Abstract
Robust statistical, genetic and functional evidence supports a role for DISC1 in the aetiology of major mental illness. Furthermore, many of its protein-binding partners show evidence for involvement in the pathophysiology of a range of neurodevelopmental and psychiatric disorders. Copy number variants (CNVs) are suspected to play an important causal role in these disorders. In this study, CNV analysis of DISC1 and its binding partners PAFAH1B1, NDE1, NDEL1, FEZ1, MAP1A, CIT and PDE4B in Scottish and Northern Swedish population-based samples was carried out using multiplex amplicon quantification. Here, we report the finding of rare CNVs in DISC1, NDE1 (together with adjacent genes within the 16p13.11 duplication), NDEL1 (including the overlapping MYH10 gene) and CIT. Our findings provide further evidence for involvement of DISC1 and its interaction partners in neuropsychiatric disorders and also for a role of structural variants in the aetiology of these devastating diseases.
Key Words: Copy number variants, DISC1, NDE1, NDEL1, Schizophrenia, Affective disorder, Intellectual disability
Introduction
Schizophrenia, schizoaffective disorder and bipolar disorder are phenotypically and genetically overlapping severe psychiatric disorders, cumulatively affecting approximately 3% of the population worldwide [1,2,3,4]. Like many other common diseases, they are complex and multifactorial, with contributions from multiple susceptibility genes and epigenetic, stochastic, and environmental factors [5]. Family, twin and adoption studies have shown that genetic factors play a major role in the development of schizophrenia [4], and also in mood disorders [6] which have estimated heritability values between 60 and 85%. The search for susceptibility genes has however been challenging, especially in mood disorders, where it has been hampered by clinical and genetic heterogeneity.
It has been shown in schizophrenia research that there is a spectrum of risk alleles from common, low penetrant forms such as single nucleotide polymorphisms (SNPs) identified from large genome-wide association studies (GWAS) to rarer, highly penetrant forms such as copy number variants (CNVs; segments of DNA with duplications and deletions) found at increased frequency in disease from a number of genome-wide CNV studies [7,8]. Whilst the majority of mental illness at the population and individual level is likely to be caused by a combination of many risk alleles, there may be some forms of illness which are due to rarer variants that are themselves sufficient to cause illness in those individuals or families expressing them. Such variants can be biologically informative because they can be used to develop molecular, cellular and animal models of psychosis to help unravel the underlying disease mechanisms.
One such rare variant, first discovered in a Scottish cytogenetic study of juvenile delinquency, has been a balanced translocation from chromosome 11 into the long arm of chromosome 1. It was found to segregate with schizophrenia, bipolar disorder and major depression in other family members with major mental illness being found in roughly half of family members carrying the translocation (log of the odds ratio = 7.1) [9,10]. Ten years later, the previously unknown gene Disrupted-In-Schizophrenia 1 (DISC1) on chromosome 1 was identified as disrupted by the translocation [11]. Since then, DISC1 has attracted much attention, not only being associated with schizophrenia but also predisposing individuals to a wide range of major mental disorders, including bipolar disorder, major depression and autism spectrum disorders (ASD), in different populations [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29]. In addition, association has been detected between DISC1 alleles or haplotypes and measurable traits related to schizophrenia and bipolar disorder, including working memory [30,31,32], cognition [22,25], reduced grey matter volume, brain activation [33] and abnormalities in hippocampal structure and function [21,34]. By conditioning a previous GWAS study on DISC1, 8 genes (LCT, CCDC141, TTN, CXCL3, KIAA1128, MED13L, MIR620 and TMC07) have been identified as associating with psychosis proneness, and these molecules predominantly link to the DISC1 pathway [35]. Through genetics, cell biology, animal modelling and neuroimaging, the DISC1 pathway is emerging as a pivotal mediator of quantitative and pathological brain dysfunction [36].
DISC1 expression in the brain is particularly high in the hippocampus during neurogenesis and remains high in the adult dentate gyrus, olfactory bulb and limbic regions [37,38], and it appears that DISC1 regulates important developmental processes such as neuronal migration, integration [39], synapse formation and neuronal stem cell maturation [38,40,41,42]. DISC1 is thus critical for neurodevelopment and normal adult neuronal function. In addition, transgenic or mutant mice with impaired DISC1 function show brain morphological changes, deficits in neural circuits, working memory impairment and behavioural traits related to schizophrenia and also bipolar disorder [43,44,45,46,47,48,49].
DISC1 acts as a large scaffold protein interacting with multiple protein partners [50,51,52], several of which have been shown to be important for cytoskeleton stability and modulation, intracellular transport, cAMP signalling, synaptic signal transduction and synaptic plasticity [53]. These DISC1 interactors are independent risk factors for major mental illness [54], making the DISC1 protein interaction network a potential target for future therapeutic intervention. Synaptic deficits and transcriptional dysregulation have recently been shown in induced pluripotent stem cell-derived forebrain neurons from patients with mutant DISC1 [55]. The so-called modelled ‘DISC1 interactome’ has been proposed to consist of 127 proteins and 158 interactions [52]. Sixty-seven of these proteins are direct DISC1 interactors and interestingly many of these DISC1 interactors also interact with one another, suggesting their involvement in common pathways, although the functional role of the majority of these potential interactors in neuronal development has yet to be shown in vivo [54]. Of these DISC1 interactors, we selected 7 that have a likely functional relation with psychiatric disorders based on evidence from the literature, namely platelet-activating factor acetylhydrolase 1b regulatory subunit 1 gene (PAFAH1B1), Nuclear distribution factor E-homolog 1 gene (NDE1), NDE-like 1 gene (NDEL1), Fasciculation and Elongation Protein Zeta-1 gene (FEZ1), Microtubule-associated Protein 1A gene (MAP1A), CITRON gene (CIT) and cAMP-hydrolysing enzyme phosphodiesterase 4 gene (PDE4B). Altogether, a substantial body of genetic and biological evidence supports a role for DISC1 and the 7 described interacting partners (summarised in table 1) in the development of psychiatric disorders.
Table 1.
Evidence supporting a role for the seven DISC1 interacting partners in the development of psychiatric disorders
| DISC1 interactor | Expression and proposed function | Disease associations and animal models |
|---|---|---|
| PAFAH1B1 | Encodes protein LIS1 which localizes to the centrosome and is a component of the microtubule-based dynein motor complex involved in neurogenesis and neuronal migration | Mutations in PAFAH1B1 cause the neuronal migration disorder lissencephaly [68,69,70] |
| Disruption of PAFAH1B1 in animal models show cerebral cortex malformations [71, 72] | ||
| NDE1 and NDEL1 | NDE1 and NDEL1 localize to the centrosome and are components of the microtubule-based dynein motor complex | Mouse studies suggest that NDE1-mediated heterochromatin replication is critical for neuronal differentiation, and loss of NDE1 function may lead to genomic neurological disorders [77] |
| NDE1 is a cytoskeletal protein that participates in essential neurodevelopmental processes, including neuronal precursor proliferation and differentiation, neuronal migration, and neurite outgrowth [73] | Familial mutations in NDE1 cause severe failure of neurogenesis and cortical lamination (microlissencephaly) [78, 79] | |
| Phosphorylated NDE1 is present at the postsynaptic density, in proximal axons, within the nucleus, and at the centrosome where it becomes substantially enriched during mitosis. Mutation of the NDE1 phosphorylation site (T131) inhibits neurite outgrowth [74] | NDE1 associated with psychosis proneness in a large Finnish birth cohort upon re-analysis of GWAS linkage data conditioned on a DISC1-associating risk haplotype [35] | |
| Abundance of the NDEL1-DISC1 complex is highest during neuronal migration in the developing cortex [75] NDE1 and NDEL1 may compete with each other for DISC1 binding [76] | Linkage found between NDE1 and schizophrenia in families with individuals carrying DISC1 risk alleles [73] – interaction between NDE1 or NDEL1 genotypes and DISC1 genotypes are associated with schizophrenia [76] | |
| NDEL1 expression is reduced in brain tissue of patients with schizophrenia [39] | NDE1 CNVs associate with a range of phenotypically different neurodevelopmental disorders including intellectual disability [80], ASD [81], attention deficit hyperactivity disorder [82], epilepsy [83] and schizophrenia [67, 80] | |
| FEZ1 | Anchors microtubules near the cell membrane and plays a role in axon bundling and axonal transport | FEZ1 mRNA expression is reduced in schizophrenia [85] Association between FEZ1 and schizophrenia was found in a Japanese cohort [86] |
| FEZ1 expression pattern closely resembles that of DISC1 and the DISC1/FEZ1 complex appears to be involved in neurite outgrowth [41] | FEZ1 knockout mice exhibit abnormal brain architecture, hyperactivity and enhanced responsiveness to psycho-stimulants [87] | |
| FEZ1 interacts with DISC1 to synergistically regulate dendritic growth of newborn neurons in the adult mouse hippocampus [84] | Epistatic interaction between FEZ1 and DISC1-associated with risk of schizophrenia [84] | |
| MAP1A | Controls microtubule polymerization and stabilization and is required for axon and dendrite development [88, 89] | Binds with post-synaptic density protein PSD-95 [90, 91] – essential for the correct membrane localization of multiple neurotransmitter receptors/synaptic proteins |
| CIT | CITRON (encoded by CIT) binds with post-synaptic density protein PSD-95 [90] | CIT has been shown to be associated with BP disorder [92] |
| PDE4B | Plays a role in intracellular signal transduction by degrading cAMP | PDE4B was found to be disrupted by a translocation breakpoint in 2 related individuals with psychosis [93] |
| Dissociation of PDE4B from DISC1 inactivates its enzymatic activity | Association has been found between SNPs in PDE4B and schizophrenia [94,95,96] | |
| PDE4B interacts with the LIS1/NDEL1/NDE1/DISC1 dynein motor complex at the centrosome and may play a role in regulating its structure and function [53] Mainly expressed in the cortex, hippocampus and striatum, brain regions implicated in memory and cognition | Specific PDE4 inhibitor rolipram shown to facilitate long-term memory and to have antidepressant and antipsychotic activity [97, 98] | |
| PDE4B knockout mice show antidepressant-like behaviour and a proliferation of neuronal cells in the hippocampus [99, 100] | ||
| Two DISC1 missense mutations in mice, which result in depressed and schizophrenic phenotypes, are located within specific PDE4B binding sites [44] | ||
In GWAS performed for schizophrenia and bipolar disorder, no SNPs in DISC1 or in any of its binding partners were found among the most significant results. Only a few SNPs with moderate significance were found, but there has been no consistency between different studies (catalogue of published GWAS: http://www.genome.gov/26525384) [56]. Current GWAS effectively capture the majority of common variants but not rare variants and structural variations, such as genomic rearrangements or CNVs, in the European population. This type of variation may contribute significantly to the inter-individual variation in susceptibility to diseases [57,58,59,60]. Genome-wide CNV studies have shown that rare CNVs are found at an increased frequency in patients with schizophrenia compared to control individuals [56,61,62,63,64,65,66,67], constituting risk factors with low prevalence but high penetrance. Therefore, in this study we test the hypothesis that CNVs disrupting or encompassing DISC1 or any of the 7 binding partners influence susceptibility to schizophrenia and bipolar disorder in the Scottish and Swedish populations and to major depressive disorder and intellectual disability (ID) in the Scottish population samples. Hereto, we used a technique named multiplex amplicon quantification (MAQ), which allows high-resolution CNV detection (kilobase range) at specific genomic regions, and thus covers small CNVs that are missed by many current genome-wide CNV detection platforms.
Subjects and Methods
Subjects
Scotland
The Scottish sample used in this study consists of a cohort of 1,293 unrelated cases [647 with schizophrenia, 213 with bipolar affective disorder, 192 with major depressive disorder and 241 with ID (ID, ID + schizophrenia and ID + ‘comorbid’] and 352 unrelated controls. The study was approved by the Multi-Centre Research Ethics Committee for Scotland, and patients gave written informed consent for the collection of DNA samples for use in genetic studies. The sample comprised Caucasian individuals contacted through the inpatient and outpatient services of hospitals in North East (nearest city: Aberdeen) and South East (nearest city: Edinburgh) Scotland. For the South East Scotland samples, a diagnosis of schizophrenia was based on information from an interview with the patient using the Schedule for Affective Disorders and Schizophrenia-Lifetime version (SADS-L) supplemented by case note review and frequently by information from medical staff, relatives and caregivers. Patients with bipolar affective disorder and unipolar depression were also identified at the Royal Edinburgh Hospital and associated hospitals, and final diagnoses, based on DSM-IV-TR criteria [101], were reached by consensus between 2 trained psychiatrists and where possible using the Operational Criteria Checklist for Psychotic Illness (OPCRIT) [102]. Patients diagnosed with ID were over 18 years of age and recruited through the Learning Disability Services of South East Scotland. To be in this service, all patients were assessed to have an IQ <70. Within the National Health Service in the UK, psychiatrists are involved in the management of these patients regardless of whether or not they have a mental illness. We did not routinely reassess IQ in this study but relied on case note records as patients in the Learning Disability Services have previously had IQ assessments by clinical psychologists. Only ID patients with an IQ between 50 and 70 were included in this study. IQ assessment used Wechsler Intelligence Scale for Children (WISC) or Wechsler Adult Intelligence Scale (WAIS; most patients were part of the Learning Disabilities service since childhood). Most patients gave consent to take part in the study; however, if incapable of giving fully informed consent, a welfare guardian or nearest relative was approached to also give consent. Initial contact was through the responsible psychiatrist. Some patients were being treated for schizophrenia and major depression and some had no history of mental illness. Diagnoses according to DSM-IV criteria were reached using the semi-structured interview Psychiatric Assessment Schedule for Adults with a Developmental Disability (PAS-ADD 10) involving the patient and a key informant. Additional clinical information was obtained from hospital case note review. The Social Communication Questionnaire-Lifetime version (SCQ-L) provided a screen for autism, and high- scoring individuals were followed up using the Autism Diagnostic Instrument-Revised (ADI-R). Initial diagnoses were confirmed through consultation between the relevant specialist clinician involved and one of the research team members who was also a specialist in the psychiatry of ID (the late Prof. Walter Muir). Ethnically matched controls from the same region were recruited through the South of Scotland Blood Transfusion Service and from hospital staff. All controls were not directly screened to exclude those with a personal or family history of psychiatric illness; however, the Blood Transfusion Service does not accept blood donations from subjects taking regular medication or with a history of major illness. Of the North East Scotland samples, all participants self-identified as born in the British Isles (95% in Scotland), diagnosis was confirmed with the OPCRIT symptom checklist [102], and all cases met the DSM-IV and International Classification of Diseases 10th Edition (ICD-10) criteria for schizophrenia. Detailed medical and psychiatric histories were collected, and a clinical interview using Structured Clinical Interview for DSM-IV (SCID) was also performed on schizophrenia cases. Control samples were volunteers recruited through general practices in North East Scotland. Practice lists were screened for potentially suitable volunteers by age and sex and by exclusion of subjects with major mental illness or use of neuroleptic medication. Volunteers who replied to a written invitation were interviewed using a short questionnaire to exclude major mental illness in themselves and first-degree relatives.
Sweden
All participants were Caucasians and originated from a geographically isolated population living in the County of Västerbotten in Northern Sweden. Two large samples were used in this study from the Swedish population. One sample consists of 909 unrelated patients (410 schizophrenia, 120 schizoaffective disorder, 314 bipolar affective disorder) and 512 unrelated control individuals. The other sample comprises 152 related patients and 227 of their first-degree relatives, all members of a large 14-generation pedigree from Västerbotten County (table 2).
Table 2.
Prevalence of CNVs in the Scottish and Swedish unrelated patient and control groups
| Group | DISC1 |
NDE1 |
NDEL1 |
CIT |
Total Del, count (%) | Total Dup, count (%) | Total CNVs, count (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Del | Dup | Del | Dup | Del | Dup | Del | Dup | ||||
| SCZ (Sc) | 1/647 | 2/647 | 1/647 | 2/647 | 0/647 | 0/647 | 0/647 | 1/647 | 2 (0.31) | 5 (0.77) | 7 (1.08) |
| SCZ (Sw) | 0/410 | 7/410 | 0/410 | 0/410 | 0/410 | 1/410 | 0/410 | 0/410 | 0 | 8 (1.95) | 8 (1.95) |
| Total SCZ | 1/1,057 | 9/1,057 | 1/1,057 | 2/1,057 | 0/1,057 | 1/1,057 | 0/1,057 | 1/1,057 | 2 (0.19) | 13 (1.23) | 15 (1.42) |
| Total SA (Sw) | 0/120 | 0/120 | 0/120 | 1/120 | 0/120 | 0/120 | 0/120 | 0/120 | 0 | 0 | 0 |
| BPD (Sc) | 0/213 | 0/213 | 0/213 | 0/213 | 0/213 | 0/213 | 0/213 | 0/213 | 0 | 0 | 0 |
| BPD (Sw) | 0/314 | 6/314 | 0/314 | 0/314 | 0/314 | 0/314 | 0/314 | 0/314 | 0 | 6 (1.91) | 6 (1.91) |
| Total BPD | 0/527 | 6/527 | 0/527 | 0/527 | 0/527 | 0/527 | 0/527 | 0/527 | 0 | 6 (1.14) | 6 (1.14) |
| Total MDD (Sc) | 0/192 | 0/192 | 0/192 | 0/192 | 0/192 | 0/192 | 0/192 | 0/192 | 0 | 0 | 0 |
| Total ID (Sc) | 0/72 | 2/72 | 0/72 | 1/72 | 0/72 | 0/72 | 0/72 | 0/72 | 0 | 3 (4.17) | 3 (4.17) |
| ID + SCZ (Sc) | 0/92 | 0/92 | 0/92 | 1/92 | 0/92 | 0/92 | 0/92 | 0/92 | 0 | 1 (1.08) | 1 (1.08) |
| ID + SCZ (Sw) | 0/24 | 0/24 | 0/24 | 0/24 | 0/24 | 0/24 | 0/24 | 0/24 | 0 | 0 | 0 |
| Total ID + SCZ | 0/116 | 0/116 | 0/116 | 1/116 | 0/116 | 0/116 | 0/116 | 0/116 | 0 | 1 (0.86) | 1 (0.86) |
| ID + comorbid (Sc) | 0/77 | 0/77 | 0/77 | 1/77 | 0/77 | 0/77 | 0/77 | 0/77 | 0 | 1 (1.30) | 1 (1.30) |
| ID + comorbid (Sw) | 0/19 | 0/19 | 0/19 | 0/19 | 0/19 | 0/19 | 0/19 | 0/19 | 0 | 0 | 0 |
| Total ID + comorbid | 0/96 | 0/96 | 0/96 | 1/96 | 0/96 | 0/96 | 0/96 | 0/96 | 0 | 1 (1.04) | 1 (1.04) |
| Control (Sc) | 1/352 | 1/352 | 0/352 | 0/352 | 0/352 | 0/352 | 0/352 | 0/352 | 1 (0.28) | 1 (0.28) | 2 (0.57) |
| Control (Sw) | 0/512 | 6/512 | 0/512 | 3/512 | 0/512 | 0/512 | 0/512 | 0/512 | 0 | 9 (1.76) | 9 (1.76) |
| Total control | 1/864 | 7/864 | 0/864 | 3/864 | 0/864 | 0/864 | 0/864 | 0/864 | 1 (0.16) | 10 (1.16) | 11 (1.27) |
Of the 8 gene regions screened, CNVs (duplications and deletions) were found in DISC1, NDE1, NDEL1 and CIT regions. No CNVs were found in PAFAH1B1, FEZ1, MAP1A or PDE4B in this study. SCZ = Schizophrenia; SA = schizoaffective disorder; BPD = bipolar disorder; MDD = major depressive disorder; Sc = Scottish; Sw = Swedish. Note: total ID + comorbid (Sc) – diagnoses with ID include BPD (n = 32), MDD (n = 26), autism (n = 17), Asperger's (n = 1) and Klinefelter's syndrome (n = 1); total ID + comorbid (Sw) – diagnoses with ID include schizoaffective disorder-bipolar type (n = 11), schizoaffective disorder-depressive type (n = 3), BPD (n = 2) and psychotic disorder not otherwise specified (n = 3). Shaded rows represent total values for Scottish and Swedish populations combined.
The unrelated patients were identified through the in-patient hospital registers in the region, and the ascertainment was carried out from 1992 to 2005. Diagnostic evaluation was performed by trained research nurses and research psychiatrists using register data, data from medical records, multiple informants, and, when deemed necessary and feasible, semi-structured interviews were used including MINI (Mini-International Neuropsychiatric Interview) [103], DIGS (The Diagnostic Interview for Genetic Studies) [104], FIGS (The Family Interview for Genetic Studies) [105] and SCAN (The Schedules for Clinical Assessment in Neuropsychiatry) [106]. The diagnoses were defined according to the DSM-IV criteria, and patients were included only if full consensus was reached by 2 research psychiatrists. To date, the patients have been followed-up for 3-13 years, giving ample opportunities for diagnostic validation and phenotypic characterization.
The 512 unrelated Swedish control individuals were recruited from the Betula study sample of 4,360 individuals representative of the general population of the region, described in detail elsewhere [107]. A prerequisite for inclusion in the Betula study was the ability to comply with a test battery consisting of a health examination, blood sample testing, interviews and self-rating scales about somatic and mental health, and an extensive examination of memory functions. For 9 Swedish control individuals carrying a CNV, in-depth clinical evaluation showed no signs of lifetime psychiatric disorders. The 152 probands of the nuclear families were initially identified through hospital registers, and the diagnostic work-up was similar to the procedure described for the unrelated cases. A detailed description of the pedigree has been previously published [108]. For the purpose of this study, we were not dependent on a final DSM-IV diagnosis (ongoing assessment) of all 227 first-degree relatives. Lifetime psychiatric disorders were evaluated in 13 first-degree relatives using a semi-structured interview based on the DIGS and FIGS. In these 13 individuals, 6 were CNV carriers and 7 were not but were a first-degree relative of a proband with a CNV. Of the 6 with a CNV, 3 had bipolar affective disorder and 3 were unaffected. Out of the 7 non-CNV carriers, 1 had bipolar disorder, 1 had schizoaffective disorder and 5 were unaffected (table 2). All subjects participated after giving written informed consent, and the study was approved by the regional Medical Ethics Committees of the Universities of Umeå and Antwerp. We also analysed the 30 HapMap trios of the CEU (Utah residents with ancestry from Northern and Western Europe) population. DNA of these 90 individuals was obtained from the Coriell Institute (Camden, N.J., USA).
Multiplex Amplicon Quantification
Genomic DNA was extracted from peripheral blood using standard methods. To explore the 8 genes for CNVs, we used MAQ [109,110]. It consists of a multiplex PCR amplification of several fluorescently labelled target and reference amplicons, followed by fragment analysis on an ABI 3730 DNA analyzer (Applied Biosystems). The comparison of normalised peak areas between patient and control individuals results in a dosage quotient (DQ) of the target amplicon. Seventy-nine target amplicons and 21 reference amplicons were amplified in three MAQ assays (online suppl. table SI; see www.karger.com/doi/10.1159/000438788 for all online suppl. material). These assays are available on request from Prof. Del-Favero (jurgen.delfavero@molgen-vib.ua.be). The MAQ reactions were performed on 20-ng genomic DNA. DQs were calculated using an in-house developed MAQ software package (MAQ-S; http://www.vibgeneticservicefacility.be/MAQ.htm). A DQ between 1.3 and 1.7 was considered indicative of a heterozygous duplication, a DQ between 0.3 and 0.7 indicative of a heterozygous deletion.
Allelic Quantification
To confirm the findings of the MAQ analysis in the Northern Swedish population samples, we used allelic quantification by genotyping SNPs in and around the duplicated amplicons using the Sequenom MassARRAY iPLEX Gold system (Sequenom, Germany). Individuals heterozygous for an SNP in a duplicated region show an allele ratio of 2:1 (relative allele frequencies 0.667:0.333) instead of the 1:1 ratio (relative allele frequencies 0.5:0.5) in a normal diploid genome. Relative quantification assays are run with the Typer Analyzer program (Typer 4.0, Sequenom) using Allelotyping mode. SNPs with the highest possible heterozygosity were selected from HapMap (www.hapmap.org) and genotyped in the individuals showing a duplication with MAQ. Primers for PCR amplification and extension primers (online suppl. table SII) were designed using the Assay Design 3.1 program (Sequenom). Genotyping of the SNPs was carried out following the protocol provided by Sequenom. Analysis and scoring were performed using the program Typer 4.0 (Sequenom).
The primer extension products of each allele are not always equally represented, so the expected relative allele frequencies when a duplication is present were calculated based on the mean relative frequency of allele A (fA) and the mean relative frequency of allele B (fB) obtained from analysis of individuals without a duplication in that specific region. Expected relative frequencies in a duplicated region would be (2fA/fB)/[1 + (2fA/fB)] and 1 - (2fA/fB)/[1 + (2fA/fB)] for duplication of the A allele or (fA/2fB)/[1 + (fA/2fB)] and (fA/2fB)/[1 + (fA/2fB)] for duplication of the B allele.
Chromosomal Microarray
For several of the Scottish samples, CNVs initially found by MAQ assay were confirmed by microarray analysis using a CytoScan 750K Array (Affymetrix UK Ltd.). Microarray analysis was also used to genotype other family members. The test DNA was referenced against a normal control dataset using the Affymetrix Chromosome Analysis Suite (ChAS) and ArrayLIMs. Further details of the array designs are available at affymetrix.com.
Results
MAQ Analysis
We explored DISC1, PAFAH1B1, NDE1, NDEL1, FEZ1, MAP1A, CIT and PDE4B for whole or partial gene deletions or duplications in the Scottish and Northern Swedish samples. In the unrelated Scottish population-based sample set, we screened 647 patients with a diagnosis of schizophrenia, 213 patients with bipolar disorder, 192 patients with major depressive disorder, 72 patients with ID, 92 patients with ID and comorbid schizophrenia, 77 patients with ID and a comorbid other psychiatric diagnosis including: 32 with bipolar affective disorder, 26 with major depressive disorder, 17 with autism, 1 with Asperger's syndrome and 1 with Klinefelter's syndrome. The analysis also included screening 352 unrelated control individuals from the Scottish population (table 2). The unrelated Swedish population-based samples included 410 patients with a diagnosis of schizophrenia, 120 patients with schizoaffective disorder, 314 patients with bipolar disorder, 24 with ID and comorbid schizophrenia, and 19 with ID plus another comorbid psychiatric diagnosis including: 11 with schizoaffective disorder bipolar type, 3 with schizoaffective disorder depressive type, 2 with bipolar affective disorder and 2 with psychotic disorder not otherwise specified (table 2). This Swedish population-based sample analysis included screening 512 unrelated control individuals. Within the Swedish sample analysis, we also screened 152 related probands from a large pedigree as well as 227 first-degree relatives of these patients from an isolated Northern Swedish population (table 3). Ninety Swedish HapMap CEU individuals were also analysed in this study. We used the accurate high-throughput MAQ method to detect whole or partial gene deletions or duplications, quantitatively amplifying several target amplicons per gene (fig. 1; online suppl. table SI). Fifty-seven target amplicons in and around the 8 genes (‘amp’, fig. 1) and 21 reference amplicons were analysed in a first screen (online suppl. table SI).
Table 3.
Prevalence of CNVs in related probands and first-degree relatives of the pedigree from the County of Västerbotten in Northern Sweden
| Group | Diagnosis | n (M:F) | First-degree relatives, n | Nuclear families, n | Mean age at onset, years | Carriers of a duplication in: |
||
|---|---|---|---|---|---|---|---|---|
| DISC1, n % | NDE1, n % | NDEL1, n % | ||||||
| Related probands (152 in total) | SCZ | 27 (18:9) | 26 | 11 | 23.2 ± 6.6 | 2 (7.41) | 0 | 0 |
| SCZ + ID | 2 (1:1) | 3 | 2 | 28.5 ± 3.5 | 0 | 0 | 0 | |
| SCA | 21 (13:8) | 23 | 9 | 21.6 ± 7.9 | 0 | 1 (4.76) | 0 | |
| SCA + ID | 1 (0:1) | 0 | 0 | 15 | 0 | 0 | 0 | |
| BPD1 | 49 (22:27) | 73 | 35 | 28.2 ± 14.0 | 2 (4.08) | 0 | 0 | |
| BPD1 + ID + autism | 1 (0:1) | 2 | 1 | 23 | 0 | 0 | 0 | |
| BPD2 | 49 (17:32) | 95 | 37 | 30.5 ± 16.8 | 1 (2.04) | 0 | 0 | |
| BPD2 + autism | 1 (1:0) | 3 | 1 | 20 | 0 | 0 | 0 | |
| BPD NOS | 1 (0:1) | 1 | 1 | 42 | 0 | 0 | 0 | |
| First-degree relativesa (227 in total) | Total sample | 227 (85:142) | n.a. | 98 | n.a. | 6 (2.64) | 0 | 0 |
| BPD1 | 2 (0:2) | n.a. | n.a. | 25.5 ± 4.9 | 1 | 0 | 0 | |
| BPD2 | 1 (0:1) | n.a. | n.a. | 14 | 1 | 0 | 0 | |
| BPD NOS | 1 (0:1) | n.a. | n.a. | 65 | 1 | 0 | 0 | |
| SCA (bipolar type) | 1 (0:1) | n.a. | n.a. | 14 | 0 | 0 | 0 | |
| Unaffected | 8 (4:4) | n.a. | n.a. | n.a. | 3 | 0 | 0 | |
Duplications were found only in DISC1, NDE1 and NDEL1. SCZ = Schizophrenia; SCA = schizoaffective disorder; BPD = bipolar disorder; NOS = not otherwise specified; n.a. = not applicable.
Diagnostic information is shown in 13 out of 227 first-degree relatives; 6 carrying a CNV; 7 not carrying a CNV but are first-degree relatives of a proband with a CNV. Of the 6 with a CNV, 3 had BPD and 3 were unaffected. Out of the 7 non-CNV carriers, 1 had BPD, 1 had SCA and 5 were unaffected.
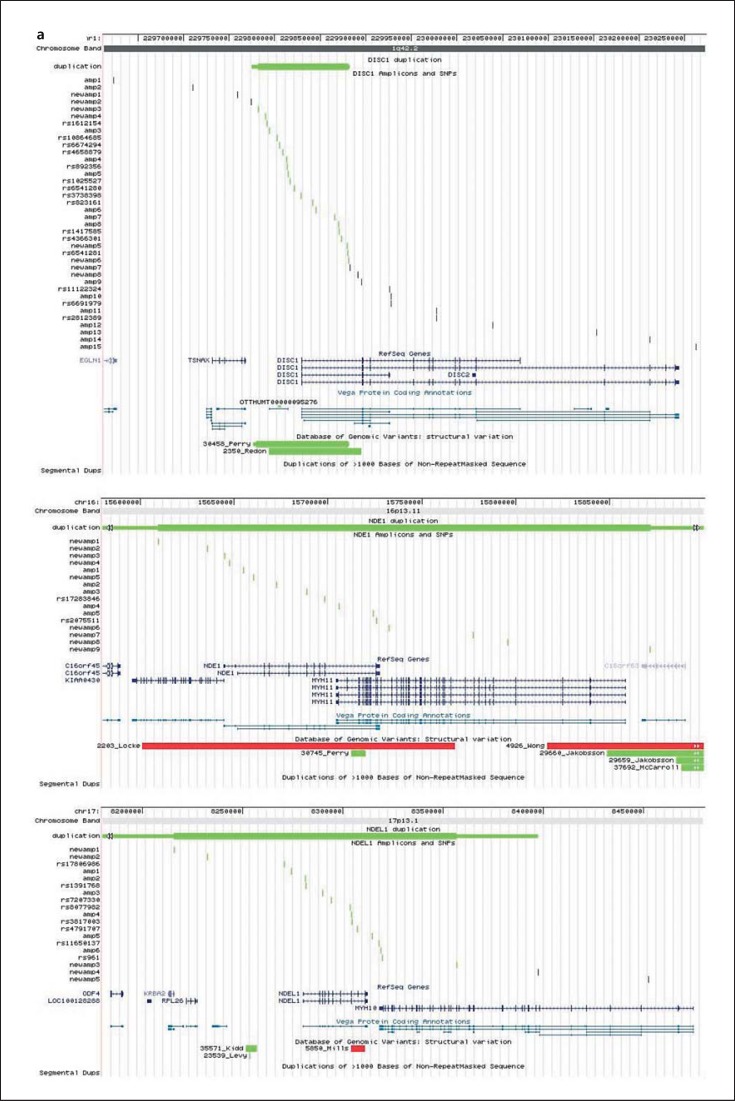
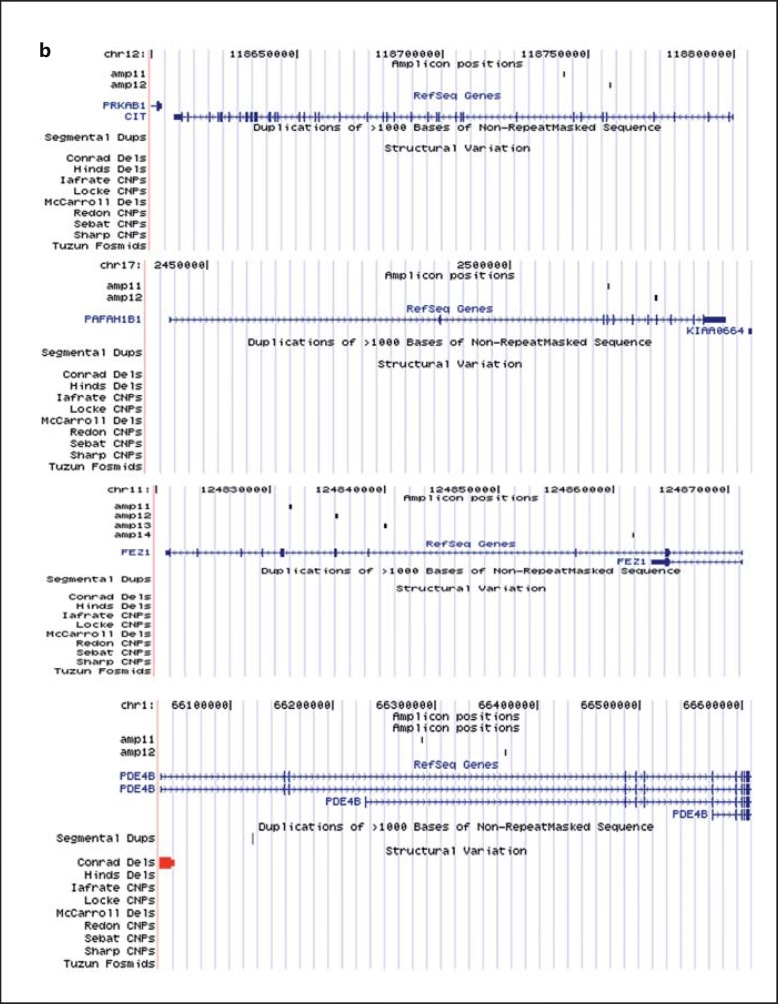
Fig. 1.
Location of the quantitatively typed MAQ amplicons and SNPs with reference to the investigated and other known genes and to structural variants in the Database of Genomic Variants. Duplications are drawn in green, deletions in red and CNVs with unknown direction in orange colour. a Detected duplications in 3 genes: DISC1, NDE1 and NDEL1. The minimum size is indicated with a thick bar, the maximal size with a thin bar and undefined boundaries are indicated with arrows. b Other genomic regions investigated included CIT, PAFAH1B, FEZ1 and PDE4B. The figure was made by adding custom tracks to the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway).
DISC1 Copy Number Variants
In both the Scottish and Swedish unrelated samples, we detected CNVs in DISC1 (table 2). In the Scottish samples, we detected one affected individual with a deletion of at least 48 kb with a minimal region of 229,904,902-229,952,723 (coordinates based on UCSC build 36) encoding intron 3 and 3 alternate exons of DISC1 (chromosome 1q42.2). This particular deletion was found only in this one Scottish male patient with a diagnosis of schizophrenia (1/647 = 0.15% in Scottish schizophrenia samples or 1/1,057 = 0.09% combining both Scottish and Swedish schizophrenia samples) and was not found in any other of the Scottish or Swedish affected samples or in any of the Scottish or Swedish controls (table 2). We found an unaffected Scottish individual who had a large deletion of at least 192 kb encoding the TSNAX and the TSNAX-DISC1 boundary region. However, the deletion in this control individual spans from 229,622,813 to 229,814,916, which is further upstream from the start of the DISC1 deletion found in the patient with schizophrenia (table 4).
Table 4.
Details on the individual CNV carriers that were identified in the Scottish samples
| ID | CNV/ cytoband | Minimal region | Size, kb | Genes | Diagnosis | Origin | Sex | AAS/AAO | Additional information |
|---|---|---|---|---|---|---|---|---|---|
| Sc11052 | Dup 1q42.2 | 229,828,419–229,830,272 | 1.9 | exon 1 DISC1 | SCZ | Sc | M | 36/28 | |
| Sc7401 | Del 1q42.2 | 229,904,902–229,952,723 | 48 | intron 3 and 3 alternate exons DISC1 | SCZ | Sc | M | 24/23 | |
| Sc11532 | Dup 1q42.2 | 229,814,616–229,845,359 | 30 | exon 1 DISC1 | SCZ | Sc | M | 34/19 | |
| Sc7473 | Dup 1q42.2 | 229,828,419–229,830,272 | 1.9 | exon 1 DISC1 | ID | Sc | F | 76/16 | mild ID |
| Sc7473 | Dup 1q42.2 | 229,814,616–229,845,359 | 30 | exon 1 DISC1 | ID | Sc | F | 76/16 | two DISC1 duplications were detected in this patient |
| Sc11986 | Dup 1q42.2 | 229,793,461–229,845,359 | 52 | exon 1 DISC1 | control | Sc | F | 59/- | |
| Sc11293 | Del 1q42.2 | 229,622,813–229,814,916 | 192 | TSNAX/TSNAX-DISC1 | control | Sc | M | 53/- | |
| Sc11120 | Del 16p13.11 | 15,654,809–15,723,967 | 69 | middle NDE1/end MYH11 overlap | SCZ | Sc | M | 30/17 | |
| Sc11034 | Dup 16p13.11 | 15,654,809–15,723,967 | 69 | middle NDE1/end MYH11 overlap | SCZ | Sc | M | 49/14 | |
| Sc6638 | Dup 16p13.11 | 15,654,809–15,723,967 | 69 | middle NDE1/end MYH11 overlap | SCZ | Sc | F | 39/37 | |
| Sc7292 | Dup 16p13.11 | 15,654,809–15,723,967 | 69 | middle NDE1/end MYH11 overlap | SCZ + ID | Sc | M | 27/21 | confirmed in GWAS |
| Sc7474 | Dup 16p13.11 | 15,654,809–15,723,967 | 69 | middle NDE1/end MYH11 overlap | ID | Sc | M | 43/12 | |
| Sc7615 | Dup 16p13.11 | 15,654,809–15,723,967 | 69 | middle NDE1/end MYH11 overlap | SCZ + ID + ASD | Sc | M | 18/14 | confirmed in GWAS |
| Sc11553 | Dup 12q24.23 | 118,741,727–118,757,944 | 16 | exons 6–9 CIT | SCZ | Sc | F | 57/18 | |
Coordinates are according to UCSC build 36 (hg18). Size is minimal CNV size. AAS = Age at sampling; AAO = age at onset; SCZ = schizophrenia; BPD = bipolar disorder; MDD = major depressive disorder; Sc = Scottish; Sw = Swedish.
In contrast to DISC1 deletions, DISC1 duplications were found to be more abundant in both the Scottish and Swedish population-based samples. Duplications of exon 1 of DISC1 were found in 5 individuals in the Scottish samples: in 2 patients with schizophrenia (2/647; 0.93%), 2 patients with ID (2/72; 2.78%) and in 1 control sample (1/352; 0.28%). In the unrelated Swedish population-based samples (table 2), duplications in DISC1 were found in 7 patients with schizophrenia (7/410; 1.7%), 5 with bipolar type 1 disorder and 1 with bipolar type 2 disorder (6/314; 1.9%) and 6 control individuals (6/512; 1.2%). In the related Swedish samples (table 3), a DISC1 duplication was detected in 2 patients with schizophrenia (2/27; 7.4%), 3 with bipolar type 1, 2 with bipolar type 2 and 1 with bipolar not otherwise specified patient (6/102; 5.8%) and in 3 unaffected relatives. On further analysis of the pedigrees of the nuclear families, the CNV did not segregate with psychiatric disease in these Swedish samples: for the affected carriers as well as for the unaffected carriers, both affected and unaffected non-carrying first-degree relatives were found. An extra 22 target amplicons in this genomic region were subsequently analysed for fine mapping of the samples (‘newamp’, fig. 1; online suppl. table SI). The duplication in DISC1 in the Swedish samples has a maximum size of 107.4 kb and a minimum size of 98.8 kb and encompasses the first exon of DISC1 (fig. 1). The DISC1 duplication in the Scottish samples also encompasses the first exon of DISC1 and has a maximum size of 52 kb and a minimum size of 1.9 kb (table 4). In the Database of Genomic Variants (DGV; http://dgv.tcag.cag, update October 2014), 5 CNVs are reported at this genomic location: variation 3334, variation 48275, variation 48276, variation 30458 and variation 2350. Variations 30458 and 2350 (fig. 1) were detected in two HapMap CEU individuals, NA11830 (mother) and NA10856 (her son) [111,112]. We analysed the 90 HapMap CEU individuals with MAQ and allelic quantification assays and also detected the DISC1 duplication in NA11830 and NA10856, suggesting that these ‘three’ duplications are in fact one recurrent duplication. With a frequency of about 1.5%, this duplication is a common variant in the Northern Swedish population. This might be a result of genetic drift in an isolated population, but the duplication may be a common variant in the general (Caucasian/European) population, supported by its detection in 2 (related) HapMap CEU individuals. In the Northern Swedish population-based sample, the partial DISC1 duplication is not associated with schizophrenia (χ2 p value = 0.70), bipolar disorder (χ2 p value = 0.44) or with both disorders together (χ2 p value = 0.53). In addition, in the nuclear families, both affected and unaffected duplication carriers were found with both affected and unaffected first-degree relatives not carrying the duplication. So the partial DISC1 duplication is most likely not related to the disease phenotypes under study. Interestingly, in one Northern Swedish schizophrenia patient, 4 copies of the DISC1 CNV were found (4 copies in total, i.e. 2 normal copies and 2 additional). DNA from the parents was not available for typing. It is possible that the extra copy present in this patient does disturb DISC1 expression or protein function. Another interesting observation is that the duplication also covers a novel processed transcript (OTTHUMT00000095276, Vega gene ID RP11-17H4.2, fig. 1). This ‘gene’ has no open reading frame, and the function of the RNA transcript is yet unknown, but it might be implicated in the regulation of expression of (a) protein coding gene(s). Four functional copies of this DNA sequence might lead to a disturbingly elevated level of this non-coding RNA and interfere with its normal regulatory function.
NDE1 Copy Number Variants
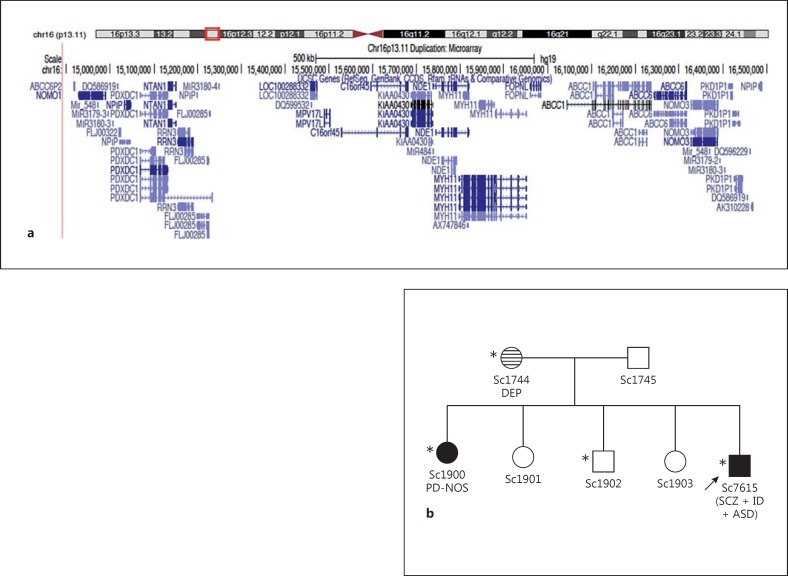
CNVs were detected in NDE1 in both the Scottish and Swedish sample sets (table 2). A deletion in NDE1 was detected by MAQ assay in one Scottish patient with a diagnosis of schizophrenia (1/647; 0.15%) and was not found in any other samples analysed in this study. Within the Scottish samples, we also found NDE1 duplications in 5 patient samples but in no Scottish controls (tables 2 and 4). Two of the Scottish patients with NDE1 duplications have a diagnosis of schizophrenia (Sc11034 and Sc6638), one has a diagnosis of ID and schizophrenia (Sc7292), one has a diagnosis of ID alone (Sc7474) and one patient Sc7615 has diagnoses of ID, autism and schizophrenia (table 4). The NDE1 duplications in 2 of the patients (Sc7292 and Sc7615) were also independently confirmed from GWAS, as they had been included in that analysis. Blood samples were subsequently obtained from the parents and all 4 siblings of Sc7615 and screened by microarray analysis using the Affymetrix CytoScan 750K array. This shows a single copy gain of the short arm of chromosome 16 at band p13.11. The duplication is approximately 1.65 Mb in size spanning 14,892,975-16,544,033 (NCBI Build 37). This 16p13.11 duplication was found to be maternally inherited and also present in two other siblings of Sc7615, one of whom has a history of psychosis (fig. 2). The mother of Sc7615 has a strong family history of schizophrenia and affective disorders. In the Swedish population sample, the duplication was detected in 1 patient with a diagnosis of schizoaffective disorder, bipolar type (1/120; 0.83%) and 3 control individuals (3/512; 0.59%) from the population-based sample, and in 1 patient with schizoaffective disorder, bipolar type from the related patients and relatives sample set (table 3). Fisher's exact test for these Swedish case-control studies results in non-significant p values of 0.27, 0.13 and 0.33, respectively. However, we believe that statistical analysis of the results of individual regions is not appropriate given how rare these CNVs are. In order to generate adequate statistical power, we would have to screen far larger sample sets than what was available. For these Swedish patients, no first-degree relative samples were available for testing, so segregation could not be verified. The duplication has a minimum size of 262 kb and encompasses NDE1 and other genes; the exact 3′ and 5′ boundaries are undefined (fig. 1).
Fig. 2.
a Microarray analysis of subject Sc1744 using the Affymetrix CytoScan 750K array shows a single copy gain of the short arm of chromosome 16 at band p13.11. The duplication is approximately 1.65 Mb in size spanning 14,892,975-16,544,033 (NCBI Build 37) and contains a number of genes. The figure was made by adding custom tracks to the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway). b Familial pedigree of Sc7615 (index individual indicated by an arrow). Samples from both parents and all 4 siblings of Sc7615 were screened by microarray. Filled symbols represent subjects affected with psychotic disorders [schizophrenia (SCZ), psychotic disorder not otherwise specified (PD-NOS)] and the horizontal-shaded symbol represents depressive disorder [depression (DEP)]. Other diagnoses include ID and ASD. The 16p13.11 duplication (carrier status denoted by an asterisk) was found to be maternally inherited and also present in two siblings of Sc7615 (Sc1900 and Sc1902). It is interesting to note that Sc1744 (the mother of Sc7615) has a strong family history of schizophrenia and affective disorders on her side of the family.
NDEL1 Copy Number Variants
A duplication in NDEL1 was found in 1 patient with a diagnosis of schizophrenia in the Swedish samples (1/410; 0.24%) but not found in any Scottish samples or in any Scottish or Swedish control individuals. It encompasses the whole gene and has a minimum size of 141.3 kb (fig. 1). The 5′ boundary is undefined; at the 3′ side, the duplication maximally extends 85 kb past exon 9.
CIT Copy Number Variants
A duplication in CIT was detected in 1 female Scottish patient with schizophrenia (1/647; 0.15%) but was not detected in the Swedish population-based sample or in any control samples from either Scotland or Sweden (tables 2 and 4). This is unsurprising given the low prevalence and de novo nature of this CNV.
No CNVs were found in PAFAH1B1, FEZ1, MAP1A or PDE4B in either the Scottish or Swedish samples screened in this study.
Allelic Quantification
Allelic quantification was used as a second independent method to confirm the 3 duplications detected in the MAQ analysis in the Swedish sample set. We selected 15 SNPs in the DISC1 duplication, 2 in the NDE1 and 8 in the NDEL1 duplication (fig. 1; online suppl. table SII). The duplications were confirmed in all the individuals.
Discussion
In this population-based CNV study, we detected duplications in DISC1, NDE1 and CIT in the Scottish sample set and in DISC1, NDE1 and NDEL1 in the Swedish unrelated samples. We found far fewer deletions, detecting only 2 in DISC1 and 1 in NDE1 in the Scottish samples. No deletions were detected in the unrelated population-based Swedish samples. Duplications may result in elevated mRNA and functional protein which might produce alterations at the cellular or pathway level. Alternatively, tandem duplications may disrupt the normal gene sequence, causing reduced expression of the gene and protein, or result in dysfunctional protein expression causing haploinsufficiency or loss-of-function.
A DISC1 duplication, encompassing the first exon, was found in patients and control individuals of the population-based sample, as well as in the related patients and their unaffected first-degree relatives and appears to be an inherited CNV. GWAS and next-generation sequencing approaches have not identified a clear association between DISC locus variants and schizophrenia, bipolar disorders and major depressive disorder, suggesting that there are no common variants at this locus to act independently to increase the risk of developing these disorders. These approaches have however raised the importance of rare genomic events such as CNVs conferring risk for major psychiatric disorders, but the presence of CNVs in DISC1 and its binding partners has received relatively little attention. Duplications in the DISC1 locus have been detected in 3 individuals (1 with schizophrenia and 2 controls) [113] and a 2-Mb duplication including 7 genes, including DISC1 and DISC2 has been described in two bothers with ASD, attention deficit hyperactivity disorder and mild ID [114]. Interestingly, we detected a deletion encoding intron 3 and 3 alternate exons of DISC1 in a Scottish patient with schizophrenia which was not found in any of the control samples. We also detected a deletion in TSNAX-DISC1 in a Scottish control sample. No deletions were observed in the Swedish samples. A 2-Mb maternally inherited deletion, encompassing TSNAX, DISC1 and DISC2, has been identified in a child with ASD and has also been shown to be associated with structural MRI changes including gliosis and bilateral cerebral white matter changes [115].
Duplications and deletions of a large region around NDE1 on chromosome 16p13.11 have previously been detected in CNV studies for autism and/or ID [81,116] and schizophrenia [62,67,117], and described as a potential risk factor for neuropsychiatric disorders. The (overlapping) CNVs probably result from non-allelic homologous recombination between chromosome 16-specific and relatively frequent low copy repeats termed LCR16 [116]. The deCODE genetics study examined 4,345 schizophrenia patients and 35,079 controls from 8 European populations for duplications and deletions at the 16p13.11 locus and found a threefold excess of duplications and deletions in schizophrenia cases compared with controls with duplications present in 0.30% of cases versus 0.09% of controls (p = 0.007) and deletions in 0.12% of cases and 0.04% of controls (p > 0.05) [117]. In the Scottish population sample in our study, we detected the duplication in 5 patients (2 with schizophrenia; 1 with schizophrenia and ID; 1 with ID alone and in 1 with schizophrenia, ID and ASD) and in no controls (0/352), and the deletion was detected in 1 patient with schizophrenia (1/647) and in no controls (0/352). It has been shown that approximately 16 protein-coding genes are expressed in this 2-Mb duplicated region (fig. 2), 10 of which have evidence of brain expression, of which 5 (ABCC1, NDE1, NOMO1, NTAN1 and PDXDC1) are thought to play a role in brain development [118].
The duplication in NDEL1 encompasses the complete gene, so potentially may cause an elevated expression of NDEL1 mRNA and intact protein. This CNV was only found in 1 patient with schizophrenia but not in unaffected individuals, so it could represent a rare but potentially pathogenic variation conferring susceptibility to schizophrenia. DNA from the parents of the patient was unavailable for typing, so we could not confirm whether this is a de novo or inherited variation. NDEL1 null mice are embryonic lethal, but mice with reduced levels of NDEL1 show neuronal migration defects resulting in abnormal organization of the cerebral cortex [119,120]. At a dosage-sensitive locus with deletion phenotypes, duplications have a high likelihood to also produce disease phenotypes [121]. While deletions of PAFAH1B1 are known to cause lissencephaly with severe brain malformations, individuals with PAFAH1B1 duplications showed mild brain malformations and moderate to severe developmental delay [122]. Duplication of NDEL1 with increased expression of the gene and protein may result in a phenotype with ‘milder’ affected brain structure and/or function such as psychiatric disorders. Interestingly, the observed duplication also encompasses part of MYH10. This gene encodes myosin heavy chain 10 non-muscle (myosin IIb) that is abundantly expressed in the brain and enriched in the post-synaptic density of neurons. It is a molecular motor that binds and contracts actin filaments and has been shown to play an essential role in axonal outgrowth, dendritic spine motility and synaptic function [123,124]. Interference with the expression of this gene or the function of its protein, maybe in combination with overexpression of NDEL1, possibly constitutes an interesting pathological mechanism implicated in schizophrenia. The low frequency of the variant nevertheless requires more patients and also more control individuals to be typed to confirm the relationship of this CNV with these disorders.
In published genome-wide CNV studies, 4 recurrent but rare deletions, at 22q11.2 (VCFS or Di George region), 1q21.1, 15q11.2 and 15q13.3, showed significant association with schizophrenia in large patient-control and trio samples [62,63]. An elegant translational study using human induced pluripotent stem cell-derived neural progenitor cells, carrying the 15q11.2 deletion, has recently shed light on how this CNV may associate with neurodevelopmental disorders [125]. As a result of haploinsufficiency of CYFIP1, a gene within 15q11.2 that encodes a subunit of the WAVE complex which regulates cytoskeletal dynamics, the neural progenitors carrying this mutation show deficits in adherens junctions and apical polarity. By engineering a similar mutation into a mouse model, they were able to demonstrate defective radial glial cell migration in the developing mouse cortex [125]. Other rare CNVs detected (almost) exclusively in patients were described as possible risk factors [61,62,63,64,65,66,67,126], supporting the hypothesis that rare variants with relatively high penetrance contribute to the aetiology of schizophrenia. In the literature, 5 of the CNVs associating with schizophrenia were duplications, raising the potential pathogenic role duplications have in disease causation: a de novo duplication at 15q13.1 spanning APBA2[61], a duplication at 7q21 including MAGI2[65], a duplication at 11p13 encompassing HIPK3[127], a de novo duplication at 12q24.23 comprising 8 exons of CIT and a de novo duplication at 14q32 spanning DICER1[64].
In conclusion, DISC1 and several of its interacting partners, which are proposed to be important for normal neuronal function, are likely candidates for involvement in schizophrenia, schizoaffective disorder and/or bipolar disorders. Our findings provide further evidence for involvement of these genes in neuropsychiatric disorders and also support the hypothesis of structural variation playing an important pathogenic role. Although the variants we have detected are rare, particularly those that are family specific, they are likely to be intractable to GWAS, which is of course optimised for the detection of common variants across populations. Variants in DISC1, which were originally implicated in a single Scottish family, have only ever reached genome-wide significance levels when studied using linkage analysis, consistent with their rare frequency and large effect size. Although exceptionally rare, they may nonetheless be biologically very informative. The consequences of these CNVs on transcript and protein level should be further investigated. The functional implication of these variants in mental illness and the mechanism of disease causation remain elusive, although the potential of investigating this in neuronal cell types derived from human induced pluripotent stem cells holds much promise.
Statement of Ethics
All human subjects participated after giving written informed consent or assent after receiving a full explanation of the study. The study was approved by the Multi-Centre Research Ethics Committee for Scotland and by the regional Medical Ethics Committees of the Universities of Umeå (Sweden) and Antwerp (Belgium).
Disclosure Statement
The authors declare that there is no conflict of interest with regard to the findings reported in this paper.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgements
We would like to thank all of the patients, relatives and control individuals who participated in the study. We are indebted to the late Prof. Walter Muir, Chair of Developmental Psychiatry and Honorary Consultant in Learning Disability Psychiatry, University of Edinburgh, who initiated these studies and whose work was dedicated to the welfare of the patients who generously participated. We are also grateful to Mrs. Pat Malloy for her assistance with DNA collection and MAQ assays screening of the Scottish samples. The Scottish sample collection was supported by a grant from the Chief Scientist Office (CSO), part of the Scottish Government Health and Social Care Directorates. This research was funded by grants from the CSO to B.S.P. (grant CZB/4/610), The Academy of Medical Sciences/Wellcome Trust to M.J. (grant R41455) and The RS Macdonald Charitable Trust (grant D21419 together with J.H.), the Swedish Research Council (grants 2003-5158 and 2006-4472), the Medical Faculty, Umeå University, and the County Councils of Västerbotten and Norrbotten, Sweden, as well as by grants from the Fund for Scientific Research Flanders (FWO-F), the Industrial Research Fund (IWT) and the Special Research Fund of the University of Antwerp, Belgium. M.J. is funded by a Wellcome Trust Clinical Research Fellowship for MB PhD graduates (R42811). We acknowledge the contribution of the personnel of the VIB Genetic Service Facility (http://www.vibgeneticservicefacility.be/) for the genetic analysis of the Swedish samples. Research nurses Gunnel Johansson, Lotta Kronberg, Tage Johansson and Lisbeth Bertilsson are thankfully acknowledged for their help and expertise. The Betula Study was funded by the Swedish Research Council (grants 345-2003-3883 and 315-2004-6977). We also acknowledge the contribution by the staff in the Betula project.
References
- 1.Berrettini W. Evidence for shared susceptibility in bipolar disorder and schizophrenia. Am J Med Genet C Semin Med Genet. 2003;123C:59–64. doi: 10.1002/ajmg.c.20014. [DOI] [PubMed] [Google Scholar]
- 2.Craddock N, O'Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32:9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry. 2008;64:89–97. doi: 10.1016/j.biopsych.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lichtenstein P, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–9. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayhan Y, et al. Animal models of gene-environment interactions in schizophrenia. Behav Brain Res. 2009;204:274–81. doi: 10.1016/j.bbr.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craddock N, Forty L. Genetics of affective (mood) disorders. Eur J Hum Genet. 2006;14:660–668. doi: 10.1038/sj.ejhg.5201549. [DOI] [PubMed] [Google Scholar]
- 7.Gershon ES, Alliey-Rodriguez N, Liu C. After GWAS: searching for genetic risk for schizophrenia and bipolar disorder. Am J Psychiatry. 2011;168:253–256. doi: 10.1176/appi.ajp.2010.10091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassett AS, Scherer SW, Brzustowicz LM. Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry. 2010;167:899–914. doi: 10.1176/appi.ajp.2009.09071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.St Clair D, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 10.Blackwood DH, et al. Schizophrenia and affective disorders - cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millar JK, et al. Genomic structure and localisation within a linkage hotspot of Disrupted In Schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol Psychiatry. 2001;6:173–178. doi: 10.1038/sj.mp.4000784. [DOI] [PubMed] [Google Scholar]
- 12.Ekelund J, et al. Chromosome 1 loci in Finnish schizophrenia families. Hum Mol Genet. 2001;10:1611–1617. doi: 10.1093/hmg/10.15.1611. [DOI] [PubMed] [Google Scholar]
- 13.Ekelund J, et al. Replication of 1q42 linkage in Finnish schizophrenia pedigrees. Mol Psychiatry. 2004;9:1037–1041. doi: 10.1038/sj.mp.4001536. [DOI] [PubMed] [Google Scholar]
- 14.Hwu HG, et al. Linkage of schizophrenia with chromosome 1q loci in Taiwanese families. Mol Psychiatry. 2003;8:445–452. doi: 10.1038/sj.mp.4001235. [DOI] [PubMed] [Google Scholar]
- 15.Curtis D, et al. Genome scan of pedigrees multiply affected with bipolar disorder provides further support for the presence of a susceptibility locus on chromosome 12q23-q24, and suggests the presence of additional loci on 1p and 1q. Psychiatr Genet. 2003;13:77–84. doi: 10.1097/01.ypg.0000056684.89558.d2. [DOI] [PubMed] [Google Scholar]
- 16.Brzustowicz LM, et al. Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am J Hum Genet. 2004;74:1057–1063. doi: 10.1086/420774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macgregor S, et al. A genome scan and follow-up study identify a bipolar disorder susceptibility locus on chromosome 1q42. Mol Psychiatry. 2004;9:1083–1090. doi: 10.1038/sj.mp.4001544. [DOI] [PubMed] [Google Scholar]
- 18.Hennah W, et al. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003;12:3151–3159. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- 19.Hamshere ML, et al. Genomewide linkage scan in schizoaffective disorder: significant evidence for linkage at 1q42 close to DISC1, and suggestive evidence at 22q11 and 19p13. Arch Gen Psychiatry. 2005;62:1081–1088. doi: 10.1001/archpsyc.62.10.1081. [DOI] [PubMed] [Google Scholar]
- 20.Cannon TD, et al. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- 21.Callicott JH, et al. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci USA. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson PA, et al. Association between genotype at an exonic SNP in DISC1 and normal cognitive aging. Neurosci Lett. 2005;389:41–45. doi: 10.1016/j.neulet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Maeda K, et al. Differential expression of disrupted-in-schizophrenia (DISC1) in bipolar disorder. Biol Psychiatry. 2006;60:929–935. doi: 10.1016/j.biopsych.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 24.Qu M, et al. Positive association of the Disrupted-in-Schizophrenia-1 gene (DISC1) with schizophrenia in the Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:266–270. doi: 10.1002/ajmg.b.30322. [DOI] [PubMed] [Google Scholar]
- 25.Palo OM, et al. Association of distinct allelic haplotypes of DISC1 with psychotic and bipolar spectrum disorders and with underlying cognitive impairments. Hum Mol Genet. 2007;16:2517–2528. doi: 10.1093/hmg/ddm207. [DOI] [PubMed] [Google Scholar]
- 26.Chen QY, et al. Case-control association study of Disrupted-in-Schizophrenia-1 (DISC1) gene and schizophrenia in the Chinese population. J Psychiatr Res. 2007;41:428–434. doi: 10.1016/j.jpsychires.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Perlis RH, et al. Family-based association study of lithium-related and other candidate genes in bipolar disorder. Arch Gen Psychiatry. 2008;65:53–61. doi: 10.1001/archgenpsychiatry.2007.15. [DOI] [PubMed] [Google Scholar]
- 28.Saetre P, et al. Association between a disrupted-in-schizophrenia 1 (DISC1) single nucleotide polymorphism and schizophrenia in a combined Scandinavian case-control sample. Schizophr Res. 2008;106:237–241. doi: 10.1016/j.schres.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 29.Chubb JE, et al. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 30.Gasperoni TL, et al. Genetic linkage and association between chromosome 1q and working memory function in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2003;116B:8–16. doi: 10.1002/ajmg.b.10757. [DOI] [PubMed] [Google Scholar]
- 31.Hennah W, et al. A haplotype within the DISC1 gene is associated with visual memory functions in families with a high density of schizophrenia. Mol Psychiatry. 2005;10:1097–1103. doi: 10.1038/sj.mp.4001731. [DOI] [PubMed] [Google Scholar]
- 32.Burdick KE, et al. DISC1 and neurocognitive function in schizophrenia. Neuroreport. 2005;16:1399–1402. doi: 10.1097/01.wnr.0000175248.25535.f6. [DOI] [PubMed] [Google Scholar]
- 33.Whalley HC, et al. Effects of a mis-sense DISC1 variant on brain activation in two cohorts at high risk of bipolar disorder or schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:343–353. doi: 10.1002/ajmg.b.32035. [DOI] [PubMed] [Google Scholar]
- 34.Di Giorgio A, et al. Association of the SerCys DISC1 polymorphism with human hippocampal formation gray matter and function during memory encoding. Eur J Neurosci. 2008;28:2129–2136. doi: 10.1111/j.1460-9568.2008.06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomppo L, et al. DISC1 conditioned GWAS for psychosis proneness in a large Finnish birth cohort. PLoS One. 2012;7:e30643. doi: 10.1371/journal.pone.0030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamiya A, Sedlak TW, Pletnikov MV. DISC1 pathway in brain development: exploring therapeutic targets for major psychiatric disorders. Front Psychiatry. 2012;3:25. doi: 10.3389/fpsyt.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer KD, Morris JA. Immunohistochemical analysis of Disc1 expression in the developing and adult hippocampus. Gene Expr Patterns. 2008;8:494–501. doi: 10.1016/j.gep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Duan X, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipska BK, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum Mol Genet. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 40.Ozeki Y, et al. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci USA. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyoshi K, et al. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 2003;8:685–694. doi: 10.1038/sj.mp.4001352. [DOI] [PubMed] [Google Scholar]
- 42.Kamiya A, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 43.Koike H, et al. Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc Natl Acad Sci USA. 2006;103:3693–3697. doi: 10.1073/pnas.0511189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clapcote SJ, et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 45.Hikida T, et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci USA. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, et al. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc Natl Acad Sci USA. 2007;104:18280–18285. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen S, et al. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pletnikov MV, et al. Enlargement of the lateral ventricles in mutant DISC1 transgenic mice. Mol Psychiatry. 2008;13:115. doi: 10.1038/sj.mp.4002144. [DOI] [PubMed] [Google Scholar]
- 49.Pletnikov MV, et al. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008;13:173–186. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- 50.Millar JK, Christie S, Porteous DJ. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun. 2003;311:1019–1025. doi: 10.1016/j.bbrc.2003.10.101. [DOI] [PubMed] [Google Scholar]
- 51.Morris DW, et al. No evidence for association of the dysbindin gene [DTNBP1] with schizophrenia in an Irish population-based study. Schizophr Res. 2003;60:167–172. doi: 10.1016/s0920-9964(02)00527-3. [DOI] [PubMed] [Google Scholar]
- 52.Camargo LM, et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 53.Bradshaw NJ, et al. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem Biophys Res Commun. 2008;377:1091–1096. doi: 10.1016/j.bbrc.2008.10.120. [DOI] [PubMed] [Google Scholar]
- 54.Bradshaw NJ, Porteous DJ. DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology. 2012;62:1230–1241. doi: 10.1016/j.neuropharm.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alaerts M, Del-Favero J. Searching genetic risk factors for schizophrenia and bipolar disorder: learn from the past and back to the future. Hum Mutat. 2009;30:1139–1152. doi: 10.1002/humu.21042. [DOI] [PubMed] [Google Scholar]
- 57.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 58.Rocha D, et al. Seventh international meeting on single nucleotide polymorphism and complex genome analysis: ‘ever bigger scans and an increasingly variable genome’. Hum Genet. 2006;119:451–456. doi: 10.1007/s00439-006-0151-z. [DOI] [PubMed] [Google Scholar]
- 59.Nadeau JH, Lee C. Genetics: copies count. Nature. 2006;439:798–799. doi: 10.1038/439798a. [DOI] [PubMed] [Google Scholar]
- 60.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 61.Kirov G, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 62.International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu B, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 65.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 66.Need AC, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirov G, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reiner O, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 69.Lo Nigro C, et al. Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome. Hum Mol Genet. 1997;6:157–164. doi: 10.1093/hmg/6.2.157. [DOI] [PubMed] [Google Scholar]
- 70.Uyanik G, et al. Location and type of mutation in the LIS1 gene do not predict phenotypic severity. Neurology. 2007;69:442–447. doi: 10.1212/01.wnl.0000266629.98503.d0. [DOI] [PubMed] [Google Scholar]
- 71.Hirotsune S, et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 72.Gambello MJ, et al. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hennah W, et al. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum Mol Genet. 2007;16:453–462. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- 74.Bradshaw NJ, et al. PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1. J Neurosci. 2011;31:9043–9054. doi: 10.1523/JNEUROSCI.5410-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brandon NJ, et al. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 76.Burdick KE, et al. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum Mol Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Houlihan SL, Feng Y. The scaffold protein Nde1 safeguards the brain genome during S phase of early neural progenitor differentiation. Elife. 2014;3:e03297. doi: 10.7554/eLife.03297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alkuraya FS, et al. Human mutations in NDE1 cause extreme microcephaly with lissencephaly [corrected] Am J Hum Genet. 2011;88:536–547. doi: 10.1016/j.ajhg.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bakircioglu M, et al. The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am J Hum Genet. 2011;88:523–535. doi: 10.1016/j.ajhg.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mefford HC, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ullmann R, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 82.Williams NM, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Kovel CG, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kang E, et al. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72:559–571. doi: 10.1016/j.neuron.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lipska BK, et al. Functional genomics in postmortem human brain: abnormalities in a DISC1 molecular pathway in schizophrenia. Dialogues Clin Neurosci. 2006;8:353–357. doi: 10.31887/DCNS.2006.8.3/blipska. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamada K, et al. Association analysis of FEZ1 variants with schizophrenia in Japanese cohorts. Biol Psychiatry. 2004;56:683–690. doi: 10.1016/j.biopsych.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 87.Sakae N, et al. Mice lacking the schizophrenia-associated protein FEZ1 manifest hyperactivity and enhanced responsiveness to psychostimulants. Hum Mol Genet. 2008;17:3191–3203. doi: 10.1093/hmg/ddn215. [DOI] [PubMed] [Google Scholar]
- 88.Noiges R, et al. Microtubule-associated protein 1A (MAP1A) and MAP1B: light chains determine distinct functional properties. J Neurosci. 2002;22:2106–2114. doi: 10.1523/JNEUROSCI.22-06-02106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Halpain S, Dehmelt L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006;7:224. doi: 10.1186/gb-2006-7-6-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang W, Benson DL. Targeting and clustering citron to synapses. Mol Cell Neurosci. 2006;31:26–36. doi: 10.1016/j.mcn.2005.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reese ML, et al. The guanylate kinase domain of the MAGUK PSD-95 binds dynamically to a conserved motif in MAP1a. Nat Struct Mol Biol. 2007;14:155–163. doi: 10.1038/nsmb1195. [DOI] [PubMed] [Google Scholar]
- 92.Lyons-Warren A, et al. Evidence of association between bipolar disorder and Citron on chromosome 12q24. Mol Psychiatry. 2005;10:807–809. doi: 10.1038/sj.mp.4001703. [DOI] [PubMed] [Google Scholar]
- 93.Millar JK, et al. Disrupted in schizophrenia 1 (DISC1): subcellular targeting and induction of ring mitochondria. Mol Cell Neurosci. 2005;30:477–484. doi: 10.1016/j.mcn.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 94.Pickard BS, et al. The PDE4B gene confers sex-specific protection against schizophrenia. Psychiatr Genet. 2007;17:129–133. doi: 10.1097/YPG.0b013e328014492b. [DOI] [PubMed] [Google Scholar]
- 95.Fatemi SH, et al. Phosphodiesterase-4A expression is reduced in cerebella of patients with bipolar disorder. Psychiatr Genet. 2008;18:282–288. doi: 10.1097/YPG.0b013e3283060fb8. [DOI] [PubMed] [Google Scholar]
- 96.Numata S, et al. Positive association of the PDE4B (phosphodiesterase 4B) gene with schizophrenia in the Japanese population. J Psychiatr Res. 2008;43:7–12. doi: 10.1016/j.jpsychires.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 97.O'Donnell BF, et al. Neural synchronization deficits to auditory stimulation in bipolar disorder. Neuroreport. 2004;15:1369–1372. doi: 10.1097/01.wnr.0000127348.64681.b2. [DOI] [PubMed] [Google Scholar]
- 98.Kanes SJ, et al. Rolipram: a specific phosphodiesterase 4 inhibitor with potential antipsychotic activity. Neuroscience. 2007;144:239–246. doi: 10.1016/j.neuroscience.2006.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Siuciak JA, et al. Behavioral and neurochemical characterization of mice deficient in the phosphodiesterase-4B (PDE4B) enzyme. Psychopharmacology (Berl) 2008;197:115–126. doi: 10.1007/s00213-007-1014-6. [DOI] [PubMed] [Google Scholar]
- 100.Zhang HT, et al. Anxiogenic-like behavioral phenotype of mice deficient in phosphodiesterase 4B (PDE4B) Neuropsychopharmacology. 2008;33:1611–1623. doi: 10.1038/sj.npp.1301537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders: DSM-IV-TR. Arlington: American Psychiatric Publishing; 2000. [Google Scholar]
- 102.McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system. Arch Gen Psychiatry. 1991;48:764–770. doi: 10.1001/archpsyc.1991.01810320088015. [DOI] [PubMed] [Google Scholar]
- 103.Lecrubier Y, et al. The M.I.N.I. International Neuropsychiatric Interview (M.I.N.I.) A Short Diagnostic Structured Interview: Reliability and Validity According to the CIDI. Eur Psychiatry. 1997;12:224–231. [Google Scholar]
- 104.Nurnberger JI Jr, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. discussion 863-864. [DOI] [PubMed] [Google Scholar]
- 105.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wing JK, et al. SCAN. Schedules for Clinical Assessment in Neuropsychiatry. Arch Gen Psychiatry. 1990;47:589–593. doi: 10.1001/archpsyc.1990.01810180089012. [DOI] [PubMed] [Google Scholar]
- 107.Nilsson L, et al. The Betula prospective cohort study: memory, health, and aging. Aging Neuropsychol Cogn. 1997;4:1–32. [Google Scholar]
- 108.Alaerts M, et al. Support for NRG1 as a susceptibility factor for schizophrenia in a northern Swedish isolated population. Arch Gen Psychiatry. 2009;66:828–837. doi: 10.1001/archgenpsychiatry.2009.82. [DOI] [PubMed] [Google Scholar]
- 109.Suls A, et al. Microdeletions involving the SCN1A gene may be common in SCN1A-mutation-negative SMEI patients. Hum Mutat. 2006;27:914–920. doi: 10.1002/humu.20350. [DOI] [PubMed] [Google Scholar]
- 110.Sleegers K, et al. APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–2983. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- 111.Perry GH, et al. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Buizer-Voskamp JE, et al. Genome-wide analysis shows increased frequency of copy number variation deletions in Dutch schizophrenia patients. Biol Psychiatry. 2011;70:655–662. doi: 10.1016/j.biopsych.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Crepel A, et al. DISC1 duplication in two brothers with autism and mild mental retardation. Clin Genet. 2010;77:389–394. doi: 10.1111/j.1399-0004.2009.01318.x. [DOI] [PubMed] [Google Scholar]
- 115.Williams JM, et al. A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am J Med Genet A. 2009;149A:1758–1762. doi: 10.1002/ajmg.a.32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hannes FD, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ingason A, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Paciorkowski AR, et al. Deletion 16p13.11 uncovers NDE1 mutations on the non-deleted homolog and extends the spectrum of severe microcephaly to include fetal brain disruption. Am J Med Genet A. 2013;161A:1523–1530. doi: 10.1002/ajmg.a.35969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shu T, et al. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 120.Sasaki S, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39(suppl):S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 122.Bi W, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Turney SG, Bridgman PC. Laminin stimulates and guides axonal outgrowth via growth cone myosin II activity. Nat Neurosci. 2005;8:717–719. doi: 10.1038/nn1466. [DOI] [PubMed] [Google Scholar]
- 124.Ryu J, et al. A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron. 2006;49:175–182. doi: 10.1016/j.neuron.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 125.Yoon KJ, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15:79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Van den Bossche MJ, et al. Rare copy number variants in neuropsychiatric disorders: specific phenotype or not? Am J Med Genet B Neuropsychiatr Genet. 2012;159B:812–822. doi: 10.1002/ajmg.b.32088. [DOI] [PubMed] [Google Scholar]
- 127.Mizuguchi T, et al. Microarray comparative genomic hybridization analysis of 59 patients with schizophrenia. J Hum Genet. 2008;53:914–919. doi: 10.1007/s10038-008-0327-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data