receptor expression are hypothesized to contribute to differences in motivated behaviors and risk of related neuropsychiatric disorders. For example, striatal D2 receptor manipulation in rodents alters motivated behaviors,8 and human studies have consistently demonstrated abnormally low striatal D2 receptor levels among individuals with addictions.9 The factors that underlie individual differences in D2 receptor expression in humans are not yet defined, but presumably both environmental and genetic factors are at work.
To address the role of genetic factors, numerous studies have used in vivo molecular imaging (positron emission tomography (PET) or single-photon emission computed tomography (SPECT)) to test whether common genetic variants influence D2 receptor binding potential (BP) in humans. However, consistent and robust genetic effects have not yet been demonstrated, and the magnitude of genetic effects is unknown. For example, among seven published studies of the single-nucleotide polymorphism (SNP) rs1800497, four4, 6, 10, 11 reported a significant association and three5, 12, 13 found no significant effect on striatal BP. There are several potential causes of such discrepancies. A genetic variant may have no true underlying effect (type I error), the true effects may be different in different studies (heterogeneity), or a true effect may be present but not consistently detected due to the low power of individual studies (type II error). This latter possibility may be particularly relevant to molecular imaging studies as their cost and complexity often necessitate small sample sizes.
Recent work has highlighted the importance of resolving such apparent discrepancies in the genetics-imaging literature. For example, despite compelling preclinical evidence and numerous human imaging studies, a recent meta-analysis of the brain-derived neurotrophic factor Val66Met polymorphism found no significant effect on hippocampal volume.14 Similarly, meta-analyses of the variable repeat polymorphism in the promoter of the serotonin transporter and amygdala activation have indicated a much smaller effect size than initially reported.15, 16 Our objective here was to perform a review and quantitative analysis of all published studies that tested genetic polymorphisms and measured human in vivo D2 receptor BP, thereby establishing associations that are known with confidence.
Materials and methods
We performed a systematic search of PubMed in May 2015 using the following search string: ‘(variant* OR polymorphism* OR genetic OR gene) AND (pet OR positron OR emission OR spect OR photon OR tomography) AND (dopamine OR d2 OR d3) AND receptor.' The titles and abstracts of all search results were reviewed to identify studies that examined genetic polymorphisms and used human D2 molecular imaging (PET or SPECT). Full text was retrieved for all potentially relevant articles. The references of full text articles were scanned to identify additional sources missed by the original search.
Meta-analysis was performed when possible for genetic variants tested in multiple independent samples. We used the ‘metafor' package (ver. 1.9–3, W. Viechtbauer, http://cran.r-project.org/web/packages/metafor) and ‘meta' package (ver. 3.7-0, G. Schwarzer, http://cran.r-project.org/web/packages/meta) with RStudio (ver. 0.97.551) within the R statistical computing environment (ver. 3.0.2, http://www.R-project.org/). The effect size was represented by the standardized mean difference (SMD), that is, the mean BP for one genotype group minus mean BP for the second genotype group, divided by Hedges' pooled s.d. Inverse-variance weighting was used to compute the pooled SMD. Fixed-effect and random-effects models were estimated. Forest plots and funnel plots were inspected for outliers and bias. Sensitivity analyses used the leave-one-out method to test for undue influence of single studies. When Cochran's Q-test for heterogeneity indicated nonsignificant heterogeneity across studies (I2<0.5 and P>0.10 as suggested17), we adopted the effect size from the fixed-effect model as our final estimate. Only BP values from anatomically defined regions of interest were used in meta-analyses. Because BP estimates from different brain regions within the same sample are highly correlated, and valid meta-analysis requires independent samples, a single estimate from each genotype group was derived (pooled mean and s.d. with inverse-variance weighting) in cases where BP estimates from multiple anatomical regions were reported. Bilateral striatum was preferred and was used for all studies except Savitz et al.11 who reported a middle caudate region of interest. To avoid the confounding effects of disease and treatment, medication-free healthy control subjects were analyzed separately from subjects with neuropsychiatric disorders. Studies with different demographic characteristics (sex and age distribution) and imaging methods (SPECT versus PET, region of interest imaged) were first analyzed together based on the assumption that these study-level variables have effects on binding measures that are similar in the two genotype groups. Although the small number of studies severely limited our power to detect statistical effects of these study-level variables on the estimated genetic effect, we did explore these potential moderators with fixed-effect meta-regression and leave-one-out sensitivity analysis.
Results
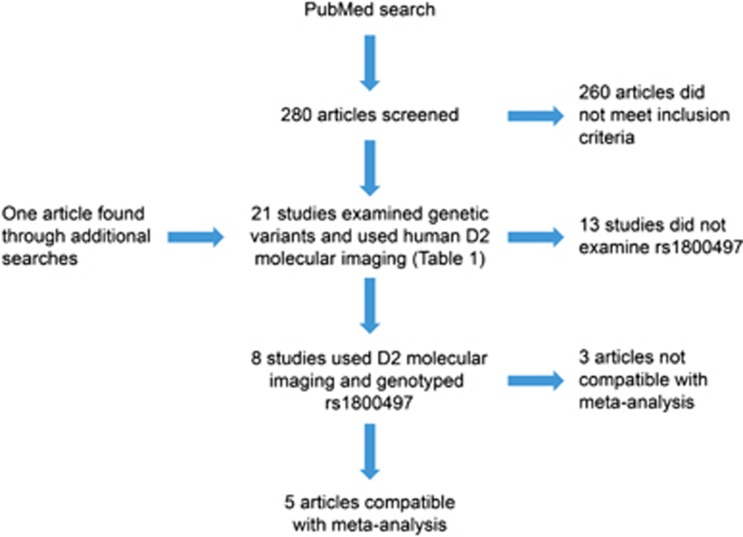
We identified 21 studies that examined genetic polymorphisms and used human D2 molecular imaging (Figure 1). The 21 studies examined 19 variants in 11 genes (Table 1). None of the investigated variants were in strong linkage disequilibrium (LD; all r2<0.8) except rs1079597 and rs1076560 (r2=0.87).
Figure 1.
Flow diagram of systematic literature search.
Table 1. Published studies of genetic polymorphisms and in vivo D2 receptor BP.
| Study | No. of subjects | Age, years, mean (s.d.) | Sex (% female) | Imaging technique | Reported effects on striatal D2 receptor BP in vivo |
|---|---|---|---|---|---|
| ANKK1 rs1800497 Glu713Lys ‘Taq1A' (missense) | |||||
| Laruelle et al.5 | 47 23a | NA | 14 | [123I] IBZM SPECT | No significant difference between genotype groups; no significant difference between healthy and schizophrenic participants |
| Pohjalainen et al.6 | 54 | 40 (17) | 39 | [11C] RAC PET | A1 carriers significantly lower than A2/A2 |
| Jönsson et al.4 | 56 | 32 (12) | 23 | [11C] RAC PET | A1 carriers significantly lower than A2/A2 |
| Brody et al.12 | 45 | 37 (11) | 29 | [11C] RAC PET | No significant difference between genotype groups; no significant association with cigarette-induced decrease in binding |
| Hirvonen et al.10 | 45 | 36 (14) | 40 | [11C] RAC PET | A1 carriers significantly lower than A2/A2; subsample of Pohjalainen et al.6 |
| Hirvonen et al.18 | 38 | 29 (7) | 0 | [11C] FLB 457 PET | A1 carriers significantly higher than A2/A2 in cerebral cortex and thalamus (extrastriatal) |
| Savitz et al.11 | 24 12b | 35 (8) 38 (11)b | 54 67b | [11C] RAC PET | A1 carriers significantly lower than A2/A2 among healthy controls; A1 carriers significantly higher than A2/A2 among depressed patients; no significant association with reward-related decrease in binding |
| Wagner et al.13 | 13 12c | ~30 ~33 | 0 | [11C] RAC PET | No significant difference between genotype groups for either healthy controls or patients with traumatic brain injury |
| COMT rs4680 Val158Met (missense) | |||||
| Brody et al.12 | 45 | 37 (11) | 29 | [11C] RAC PET | No significant difference between genotype groups for baseline binding; significantly greater cigarette-induced decrease in binding among Val/Val subjects relative to Met carriers |
| Hirvonen et al.19 | 45 | 36 (14) | 40 | [11C] RAC PET | No significant difference between genotype groups |
| Hirvonen et al.19 | 38 | 29 (7) | 0 | [11C] FLB 457 PET | No significant difference between genotype groups (extrastriatal) |
| Boot et al.20 | 15d | NA | 60d | [123I] IBZM SPECT | Met hemizygotes significantly lower than Val hemizygotes (all participants with 22q11 deletion syndrome) |
| DAT (SLC6A3) 40-bp VNTR polymorphism (3′ untranslated region) | |||||
| Brody et al.12 | 45 | 37 (11) | 29 | [11C] RAC PET | No significant difference between genotype groups for baseline binding; significantly greater cigarette-induced decrease in binding among 9-repeat allele carriers compared with 10-repeat homozygotes |
| Wagner et al.13 | 13 12c | NA | 0 | [11C] RAC PET | No significant difference between genotype groups in healthy controls; 10-repeat homozygotes marginally higher than 9-repeat carriers among patients with traumatic brain injury |
| DRD2 rs1079597e ‘Taq1B' (intronic) | |||||
| Laruelle et al.5 | 47 23a | NA | 14 | [123I] IBZM SPECT | No significant difference between genotype groups; no significant difference between healthy and schizophrenic participants |
| Jönsson et al.4 | 56 | 32 (12) | 23 | [11C] RAC PET | B1 carriers significantly lower than B2/B2 |
| DRD2 rs1076560e G>T (intronic) | |||||
| Bertolino et al.21 | 32 | NA | NA | [123I] IBZM SPECT | T-allele carriers significantly lower than G/G; binding values not reported |
| Taurisano et al.22 | 26 | 23 (3) | 50 | [123I] IBZM SPECT | Main effect of genotype not reported; significant interaction between genotype and schizotypy; binding values not reported |
| DRD2 rs6277 C957T (synonymous) | |||||
| Hirvonen et al.23, 24 | 45 | 36 (14) | 40 | [11C] RAC PET | T allele associated with significantly higher BP |
| Hirvonen et al.10 | 45 | 36 (14) | 40 | [11C] RAC PET | T allele associated with significantly lower affinity (Kd); no significant association with receptor density (Bmax); subsample of Hirvonen et al.23, 24 |
| Hirvonen et al.18 | 38 | 29 (7) | 0 | [11C] FLB 457 PET | T allele associated with significantly lower BP in cerebral cortex and thalamus (extrastriatal) |
| DRD2 rs1801028 Ser311Cys (missense) | |||||
| Pohjalainen et al.25 | 49 | 41 (18) | 33 | [11C] RAC PET | No significant difference between genotype groups |
| DRD2 rs1799732 −141C ins/del (upstream) | |||||
| Jönsson et al.4 | 56 | 32 (12) | 23 | [11C] RAC PET | Deletion associated with significantly higher binding |
| Hirvonen et al.18 | 38 | 29 (7) | 0 | [11C] FLB 457 PET | No significant difference between genotype groups (only extrastriatal regions measured) |
| DRD2 (CA)n repeat polymorphism (intron 2) | |||||
| Jönsson et al.4 | 56 | 32 (12) | 23 | [11C] RAC PET | No significant difference between genotype groups |
| DRD2 rs4274224 (intronic) and rs12364283 (upstream) | |||||
| Peciña et al.26 | 52 | 27 (5) | 58 | [11C] RAC PET | No significant difference between genotype groups for baseline binding for either variant; significantly lower pain-induced decrease in binding among rs4274224 A/G heterozygotes relative to A/A or G/G homozygotes |
| DRD3 rs6280 Ser9Gly (missense) | |||||
| Savitz et al.27 | 26 10b | 34 (8) 38 (11)b | 58 80b | [11C] RAC PET | No significant difference between genotype groups for baseline binding; Gly allele associated with significantly greater reward-induced decrease in binding |
| DRD4 48-bp VNTR polymorphism (exon 3) | |||||
| Brody et al.12 | 45 | 37 (11) | 29 | [11C] RAC PET | No significant difference between genotype groups for baseline binding; significantly greater cigarette-induced decrease in binding among subjects with fewer than seven repeats |
| HTR2C rs6318 Cys23Ser (missense) | |||||
| Mickey et al.28 | 54 | 27 (5) | 60 | [11C] RAC PET | No significant difference between genotype groups for baseline binding; significantly greater pain-induced decrease in binding among Ser allele carriers |
| LEP rs12706832 (intronic) and rs3828942 (intronic) | |||||
| Burghardt et al.29 | 50 | 26 (5) | 56 | [11C] RAC PET | No significant difference between genotype groups for baseline binding for either variant; significantly greater pain-induced decrease in binding among rs12706832 G/G homozygotes |
| OPRM1 rs1799971 A118G Asn40Asp (missense) | |||||
| Domino et al.30 | 20f | 26 (5)f | 0f | [11C] RAC PET | Differences between genotype groups not reported for baseline binding; significantly greater cigarette-induced decrease in binding among G-allele carriers relative to A/A homozygotes |
| OXT rs4813625 (upstream) | |||||
| Love et al.31 | 55 | 27 (5) | 58 | [11C] RAC PET | No significant difference between genotype groups for baseline binding; among females only, significantly greater pain-induced decrease in binding among C-allele carriers compared with G/G |
| PER2 VNTR polymorphism (intron 3) | |||||
| Shumay et al.32 | 52 | 35 (9) | 17 | [11C] RAC PET | Carriers of 3-repeat or rare alleles significantly lower than 4-repeat homozygotes |
Abbreviations: BP, binding potential; bp, base pair; FLB 457, (S)-5-bromo-N-((1-ethyl-2-pyrrolidinyl)methyl)-2,3-dimethoxybenzamide; IBZM, iodobenzamide; NA, not available; PET, positron emission tomography; RAC, raclopride; SPECT, single-photon emission computed tomography; VNTR, variable-number tandem repeat.
All data are from healthy adult subjects unless otherwise specified.
Participants with schizophrenia.
Participants with major depressive disorder.
Participants with traumatic brain injury.
Participants with 22q11 deletion syndrome.
rs1079597 and rs1076560 are in linkage disequilibrium (r2=0.87, D′=0.93, northern and western European ancestry).
Participants with tobacco dependence, otherwise healthy.
The most commonly studied variant was rs1800497, a SNP within the ankyrin repeat and kinase domain containing 1 (ANKK1) gene and 10 kb downstream from the dopamine D2 receptor (DRD2) gene. This missense mutation (C>T) is predicted to change a glutamate residue to a lysine residue at position 713 (Glu713Lys) within the 11th ankyrin repeat of the ANKK1 protein.33 This variant is often referred to as the ‘Taq1A' polymorphism (after a restriction enzyme initially used to detect it) with the minor Lys713 allele denoted ‘A1' and the major Glu713 allele denoted ‘A2'.34
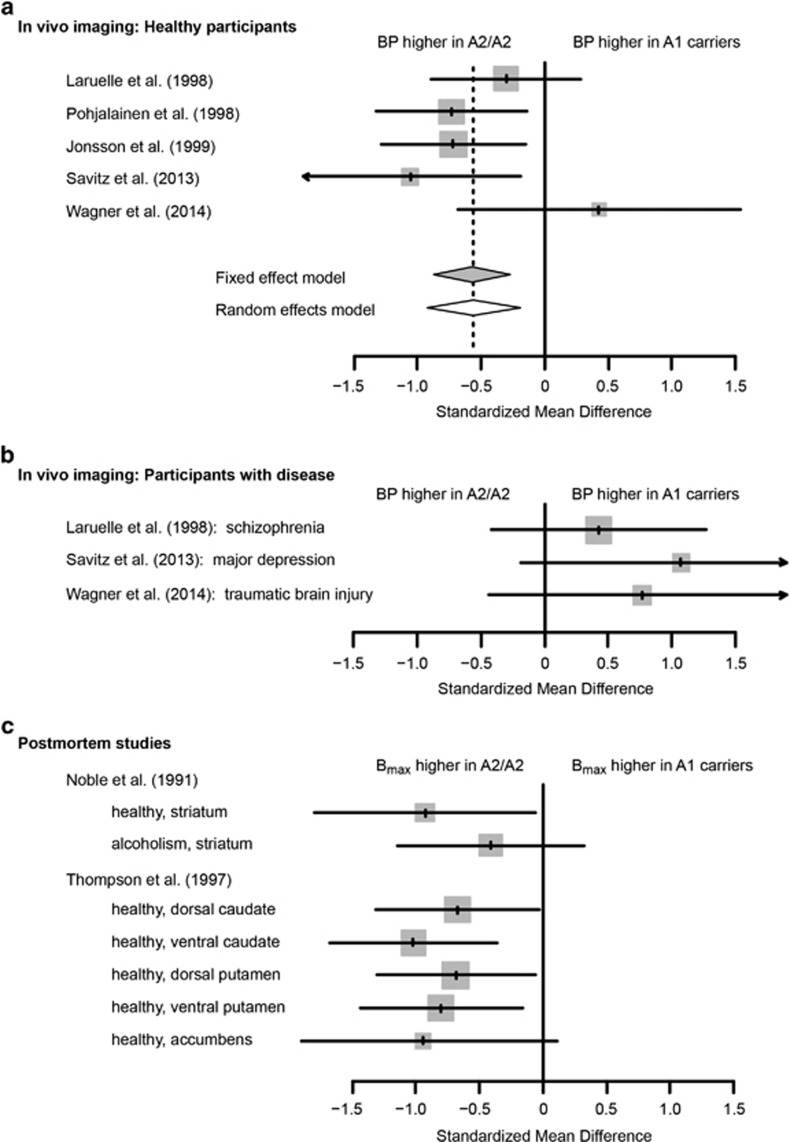
Five of the eight Taq1A studies were compatible with meta-analysis. One article was not compatible because baseline BP values were not reported.12 A second article was incompatible because only extrastriatal BP was quantified.18 A third study10 was excluded because it included subjects from a previously reported sample.6 The remaining five studies included 194 healthy participants. Striatal BP was significantly and robustly lower among healthy A1-allele carriers relative to healthy A2 homozygotes (Figure 2a). The weighted SMD was −0.57 (95% confidence interval (95% CI)=(−0.87, −0.27), P=0.0002) under the fixed-effect model and −0.56 (95% CI=(−0.93, −0.19), P=0.003) under the random-effects model. Sensitivity analysis showed that the results were robust when individual studies were omitted (all P<0.005) indicating that no single study unduly influenced the results. We found no evidence of heterogeneity across studies (Q=5.7, degrees of freedom (df)=4, I2=0.29, P=0.22). Likewise, a funnel plot did not suggest heterogeneity or bias (data not shown). Thus, our results are consistent with a single underlying effect of the Taq1A polymorphism across studies.
Figure 2.
Forest plots of effect size for rs1800497 (Taq1A) and D2 receptor binding. Each genotype group comparison is represented by a gray square and horizontal error bars (mean and 95% confidence intervals). Square size is proportional to study weight. The solid vertical line represents the null hypothesis (no effect of the genetic variant). (a) In vivo imaging studies of healthy participants. Diamonds represent means and 95% confidence intervals for the fixed-effect model (filled diamond) and random-effects model (open diamond). The dashed vertical line is the fixed-effect weighted mean. (b) In vivo imaging studies of participants with disease. (c) Postmortem studies. BP, binding potential; Bmax, total number of receptors.
Meta-regression and sensitivity analyses were used to explore the potential influence of study imaging modality (PET versus SPECT), brain region of interest, sex distribution and mean age on the Taq1A findings. We considered these exploratory analyses, as our power to detect effects of study-level variables across five studies was very limited. Meta-regression suggested that sex distribution may be a moderator of the Taq1A effect (β=0.024, s.e.m.=0.011, Q=4.4, df=1, P=0.035) with stronger genetic effects evident in studies with a greater proportion of females. Meta-regression revealed no significant effects of imaging modality (P=0.48), brain region (P=0.27) or mean subject age (P=0.38). Finally, fixed-effect analyses that excluded the SPECT study5 or the study that quantified a middle caudate region11 produced similar estimates of effect size (SMD=−0.66 and −0.50, P=0.0002 and 0.002, respectively).
Three of the Taq1A in vivo imaging studies5, 11, 13 quantified striatal BP in participants with disease. Laruelle et al.5 reported data from 23 subjects with schizophrenia, Savitz et al.11 reported data from 12 subjects with major depressive disorder and Wagner et al.13 reported data from 12 subjects with traumatic brain injury. In all cases, participants were free of dopaminergic medications for at least 3 weeks, and individuals with recent alcohol or drug dependence were excluded. In contrast to healthy participants, striatal BP in these subjects tended to be higher (rather than lower) among A1-allele carriers relative to A2 homozygotes (Figure 2b). The effect sizes for each of these three patient groups were not significantly different from 0, but they did differ significantly from the above-calculated weighted mean effect size for healthy controls (95% CI for between-group differences: (0.1, 1.9), (0.4, 2.9) and (0.1, 2.6), respectively). This finding suggested that for each of these diagnostic groups, the disease moderated the effect of the Taq1A polymorphism on D2 receptor BP. Meta-regression including healthy and disease samples confirmed that disease status was a significant moderator (Q=12.0, df=1, P=0.0005).
In addition to the eight in vivo imaging studies, two studies of postmortem brain have reported binding density (Bmax) versus Taq1A status,2, 3 so we extracted effect sizes from these studies as well. Striatal binding was generally lower among healthy A1-allele carriers relative to A2 homozygotes (Figure 2c) and the magnitude of this difference was consistent with in vivo measurements from healthy participants (Figure 2a).
The intronic DRD2 SNP rs1079597 was examined in two comparable, independent, molecular imaging studies.4, 5 This variant, which is in moderate LD with the Taq1A variant (r2=0.70), is sometimes called the ‘Taq1B' polymorphism, with the minor allele denoted ‘B1' and the major allele denoted ‘B2'.35, 36 The first study5 reported no significant effect of this polymorphism on D2 binding but the second study4 reported significantly lower striatal BP among healthy B1-allele carriers relative to healthy B2 homozygotes. Although this apparent discrepancy has been interpreted as a replication failure,4 the effect sizes are actually consistent with each other (mean=−0.40, 95% CI=(−1.02, 0.21); and mean=−0.72, 95% CI=(−1.30, 0.15), respectively) and the weighted mean effect size from the two samples is −0.58 (95% CI=(−1.00, −0.15), P=0.008). Thus, the Taq1A and Taq1B polymorphisms appear to have a similar magnitude of association with D2 receptor BP in healthy participants.
Catechol-O-methyltransferase (COMT) is an enzyme that degrades dopamine, so genetic variation in the COMT gene has been hypothesized to affect dopamine receptor availability. The effect of the COMT missense variant rs4680 (Val158Met) on striatal D2 receptor BP was examined in three independent molecular imaging studies. One study of 45 cigarette smokers found no significant effect, but reported only a lower bound on significance (P>0.25).12 A second study of 45 healthy participants also reported no significant effect (SMD=0.02, 95% CI=(−0.59, 0.63)).19 A third study of 15 individuals with 22q11 deletion syndrome reported lower binding associated with the Met158 allele (SMD=1.38, 95% CI=(0.17, 2.60)).20 Thus, the available evidence does not support an effect of this polymorphism on D2 receptor BP in diploid individuals, but the genetic effects may be stronger among individuals with 22q11 deletion syndrome, who have only one copy of COMT.
Two independent molecular imaging studies examined a 3′ variable-number tandem repeat polymorphism in the dopamine transporter gene (SLC6A3). No significant differences between genotype groups were found among healthy controls13 or cigarette smokers.12 Among patients with traumatic brain injury, 10-repeat homozygotes showed marginally higher D2 receptor binding than 9-repeat carriers (P=0.052).13
For the remaining 15 genetic variants identified in the literature search, we found no other replications in which two or more comparable, independent, studies of striatal D2 receptors tested the same variant. Significant associations with baseline (resting) striatal D2 receptor BP were reported for 4 of the 15 variants. Three of the 4 variants reported were in the DRD2 gene: the intronic SNP rs1076560 (ref. 21), the synonymous SNP rs6277 (ref. 23) and the upstream single-nucleotide insertion rs1799732 (ref. 4). Finally, Shumay et al. studied an intronic variable-number tandem repeat polymorphism in the circadian gene PER2 based on preclinical evidence linking striatal dopamine release with circadian rhythms and PER2. They reported that fewer repeats were associated with lower baseline D2 receptor BP.32
Several genetic association studies used the radiotracer [11C] raclopride, which is displaceable by endogenous dopamine, in combination with a behavioral challenge intended to induce striatal dopamine release (cigarettes, pain and reward task). Significant associations were reported for polymorphisms in genes directly involved in dopamine signaling, viz., the dopamine transporter,12 D2 receptor,26 D3 receptor27 and D4 receptor.12 In addition, significant associations were found for variants in genes for the serotonin 2C receptor28 and mu-opioid receptor30, 37—both known to regulate dopamine release via their expression in ventral tegmental interneurons—and for variants in the genes for leptin29 and oxytocin,31 which act at their respective receptors within the mesoaccumbal and nigrostriatal pathways.
Discussion
This systematic review and meta-analysis has produced several principal findings. First, variants in DRD2 and 10 other genes have been tested for effects on in vivo striatal D2 receptor BP, but only 4 variants (rs1800497, rs1079597, rs4680 and SLC6A3 variable-number tandem repeat) have been examined in two or more comparable, independent samples. Second, despite apparent discrepancies in the literature, the ANKK1 Taq1A polymorphism (rs1800497) is robustly associated with D2 receptor BP in healthy humans. Third, the DRD2 variant rs1079597 has effects on D2 receptor BP that are similar to rs1800497. Fourth, several examples were identified in which the presence of a neuropsychiatric syndrome modified the effects of genetic variants on D2 receptor BP.
Our study has notable limitations. The first type of limitation is related to interpretation of in vivo receptor binding measures. The PET and SPECT studies examined here estimated a quantity proportional to Bmax/Kd, where Bmax is the total number of available receptor sites and 1/Kd is the affinity of the radiotracer for the receptor. Consequently, group differences may arise from a difference in Bmax, a difference in Kd or both. In addition, because raclopride is displaceable by endogenous dopamine in vivo, Bmax reflects the number of receptor sites not occupied by endogenous ligand rather than the total number of receptor sites.38 In other words, BP is sensitive to endogenous dopaminergic tone, which may be an additional source of variance. Furthermore, raclopride is known to bind D2 receptors in both the G-protein-coupled (‘high' or active) state and the non-coupled (‘low' or inactive) state, so genetic influences on the proportion of receptors in the high versus low state cannot be distinguished.39 It is notable that different tracers or modeling methods used across different laboratories will yield different absolute BP estimates (for example, SPECT versus PET, or reference region versus arterial input function). BP estimates may also be altered by between-study differences in how the striatum was defined anatomically, and by striatal atrophy in disease groups. We believe that such specific methodological factors are unlikely to substantially influence our analyses of genetic effect sizes because such factors are likely to influence genotype groups equally.
The second type of limitation is related to systematic review and meta-analysis methodology. With the exception of rs1800497, the paucity of replications prevented reliable quantification of effect size for most genetic variants. In several cases, studies could not be included in meta-analyses because effect sizes (or data sufficient to compute effect sizes) were not reported. We used heterogeneity tests, funnel plots and sensitivity analyses to evaluate for bias and inconsistency across studies, but the utility of these approaches is limited when examining only five studies. Meta-analysis methodology is incapable of answering questions about (or controlling for) potentially interesting individual-level variables such as sex, age, race/ancestry, body mass index or striatal anatomy. Therefore, we were unable to determine with confidence whether these variables moderated the effect of rs1800497 on in vivo D2 receptor binding. If protocols for sharing of molecular imaging data are developed, future mega-analysis40 of individual participant data could address these and other interesting questions. Finally, relevant data may have been unreported due to negative-results reporting bias (that is, the ‘file drawer problem') and our meta-analysis may have overlooked relevant articles.
Our most robust finding is the effect of the ANKK1 Taq1A variant (rs1800497) on striatal BP in healthy participants. BP was lower among carriers of the minor allele (‘A1' or Lys713) relative to A2 homozygotes, with a SMD of −0.57 and 95% CI of (−0.87, −0.27). Furthermore, the findings from in vivo molecular imaging studies were consistent with effect sizes for Bmax values measured in two postmortem studies. It is instructive to compare this effect size of 0.57 with other, commonly encountered, individual differences. Human males are typically taller and heavier than females, but the sexes overlap substantially. Among US adults, the SMD in height between men and women is ~1.2 and that for weight is ~0.4,41 so we can conclude that the Taq1A polymorphism is more strongly associated with D2 receptor BP than sex is with weight, but less so than sex is with height. It is also instructive to consider the proportion of the observed variance in BP that is explained by the Taq1A variant, as there are presumably numerous genetic and environmental factors that influence D2 receptor BP. For a SMD d with group sizes n1 and n2, the equivalent Pearson correlation is r=d/(d2+c)1/2, where c=(n1+n2)2/n1n2 (ref. 17). In the case of the Taq1A variant, d=0.57, n1=73 and n2=121, so the equivalent r=0.266 and r2=0.071. This indicates that the proportion of variance in striatal BP that is explained by rs1800497 is ~7%, with a 95% CI of (1.7%, 15%). With an effect size of this magnitude, it is not surprising that some molecular imaging studies did not detect an effect of the Taq1A variant on D2 receptor BP. The largest study reviewed here included just 56 healthy participants. This sample size has only 50–60% power to detect a true effect size of 0.57 (two-tailed test, α=0.05), whereas a sample of ~200 subjects provides >95% power.42
Several large studies have reported associations of the A1 allele of rs1800497 with poorer cognitive function. A study of ~2000 older adults linked the A1 allele with lower general cognitive ability43 and a study of ~500 patients linked the A1 allele with poorer cognitive outcomes after traumatic brain injury.44 A recent meta-analysis suggested an association of the A1 allele with attention deficit hyperactivity disorder, although the authors observed significant unexplained heterogeneity across studies,45 possibly related to differences in diagnostic procedures or clinical subtypes. A recent study of ~1300 adolescents showed that A1-allele carriers had poorer performance on visuospatial working memory tasks.46 That study also measured striatal activation using functional magnetic resonance imaging during a reward task and found no significant main effects of the Taq1A polymorphism, but the authors did detect an interaction whereby working memory performance was associated with striatal activation only among A1 carriers.46 This finding is consistent with the idea that lower D2 receptor expression among A1 carriers increases the dependence of working memory performance on striatum-based motivational circuitry.47 Taken together, these studies suggest that lower expression of D2 receptors among A1 carriers confers poorer cognitive function, although this model is yet to be directly tested.
Although the association of rs1800497 with neurocognitive function has been well studied, remarkably little is known about the impact of this variant at the molecular, cellular or circuit level. This SNP is predicted to cause a missense mutation in ANKK1 (Glu713Lys) and lies within 10 kb of DRD2.33 To our knowledge, no in vitro mechanistic studies have yet examined the effects of rs1800497 on the expression or molecular function of DRD2 or ANKK1 proteins. ANKK1 and DRD2 mRNA are both expressed in human striatum (Allen Human Brain Atlas, http://human.brain-map.org),48 and expression of ANKK1 in rodents appears to be regulated by dopamine.49, 50 To our knowledge, direct regulation of D2 receptors by ANKK1 has not been shown. Although a causal link between rs1800497 and D2 receptor density or affinity appears plausible, it is equally plausible that another polymorphism in LD with rs1800497 is the causal variant. For example, rs1800497 is in LD with the ANKK1 missense SNP rs7118900, which is predicted to create a new ANKK1 phosphorylation site and which has been associated with differential subcellular ANKK1 expression patterns in vitro.49, 51 It is also in LD with the intronic DRD2 SNP rs1079597 (described above) and with the intronic DRD2 SNPs rs2283265 and rs1076560, which were reported to alter DRD2 RNA splicing.52 Given the clear association of rs1800497 with D2 receptor BP in humans that we have demonstrated here, and the multiple potentially causal SNPs in LD with rs1800497, further in vitro and in vivo mechanistic studies are warranted.
Our analyses suggest that disease status and sex may moderate the genetic effect of rs1800497 on D2 receptor BP. The robust association of the minor allele with lower D2 receptor BP that we observed among healthy subjects was not present in samples with schizophrenia, depression or traumatic brain injury (TBI) (Figure 2b). Similarly, exploratory analysis suggested that the genetic effect of rs1800497 was weaker in studies that included a greater proportion of males. Similar disease-by-genotype and sex-by-genotype interactions have been reported in cognitive outcomes after TBI. The rs1800497 minor allele was associated with poorer cognition among predominantly mild TBI44 but with better cognition among a more severe TBI sample.53 Along the same lines, rs1800497 was a significant predictor of cognitive performance among females with TBI, but not among males with TBI.54 Taken together with our finding that the effect of rs1800497 on D2 receptor BP is stronger in studies with a greater proportion of females, these findings suggest that this polymorphism may be more penetrant in females. Although the neurobiological basis of this sex difference is unclear, sex steroids may have a role, given evidence in rodents that estrogens and testosterone influence D2 receptor expression.55, 56, 57
Our findings carry implications for future studies of the D2 receptor. First, based on the robust evidence that the ANKK1 polymorphism rs1800497 is associated with D2 receptor binding in humans, future studies are warranted to determine whether rs1800497 is the causal variant (versus another polymorphism in LD), and to elucidate the molecular and cellular basis of this association. Second, our findings highlight the possibility that other variants of similar effect size exist. Adequately powered human in vivo molecular imaging studies are warranted to identify the most influential variants. Third, genetic variation, sex and disease may interact to influence D2 receptor BP in complex ways, so the design and analysis of future studies that examine any one of these variables should consider effects of the other two as well. Such effects could be especially important for sexually dimorphic diseases (for example, depression and attention deficit hyperactivity disorder). Our findings also raise the possibility of interactions with other variables known to influence D2 receptor BP (for example, age and body mass index). Ultimately, we expect that a fuller description of genetic and environmental factors that influence D2 receptor levels and related neurocognitive functions will improve our understanding of neuropsychiatric disease risk, pharmacological response and clinically relevant outcomes.
Acknowledgments
BJM was supported by the National Institute of Mental Health (K23 MH 092648) and the Taubman Medical Research Institute.
Footnotes
During the 1-year period before publication, BJM was a paid consultant to Alkermes, on a topic unrelated to this work. BSG declares no conflict of interest.
References
- Beaulieu JM, Gainetdinov RR. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev 2011; 63: 182–217. [DOI] [PubMed] [Google Scholar]
- Noble EP, Blum K, Ritchie T, Montgomery A, Sheridan PJ. Allelic association of the D2 dopamine receptor gene with receptor-binding characteristics in alcoholism. Arch Gen Psychiatry 1991; 48: 648–654. [DOI] [PubMed] [Google Scholar]
- Thompson J, Thomas N, Singleton A, Piggott M, Lloyd S, Perry EK et al. D2 dopamine receptor gene (DRD2) Taq1 A polymorphism: reduced dopamine D2 receptor binding in the human striatum associated with the A1 allele. Pharmacogenetics 1997; 7: 479–484. [DOI] [PubMed] [Google Scholar]
- Jönsson EG, Nothen MM, Grunhage F, Farde L, Nakashima Y, Propping P et al. Polymorphisms in the dopamine D2 receptor gene and their relationships to striatal dopamine receptor density of healthy volunteers. Mol Psychiatry 1999; 4: 290–296. [DOI] [PubMed] [Google Scholar]
- Laruelle M, Gelernter J, Innis RB. D2 receptors binding potential is not affected by Taq1 polymorphism at the D2 receptor gene. Mol Psychiatry 1998; 3: 261–265. [DOI] [PubMed] [Google Scholar]
- Pohjalainen T, Rinne JO, Nagren K, Lehikoinen P, Anttila K, Syvalahti EK et al. The A1 allele of the human D2 dopamine receptor gene predicts low D2 receptor availability in healthy volunteers. Mol Psychiatry 1998; 3: 256–260. [DOI] [PubMed] [Google Scholar]
- Farde L, Hall H, Pauli S, Halldin C. Variability in D2-dopamine receptor density and affinity: a PET study with [11C]raclopride in man. Synapse 1995; 20: 200–208. [DOI] [PubMed] [Google Scholar]
- Trifilieff P, Feng B, Urizar E, Winiger V, Ward RD, Taylor KM et al. Increasing dopamine D2 receptor expression in the adult nucleus accumbens enhances motivation. Mol Psychiatry 2013; 18: 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Tomasi D. Addiction circuitry in the human brain. Annu Rev Pharmacol Toxicol 2012; 52: 321–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvonen MM, Laakso A, Nagren K, Rinne JO, Pohjalainen T, Hietala J. C957T polymorphism of dopamine D2 receptor gene affects striatal DRD2 in vivo availability by changing the receptor affinity. Synapse 2009; 63: 907–912. [DOI] [PubMed] [Google Scholar]
- Savitz J, Hodgkinson CA, Martin-Soelch C, Shen PH, Szczepanik J, Nugent AC et al. DRD2/ANKK1 Taq1A polymorphism (rs1800497) has opposing effects on D2/3 receptor binding in healthy controls and patients with major depressive disorder. Int J Neuropsychopharmacol 2013; 16: 2095–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody AL, Mandelkern MA, Olmstead RE, Scheibal D, Hahn E, Shiraga S et al. Gene variants of brain dopamine pathways and smoking-induced dopamine release in the ventral caudate/nucleus accumbens. Arch Gen Psychiatry 2006; 63: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AK, Scanlon JM, Becker CR, Ritter AC, Niyonkuru C, Dixon CE et al. The influence of genetic variants on striatal dopamine transporter and D2 receptor binding after TBI. J Cereb Blood Flow Metab 2014; 34: 1328–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrisberger F, Smieskova R, Schmidt A, Lenz C, Walter A, Wittfeld K et al. BDNF Val66Met polymorphism and hippocampal volume in neuropsychiatric disorders: a systematic review and meta-analysis. Neurosci Biobehav Rev 2015; 55: 107–118. [DOI] [PubMed] [Google Scholar]
- Murphy SE, Norbury R, Godlewska BR, Cowen PJ, Mannie ZM, Harmer CJ et al. The effect of the serotonin transporter polymorphism (5-HTTLPR) on amygdala function: a meta-analysis. Mol Psychiatry 2013; 18: 512–520. [DOI] [PubMed] [Google Scholar]
- Bastiaansen JA, Servaas MN, Marsman JB, Ormel J, Nolte IM, Riese H et al. Filling the gap: relationship between the serotonin-transporter-linked polymorphic region and amygdala activation. Psychol Sci 2014; 25: 2058–2066. [DOI] [PubMed] [Google Scholar]
- Borenstein M, Hedges LV, Higgins JPT, Rothstein HR. Introduction to Meta-Analysis. John Wiley & Sons: West Sussex, UK, 2009. [Google Scholar]
- Hirvonen MM, Lumme V, Hirvonen J, Pesonen U, Nagren K, Vahlberg T et al. C957T polymorphism of the human dopamine D2 receptor gene predicts extrastriatal dopamine receptor availability in vivo. Prog Neuropsychopharmacol Biol Psychiatry 2009; 33: 630–636. [DOI] [PubMed] [Google Scholar]
- Hirvonen MM, Nagren K, Rinne JO, Pesonen U, Vahlberg T, Hagelberg N et al. COMT Val158Met genotype does not alter cortical or striatal dopamine D2 receptor availability in vivo. Mol Imaging Biol 2010; 12: 192–197. [DOI] [PubMed] [Google Scholar]
- Boot E, Booij J, Zinkstok JR, Baas F, Swillen A, Owen MJ et al. COMT Val(158) met genotype and striatal D(2/3) receptor binding in adults with 22q11 deletion syndrome. Synapse 2011; 65: 967–970. [DOI] [PubMed] [Google Scholar]
- Bertolino A, Taurisano P, Pisciotta NM, Blasi G, Fazio L, Romano R et al. Genetically determined measures of striatal D2 signaling predict prefrontal activity during working memory performance. PloS One 2010; 5: e9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurisano P, Romano R, Mancini M, Giorgio AD, Antonucci LA, Fazio L et al. Prefronto-striatal physiology iss associated with schizotypy and is modulated by a functional variant of DRD2. Front Behav Neurosci 2014; 8: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvonen M, Laakso A, Nagren K, Rinne JO, Pohjalainen T, Hietala J. C957T polymorphism of the dopamine D2 receptor (DRD2) gene affects striatal DRD2 availability in vivo. Mol Psychiatry 2004; 9: 1060–1061. [DOI] [PubMed] [Google Scholar]
- Hirvonen M, Laakso A, Nagren K, Rinne JO, Pohjalainen T, Hietala J. Corrigendum: C957T polymorphism of the dopamine D2 receptor (DRD2) gene affects striatal DRD2 availability in vivo. Mol Psychiatry 2005; 10: 889. [DOI] [PubMed] [Google Scholar]
- Pohjalainen T, Cravchik A, Gejman PV, Rinne J, Nagren K, Syvalahti E et al. Antagonist binding characteristics of the Ser311—>Cys variant of human dopamine D2 receptor in vivo and in vitro. Biochem Biophys Res Commun 1997; 232: 143–146. [DOI] [PubMed] [Google Scholar]
- Pecina M, Mickey BJ, Love T, Wang H, Langenecker SA, Hodgkinson C et al. DRD2 polymorphisms modulate reward and emotion processing, dopamine neurotransmission and openness to experience. Cortex 2013; 49: 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitz J, Hodgkinson CA, Martin-Soelch C, Shen PH, Szczepanik J, Nugent A et al. The functional DRD3 Ser9Gly polymorphism (rs6280) is pleiotropic, affecting reward as well as movement. PloS One 2013; 8: e54108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickey BJ, Sanford BJ, Love TM, Shen PH, Hodgkinson CA, Stohler CS et al. Striatal dopamine release and genetic variation of the serotonin 2C receptor in humans. J Neurosci 2012; 32: 9344–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghardt PR, Love TM, Stohler CS, Hodgkinson C, Shen PH, Enoch MA et al. Leptin regulates dopamine responses to sustained stress in humans. J Neurosci 2012; 32: 15369–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domino EF, Evans CL, Ni L, Guthrie SK, Koeppe RA, Zubieta JK. Tobacco smoking produces greater striatal dopamine release in G-allele carriers with mu opioid receptor A118G polymorphism. Prog Neuropsychopharmacol Biol Psychiatry 2012; 38: 236–240. [DOI] [PubMed] [Google Scholar]
- Love TM, Enoch MA, Hodgkinson CA, Pecina M, Mickey B, Koeppe RA et al. Oxytocin gene polymorphisms influence human dopaminergic function in a sex-dependent manner. Biol Psychiatry 2012; 72: 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumay E, Fowler JS, Wang GJ, Logan J, Alia-Klein N, Goldstein RZ et al. Repeat variation in the human PER2 gene as a new genetic marker associated with cocaine addiction and brain dopamine D2 receptor availability. Transl Psychiatry 2012; 2: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: a novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Hum Mutat 2004; 23: 540–545. [DOI] [PubMed] [Google Scholar]
- Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P et al. Allelic association of human dopamine D2 receptor gene in alcoholism. JAMA 1990; 263: 2055–2060. [PubMed] [Google Scholar]
- Castiglione CM, Deinard AS, Speed WC, Sirugo G, Rosenbaum HC, Zhang Y et al. Evolution of haplotypes at the DRD2 locus. Am J Hum Genet 1995; 57: 1445–1456. [PMC free article] [PubMed] [Google Scholar]
- Hauge XY, Grandy DK, Eubanks JH, Evans GA, Civelli O, Litt M. Detection and characterization of additional DNA polymorphisms in the dopamine D2 receptor gene. Genomics 1991; 10: 527–530. [DOI] [PubMed] [Google Scholar]
- Pecina M, Love T, Stohler CS, Goldman D, Zubieta JK. Effects of the Mu opioid receptor polymorphism (OPRM1 A118G) on pain regulation, placebo effects and associated personality trait measures. Neuropsychopharmacology 2015; 40: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laruelle M. Imaging synaptic neurotransmission with in vivo binding competition techniques: a critical review. J Cereb Blood Flow Metab 2000; 20: 423–451. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Strange PG. Mechanisms of inverse agonist action at D2 dopamine receptors. Br J Pharmacol 2005; 145: 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Schwander K, Arnett DK, Kardia SL, Rankinen T, Bouchard C et al. An empirical comparison of meta-analysis and mega-analysis of individual participant data for identifying gene-environment interactions. Genet Epidemiol 2014; 38: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell MA, Fryar CD, Ogden CL, Flegal KM. Anthropometric reference data for children and adults: United States, 2003–2006. Natl Health Stat Rep 2008; 10: 1–48. [PubMed] [Google Scholar]
- Kraemer HC, Thiemann S. How Many Subjects? Statistical Power Analysis in Research. Sage Publications: Newbury Park, CA, USA, 1987. [Google Scholar]
- Bolton JL, Marioni RE, Deary IJ, Harris SE, Stewart MC, Murray GD et al. Association between polymorphisms of the dopamine receptor D2 and catechol-o-methyl transferase genes and cognitive function. Behav Genet 2010; 40: 630–638. [DOI] [PubMed] [Google Scholar]
- Yue JK, Pronger AM, Ferguson AR, Temkin NR, Sharma S, Rosand J et al. Association of a common genetic variant within ANKK1 with six-month cognitive performance after traumatic brain injury. Neurogenetics 2015; 16: 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YQ, Qiao L, Xue XD, Fu JH. Association between ANKK1 (rs1800497) polymorphism of DRD2 gene and attention deficit hyperactivity disorder: a meta-analysis. Neurosci Lett 2015; 590: 101–105. [DOI] [PubMed] [Google Scholar]
- Nymberg C, Banaschewski T, Bokde AL, Buchel C, Conrod P, Flor H et al. DRD2/ANKK1 polymorphism modulates the effect of ventral striatal activation on working memory performance. Neuropsychopharmacology 2014; 39: 2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderqvist S, Matsson H, Peyrard-Janvid M, Kere J, Klingberg T. Polymorphisms in the dopamine receptor 2 gene region influence improvements during working memory training in children and adolescents. J Cogn Neurosci 2014; 26: 54–62. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenicka J, Quinones-Lombrana A, Espana-Serrano L, Alvira-Botero X, Kremer L, Perez-Gonzalez R et al. The ANKK1 gene associated with addictions is expressed in astroglial cells and upregulated by apomorphine. Biol Psychiatry 2010; 67: 3–11. [DOI] [PubMed] [Google Scholar]
- Ponce G, Quinones-Lombrana A, Martin-Palanco NG, Rubio-Solsona E, Jimenez-Arriero MA, Palomo T et al. The addiction-related gene Ankk1 is oppositely regulated by D1R- and D2R-like dopamine receptors. Neurotox Res 2015. [DOI] [PubMed]
- Garrido E, Palomo T, Ponce G, Garcia-Consuegra I, Jimenez-Arriero MA, Hoenicka J. The ANKK1 protein associated with addictions has nuclear and cytoplasmic localization and shows a differential response of Ala239Thr to apomorphine. Neurotox Res 2011; 20: 32–39. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bertolino A, Fazio L, Blasi G, Rampino A, Romano R et al. Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci USA 2007; 104: 20552–20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failla MD, Myrga JM, Ricker JH, Dixon CE, Conley YP, Wagner AK. Posttraumatic brain injury cognitive performance is moderated by variation within ANKK1 and DRD2 genes. J Head Trauma Rehabil 2015; 30: E54–E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrga JM, Failla MD, Ricker JH, Dixon CE, Conley YP, Arenth PM et al. A dopamine pathway gene risk score for cognitive recovery following traumatic brain injury: methodological considerations, preliminary findings, and interactions with sex. J Head Trauma Rehabil 2015. [DOI] [PubMed]
- Al Sweidi S, Morissette M, Rouillard C, Di Paolo T. Estrogen receptors and lesion-induced response of striatal dopamine receptors. Neuroscience 2013; 236: 99–109. [DOI] [PubMed] [Google Scholar]
- Purves-Tyson TD, Owens SJ, Double KL, Desai R, Handelsman DJ, Weickert CS. Testosterone induces molecular changes in dopamine signaling pathway molecules in the adolescent male rat nigrostriatal pathway. PloS One 2014; 9: e91151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvari M, Deli L, Kocsis P, Mark L, Maasz G, Hrabovszky E et al. Estradiol and isotype-selective estrogen receptor agonists modulate the mesocortical dopaminergic system in gonadectomized female rats. Brain Res 2014; 1583: 1–11. [DOI] [PubMed] [Google Scholar]