Abstract
Comprehensive characterization of protein glycosylation is critical for understanding the structure and function of glycoproteins. However, due to the complexity and heterogeneity of glycoprotein conformations, current glycoprotein analyses focus mainly on either the de-glycosylated glycosylation site (glycosite)-containing peptides or the released glycans. Here, we describe a chemoenzymatic method called solid phase extraction of N-linked glycans and glycosite-containing peptides (NGAG) for the comprehensive characterization of glycoproteins that is able to determine glycan heterogeneity for individual glycosites in addition to providing information about the total N-linked glycan, glycosite-containing peptide and glycoprotein content of complex samples. The NGAG method can also be applied to quantitatively detect glycoprotein alterations in total and site-specific glycan occupancies.
Protein glycosylation is one of the most common and important protein modifications1,2. It plays fundamental roles in many biological processes and in the pathological progression of many diseases, and glycoproteins are a major class of current therapeutic targets and clinical biomarkers3. Protein glycosylation is also the most structurally complicated and diverse type of protein modification1. Each glycoprotein can contain multiple glycosites, and each glycosite can be modified by multiple different glycans1. Glycoprotein characterization often entails determining the fraction of protein that is glycosylated, where the glycosites are located, the occupancy of each glycosite and which glycan structures are associated with each glycosite4–6. The complexity of glycosylation heterogeneity poses great challenges for the identification of specific glycosylation patterns that are associated with different biological functions and diseases.
In the last decade, mass spectrometry (MS)-based glycoproteomics and glycomics have become emerging technologies for the large-scale characterization of glycoproteins4–7. These methods include (i) physical-chemical methods such as chromatography8,9, peptide sequencing10 and electrophoresis11; (ii) methods using affinity-based reagents such as lectins and antibodies5,12,13; and (iii) tagging glycan moieties using enzymes14, chemicals4,15 or metabolic labeling16. Recent developments in MS-based technologies also enable intact glycopeptide analysis17–25. These techniques have provided valuable insights into the interdependence of glycoprotein structure and protein function. However, these technologies focus mainly on the analysis of either released glycans or deglycosylated peptides, or concentrate on glycopeptides, and lack the capability to simultaneously analyze all relevant parameters such as total glycans, glycoproteins and their glycosites, as well as site-specific glycosylation, especially in complex biological or clinical samples.
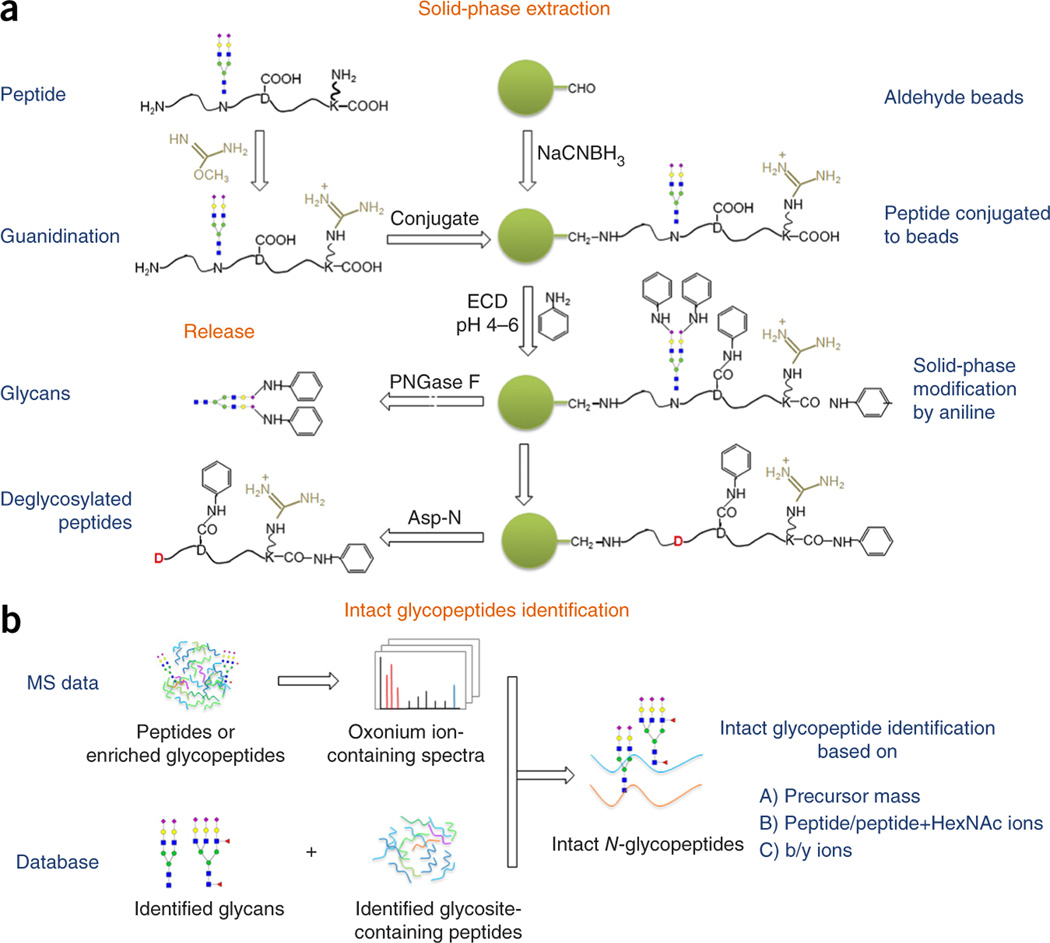
In this study, we developed a chemoenzymatic method termed solid phase extraction of NGAG for the comprehensive characterization of N-linked glycoproteins. The NGAG method includes seven steps (Fig. 1a and Supplementary Fig. 1). (i) Protein samples were digested into peptides, followed by guanidination to block the ε-amino groups on the side chain of lysine residues26. (ii) Peptides were covalently conjugated to an aldehyde-functionalized solid support through their N termini (α-amino groups). (iii) Carboxyl groups of aspartic acid (D), glutamic acid (E), and peptide C termini were modified with aniline. At the same time sialic acids on the sialylated glycopeptides were also modified by aniline to facilitate mass spectrometric detection of sialylated glycans15. (iv) N-linked glycans were released from the solid support by PNGase F digestion, in parallel with conversion of asparagine residues at N-linked glycosites to aspartic acid residues by deamination. (v) Glycosite-containing peptides with aspartic residues at N-glycosites were specifically released from solid phase by Asp-N digestion. Asp-N specifically cleaves peptide bonds N-terminal to aspartic acid residues. However, because all original aspartic acid residues of the peptides were modified with esterification, only newly generated aspartic acid residues at the N-glycosites after N-glycan release were cleaved by Asp-N. (vi) The released N-glycans and glycosite-containing peptides were identified by MS. (vii) The intact N-glycopeptides were then reconstructed by assigning the oxonium ions-containing MS/MS spectra extracted from global proteomic or enriched glycopeptide data to the glycopeptide candidate database, which was comprised of all possible combinations of identified glycans and glycosite-containing peptides (Fig. 1b).
Figure 1.
Workflow of solid-phase extraction of NGAG. (a) Principles of the NGAG method and chemical modifications involved in the isolation of N-glycans and glycosite-containing peptides. (b) Identification of intact glycopeptides from proteomic or enriched glycopeptide data using glycan and glycosite-containing peptide databases.
First, we tested our method on bovine fetuin and determined glycan content, glycosites, total glycoproteins and site-specific glycosylation in a single experiment (Supplementary Results and Discussion, Supplementary Figs. 2 and 3, and Supplementary Tables 1 and 2).
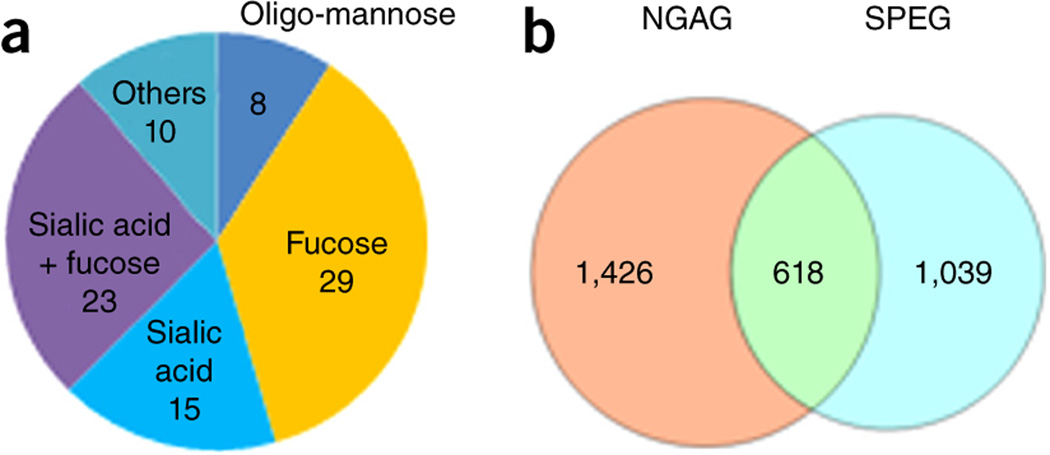
To evaluate the applicability of the NGAG method for the comprehensive analysis of N-glycoproteins from complex samples, we applied the NGAG method to analyze the influence of the N-linked glycosylation inhibitor tunicamycin on the glycosylation patterns of glycoproteins from OVCAR-3 ovarian cancer cells. Briefly, N-linked glycans and glycosite-containing peptides were extracted separately from either unlabeled OVCAR-3 cells with/without tunicamycin treatment (1 µM for 48 h) or Stable Isotope Labeling by Amino acids in Cell culture (SILAC)-labeled cells, using the NGAG method. We identified N-glycans with 85 different compositions from unlabeled OVCAR-3 cells ± tunicamycin by matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF)-MS and liquid chromatography (LC)-MS/MS (Supplementary Figs. 4 and 5 and Supplementary Table 3), of which 52 contained fucose, 38 contained sialic acid, 23 contained both fucose and sialic acid residues and 8 were of the oligo-mannose type (Fig. 2a). Glycosite-containing peptides from each unlabeled cell population ± tunicamycin were mixed with an equivalent amount of heavy labeled glycosite-containing peptides isolated from heavy SILAC-labeled cells, and each glycosite-containing peptide mixture was fractionated into 12 fractions by basic reversed-phase liquid chromatography (bRPLC) before LC-MS/MS analysis of each fraction. From these two samples, we identified 2,044 unique glycosite-containing peptides with an N-X-S/T motif at their N termini at a 1% false-discovery rate (FDR) (Fig. 2b and Supplementary Table 4).
Figure 2.
Extraction and identification of N-linked glycans and glycosite-containing peptides from OVCAR-3 cells using the NGAG method. (a) Compositions of the 85 identified N-glycans in OVCAR-3 cells. (b) Glycosite-containing peptides identified from OVCAR-3 cells using the NGAG and SPEG methods.
We previously reported a hydrazide chemistry method for the identification of N-linked glycosites using mass spectrometric analysis of PNGase F released peptides from immobilized glycopeptides by glycans (SPEG)4,27. To determine whether there were differences in the glycosites identified using NGAG from those by the SPEG method, an equivalent amount of cell lysate–derived proteins and the same LC-MS/MS analysis method as used in the NGAG method were also used for glycosite analysis by the SPEG method. This resulted in identification of 1,657 unique N-glycosite-containing peptides (Supplementary Table 5). Among these glycosite-containing peptides, 618 overlapped with glycosites identified using the NGAG method. In total, 3,083 unique N-glycosite-containing peptides from 1,473 glycoproteins were identified from OVCAR-3 cells using the SPEG and NGAG methods (Fig. 2b and Supplementary Table 6).
We next used the identified glycans and glycosite-containing peptides to construct a sample-specific database (consisting of all possible combinations of identified glycans and glycosite-containing peptides) for assignment of the tandem MS spectra of intact glycopeptides. The intact glycopeptides were enriched from the tryptic peptides of OVCAR-3 cells using a hydrophilic interaction column (HILIC) and analyzed by LC-MS/MS. The oxonium ion-containing MS/MS spectra were extracted and assigned to intact glycopeptides in the database using the precursor mass matching option in the GPQuest software28. With filtering based on the presence of peptide+HexNAc and/or peptide ions, as well as ≥7 observed b and y ions (1% FDR), we could assign 4,562 oxonium ion–containing spectra to 1,562 unique glycopeptides (containing 518 glycosites and 81 glycans, Supplementary Table 7).
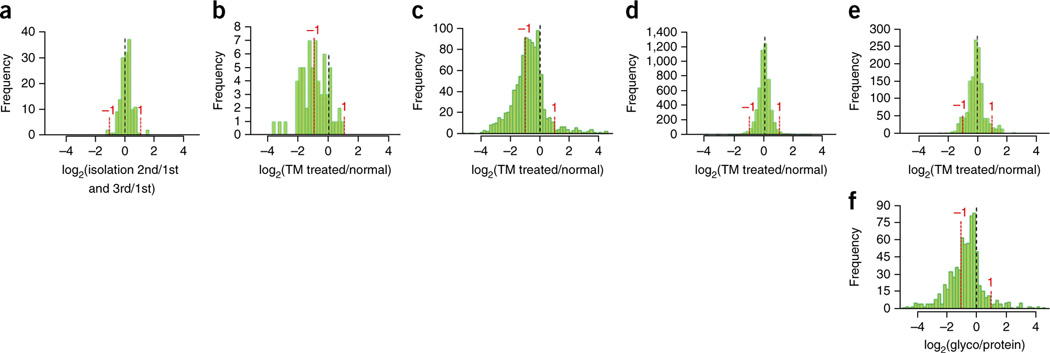
The NGAG method was then used to determine quantitative glycoprotein changes in cells treated with tunicamycin. We first determined the precision of glycoprotein quantitation for complex samples using NGAG. Among the glycans and glycosite-containing peptides of OVCAR-3 cells quantified from triplicate NGAG isolations, 97.3% of glycans (Fig. 3a) and 95.4% of glycosite-containing peptides (Supplementary Results and Discussion and Supplementary Figs. 6–9) varied less than twofold. Therefore, a twofold change in either direction was used as a reasonable cutoff for detecting glycan and glycosite-containing peptide alterations between tunicamycin-treated and untreated OVCAR-3 cells.
Figure 3.
Quantification of glycans and glycosite-containing peptides in tunicamycin-treated OVCAR-3 cells. The distribution of the log2(+tunicamycin/−tunicamycin ratios) was used to show the alteration of glycosylation in tunicamycin-treated cells (x axis). Y axis indicates the number of glycans, peptides or proteins at each alteration range. (a) Glycans quantified among triplicate isolations of untreated cells using a label-free approach. (b) Glycans quantified between tunicamycin-treated and untreated OVCAR-3 cells using a label-free approach. (c) Glycosite-containing peptides quantified between tunicamycin-treated and untreated cells using the NGAG method. (d) Total protein quantification from proteomic data. (e) Total glycoprotein quantification using nonglycosylated peptides identified from proteomic data. (f) Distribution of the log2(glycosite/protein) in tunicamycin-treated cells. The dotted lines indicate the unchanged glycans (a, b), glycosites (c), total proteins (d), total glycoproteins (e) or glycosite occupancies (f) (log2 = 0).
The quantification results indicated that 31 out of 64 quantifiable glycans (48.4%) and 359 out of 998 quantifiable glycosite-containing peptides (36%) isolated by the NGAG method decreased more than twofold in the tunicamycin-treated cells (Fig. 3b,c). These data indicated that the levels of both N-glycans and glycosite-containing peptides decreased in tunicamycin-treated OVCAR-3 cells. To determine whether the decrease of glycosite-containing peptides occurred at the protein expression or glycosylation occupancy levels, we identified and quantified the global proteins from the same treated cells. We identified 87,247 unique peptides from 7,764 protein groups, and the relative abundance of 96.8% (5,953/6,149) of these proteins changed less than twofold after tunicamycin treatment (Fig. 3d). Using only nonglycosylated peptides for quantification, we found that the relative abundances of the majority of glycoproteins (92.4%, 837/906) were also within a twofold change (Fig. 3e). However, when we quantified the N-glycosite-containing peptides, we found that 559 of these peptides exhibited greater than twofold decreases (ratio < 0.5) in addition to the changes in total glycoprotein levels (Fig. 3f). These data indicate that the decrease of glycosite-containing peptides in tunicamycin-treated cells was mainly caused by reduced glycan occupancy (stoichiometry) of the glycosites. This result is consistent with the known inhibitory role of tunicamycin to N-linked glycosylation. The results also showed that the extent of glycosylation alteration could depend on both glycosites and glycans as different glycans and glycosites exhibited different changes (Fig. 3b,c).
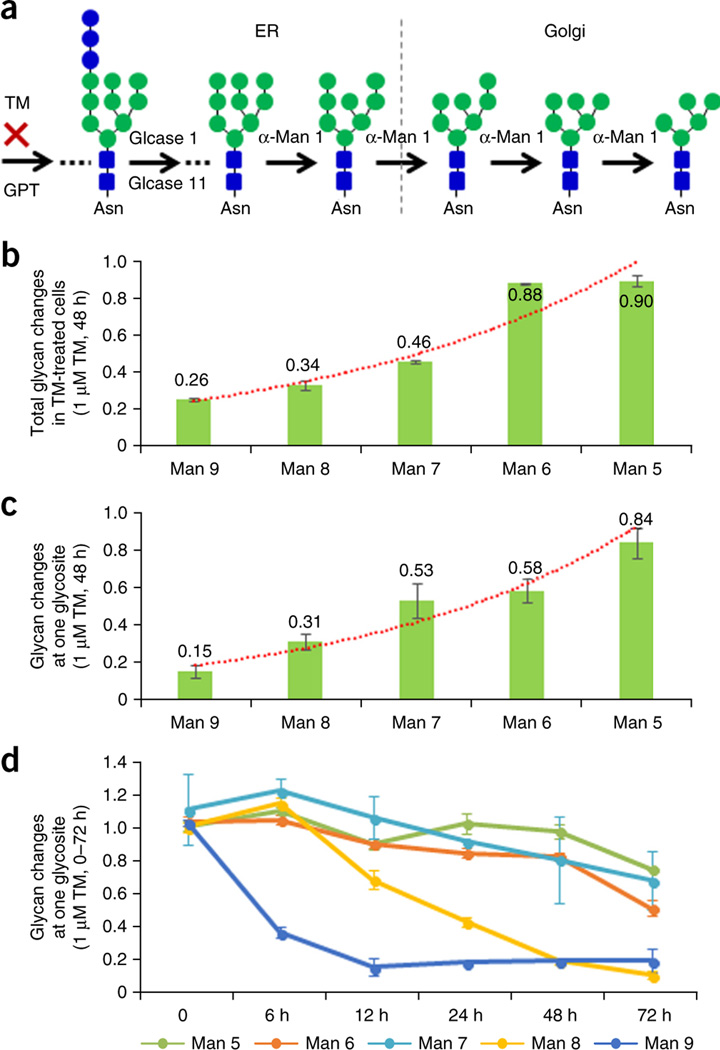
Diverse glycan alterations were observed in tunicamycin-treated OVCAR-3 cells at the total glycan level (Fig. 3b). To further investigate the pattern of glycan alteration in tunicamycin-treated cells, we focused on the five oligo-mannose glycans (Man5–Man9) for further analysis. Protein glycosylation initiates with the transfer of the glycan precursor Glc3Man9GlcNAc2 from dolichol pyrophosphate to Asn-X-Ser/Thr in newly synthesized glycoproteins29. The glycan precursor is trimmed to Man9, then to Man8 and gradually down to Man 5 (ref. 29; Fig. 4a). Our glycomic results showed that five oligo-mannose glycans were differentially altered with tunicamycin treatment, and the alteration of these five oligo-mannose glycans (Man9–Man5) was apparently associated with the location of the glycan in the N-glycan biosynthesis pathway (Fig. 4b). The trend in the tunicamycin inhibition was reflected by a readily observable decrease at Man9 (0.26-fold), to a gradual lesser degree of change at Man5 (0.90-fold) (Fig. 4b). To determine whether this phenomenon could be observed when glycans were attached to a specific glycosite, we performed a site-specific glycosylation analysis using the intact glycopeptide quantification data from the global proteomic data. Our results showed that not all glycosites exhibited this alteration trend. This indicated that alterations of glycosylation occurred in a glycosite-specific manner. Therefore, we reasoned that the observed overall glycan changes might be mainly reflected by the high abundance of glycoproteins.
Figure 4.
Differential alterations of oligo-mannose glycans in tunicamycin-treated OVCAR-3 cells. (a) Glycosylation pathway of oligo-mannose glycan synthesis. (b) Total oligo-mannose glycan changes in OVCAR-3 cells after 48 h of tunicamycin treatment. (c) Site-specific oligo-mannose glycan changes at endoplasmin glycosite Asn217 in OVCAR-3 cells after 48 h of tunicamycin treatment. (d) The dynamics of oligo-mannose glycan changes at endoplasmin glycosite Asn217 in OVCAR-3 cells treated with tunicamycin. Error bars, means ± s.d.
We then selected glycosite Asn217 of Endoplasmin, which had the highest number of spectra at both the glycosite-containing peptide and the intact glycopeptide levels. This glycosite underwent a dramatic decrease at its glycosite-containing peptide level (0.54-fold) with tunicamycin treatment. Five oligo-mannose glycans at the Endoplasmin glycosite Asn217 in tunicamycin-treated OVCAR-3 cells exhibited a similar alteration trend as observed at the total glycan level (Fig. 4c).
To further investigate the dynamics of the glycosylation changes, OVCAR-3 cells were treated with 1 µM tunicamycin for six different time periods (0, 6, 12, 24, 48 and 72 h). The proteins from the six time points plus repeated 0 and 48 h time points were labeled using 8-plex iTRAQ reagents for quantifying the intact glycopeptides at Endoplasmin glycosite Asn217 at each time point. The results showed that the amount of the Man9 glycan rapidly decreased to 37.2% of its baseline abundance after 6 h of tunicamycin treatment and to 15.7% in 12 h. The abundance of the Man8 glycan slowly decreased to 43.8% in 24 h and to 10.8% after 72 h of tunicamycin treatment, whereas the rate of decrease of the other three oligo-mannose glycans (Man5–Man7) was much slower compared to the Man9’s and Man8’s (Fig. 4d). These dynamic alteration results showed that (i) different glycans were altered at different rates, and (ii) that the closer to the reaction that was inhibited by tunicamycin a glycan was located in the N-glycan synthesis pathway, the faster the glycosylation changes occurred.
The observed glycan inhibition pattern by tunicamycin treatment can be explained by the following possible mechanism: the cells usually have some reserves of N-glycan precursor and processing glycoproteins in the endoplasmic reticulum. When the N-glycosylation pathway is inhibited, these reserves can be used to compensate for the effects of the inhibition. However, these reserves may only last for a short time. When the precursor glycans are depleted, the abundance of Man9 begins to decrease. Following the same mechanism, when Man9 is depleted, Man8 starts to decrease, and so on. To a certain degree, the rate of decrease of each oligo-mannose glycan with tunicamycin treatment could reflect the consumption rates of the glycan in the cells.
Compared to previous work8–25, our NGAG method allows the simultaneous large-scale profiling and monitoring of N-linked glycans, glycosites, glycoproteins and site-specific glycosylation in a single method (Supplementary Table 8). In tunicamycin-treated cells, we show that our method detected not only the overall reduction of total glycans and glycosite occupancy, but also the changes in site-specific glycosylation. Such a comprehensive and integrated characterization of protein glycosylation is complementary to genomic, transcriptomic and metabolomic data and is crucial for a complete understanding of the consequences perturbing biological systems.
ONLINE METHODS
OVCAR-3 cell culture and tunicamycin treatment
Human ovarian carcinoma cell line OVCAR-3 (ATCC, HTB161) was obtained from ATCC (Rockville, MD). The cell line was tested for mycoplasma contamination by ATCC before being used in experiments. A total of 32 dishes (15 cm × 15 cm) of OVCAR-3 cells were cultured in RPMI 1640 medium with 10% FBS to 50% confluence, and half of the dishes were treated with 1 µM tunicamycin for 48 h to inhibit N-glycosylation. In order to analyze the glycoprotein changes in tunicamycin-treated cells, the OVCAR-3 cells were also treated with 1 µM tunicamycin in triplicate and collected at six different time points (0, 6, 12, 24, 48 and 72 h). Separate dishes of OVCAR-3 cells were cultured in SILAC RPMI 1640 medium containing heavy isotope-labeled 13C6-l-lysine and 13C615N4-l-arginine (Cambridge Isotope Laboratories, Andover, MA) supplemented with 10% dialyzed FBS (diFBS) (Invitrogen, Carlsbad, CA) to generate heavy SILAC-labeled (K6, R10) OVCAR-3 cells. The cells were cultured for approximately ten doublings in the SILAC medium to ensure complete labeling31. After removing the medium, the cells were washed five times with phosphate buffered saline (PBS, pH 7.4) buffer and then lysed directly with 8 M urea/1 M NH4HCO3 solution32. Lysates were briefly sonicated until the solutions were clear. Protein concentrations were determined by BCA protein assay reagent (Thermo Scientific, Fair Lawn, NJ).
Protein digestion
Fetuin, and proteins from normal, tunicamycin-treated and heavy SILAC-labeled OVCAR-3 cells were denatured in 8 M urea/1 M NH4HCO3 buffer, reduced by 10 mM TCEP at 37 °C for 1 h and alkylated by 15 mM iodoacetamide at room temperature in the dark for 30 min. The solutions were diluted fivefold with deionized water. Then sequencing grade trypsin (Promega, Madison, WI; protein: enzyme, 50:1, w/w) was added to the samples and incubated at 37 °C overnight with shaking. Samples were centrifuged at 13,000g for 10 min to remove any particulate matter and purified by a C18 solid-phase extraction. Peptides were eluted from the C18 column in 60% ACN/0.1% TFA, and the peptide concentrations were measured by BCA reagent.
Two hundred microgram of peptides from normal and tunicamycin-treated OVCAR-3 cells were mixed with an equivalent amount of heavy SILAC peptides, and the combined samples were further purified by SCX column (Glygen, Columbia, MD) for proteomic analysis.
Extraction of N-linked glycans and glycosite-containing peptides using the NGAG method
One milligram of peptides (400 µl) from fetuin, 3 mg of peptides from normal and tunicamycin-treated OVCAR-3 cells, or 6 mg of peptides from heavy SILAC (K6R10)-labeled OVCAR-3 cells were used for extraction of N-linked glycans and glycosite-containing peptides using the NGAG method. The guanidination of lysine on tryptic peptides was performed according to the Sigma ProteoMass guanidination protocol (Sigma-Aldrich, St. Louis, Missouri) by adding 400 µl of 2.85 M aqueous ammonia and 400 µl of 0.6M O-methylisourea into the peptide solution (final pH = 10.5) and incubating at 65 °C for 30 min (ref. 26) (the volumes and concentrations of the solutions and reagents described here and below were for 1 mg peptides; the volumes and concentrations can be adjusted according to the actual volumes and amounts of the peptides). The reactions were stopped by adding 1.5 ml 10% TFA to adjust the pH to <3. The samples were desalted by C18 column and eluted in 400 µl 60% ACN/0.1% TFA.
The pH of the peptide solution was adjusted to 7.4 by adding 400 µl of 10× PBS buffer (pH 7.4). The peptides were incubated with 1 ml equilibrated AminoLink resin (50% slurry, Pierce, Rockford, IL) in the presence of 50 mM NaCNBH3 at room temperature for at least 2 h or overnight with shaking. The remaining aldehyde groups on the resin were blocked by 1 M Tris-HCl solution (pH 7.4) in the presence of 50 mM NaCNBH3 at room temperature for 30 min. The resin was washed three times with water. All carboxyl groups of the peptides, including the sialic acid of glycans, aspartic acid (D) and glutamic acid (E) residues and C termini of peptides, were modified by aniline by incubating beads with 1 M aniline and 0.5 M 1-ethyl-3-(3-dimethylamin-opropyl)carbodiimide(EDC) in 0.2 M 2-(N-morpholino)ethanesulfonic acid (MES) buffer (~pH 5) at room temperature for 6 h or overnight. The EDC solution was added three times (2–4 h intervals) to completely modify the carboxyl groups. The pH of the solution was checked frequently to ensure that it was 4–6 (refs. 33,34).
The resin was washed three times each with 50% ACN and 100 mM NH4HCO3 solution, and resuspended in 100 µl of 100 mM NH4HCO3 solution. One microgram of Asp-N was added and incubated with resin overnight at 37 °C with shaking to remove all possibly unlabeled D/E containing peptides. The resin was washed three times each with 50% ACN, 1.5 M NaCl, H2O, and 0.1 M NH4HCO3 solution. The N-glycans were released from the resin by 2 µl PNGase F (New England Biolab, Beverly, MA) in 200 µl 25 mM NH4HCO3 solution at 37 °C for 3 h or overnight with shaking. The resin was washed twice with 200 µl deionized water. The N-glycans were collected in the supernatant/wash solutions and purified by Carbograph column (Grace, Deerfield, IL) before being dried by vacuum35. The resin was washed three times each with 50% ACN, 1.5 M NaCl, H2O and 0.1 M NH4HCO3 solution. The former N-glycopeptides were released from the resin by 1 µg Asp-N in 200 µl 25 mM NH4HCO3 solution at 37 °C overnight with shaking. The resin was washed twice with 200 µl 50% ACN and the glycosite-containing peptides were collected in the supernatant/wash solutions.
The glycosite-containing peptide samples from the normal and tunicamycin-treated OVCAR-3 cells were mixed with the same amount of glycosite-containing peptides from the heavy SILAC-labeled cells. The mixtures were dried by vacuum. The glycosite-containing peptides were resuspended in 20 µl 0.2% formic acid (pH < 3) for LC-MS/MS analysis directly or in 50 µl 0.2% FA (pH < 3) for high-performance (HP)LC fractionation.
To determine the reproducibility of the NGAG method, N-linked glycans and glycosite-containing peptides were extracted in triplicate from different amounts of fetuin (100 µg, 100 µg, 200 µg, 300 µg) or the same amount of OVCAR-3 cells (0.5 mg) using NGAG as described above. The glycans (after being purified) and glycosite-containing peptides were directly analyzed by LC-MS/MS.
N-linked glycosite-containing peptide extraction by SPEG method
N-linked glycosite-containing peptide isolation by the Solid Phase Extraction of Glycopeptide method (SPEG, based on hydrazide chemistry) was performed as described previously27 with minor modification36. Briefly, three milligram of peptides from normal and tunicamycin-treated OVCAR-3 cells were mixed with an equivalent amount of SILAC heavy peptides before SPEG. Glycopeptides in the solution were oxidized by 10 mM NaIO4 solution at room temperature in the dark for 1 h. The samples were diluted with 0.1% TFA to 5% ACN and cleaned by C18 column, and the elution solution was collected directly into equilibrated hydrazide beads (300 µl of 50% slurry for each sample) and incubated with 100 mM aniline at room temperature for 1 h (ref. 36). The beads were washed 3 times each with 50% ACN, 1.5 M NaCl, water and 100 mM NH4HCO3 buffer. Former N-glycopeptides were released via 3 µl PNGase F (New England Biolabs, Beverly, MA) in 25 mM NH4HCO3 buffer at 37 °C overnight with shaking. Glycosite-containing peptides were collected in supernatants/wash solutions (200 µl of 50% ACN), and they were dried by vacuum and resuspended in 50 µl 0.2% FA (pH < 3) for HPLC fractionation.
Enrichment of intact glycopeptides using hydrophilic interaction column (HILIC)
The intact glycopeptides were enriched from OVCAR-3 cells using a HILIC enrichment method37. Briefly, the tryptic peptides from normal OVCAR-3 cells were purified by C18 and were enriched in 45% ACN/0.1% TFA. The peptide solution (~4 mg peptides) was diluted by 95% ACN/0.1% TFA to a final solvent composition of 80% ACN/0.1% TFA. The HILIC column (SeQuant, Southborough, MA) was washed twice each by 0.1% TFA and 80% ACN/0.1% TFA, followed by sample loading and three washes with 80% ACN/0.1% TFA. The glycopeptides bound to the column were eluted in 0.4 ml of 0.1% TFA solution. The samples were dried by SpeedVac and resuspended in 50 µl of 0.2% FA for high-pH RPLC fractionation.
iTRAQ labeling
Equal amounts of tryptic peptides from normal and triplicate tunicamycin-treated OVCAR-3 cells (set 1), and tunicamycin-treated cells with different time points (set 2) were labeled by 8-plex iTRAQ reagents according to the manufacturer’s protocols for intact glycopeptide quantification (set 1: normal: iTRAQ 113 and 117; #1 treated cells: iTRAQ 114 and 118; #2 treated cells: iTRAQ 115 and 119; #3 treated cells: iTRAQ 116 and 121. Set 2: 0 h tunicamycin treatment: iTRAQ 113 and 119; 6 h treatment: iTRAQ 114; 12 h treatment: iTRAQ 115; 24 h treatment: iTRAQ 116; 48 h treatment: iTRAQ 117 and 121; 72 h treatment: iTRAQ 118). The glycosite-containing peptides isolated from different amounts of fetuin were labeled by 4-plex iTRAQ reagents (three sets). The iTRAQ-labeled samples were pooled and purified by SCX and C18 columns for further analysis.
Analysis of glycans using mass spectrometry
N-glycans extracted from bovine fetuin and OVCAR-3 cells using NGAG were analyzed by an Axima MALDI Resonance mass spectrometer (Shimadzu). Dried N-glycans were resuspended in 20 µl deionized water followed by spotting 1 µl glycan sample with 1 µl 2,5-dihydroxybenzoic acid/N,N-dimethylaniline (DHB/DMA) matrix (100 mg/ml DHB, 2% DMA in 50% acetonitrile, 0.1 mM NaCl) onto a MALDI plate15,35. The laser power was set to 100 for two shots each in 100 locations per spot. The average MS spectra (200 profiles) were used for glycan assignment by GlycoMod software (http://web.expasy.org/glycomod/) and comparison to human glycan database in CFG website (http://www.functionalglycomics.org/) as well as previously analyzed in our laboratory35,38. The possible glycan structures of the identified glycan compositions were generated by GlycoWorkbeach39. The identified glycans were further verified by LC-MS/MS (Q-Exactive mass spectrometer, Thermo Fisher Scientific), in which the glycan isotopic mass is more accurate (<10 p.p.m.) than MALDI-TOF-MS, and the glycans with different numbers of sialic acids usually have different retention times on C18 column (due to different number of aniline modifications on the sialic acid). The detailed LC-MS/MS setting is described below. The glycans were directly injected to the analytical C18 column without going through the trap column, and the MS signals were collected from the very beginning of the sample injection in order to detect the underivatived glycans (the neutral glycans that don’t have aniline-labeled sialic acids). The glycans identified from the different samples were quantified by a label-free quantification method based on their peak area from LC-MS/MS data using SIEVE software (Thermo Fisher Scientific), and the total ion current (TIC) was used for normalization.
Peptide fractionation using high-pH RPLC
All SILAC light and heavy mixed tryptic peptides (100 µg), iTRAQ-labeled peptides, glycosite-containing peptides (~60 µg), and intact glycopeptides (~40 mg) from OVCAR-3 cells extracted by NGAG and SPEG were injected by a 1220 Series HPLC (Agilent Technologies, Inc., CA) into a Zorbax Extend-C18 analytical column containing 1.8 µm particles at a flow rate of 0.2 ml/min. The mobile-phase A consisted of 10 mM ammonium formate (pH 10) and B consisted of 10 mM ammonium formate and 90% ACN (pH 10). Sample separation was accomplished using the following linear gradient: 0–2% B, 10 min; 2–8% B, 5 min; 8–35% B, 85 min; 35–95% B, 5 min; 95–95% B, 15 min. Peptides were detected at 215 nm and 96 fractions were collected along with the LC separation in a time-based mode from 16 to 112 min. The 96 fractions of tryptic peptide samples were concatenated into 24 fractions by combining fractions 1, 25, 49, 73; 2, 26, 50, 74, etc. The 96 fractions of the glycosite-containing peptide samples were concatenated into 12 fractions by combining fractions 1, 13, 25, 37, 49, 61, 73, 85; 2, 14, 26, 38, 50, 62, 74, 86, etc. The samples were then dried in a Speed-Vacuum and stored at −80 °C until LC-MS/MS analysis.
LC-MS/MS analysis
The tryptic peptide and N-linked glycosite-containing peptide samples underwent one LC-MS/MS run per sample (or fraction) on a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Peptides were separated on a Dionex Ultimate 3000 RSLC nano system with a 75 µm × 15 cm Acclaim PepMap100 separating column protected by a 2-cm guard column (Thermo Scientific). The mobile phase flow rate was 300 nl/min and consisted of 0.1% formic acid in water (A) and 0.1% formic acid 95% acetonitrile (B). The gradient profile was set as follows: 4–7% B for 5 min, 7–40% B for 90 min, 40–90% B for 5 min, 90% B for 10 min and equilibrated in 4% B for 10 min. MS analysis was performed using a Q-Exactive mass spectrometer. The spray voltage was set at 2.2 kV. Orbitrap MS1 spectra (AGC 1 × 106) were collected from 400–1,800 m/z at a resolution of 60 K followed by data-dependent higher-energy collisional dissociation tandem mass spectrometry (HCD MS/MS) (resolution 7,500, collision energy 45%, activation time 0.1 ms) of the 20 most abundant ions using an isolation width of 2.0 Da. Charge state screening was enabled to reject unassigned and singly charged ions. A dynamic exclusion time of 25s was used to discriminate against previously selected ions. For tryptic peptides, 100 m/z was set as the fixed first mass in MS/MS fragmentation to include all oxonium ions of glycopeptides.
Database search
All LC-MS/MS data from human and bovine resources were searched against RefSeq human protein databases40 (downloaded from NCBI website July 29, 2013) and bovine fetuin sequence by MaxQuant41 (v1.3.0.5), respectively. For global proteome data (tryptic peptide), the search parameters were set as follows: up to two missed cleavage were allowed for trypsin digestion, 20 p.p.m. and 6 p.p.m. precursor mass tolerance for first and main search, respectively; carbamidomethylation (C) was set as a static modification and oxidation (M) was set as a dynamic modification; two modifications with “Arg 10” and “Lys6” were selected as heavy labels for mixed peptides from OVCAR-3 cells; five modifications per peptide and a minimum of six amino-acid length were considered for peptide identification. All other settings were set as default values and the results were filtered with a 1% FDR. For SPEG glycosite-containing peptides data, deamination (N) was added as one additional dynamic modification.
To search LC-MS/MS data of glycosite-containing peptides extracted by NGAG, both human and bovine fetuin databases were first modified by replacing all potential N-glycosylation sites (N#-X-S/T motif, X is any amino acid except proline) with “U” (not existing in normal protein sequences) to facilitate the search parameter setting and N-linked glycosite-containing peptide identification. Then an enzyme “trypsin + U” was created and added to the enzyme list, which was assigned to specifically cleave peptide bonds at the C-terminal side of lysine/arginine (no cleavage at KP and RP) and at the N-terminal side of “U” (168.9642Da). The MaxQuant search parameters were set as follows: up to two missed cleavages were allowed for “trypsin+U” digestion, 20 p.p.m. and 6 p.p.m. precursor mass tolerance for first and main search, respectively; carbamidomethylation (C, +57.0215 Da) was set as a static modification; U->D (U, −53.9373Da), U->N (U that is not at the N termini, −54.9213 Da), guanidination (K, +42.0218 Da), aniline label (D and E that are not at the N termini, protein C termini and K and R that are at any C termini, +75.0473 Da), two aniline labels (D and E that are at the protein C termini, +150.0946 Da), guanidination + aniline label (K at any C termini, +117.0691 Da) and oxidation (M, +15.9949 Da) were set as dynamic modifications; six modifications per peptide and a minimum of six amino-acid length were considered for peptide identification. All other settings were set as default values, and the results were filtered with a 1% FDR. For glycosite-containing peptide data from the OVCAR-3 cell line, which included SILAC peptides, K6+guanidination label (K, +48.0419 Da), K6+guanidination+aniline label (K at any C termini, +123.0892 Da) and R10+aniline label (R at any C termini, +85.0555 Da) were added as additional dynamic modifications based on the parameters used for the global proteome data described above. Two modifications with “Arg 10” and “Lys6” were selected as heavy labels.
Intact glycopeptide analysis
In order to allow Q-Exactive mass spectrometer to select intact glycopeptide peaks for MS/MS analysis, we set “peptide match” as “off”. We also excluded the precursor at charge +1 and +2 for MS/MS, because the intact glycopeptides usually have masses large enough to present as multiple charge states. In order to detect oxonium ions of glycopeptides, we set 100 m/z as the fixed first mass in MS/MS fragmentation, and optimized the MS/MS fragmentation energy to generate the MS/MS spectra of glycopeptides that contained peptide/peptide+HexNAc fragment ions. These fragment ions facilitate the selection of tandem spectra from intact glycopeptides.
The precursor mass matching approach in the GPQuest software was developed for this study and used to identify intact glycopeptides28. Briefly, the proteomic raw data were converted to ‘mzXML’ format using Trans-Proteomic Pipeline (TPP)42, and then to ‘matlab’ format using GPQuest28. The oxonium ion-containing MS/MS spectra were extracted using oxonium ion HexNAc_204.087 Da and one of the other oxonium ions (including 138.055 Da, 163.061 Da, 168.066 Da, 274.093 Da, 292.103 Da and 366.140 Da) within 50 p.p.m. Note that the oxonium ions were only matched from the top five fragment ions of the MS/MS spectra, as the oxonium ions usually have the highest intensities among fragment ions of N-glycopeptides in HCD fragmentation mode. The oxonium ion-containing spectra were matched to the N-glycopeptide candidate database (comprising the identified N-glycan and glycosite-containing peptides) to assign all glycopeptide candidates of the spectra based on their precursor masses within a 10 p.p.m. mass error. The existence of a minimum of two peptide or peptide+HexNAc ions (charge 1+ and 2+, 50 p.p.m. mass error) in MS/MS spectra was used as a filter to determine the glycosite-containing peptide and glycan compositions of the glycopeptides. The quantification information of assigned glycopeptide spectra (based on their MS/MS scan numbers) were obtained from “allpeptide.txt” files from MaxQuant results (SILAC-labeled samples). The b and y ion information from peptide portion of the matched glycopeptides were used for validation.
For iTRAQ-labeled samples, the oxonium ions were matched from the top 20 fragment ions of the MS/MS spectra. The oxonium ion-containing spectra were matched to the N-glycopeptide candidate database (comprising the identified N-glycan and glycosite-containing peptides) to assign all glycopeptide candidates of the spectra based on their precursor masses within a 10 p.p.m. mass error. Then only glycopeptides that contained >30% of the theoretical b and y ions of the peptide portion were included in the final list of identified intact glycopeptides. The quantification information of assigned glycopeptide spectra (based on their MS/MS scan numbers) were obtained from the intensities of the iTRAQ reporter ions.
FDR estimation of intact glycopeptide identification
The false-discovery rate (FDR) of the assignments of spectra to intact glycopeptides of fetuin was estimated using a glycopeptide database established from human immunodeficiency virus (HIV) GP120 glycopeptides that contains 116 N-linked glycans and 17 glycosite-containing peptides (116 × 17 = 1,972 glycopeptide candidates)23. Using the same strategy and filters as fetuin glycopeptide identification, only one oxonium ion-containing MS/MS spectrum from triplicate global proteomic analyses was randomly matched to a HIV glycopeptide, which indicated an estimated FDR of 0.1%.
To estimate the FDR of glycopeptide assignment, the decoy peptide database was built by combining the amino acids from all identified glycosite-containing peptides, shuffling them, and separating them into decoy peptide sequences with the same length as the target database. The decoy database was then merged with the target database resulting in a total of 6,166 peptides in the peptide database. The glycopeptide candidate database was then created based on identified glycans with identified and decoy peptides. Using the same match strategy and filters, the oxonium ion-containing spectra were matched to the glycopeptide candidate database and further filtered by the number of matched b and y ions. The FDR of spectrum assignment was estimated using the percentage of spectra that matched to the decoy glycopeptides.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health, National Cancer Institute, Clinical Proteomic Tumor Analysis Consortium (U24CA160036), the Early Detection Research Network (EDRN, U01CA152813 and U24CA115102) and R01CA112314, and by the National Institutes of Health, National Heart, Lung, and Blood Institute Programs of Excellence in Glycosciences (PEG, P01HL107153) and the Johns Hopkins Proteomics Center (N01-HV-00240).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
S.S. and H.Z. prepared the manuscript with contributions from all co-authors; S.S. conducted most experimental and data analyses with support from other co-authors; P.S. performed part of mass spectrometric analyses; P.S. and W.Y. contributed to part of data analyses; S.T.E. developed software for intact glycopeptide analysis; N.T. and N.H. cultured cells; S.Y. and P.A. contributed to part of the glycan analysis experiments; L.C. assisted in some of sample preparation experiments; D.W.C. and Z.Z. provided additional research support.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 2010;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 3.Drake PM, et al. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin. Chem. 2010;56:223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 5.Kaji H, et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson J, et al. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods. 2009;6:809–811. doi: 10.1038/nmeth.1392. [DOI] [PubMed] [Google Scholar]
- 7.Jensen PH, Karlsson NG, Kolarich D, Packer NH. Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 2012;7:1299–1310. doi: 10.1038/nprot.2012.063. [DOI] [PubMed] [Google Scholar]
- 8.Wada Y, Tajiri M, Yoshida S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal. Chem. 2004;76:6560–6565. doi: 10.1021/ac049062o. [DOI] [PubMed] [Google Scholar]
- 9.Yu L, Li X, Guo Z, Zhang X, Liang X. Hydrophilic interaction chromatography based enrichment of glycopeptides by using click maltose: a matrix with high selectivity and glycosylation heterogeneity coverage. Chem. Eur. J. 2009;15:12618–12626. doi: 10.1002/chem.200902370. [DOI] [PubMed] [Google Scholar]
- 10.Paxton RJ, Mooser G, Pande H, Lee TD, Shively JE. Sequence analysis of carcinoembryonic antigen: identification of glycosylation sites and homology with the immunoglobulin supergene family. Proc. Natl. Acad. Sci. USA. 1987;84:920–924. doi: 10.1073/pnas.84.4.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu T, et al. Capillary electrophoresis-electrospray mass spectrometry for the characterization of high-mannose-type N-glycosylation and differential oxidation in glycoproteins by charge reversal and protease/glycosidase digestion. Anal. Chem. 2001;73:5875–5885. doi: 10.1021/ac0106748. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, et al. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat. Methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 13.Zielinska DF, Gnad F, Wiśniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Wang H, Tang H, Yang P. Endoglycosidase-mediated incorporation of 18O into glycans for relative glycan quantitation. Anal. Chem. 2011;83:4975–4981. doi: 10.1021/ac200753e. [DOI] [PubMed] [Google Scholar]
- 15.Shah P, et al. Mass spectrometric analysis of sialylated glycans with use of solid-phase labeling of sialic acids. Anal. Chem. 2013;85:3606–3613. doi: 10.1021/ac3033867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breidenbach MA, et al. Targeted metabolic labeling of yeast N-glycans with unnatural sugars. Proc. Natl. Acad. Sci. USA. 2010;107:3988–3993. doi: 10.1073/pnas.0911247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott NE, et al. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M000031-MCP201. M000031-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bern M, Kil YJ, Becker C. Byonic: advanced Peptide and protein identification software. Current Protocols in Bioinformatics. 2012 doi: 10.1002/0471250953.bi1320s40. 13.20.11–13.20.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolarich D, Jensen PH, Altmann F, Packer NH. Determination of site-specific glycan heterogeneity on glycoproteins. Nat. Protoc. 2012;7:1285–1298. doi: 10.1038/nprot.2012.062. [DOI] [PubMed] [Google Scholar]
- 20.Lattová E, Kapková P, Krokhin O, Perreault H. Method for investigation of oligosaccharides from glycopeptides: direct determination of glycosylation sites in proteins. Anal. Chem. 2006;78:2977–2984. doi: 10.1021/ac0519918. [DOI] [PubMed] [Google Scholar]
- 21.Pompach P, Chandler KB, Lan R, Edwards N, Goldman R. Semi-automated identification of N-Glycopeptides by hydrophilic interaction chromatography, nano-reverse-phase LC-MS/MS, and glycan database search. J. Proteome Res. 2012;11:1728–1740. doi: 10.1021/pr201183w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker BL, et al. Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. J. Proteome Res. 2013;12:5791–5800. doi: 10.1021/pr400783j. [DOI] [PubMed] [Google Scholar]
- 23.Yang W, et al. Glycoform analysis of recombinant and human immunodeficiency virus envelope protein gp120 via higher energy collisional dissociation and spectral-aligning strategy. Anal. Chem. 2014;86:6959–6967. doi: 10.1021/ac500876p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S-W, Pu T-H, Viner R, Khoo K-H. Novel LC-MS2 product dependent parallel data acquisition function and data analysis workflow for sequencing and identification of intact glycopeptides. Anal. Chem. 2014;86:5478–5486. doi: 10.1021/ac500945m. [DOI] [PubMed] [Google Scholar]
- 25.Wu S-W, Liang S-Y, Pu T-H, Chang F-Y, Khoo K-H. Sweet-Heart - an integrated suite of enabling computational tools for automated MS2/MS3 sequencing and identification of glycopeptides. J. Proteomics. 2013;84:1–16. doi: 10.1016/j.jprot.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 26.Brancia FL, Oliver SG, Gaskell SJ. Improved matrix-assisted laser desorption/ionization mass spectrometric analysis of tryptic hydrolysates of proteins following guanidination of lysine-containing peptides. Rapid Commun. Mass Spectrom. 2000;14:2070–2073. doi: 10.1002/1097-0231(20001115)14:21<2070::AID-RCM133>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 27.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat. Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toghi Eshghi S, Shah P, Yang W, Li X, Zhang H. GPQuest: A Spectral Library Matching Algorithm for Site-Specific Assignment of Tandem Mass Spectra to Intact N-glycopeptides. Anal. Chem. 2015;87:5181–5188. doi: 10.1021/acs.analchem.5b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freeze H, Elbein A. In: Essentials of Glycobiology. Varki A, et al., editors. Cold Spring Harbor Laboratory Press; 2009. p. 66. [PubMed] [Google Scholar]
- 30.Vizcaíno JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 31.Ong S-E, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat. Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 32.Sun S, Zhou J-Y, Yang W, Zhang H. Inhibition of protein carbamylation in urea solution using ammonium-containing buffers. Anal. Biochem. 2014;446:76–81. doi: 10.1016/j.ab.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panchaud A, et al. ANIBAL, stable isotope-based quantitative proteomics by aniline and benzoic acid labeling of amino and carboxylic groups. Mol. Cell. Proteomics. 2008;7:800–812. doi: 10.1074/mcp.M700216-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Schilling O, Barré O, Huesgen PF, Overall CM. Proteome-wide analysis of protein carboxy termini: C terminomics. Nat. Methods. 2010;7:508–511. doi: 10.1038/nmeth.1467. [DOI] [PubMed] [Google Scholar]
- 35.Yang SJ, Li Y, Shah PK, Zhang H. Glycomic analysis using glycoprotein immobilization for glycan extraction. Anal. Chem. 2013;85:5555–5561. doi: 10.1021/ac400761e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun S, et al. Analysis of N-glycoproteins using genomic N-glycosite prediction. J. Proteome Res. 2013;12:5609–5615. doi: 10.1021/pr400575f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hägglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 2004;3:556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- 38.Yang SJ, Zhang H. Glycan analysis by reversible reaction to hydrazide beads and mass spectrometry. Anal. Chem. 2012;84:2232–2238. doi: 10.1021/ac202769k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ceroni A, et al. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 2008;7:1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 40.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 42.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005;1:0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.