Abstract
The muscular dystrophies (MDs) represent a diverse collection of inherited human disorders, which affect to varying degrees skeletal, cardiac, and sometimes smooth muscle (Emery, 20021). To date, more than 50 different genes have been implicated as causing one or more types of MD (Bansal et al., 20032). In many cases, invaluable insights into disease mechanisms, structure and function of gene products, and approaches for therapeutic interventions have benefited from the study of animal models of the different MDs (Arnett et al., 20093). The large number of genes that are associated with MD and the tremendous number of animal models that have been developed preclude a complete discussion of each in the context of this review. However, we summarize here a number of the more commonly used models together with a mixture of different types of gene and MD, which serves to give a general overview of the value of animal models of MD for research and therapeutic development.
I. Dystrophin
Duchenne muscular dystrophy (DMD) and the milder Becker MD (BMD) are caused by dystrophin gene mutations.4,5 DMD/BMD is thought to be the most common form of muscular dystrophy (MD), and the most common lethal genetic disorder of children.6 Full-length dystrophin is expressed in all muscles, while shorter, truncated isoforms are expressed in many other tissues.7 Dystrophin is part of a large complex of integral and peripheral proteins that link the γ-actin cytoskeleton to the extracellular matrix via the dystrophin–glycoprotein complex (DGC) (Fig. 1).8 Studies of dystrophin and DMD/BMD have been greatly facilitated by the availability of numerous murine and canine models of DMD.9
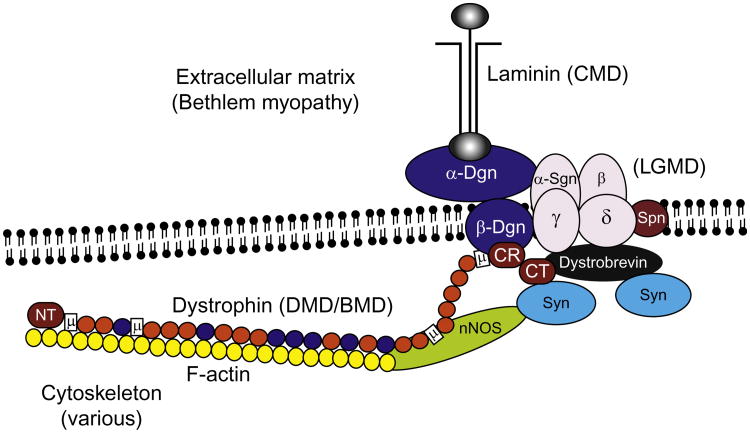
Fig. 1.
The dystrophin–glycoprotein complex (DGC) contains many proteins that, when not expressed, lead to various forms of muscular dystrophy. Shown is the organization of the DGC in skeletal muscle. Dystrophin is defective in DMD and BMD; α-, β-, γ-, and δ- sarcoglycans (Sgn) are defective in four different types of LGMD; laminin α2 is defective in one form of congenital muscular dystrophy; collagen VI in the extracellular matrix is defective in Bethlem myopathy and Ullrich congenital muscular dystrophy; while numerous other cytosolic- and membrane-associated proteins lead to other MDs. Other abbreviations include Syn, syntrophin; SPN, sarcospan; Dgn, dystroglycan; nNOS, neuronal nitric oxide synthase. Dystrophin domains listed include the N-terminal actin-binding domain (NT), various hinges (H), a WW domain (W), a cysteine-rich domain (CR), and a C-terminal domain (CT). Orange and red balls in dystrophin represent spectrin-like repeats, orange ones are acidic, while blue ones are basic and contribute to actin binding.
A. Mouse Models for Duchenne Muscular Dystrophy
Mouse models have been used extensively to elucidate the pathogenic mechanism of DMD and have been indispensable in the development of therapeutic approaches. Many mouse models with mutations in the DMD gene have been identified. Among these models, the mdx mouse is the most commonly used to analyze dystrophin gene expression and function. The mdx mouse contains a premature stop codon in exon 23 that leads to loss of full-length dystrophin, although smaller isoforms are still expressed.10–12 Although mdx skeletal muscle shows moderate signs of dystrophy, young mice exhibit little weakness and live 80% as long as controls, significantly more than that of DMD patients.13 This mild, yet progressive phenotype has been attributed in part to compensatory upregulation of utrophin.14,15 Histological examination of mdx muscle during various stages of development reveals that muscle fiber necrosis and cellular infiltration begin at approximately 3 weeks of age. This is followed by a crises period that peaks at approximately 4–6 weeks of age and is characterized by the presence of extensive necrosis, regenerating muscle fibers with centrally located nuclei, and elevated levels of serum creatine kinase (CK).10,16 After 12 weeks, the cycles of necrosis and regeneration begin to slow, although necrotic myofibers are present for the remainder of their lifespan. The fibrosis and infiltration of inflammatory cells in skeletal and cardiac muscle of the mdx are much milder than that observed in DMD patients.17,18 In contrast, the mdx mouse diaphragm exhibits severe pathological changes and functional deficit comparable to that of DMD limb muscle.19,20
Four additional strains of mdx mice, mdx2cv–5cv, have been generated with N-ethylnitrosourea chemical mutagenesis.21 All these strains have point mutations that lead to loss of full-length dystrophin isoforms. The relative location of these mutations results in a series of mdx mouse mutants that vary in their expression of different dystrophin isoforms, such that these mice should be useful for analysis of the role of the various isoforms.12 Despite their differences, all five mdx strains display essentially identical muscle pathology as mdx mice, although additional phenotypes have been observed.21–23 In the mdx2cv mouse, a single base change in the splice acceptor site of dystrophin intron 42 generates multiple transcripts due to aberrant splicing that eliminates the normal open reading frame. The mdx3cv allele arises from a mutant splice acceptor site in intron 65,12,21,22 leading to severe loss of most dystrophin isoforms, including Dp71. Of the five original mdx strains, only the mdx3cv mice accumulate detectable levels of dystrophin-positive muscle fibers, although the expressed dystrophin lacks a key functional domain and is largely nonfunctional.22–24 The mutation in the mdx4cv mouse is a point mutation that creates a stop codon in exon 53, whereas in the mdx5cv allele, a transversion mutation in exon 10 creates a new splice donor that generates a frame-shifting deletion in the mRNA. Both the mdx4cv and mdx5cv strains display a low background of reverent dystrophin-containing fibers, making them particularly useful strains in gene transfer studies exploring the feasibility of DMD therapy.23,25,26
In the mdxβgeo mouse, a β-galactosidase/neomycin phosphotransferase (βgeo) reporter gene is inserted 3′ to exon 63 of the dystrophin gene, affecting translation of dystrophin isoforms, including Dp71.27 These mice develop a dilated esophagus, cardiac hypertrophy, and the same skeletal muscle pathology as mdx mice. The lacZ reporter gene of mdxβgeo allows detection of dystrophin expression by staining for β-galactosidase activity.
Another mutant mouse known as mdx52 was generated by a homologous recombination technique.28 In this model, exon 52 of the dystrophin gene is disrupted, and dystrophin along with shorter dystrophin isoforms (Dp140 and Dp260) are absent. This exon was targeted to replicate the most common out-of-frame deletion mutation that occurs in 13% of DMD patients.29–31 Recently, antisense-mediated skipping of exon 51 using antisense oligonucleotides (AOs) to restore the reading frame of the dystrophin gene was found to be successful in some patients with DMD.32–34 Exon 51 skipping using AO in mdx52 mice successfully converted an out-of-frame mutation into an in-frame mutation, resulting in amelioration of the dystrophic pathology and improved muscle function.35
Hanaoka and colleagues generated a new DMD mouse model known as DMD-null. In this model, the entire genomic region of the DMD gene on mouse chromosome X was deleted using a Cre-loxP recombination system.36 These DMD-null males exhibited some behavioral abnormality, sterility, and pathological features very similar to those of the mdx mice. As the production of all dystrophin isoforms is prevented in the DMD-null mouse, this model system may be used to facilitate studies to clarify the function of individual dystrophin isoforms.
In efforts to make the mdx muscle phenotype more similar to that of patients, several additional mutations have been crossed onto the mdx background to generate double knockouts (DKOs). The most widely used is a dystrophin:utrophin DKO (mdx:utrn−/−).37,38, These mice display a severe phenotype and live on average only about 3 months. The severity of the phenotype supports the concept that utrophin upregulation in dystrophic muscles partially compensates for the absence of dystrophin. Further, the mdx:utrn−/− have proved useful in gene therapy studies, where the phenotype can be largely eliminated by muscle-specific expression of utrophin, miniutrophin, or mini- or micro-dystrophin.25,26,37–40 Two newer DKO strains have recently been generated that not only resulted in a several phonotype but also rely on mutations that appear to faithfully reproduce the human condition. The first is a cross between mdx and a mutation of cytidine monophosphate–sialic acid hydroxylase (Cmah).41 Cmah is inactive in humans due to a mutation, and it prevents glycosylation with N-glycolylneuraminic acid. The second is an mdx:mTR cross that effectively lacks telomerase activity.42 This cross becomes severe due to the impaired ability of myogenic stem cells to support ongoing muscle regeneration.
B. Canine Models for DMD
Clinical diagnosis of MD has been observed in the dog for several decades. Identifying a particular MD with broad symptoms (i.e., weakness and intolerance to exercise) has proven difficult. With the increasing availability of research tools, more human MDs have found a pathologically similar model in canines. Yet, the infrequency of these diagnoses and the difficulties in identifying and propagating the mutations by breeding have hindered multiple MD strains from being used as valuable research tools. One MD that has been successfully generated and utilized throughout the world is the X-linked dystrophin-deficient dog. Spontaneous mutations causing dystrophinopathy have been identified in several breeds of dog.43–45 Multiple colonies of the golden retriever muscular dystrophy dog (GRMD) have been created and is the most extensively studied breed for this model, but it is not the only one.9,46 Due to prior use in research and its smaller size, the GRMD mutation has been bred onto the beagle background.47,48 Severe symptoms commonly appear at 6 months of age in the GRMD dog, but unlike the mdx mouse, the degree of severity and time of progression are quite variable. Pathology has been observed in utero and massive necrosis can result in neonatal death.46 Compounded by the increased cost of maintaining a larger species, the GRMD model can be problematic. However, use of the GRMD model has gained much emphasis with its more clinically similar pathology than the mdx mouse model.
II. The Sarcoglycans
As a clinical group, the MDs were first appreciated in the mid-1800s with a detailed description of DMD.49,50 Approximately a century following, another group of clinically comparable progressive MDs distinct from DMD/BMD muscular dystrophy emerged which were found to follow an autosomal recessive mode of inheritance. Given their phenotypic similarity, these diseases were originally termed Duchenne-like autosomal recessive MD or severe childhood autosomal recessive MD.51 Several decades later, the discovery of mutations in the dystrophin gene as the genetic cause of DMD,52 subsequent description of the DGC,53 and demarcation of the striated muscle transmembrane sarcoglycan complex paved the way for initial characterization of the sarcoglycanopathies. Four genetically related MDs arose with pathological mutations in genes for alpha-sarcoglycan (α-SG),54 beta-sarcoglycan (β-SG),55 gamma-sarcoglycan (γ-SG),56 and delta-sarcoglycan (δ-SG)57 defining these distinct autosomal recessive MDs. Given that these disorders primarily affected proximal muscles around the scapular and pelvic girdles, and elevation of the genetic classification of limb-girdle muscular dystrophies (LGMD), there was the assignment of LGMD2C-F for the genes representing γ-SG, α-SG, β-SG, and δ-SG, respectively.
A. δ-Sarcoglycan and LGMD2F
The BIO14.6 hamster strain was established in 1962 and is recognized as the first animal model of sarcoglycanopathies.58 However, it was not until after the discovery of the causative mutation in LGMD2F patients59 that a large deletion in the 5′ portion of the δ-SG gene was identified.60 The hamsters die prematurely (∼ 8 months) from progressive myocardial necrosis and ultimately heart failure. Compared with skeletal muscle, damage to cardiac muscle occurs early (∼ 5 weeks), consistent with a cardiomyopathic phenotype with decreased stroke volume, cardiac output, and increased ventricular hypertrophy.61 As the original BIO14.6 strain was identified, there have been at least three additional strains developed: the J2-NK, the CHF147 (formerly VMX7.1), and the TO-2 strain. Differences, such as the expression profile of individual sarcoglycan isoforms, have been observed in the TO-2 as compared to the BIO14.6 model. For example, in the BIO14.6 muscles, residual protein levels of α-SG and γ-SG can be detected on the sarcolemma, while in the TO-2, there is a complete absence of these isoforms. Further, the TO-2 has a shorter lifespan with a tendency to develop a more severe cardiomyopathy displaying a necrotic mode of pathology, while the BIO14.6 model displays a hypertrophic cardiomyopathy.62,63 Mice deficient in δ-SG have also been generated with subsequent complete loss of the entire SG complex,64,65a and as with the BIO14.6 hamster, these mice display classical histological findings of dystrophy inclusive of muscle degeneration, necrosis, inflammation, fibrosis, and reduced survival. However, muscle mass, peak twitch tension (P1) and peak tetanic tension (P0), and specific force (P0/CSA) do not differ from wild-type muscle with the latter being in contrast to the BIO14.6 hamster65b suggesting species-specific variations in pathophysiology. Similar to the BIO14.6 hamster model, these mice develop cardiomyopathy with a relatively early onset (∼ 8 weeks) with areas of fibrosis. By 32 weeks, a detrimental change in pathology is observed with concurrent reduction in ejection fraction, and evidence of right ventricular dilation suggestive of pulmonary dysfunction.65c Thus, similar to the hamster model, the δ-SG-null mouse reproduces many of the pathophysiological features of the human disease, providing a useful preclinical research model.
B. γ-Sarcoglycan and LGMD2C
Mice with a targeted deletion of γ-SG also exhibit progressive MD and cardiomyopathy with a subsequent destabilization of β-SG and δ-SG while α-SG is highly reduced.66 Like the δ-SG-null hamster, cardiomyopathy in γ-SG-null mice may be dilated or hypertrophic.62,66 Similar to findings in LGMD2C patients, membrane disruptions are a common feature of mice deficient for γ-SG as evidenced by Evans blue dye uptake and elevation of the muscle-specific enzyme M-CK in serum.66,67 Importantly, in striated muscle, the expression of dystrophin, dystroglycan, and laminin appears unaltered with the loss of γ-SG when evaluated by immunostaining.64 Thus, the mechanical linkage between the extracellular matrix and the intracellular cytoskeleton appears to be maintained in the absence of γ-SG. Further, γ-SG-deficient mice show normal resistance to mechanical strain and no evidence of contraction-induced injury after exercise when compared to its wild-type counterparts.66 These results are similar to the BIO14.6 hamster and in contrast to muscles of α-SG, β-SG, and δ-SG-null mice (see below). Considering the sarcoglycans are secondarily reduced in dystrophin-deficient mice, this advocates that nonmechanical properties contribute to the pathophysiology of MD in genetically varied forms of disease.
C. β-Sarcoglycan and LGMD2E
Mice deficient for β-SG have also been generated which, like δ-SG-null mice, display MD with large areas of necrosis/fibrosis and cardiomyopathy.68,69 As an indicator of acute myocardial necrosis preceding that of fibrotic lesion formation, serum levels of the cardiac-specific troponin-I have been found elevated in these mice.65a,68,70 Additionally, like δ-SG-null mice, β-SG-null mice show severely reduced expression of the entire sarcoglycan complex inclusive of sarcospan, increased muscle mass, decreased specific force generation, myofiber cross-sectional area, and resistance to contraction-induced injury.68,69,71 In support of the former, coimmunoprecipitation studies have shown a strong association between β-SG and δ-SG, proposed to form a core complex enabling delivery and retention of the sarcoglycan complex to the sarcolemma.72 Correlative support of this hypothesis can be found in human patients with either β-SG or δ-SG mutations, where there is a strong reduction or complete absence of the entire sarcoglycan complex.73–75
D. α-Sarcoglycan and LGMD2D
Mice lacking α-SG have been generated by targeted disruption of the α-SG gene.76 These mice show a complete loss of the sarcoglycan complex, demonstrate a progressive MD, and like LGMD2D patients present with an absence of cardiomyopathy.76 Like the γ-, β-, and δ-SG-null mice, the α-SG-null mice demonstrate membrane permeability defects as evidenced by Evans blue dye uptake and elevated serum CK. Given this similarity across dystrophic models, it remains unclear whether or not changes in increased membrane permeability are causative for muscle degeneration or rather a result of it. A further interesting aspect of α-SG-null mice that differs from the γ-SG-null mice is an apparent decrease in expression of α-dystroglycan (αDG) and dystrophin with an inherent increase in expression of utrophin at the sarcolemma.76 The molecular basis for these differences has not been established, though it is tempting to speculate that the residual amount of α-SG present in the γ-SG-null could be sufficient to allow normal levels of dystrophin and dystroglycan. In the α-SG-null mice, there are notable muscle-specific differences in contractile properties. For example, in hypertrophic slow-twitch soleus muscle, there is a demonstrated increase in maximal force values and relatively unchanged specific force relative to wild-type controls.76 However, in fast-twitch extensor digitorum longus (EDL) muscles, maximal force values are unchanged while specific force is significantly lower than wild-type controls.76 In diaphragm of α-SG-null mice, there is a significant reduction in maximal force, coupling between longitudinal and transverse properties, and muscle extensibility in the direction of muscle fibers and transverse to fibers.77 Thus, as a whole, the sarcoglycan complex may also provide a complex mechanical function in the diaphragm by influencing the properties of muscle stiffness, muscle viscoelasticity, and modulation of the contractile properties.
E. Canine Models of Sarcoglycan Deficiency
Three breeds of dogs have also been reported with deficiencies in expression of the sarcoglycan complex in skeletal muscle.78,79 Where the mutation lies in each of these cases has yet to be determined, but as discussed above for both rodents and humans, a deficiency in one sarcoglycan can destabilize the subcomplex made by all four of these isoforms.
III. Calcium and Dystrophic Pathology
A. Calcium Contributes to the Pathology of Dystrophic Skeletal Muscles
Calcium plays a dual role in muscle function, serving as a signaling molecule as well as a regulator of muscle contraction and relaxation.80 To effectively regulate cytosolic [Ca2+], muscle fibers contain a complex system of channels, pumps, and buffering proteins, each dedicated to mobilize, transport, or sequester Ca2+.80 In dystrophin-deficient muscle, all three components, that is, channels, pumps, and buffering proteins, appear to be dysfunctional. Ca2+ channels in the sarcolemma of mdx mice exhibit increased activity,81,82 while Ca2+ buffering proteins83,84 and ATPase pumps in the sarcoplasmic reticulum85,86 demonstrate an impaired ability to sequester Ca2+. Together, the compromised Ca2+-cycling system in mdx muscles results in an elevated level of resting intracellular [Ca2+].87–89 Accumulation of Ca2+ is particularly deleterious to the survivability of muscle fibers, as Ca2+ activates a variety of free radical and enzymatic processes that target the cytoskeleton, sarcolemma, and mitochondria. Left unchecked, these processes may ultimately result in the necrosis of the muscle fiber.90
Investigators have gained valuable insight into the relationship between calcium and MD by developing several mouse models with altered calcium handling properties. One class of calcium handling proteins to be overexpressed is the transient receptor potential canonical (TRP) channels that facilitate the entry of Ca2+ into muscle fibers. Overexpression of TRP channels in wild-type mice causes skeletal muscles to exhibit increased fibrosis, immune cell infiltration, fatty tissue, and continuous cycles of muscle regeneration. This mouse model possesses many of the pathological characteristics of dystrophic mice and demonstrates that calcium misregulation alone is sufficient to trigger a MD-like phenotype. In a related set of experiments, investigators were able to partially correct the pathology of dystrophic mdx mice by overexpressing nonfunctional mutant TRP channels.91,92 The rescue effect stems from the suppression of endogenous TRP channel activity elicited by the dominant-negative mutant TRP channels.
B. Dysferlin Gene Mutations
In addition to overactive calcium channels, compromised membrane-repair processes are also capable of eliciting MD. Dysferlin is a protein that augments membrane repair2,93,94 and deficiencies have been linked to LGMD2B and Miyoshi myopathy.95,96 In mouse models that lack dysferlin, skeletal muscles suffer from a progressive dystrophy and display altered morphology, muscle necrosis, macrophage infiltration, and fatty tissue replacement.2,97 Without dysferlin, lesions in the membrane that may occur during contractile activity remain unrepaired, allowing calcium to rush into the muscle fiber. The uncontrolled influx of calcium into the cytosol is likely to overwhelm the endogenous calcium handling apparatus and trigger gradual muscle fiber necrosis. Studies on these mouse models suggest that imbalances in cytosolic calcium levels are sufficient to induce progressive MD, independent of the absence or presence of structural proteins such as dystrophin and the DGC. Other recent studies also suggest that dysferlin may be a component of T-tubules, but whether it regulates calcium flux there is not clear.93,98
C. Calpain 3 and LGMD2A
Mutations in calpain 3 gene cause LGMD2A, one of the most prevalent forms of the disease.99–101 Calpain 3 is a calcium-dependent nonlysosomal protease involved primarily in regulating sarcomere formation and remodeling.102 Additionally, calpain 3 serves structural functions as a component of the skeletal muscle triad103 and the dysferlin complex.104 Although calpain 3 has been found in many subcellular locations, the majority of it is anchored to titin, a giant sarcomeric protein that acts as a molecular ruler and a template for sarcomere assembly during muscle cell development.105–107 As mutations in titin result in LGMD2J and a secondary deficiency of calpain 3,108 mouse models for both forms of the disease will be discussed together.
Two calpain 3 knockout mouse models have been created. Richard et al. generated mice with disrupted proteolytic site of calpain 3 by substituting exons 2 and 3 with a neoR cassette using homologous recombination.109 Skeletal muscles from the transgenic mice showed 80% decrease in calpain 3 mRNA and a complete absence of calpain 3 protein. Phenotypically, the muscles revealed regions of focal necrosis and apoptosis of nuclei associated with perturbation of NF-kB/IkBα as well as disruption of sarcolemmal integrity. Spencer's group took a different approach and generated calpain 3-deficient mice by utilizing a gene trap retroviral vector that introduced premature stop signals that completely abolished mRNA and protein expression.110 The phenotype more closely resembled LGMD2A111,112 as it showed muscle necrosis in combination with fiber atrophy. The studies utilizing these mice indicated that calpain 3 participates in remodeling of the sarcomere by promoting ubiquitination of its ligands.102 Further, calpain 3-deficient mice were shown to have reduced calcium release in fibers due to perturbations in the triad-associated complex.103
Although most cases of LGMD2A result from mutations in the proteolytic region of calpain 3, pathogenic mutations are present along the entire sequence of calpain 3 gene. Mice expressing a proteolytically inactive mutant form (C129S) that retains its conformational properties enabled dissecting enzymatic and structural functions of calpain 3. Tagawa et al. generated transgenic mice expressing the mutated protein on the wild-type background.113 Despite low expression levels, the aged mice showed signs of a mild myopathy, including reduced motor functions, as well as the presence of centrally nucleated nuclei, and lobulated and split fibers. In a more recent study,114 analyses of C129S knock-in mice revealed progressive MD associated with inefficient redistribution of the mutant protein in stretched sarcomeres. Nevertheless, the mice showed a less severe phenotype than calpain 3 knockout mice, presumably due to preserved calpain 3 structural functions in the sarcoplasmic reticulum.103,115 Another study showed that impaired titin–calpain 3 interaction results in reduced concentration of calpain 3 in the myofibrillar fraction along with its rapid degradation.116 Here, generated transgenic mice expressed mutant cal-pains known to result in LGMD2A with preserved proteolytic function but mutated titin-binding site on the caveolin 3 knockout background.
D. Titin and LGMD2J
Mutations in the last 2 coding exons of titin lead to a mild late onset dominant tibial muscular dystrophy and a severe recessive LGMD2J. The knock-in mice carrying the most common mutation (FINmaj) recapitulate the human pathology, with heterozygotes showing a very mild phenotype and homozygotes developing a progressive MD.117 This mouse model reveals also a secondary reduction in calpain 3 expression levels. Importantly, when the mice were bred onto the calpain 3-deficient background, the pathology of the FINmaj knock-in mice was attenuated, indicating that disrupted regulation of calpain 3 is responsible for the pathology of LGMD2J. It is also important to note that the pathology of another mouse model for titinopathy, severely affected MD with myositis (mdm), was not linked to deregulation of calpain 3.118 The Mdm causing deletion of 83 amino acids in the N2A region of titin arose spontaneously in the Jackson laboratory in the early 1980s.119 The mutation disrupts the calpain 3 binding site in the N2A region of titin. Despite that, mdm/calpain 3-deficient mice showed no change in the mdm pathology.
IV. α-Actinin
The α-actinins are a family of actin-binding proteins that are structurally related to dystrophin. Of the four members in the α-actinin family, only the sarcomeric isoforms α-actinin-2 and α-actinin-3 (Actn3) have been shown to bind dystrophin and other Z-disk associated proteins at the sarcomere,120 while the cytoskeletal isoforms α-actinin-1 and α-actinin-4 are involved in cytoskeleton organization and cellular attachment to the extracellular matrix at focal adhesions.121 As α-actinin-2 is the sole sarcomeric α-actinin isoform expressed in the heart, mutations in α-actinin-2 result in hypertrophic cardiomyopathy.122 In contrast, complete deficiency of α-actinin-3 occurs in 16% of the normal human population123 and is associated with reduced muscle strength and power in elite athletes124–127 and nonathletes.128,129 Phenotypic analysis of the Actn3 knockout mouse showed that absence of α-actinin-3 alters the structural, metabolic, and physiological characteristics of fast-twitch muscle fibers toward that of slow-twitch fibers,130 suggesting that α-actinin-3 expression is essential for optimal rapid power generation in muscles of humans and mice. Recently, α-actinin-3 deficiency has also been associated with increased susceptibility to contraction-induced injury.131 Work is ongoing to examine the influence of ACTN3 genotype on muscle disease phenotypes by crossing the Actn3 knockout mouse with mouse models of muscle disease, and to discover the therapeutic potential of α-actinin-3 overexpression by generation of Actn3 transgenic models.
V. Fukutin-Related Protein and Dystroglycan Processing
Fukutin-related protein (FKRP) is a putative glycosyltransferase and is one of many that glycosylate αDG, allowing it to stabilize the sarcomere at the basal lamina via binding to laminin α2. Mutations in FKRP cause a broad spectrum of clinical phenotypes with variable disease severity. The most severe forms result in structural brain abnormalities (Walker–Warburg syndrome, WWS and muscle–eye–brain disease, MEB)132 and an inabiity to walk (MDC1C).133 FKRP mutations also result in LGMD2I, one of the most common forms of LGMD in childhood. LGMD2I itself is variable—one version has an early onset, Duchenne-like phenotype (severe LGMD2I) and the other a milder, slower progression (mild LGMD2I). Clinical severity has been correlated to depletion of glycosylated αDG and secondary reduction in laminin α2 as a consequence of reduced FKRP expression.134,135
The effect of FKRP downregulation has been explored in zebrafish136 and mice137 injected with AOs. While mouse models based on patient FKRP mutations that result in MEB (Y307N),138 WWS (E310del), and MDC1C (P448L)139 have also been generated, only those retaining the neomycin cassette succeeded in reducing Fkrp transcript levels and recapitulating the severity of the disease phenotypes.138,139 Of the three mouse models, the E310del was embryonically lethal, FKRP-neo-Y307N resulted in perinatal lethality, while some FKRP-neo-P448L mutant mice survived to 10 months. The difference in lifespan of these models could be associated with the level of Fkrp mRNA/protein present. Homozygous E310del mice completely lack functional FKRP due to deletion of the C-terminus of FKRP that contains a consensus D × D motif commonly found in glycosyltransferases, suggesting that the presence of at least some FKRP activity is critical for embryonic development.139 In contrast, the FKRP-neo-Y307N homozygotes expressed approximately 40% of Fkrp mRNA compared to controls,138 while FKRP-neo-P448L homozygotes expressed approximately 55%.139 This is consistent with results from knockdown of Fkrp transcript using RNA interference, which showed that the greater reductions of Fkrp mRNA (up to 75%) using dual shRNA cassettes induce overt dystrophic pathology, compared with the lower reductions from using single shRNA cassettes.137 However, as shRNA expression by itself can cause toxicity, it remains unclear how much of the phenotype was FKRP specific.140 Differences in disease severity between the latter two models could also be attributed to laminin α2 localization, which was reduced in FKRP-neo-Y307N mutant muscles but was normal in FKRP-neo-P448L mutants. Interestingly, disease severity may not be correlated with the level of glycosylated αDG expressed, as some expression of glycosylated αDG was detected in FKRP-neo-Y307N muscles by Western blot but not in FKRP-neo-P448L muscles. Taken together, these results suggest that FKRP could have other unknown functions, for example, in regards to laminin α2 localization, and that factors other than glycosylation of αDG may be important for modulating disease severity.
VI. Desmin
Desminopathy is primarily (∼ 80%) a dominantly inherited form of myofibrillar myopathy that leads to cardiac conduction disease, cardiomyopathy, and skeletal muscle weakness.141,142 The initial symptoms can arise before 10 years of age but are more often reported in the 30s.141,142 Symptoms usually begin with cardiac conduction disease and progress to cardiomyopathy with severe respiratory distress. There is currently no specific treatment for desminopathies to prevent the progressing cardiomyopathy and deterioration of the diaphragm. Desmin is the primary intermediate filament protein of muscle that connects the sarcomeres to the sarcolemma and organelles.143 In the heart, desmin is particularly concentrated in the conduction system, at costameres, and at the desmosome in intercalated disks. Desmin-null mice develop a severe dilated cardiomyopathy from approximately 5 days of age that eventually leads to widespread cardiomyocyte degeneration, fibrosis, and calcification.144–147 Abnormal mitochondria distribution, proliferation, and swelling are thought to be a central pathologic feature of desmin knockout cardiomyocytes and desmin-related cardiomyopathy.148–150 The cardiomyocytes are also structurally abnormal in that the sarcomeres do not align and do not attach appropriately to the intercalated disks.144–147 Desmin knockout mice develop a mild cardiac conduction disease probably owing to the disruption of desmosomes in the conduction system.151 There are two transgenic mouse models that mimic dominantly inherited desminopathy in humans. The first is a 7-amino acid deletion (R173-E179) that leads to chimeric intracellular aggregates, not normally seen in desmin-null mice.152 The second transgenic mouse is a L345P mutation that significantly alters mitochondrial morphology and function that is associated with altered Ca2+ handling.153 Together, these transgenic mice suggest that different mutations in desmin can lead to strikingly different phenotypes.
VII. Laminin α2
Laminin α2-deficient mouse models have significantly expanded our understanding of congenital MD type 1A (MDC1A). Models derive from the dystrophic dy/dy mouse line whose pathology originates from mutations in the LAMA2 gene.154,155 Differences in site and type of mutation determines residual laminin α2 expression, and by extension, disease severity.156–158 The dy/dy mouse possesses a moderate fibrotic and dystrophic phenotype, coincident with reduced laminin α2 expression.156,159 The mutation responsible for the dy/dy phenotype is unknown; however, additional mouse models with targeted mutation (dy2J/dy2J) or removal (dyW/dyW) of the LAMA2 N-terminal domain show minor and severe dystrophic phenotypes, respectively.156,160,161 Importantly, the severe dystrophic phenotype observed in dyW/dyW closely resembles that of dy3K/dy3K mice where laminin α2 expression is completely abolished.156,162 While the secondary mechanisms governing MDC1A pathogenesis remain unclear, these models underscore the connection between laminin α2 expression and disease severity and will likely prove invaluable in the design of novel therapeutic strategies.
A congenital MD was found in a mixed breed dog that had an absence of the laminin isoform α2, which is predominant in, but not specific to, skeletal muscle.163 Mutations causing laminin α2 deficiency result in central nervous system defects in humans and peripheral nerve defects in dy mice. It will be interesting to see if similar abnormalities are found in laminin α2-deficient dogs.
VIII. Collagen VI
Collagens are secreted triple-helical extracellular matrix proteins that maintain integrity across many tissues.164,165 Collagen VI forms a heterotrimer composed of three alpha chains encoded by the COL6A1, COL6A2, and COL6A3 genes. The microfibrils generated upon assembly of collagen VI molecules have been implicated in the maintenance of muscle function and provide an important link between muscle cells and the surrounding matrix through a variety of molecular interactions.166–168 Mutations in the three collagen VI alpha chains give rise to autosomal dominant Bethlem myopathy (BM)169–172 as well as the more severe usually autosomal recessive Ullrich congenital muscular dystrophy (UCMD).173,174 BM is characterized by later onset, proximal muscle weakness with joint contractures, especially of the elbow, finger, and ankle, and slow progression, while UCMD typically manifests early with rapid progression of symptoms that include muscle weakness and hypotonia, joint contractures, and hyperelastic distal joints. Respiratory failure is common in later stages of UCMD and can result in early death if untreated. In both BM and UCMD, intelligence is normal and serum CK levels are normal or slightly elevated.
Investigation of collagen function led to generation of collagen VI-deficient mice through targeted disruption of the col6a1 gene, thus abolishing assembly of the triple helix.175 The resulting phenotype strongly resembled BM, and these mice became a useful animal model for studying the pathophysiology of both BM and UCMD.167 Histology and Evans blue dye staining revealed early onset focal necrosis, fiber size variation, and mild central nucleation, all indicators of skeletal muscle pathology. Locomotor performance is not severely affected, though col6a1-deficient mice exhibit muscle weakness, and the condition progresses very slowly, similar to BM. However, the diaphragm is more severely affected than other muscle groups, similar to UCMD.167,176,177 Col6a1-deficient mice also reflect the dominant character of BM, as heterozygotes show signs of myopathy, though with milder defects.175
The mouse model of collagen VI deficiencies has revealed important mechanistic insights into the pathophysiology of BM and UCMD, including the involvement of mitochondrial dysfunction and apoptosis,167,176 but the relationship between these abnormalities and collagen mutations is still poorly understood. Nevertheless, col6a1-deficient mice have allowed testing of specific therapeutic interventions such as cyclosporine A and other pharmacological treatments to target the mitochondrial permeability transition pore.167,178 While the severity of the deficits observed in UCMD is not captured by these mice, zebrafish models have been reported with early onset, severe myopathy that could be useful for initial screening of novel therapies, and further investigating the molecular etiology of both BM and UCMD.179
IX. D4Z4 Repeats and FSHD
Facioscapulohumeral muscular dystrophy (FSHD) is caused by macrosatellite DNA contraction in the D4Z4 region of telomeric 4q35.180 The molecular events following DNA contraction include epigenetic events resulting in chromatin hypomethylation, chromatin relaxation, and expression of a highly conserved double homeobox retrogene, DUX4, embedded within the repeats.181 Recent studies suggest that disease arises only when DUX4 expression occurs in the context of an intact polyadenylation site following the last repeat on the pathogenic alleles and presents a unifying model for FSHD disease mechanism.182,183
Controversy surrounds the involvement of another gene proximal to 4q35, FRG1. Transgenic mouse studies show that overexpression of FRG1 causes an MD similar to FSHD.184 Disease severity increases with an increase in FRG1 expression. To date, this is the only model that recapitulates some of the disease features, such as a degenerative muscle phenotype more pronounced in muscles similarly affected in FSHD patients, increased central nuclei, abnormal fiber morphology with increased cross-sectional area, decreased muscle size, kyphosis, and weakness.
Attempts to produce a transgenic mouse line expressing DUX4 have not been successful, but engineering a mouse line that expresses a low level of DUX4 in adult mice to recapitulate DUX4 levels in FSHD patient muscle may prove a difficult task. DUX4 expression in muscle cells in culture results in cell death accompanied by an increased sensitivity to oxidative stress which supports the notion that DUX4 plays a role in early development but is inactivated in the adult. However, because it is generally agreed that overexpression of a gene or genes is the cause of FSHD, therapy development is now underway directed toward eliminating toxicity by targeting mRNA for destruction. Muscle structural and histological changes associated with FRG1 overexpression in the FRG1 transgenic mouse were improved with delivery of constitutively expressed RNA hairpins to direct RNA interference.140,185 More importantly, functional rescue of muscles, as measured by endurance in treadmill running studies, was possible with AAV6 short hairpin RNA intravascular systemic delivery after the onset of disease.140 These studies serve as a proof-of-principle for therapeutic antisense approaches for FSHD and other dominant muscle diseases.
X. DMPK and Myotonic Dystrophy
Myotonic dystrophy (DM1) is caused by a microsatellite CTG repeat expansion in the 3′UTR of the dystrophia myotonica protein kinase gene (DMPK).186 Transcripts with expanded repeats accumulate in the nuclei of cells and alter the availability of proteins involved in the regulation of alternative splicing during development.187,188 Important protein families for developmental splicing regulation that are affected by the DMPK mRNA repeat expansions are the muscleblind-like (MBNL) and CUGBP/Elav-like family (CELF) proteins.187,189,190 Disruption of splicing patterns to produce embryonic transcript profiles for a host of unrelated mRNAs results in a disease phenotype with seemingly unrelated features, such as muscle stiffness (myotonia), hyperinsulinemia, gastrointestinal insufficiencies, and cognitive impairment.186
Approximately 20 transgenic mouse lines have been generated to dissect the molecular pathology resulting from the toxic gain-of-function mRNA expression and to produce models for testing potential therapies for DM1 (reviewed in detail191a). The models best suited for therapy testing are based on either expressing the CTG repeat expansion or altering the levels of MBNL1 and CUGBP1. One of the most informative and useful transgenic lines, the HSALR line, was generated from expression of CTG250 from the human α-skeletal actin gene placed in the HSA 3′UTR.191b The HSALR mice display profound myotonia and subsequent changes in MBNL levels that lead to altered splicing profiles. Similar to the human disease, MBNL1 can be found in nuclear accumulations of the protein with the CUG250 repeat HSA mRNA. Notably, CUGBP1 levels are not affected, and the muscle-wasting phenotype of DM1 is not reproduced in this transgenic mouse line. Additionally, expression of HSALR is limited to muscle and therefore cannot contribute to the effects in multiple organ systems associated with the human condition. Regardless of the restriction of expression of the HSALR mRNA in muscle, the HSALR mouse has been used for testing antisense therapeutic approaches and has resulted in studies showing elimination of myotonia and reversal of splicing defects with intramuscular delivery of AOs targeting the CUG repeats.192
Another notable transgenic line contains a large fragment of the human DMPK locus with an unstable expanded CTG repeat that has become larger with increasing generations to reach 1000–1800 repeats.193 The DM500 mice display multisystemic attributes of human DM1, including splicing defects in the muscle, heart, CNS, myotonia, muscle weakness, glucose metabolism defects, and tau distribution abnormalities in the brain. The requirement for homozygosity in these mice suggests that the level of mutant DMPK mRNA expression is below a threshold necessary for pronounced disease and makes these mice expensive and difficult for testing potential therapies.
A third model allowed conditional tissue-specific expression of a 960 repeat expansion in the context of the last exon of DMPK where the expansion normally is found.194 These mice recapitulate all muscle features of DM1, including muscle wasting, highlighting the importance of the chromosomal context for a more complete disease phenocopy. Other models focus on manipulation of the levels of the MBNL and CLEF protein family members and have limited potential for therapeutic testing related to manipulation of their expression levels. MBNL1 knockout mice recapitulate the splicing defects and myotonia observed with toxic repeat expression.195 CUGBP1 overexpressing mice display a severe muscle-wasting phenotype not seen in repeat expansion-based models lacking the DMPK chromosomal context, which correlates CUGBP1 upregulation with a muscle-wasting phenotype.196 Despite the numbers of transgenic mice made to reproduce the many features of DM1, there remain only two lines that have been used for testing potential therapies: the HSALR model and the DM500 model (500 repeats in large chrosomal fragment from the DMPK locus). This situation highlights the difficulties often associated with attempts to recapitulate human disease in the mouse with gene insertion, especially related to unstable repetitive and ultimately toxic DNA.
XI. Conclusions
The large and growing number of animal models available to study MD has proven to be an indispensible resource for understanding the pathophysiology of these disorders and for testing experimental approaches to therapy.197 However, the genes for a number of forms of MD have not yet been identified and there do not yet exist corresponding animal models, and even those genes that have been found have not all been mutated in animals. Developing these newer models is an important goal, especially for the rare MDs where few patient samples are available. In such cases, therapies being developed for other disorders could potentially be quickly tested in animal models for safety prior to therapeutically oriented clinical trials.
Acknowledgments
Supported by grants from the Muscular Dystrophy Association (to G. L. O., G. B. B., J. R. C., and J. S. C.), by NIH Grant AR044533 (to J. S. C.), and by a grant from the Pacific Northwest Friends of FSHD (to J. R. C.).
References
- 1.Emery AE. The muscular dystrophies. Lancet. 2002;359:687–95. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 2.Bansal D, Miyake K, Vogel SS, Groh S, Chen CC, Williamson R, et al. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–72. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 3.Arnett AL, Chamberlain JR, Chamberlain JS. Therapy for neuromuscular disorders. Curr Opin Genet Dev. 2009;19:290–7. doi: 10.1016/j.gde.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel LM, et al. Analysis of deletions in DNA from patients with Becker and Duchenne muscular dystrophy. Nature. 1986;322:73–7. doi: 10.1038/322073a0. [DOI] [PubMed] [Google Scholar]
- 5.Monaco AP, Neve RL, Coletti-Feener C, Bertelson CJ, Kurnit DM, Kunkel LM. Isolation of candidate cDNA clones for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–50. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 6.Emery AE, Muntoni F. Duchenne muscular dystrophy. 3rd. Oxford: Oxford University Press; 2003. [Google Scholar]
- 7.Abmayr S, Chamberlain J. The structure and function of dystrophin. In: Winder SJ, editor. Molecular mechanisms of muscular dystrophies. Georgetown: Landes Biosciences; 2006. pp. 14–34. [Google Scholar]
- 8.Ervasti JM. Structure and function of the dystrophin-glycoprotein complex. In: Winder SJ, editor. Molecular mechanisms of muscular dystrophies. Georgetown: Landes Biosciences; 2006. pp. 1–13. [Google Scholar]
- 9.Banks GB, Chamberlain JS. The value of mammalian models for Duchenne muscular dystrophy in developing therapeutic strategies. Curr Top Dev Biol. 2008;84:431–53. doi: 10.1016/S0070-2153(08)00609-1. [DOI] [PubMed] [Google Scholar]
- 10.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81:1189–92. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–80. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 12.Im WB, Phelps SF, Copen EH, Adams EG, Slightom JL, Chamberlain JS. Differential expression of dystrophin isoforms in strains of mdx mice with different mutations. Hum Mol Genet. 1996;5:1149–53. doi: 10.1093/hmg/5.8.1149. [DOI] [PubMed] [Google Scholar]
- 13.Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21:2195–204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 14.Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360:588–91. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- 15.Takemitsu M, Ishiura S, Koga R, Kamakura K, Arahata K, Nonaka I, et al. Dystrophin-related protein in the fetal and denervated skeletal muscles of normal and mdx mice. Biochem Biophys Res Commun. 1991;180:1179–86. doi: 10.1016/s0006-291x(05)81320-8. [DOI] [PubMed] [Google Scholar]
- 16.Coulton GR, Curtin NA, Morgan JE, Partridge TA. The mdx mouse skeletal muscle myopathy: II. Contractile properties. Neuropathol Appl Neurobiol. 1988;14:299–314. doi: 10.1111/j.1365-2990.1988.tb00890.x. [DOI] [PubMed] [Google Scholar]
- 17.Dangain J, Vrbova G. Muscle development in mdx mutant mice. Muscle Nerve. 1984;7:700–4. doi: 10.1002/mus.880070903. [DOI] [PubMed] [Google Scholar]
- 18.Tanabe Y, Esaki K, Nomura T. Skeletal muscle pathology in X chromosome-linked muscular dystrophy (mdx) mouse. Acta Neuropathol. 1986;69:91–5. doi: 10.1007/BF00687043. [DOI] [PubMed] [Google Scholar]
- 19.Stedman HH, Sweeney HL, Shrager JB, Maguire HC, Panettieri RA, Petrof B, et al. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature. 1991;352:536–9. doi: 10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- 20.Lynch GS, Rafael JA, Hinkle RT, Cole NM, Chamberlain JS, Faulkner JA. Contractile properties of diaphragm muscle segments from old mdx and old transgenic mdx mice. Am J Physiol. 1997;272:C2063–8. doi: 10.1152/ajpcell.1997.272.6.C2063. Cell Physiol. 41. [DOI] [PubMed] [Google Scholar]
- 21.Chapman VM, Miller DR, Armstrong D, Caskey CT. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proc Natl Acad Sci USA. 1989;86:1292–6. doi: 10.1073/pnas.86.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox GA, Phelps SF, Chapman VM, Chamberlain JS. New mdx mutation disrupts expression of muscle and nonmuscle isoforms of dystrophin. Nat Genet. 1993;4:87–93. doi: 10.1038/ng0593-87. [DOI] [PubMed] [Google Scholar]
- 23.Danko I, Chapman V, Wolff JA. The frequency of revertants in mdx mouse genetic models for Duchenne muscular dystrophy. Pediatr Res. 1992;32:128–31. doi: 10.1203/00006450-199207000-00025. [DOI] [PubMed] [Google Scholar]
- 24.Rafael JA, Cox GA, Corrado K, Jung D, Campbell KP, Chamberlain JS. Forced expression of dystrophin deletion constructs reveals structure-function correlations. J Cell Biol. 1996;134:93–102. doi: 10.1083/jcb.134.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gregorevic P, Allen JM, Minami E, Blankinship MJ, Haraguchi M, Meuse L, et al. rAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic mice. Nat Med. 2006;12:787–9. doi: 10.1038/nm1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odom GL, Gregorevic P, Allen JM, Finn E, Chamberlain JS. Microutrophin delivery through rAAV6 increases lifespan and improves muscle function in dystrophic dystrophin/ utrophin-deficient mice. Mol Ther. 2008;16:1539–45. doi: 10.1038/mt.2008.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wertz K, Fuchtbauer EM. Dmd(mdx-beta geo): a new allele for the mouse dystrophin gene. Dev Dyn. 1998;212:229–41. doi: 10.1002/(SICI)1097-0177(199806)212:2<229::AID-AJA7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 28.Araki E, Nakamura K, Nakao K, Kameya S, Kobayashi O, Nonaka I, et al. Targeted disruption of exon 52 in the mouse dystrophin gene induced muscle degeneration similar to that observed in Duchenne muscular dystrophy. Biochem Biophys Res Commun. 1997;238:492–7. doi: 10.1006/bbrc.1997.7328. [DOI] [PubMed] [Google Scholar]
- 29.Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30:293–9. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 30.Helderman-van den Enden AT, Straathof CS, Aartsma-Rus A, den Dunnen JT, Verbist BM, Bakker E, et al. Becker muscular dystrophy patients with deletions around exon 51; a promising outlook for exon skipping therapy in Duchenne patients. Neuromuscul Disord. 2010;20:251–4. doi: 10.1016/j.nmd.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–28. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goemans NM, Tulinius M, van den Akker JT, Burm BE, Ekhart PF, Heuvelmans N, et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med. 2011;364:1513–22. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 33.van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–86. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 34.Chamberlain JR, Chamberlain JS. Muscling in: gene therapies for muscular dystrophy target RNA. Nat Med. 2010;16:170–1. doi: 10.1038/nm0210-170. [DOI] [PubMed] [Google Scholar]
- 35.Aoki Y, Nakamura A, Yokota T, Saito T, Okazawa H, Nagata T, et al. In-frame dystrophin following exon 51-skipping improves muscle pathology and function in the exon 52-deficient mdx mouse. Mol Ther. 2010;18:1995–2005. doi: 10.1038/mt.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kudoh H, Ikeda H, Kakitani M, Ueda A, Hayasaka M, Tomizuka K, et al. A new model mouse for Duchenne muscular dystrophy produced by 2.4 Mb deletion of dystrophin gene using Cre-loxP recombination system. Biochem Biophys Res Commun. 2005;328:507–16. doi: 10.1016/j.bbrc.2004.12.191. [DOI] [PubMed] [Google Scholar]
- 37.Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, et al. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90:717–27. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- 38.Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. 1997;90:729–38. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 39.Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM, et al. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4:1441–4. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 40.Rafael JA, Tinsley JM, Potter AC, Deconinck AE, Davies KE. Skeletal muscle-specific expression of a utrophin transgene rescues utrophin-dystrophin deficient mice. Nat Genet. 1998;19:79–82. doi: 10.1038/ng0598-79. [DOI] [PubMed] [Google Scholar]
- 41.Chandrasekharan K, Yoon JH, Xu Y, deVries S, Camboni M, Janssen PM, et al. A human-specific deletion in mouse Cmah increases disease severity in the mdx model of Duchenne muscular dystrophy. Sci Transl Med. 2010;2:42ra54. doi: 10.1126/scitranslmed.3000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–71. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper BJ, Winand NJ, Stedman H, Valentine BA, Hoffman EP, Kunkel LM, et al. The homologue of the Duchenne locus is defective in X-linked muscular dystrophy of dogs. Nature. 1988;334:154–6. doi: 10.1038/334154a0. [DOI] [PubMed] [Google Scholar]
- 44.Partridge TA. Models of dystrophinopathy, pathological mechanisms and assessment of therapies. In: Brown SC, Lacy JA, editors. Dystrophin: gene, protein and cell biology. Cambridge: Cambridge University Press; 1997. pp. 310–31. [Google Scholar]
- 45.Schatzberg SJ, Olby NJ, Breen M, Anderson LV, Langford CF, Dickens HF, et al. Molecular analysis of a spontaneous dystrophin ‘knockout’ dog. Neuromuscul Disord. 1999;9:289–95. doi: 10.1016/s0960-8966(99)00011-5. [DOI] [PubMed] [Google Scholar]
- 46.Valentine BA, Winand NJ, Pradhan D, Moise NS, de Lahunta A, Kornegay JN, et al. Canine X-linked muscular dystrophy as an animal model of Duchenne muscular dystrophy: a review. Am J Med Genet. 1992;42:352–6. doi: 10.1002/ajmg.1320420320. [DOI] [PubMed] [Google Scholar]
- 47.Shimatsu Y, Katagiri K, Furuta T, Nakura M, Tanioka Y, Yuasa K, et al. Canine X-linked muscular dystrophy in Japan (CXMDJ) Exp Anim. 2003;52:93–7. doi: 10.1538/expanim.52.93. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Kuhr CS, Allen JM, Blankinship M, Gregorevic P, Chamberlain JS, et al. Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Mol Ther. 2007;15:1160–6. doi: 10.1038/sj.mt.6300161. [DOI] [PubMed] [Google Scholar]
- 49.Meryon E. On granular and fatty degeneration of the voluntary muscles. Med Chir Trans. 1852;35(73–84):1. [PMC free article] [PubMed] [Google Scholar]
- 50.Duchenne GB. Récherches sur la paralysie musculaire pseudohypertrophique ou paralysie myo-sclerosique. Arch Gen Med. 1868;11:5–25. [Google Scholar]
- 51.Kloepfer HW, Talley C. Autosomal recessive inheritance of Duchennetype muscular dystrophy. Ann Hum Genet. 1958;22:138–43. doi: 10.1111/j.1469-1809.1957.tb01928.x. [DOI] [PubMed] [Google Scholar]
- 52.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–28. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 53.Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–31. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- 54.Roberds SL, Leturcq F, Allamand V, Piccolo F, Jeanpierre M, Anderson RD, et al. Missense mutations in the adhalin gene linked to autosomal recessive muscular dystrophy. Cell. 1994;78:625–33. doi: 10.1016/0092-8674(94)90527-4. [DOI] [PubMed] [Google Scholar]
- 55.Bonnemann CG, Modi R, Noguchi S, Mizuno Y, Yoshida M, Gussoni E, et al. Beta-sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat Genet. 1995;11:266–73. doi: 10.1038/ng1195-266. [DOI] [PubMed] [Google Scholar]
- 56.Noguchi S, McNally EM, Ben Othmane K, Hagiwara Y, Mizuno Y, Yoshida M, et al. Mutations in the dystrophin-associated protein gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science. 1995;270:819–22. doi: 10.1126/science.270.5237.819. [DOI] [PubMed] [Google Scholar]
- 57.Nigro V, de Sa Moreira E, Piluso G, Vainzof M, Belsito A, Politano L, et al. Autosomal recessive limb-girdle muscular dystrophy, LGMD2F, is caused by a mutation in the delta-sarcoglycan gene. Nat Genet. 1996;14:195–8. doi: 10.1038/ng1096-195. [DOI] [PubMed] [Google Scholar]
- 58.Homburger F, Baker JR, Nixon CW, Wilgram G. New hereditary disease of Syrian hamsters. Primary, generalized polymyopathy and cardiac necrosis. Arch Intern Med. 1962;110:660–2. doi: 10.1001/archinte.1962.03620230106015. [DOI] [PubMed] [Google Scholar]
- 59.Nigro V, Piluso G, Belsito A, Politano L, Puca AA, Papparella S, et al. Identification of a novel sarcoglycan gene at 5q33 encoding a sarcolemmal 35 kDa glycoprotein. Hum Mol Genet. 1996;5:1179–86. doi: 10.1093/hmg/5.8.1179. [DOI] [PubMed] [Google Scholar]
- 60.Nigro V, Okazaki Y, Belsito A, Piluso G, Matsuda Y, Politano L, et al. Identification of the Syrian hamster cardiomyopathy gene. Hum Mol Genet. 1997;6:601–7. doi: 10.1093/hmg/6.4.601. [DOI] [PubMed] [Google Scholar]
- 61.Awad SS, Welty JD. Comparisons of hemodynamics throughout the life span of the Bio 14.6 cardiomyopathic with the F1B normal hamster. Comp Biochem Physiol A Comp Physiol. 1990;97:487–91. doi: 10.1016/0300-9629(90)90115-9. [DOI] [PubMed] [Google Scholar]
- 62.Sakamoto A, Ono K, Abe M, Jasmin G, Eki T, Murakami Y, et al. Both hypertrophic and dilated cardiomyopathies are caused by mutation of the same gene, delta-sarcoglycan, in hamster: an animal model of disrupted dystrophin-associated glycoprotein complex. Proc Natl Acad Sci USA. 1997;94:13873–8. doi: 10.1073/pnas.94.25.13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakamoto A. Molecular pathogenesis of severe cardiomyopathy in the TO-2 hamster. Exp Clin Cardiol. 2003;8:143–6. [PMC free article] [PubMed] [Google Scholar]
- 64.Hack AA, Groh ME, McNally EM. Sarcoglycans in muscular dystrophy. Microsc Res Tech. 2000;48:167–80. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<167::AID-JEMT5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 65a.Coral-Vazquez R, Cohn RD, Moore SA, Hill JA, Weiss RM, Davisson RL, et al. Disruption of the sarcoglycan-sarcospan complex in vascular smooth muscle: a novel mechanism for cardiomyopathy and muscular dystrophy. Cell. 1999;98:465–74. doi: 10.1016/s0092-8674(00)81975-3. [DOI] [PubMed] [Google Scholar]
- 65b.Burbach JA, Schlenker EH, Johnson JL. Morphometry, histochemistry, and contractility of dystrophic hamster diaphragm. Am J Physiol. 1987;253:R275–84. doi: 10.1152/ajpregu.1987.253.2.R275. [DOI] [PubMed] [Google Scholar]
- 65c.Wansapura JP, Millay DP, Dunn RS, Molkentin JD, Benson DW. Magnetic resonance imaging assessment of cardiac dysfunction in delta-sarcoglycan null mice. Neuromuscul Disord. 2011;21:68–73. doi: 10.1016/j.nmd.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hack AA, Ly CT, Jiang F, Clendenin CJ, Sigrist KS, Wollmann RL, et al. Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J Cell Biol. 1998;142:1279–87. doi: 10.1083/jcb.142.5.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ben Hamida M, Ben Hamida C, Zouari M, Belal S, Hentati F. Limb-girdle muscular dystrophy 2C: clinical aspects. Neuromuscul Disord. 1996;6:493–4. doi: 10.1016/s0960-8966(96)00395-1. [DOI] [PubMed] [Google Scholar]
- 68.Durbeej M, Cohn RD, Hrstka RF, Moore SA, Allamand V, Davidson BL, et al. Disruption of the beta-sarcoglycan gene reveals pathogenetic complexity of limb-girdle muscular dystrophy type 2E. Mol Cell. 2000;5:141–51. doi: 10.1016/s1097-2765(00)80410-4. [DOI] [PubMed] [Google Scholar]
- 69.Araishi K, Sasaoka T, Imamura M, Noguchi S, Hama H, Wakabayashi E, et al. Loss of the sarcoglycan complex and sarcospan leads to muscular dystrophy in beta-sarcoglycan-deficient mice. Hum Mol Genet. 1999;8:1589–98. doi: 10.1093/hmg/8.9.1589. [DOI] [PubMed] [Google Scholar]
- 70.Cohn RD, Durbeej M, Moore SA, Coral-Vazquez R, Prouty S, Campbell KP. Prevention of cardiomyopathy in mouse models lacking the smooth muscle sarcoglycan-sarcospan complex. J Clin Invest. 2001;107:R1–7. doi: 10.1172/JCI11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Durbeej M, Campbell KP. Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr Opin Genet Dev. 2002;12:349–61. doi: 10.1016/s0959-437x(02)00309-x. [DOI] [PubMed] [Google Scholar]
- 72.Chan YM, Bonnemann CG, Lidov HG, Kunkel LM. Molecular organization of sarcoglycan complex in mouse myotubes in culture. J Cell Biol. 1998;143:2033–44. doi: 10.1083/jcb.143.7.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duggan DJ, Manchester D, Stears KP, Mathews DJ, Hart C, Hoffman EP. Mutations in the delta-sarcoglycan gene are a rare cause of autosomal recessive limb-girdle muscular dystrophy (LGMD2) Neurogenetics. 1997;1:49–58. doi: 10.1007/s100480050008. [DOI] [PubMed] [Google Scholar]
- 74.Klinge L, Dekomien G, Aboumousa A, Charlton R, Epplen JT, Barresi R, et al. Sarcoglycanopathies: can muscle immunoanalysis predict the genotype? Neuromuscul Disord. 2008;18:934–41. doi: 10.1016/j.nmd.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 75.Vainzof M, Passos-Bueno MR, Canovas M, Moreira ES, Pavanello RC, Marie SK, et al. The sarcoglycan complex in the six autosomal recessive limb-girdle muscular dystrophies. Hum Mol Genet. 1996;5:1963–9. doi: 10.1093/hmg/5.12.1963. [DOI] [PubMed] [Google Scholar]
- 76.Duclos F, Straub V, Moore SA, Venzke DP, Hrstka RF, Crosbie RH, et al. Progressive muscular dystrophy in alpha-sarcoglycan-deficient mice. J Cell Biol. 1998;142:1461–71. doi: 10.1083/jcb.142.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patel ND, Jannapureddy SR, Hwang W, Chaudhry I, Boriek AM. Altered muscle force and stiffness of skeletal muscles in alpha-sarcoglycan-deficient mice. Am J Physiol Cell Physiol. 2003;284:C962–8. doi: 10.1152/ajpcell.00326.2002. [DOI] [PubMed] [Google Scholar]
- 78.Deitz K, Morrison JA, Kline K, Guo LT, Shelton GD. Sarcoglycan-deficient muscular dystrophy in a Boston Terrier. J Vet Intern Med. 2008;22:476–80. doi: 10.1111/j.1939-1676.2008.0080.x. [DOI] [PubMed] [Google Scholar]
- 79.Schatzberg S. Proceedings of the American College of Veterinary Internal Medicine, 22nd annual forum, Dallas, TX. 2003 [Google Scholar]
- 80.Berchtold MW, Brinkmeier H, Muntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiol Rev. 2000;80:1215–65. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- 81.Turner PR, Fong PY, Denetclaw WF, Steinhardt RA. Increased calcium influx in dystrophic muscle. J Cell Biol. 1991;115:1701–12. doi: 10.1083/jcb.115.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vandebrouck C, Martin D, Colson-Van SM, Debaix H, Gailly P. Involvement of TRPC in the abnormal calcium influx observed in dystrophic (mdx) mouse skeletal muscle fibers. J Cell Biol. 2002;158:1089–96. doi: 10.1083/jcb.200203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Culligan K, Banville N, Dowling P, Ohlendieck K. Drastic reduction of calsequestrin-like proteins and impaired calcium binding in dystrophic mdx muscle. J Appl Physiol. 2002;92:435–45. doi: 10.1152/japplphysiol.00903.2001. [DOI] [PubMed] [Google Scholar]
- 84.Dowling P, Doran P, Ohlendieck K. Drastic reduction of sarcalumenin in Dp427 (dystrophin of 427 kDa)-deficient fibres indicates that abnormal calcium handling plays a key role in muscular dystrophy. Biochem J. 2004;379:479–88. doi: 10.1042/BJ20031311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Divet A, Huchet-Cadiou C. Sarcoplasmic reticulum function in slow- and fast-twitch skeletal muscles from mdx mice. Pflugers Arch. 2002;444:634–43. doi: 10.1007/s00424-002-0854-5. [DOI] [PubMed] [Google Scholar]
- 86.Divet A, Lompre AM, Huchet-Cadiou C. Effect of cyclopiazonic acid, an inhibitor of the sarcoplasmic reticulum Ca-ATPase, on skeletal muscles from normal and mdx mice. Acta Physiol Scand. 2005;184:173–86. doi: 10.1111/j.1365-201X.2005.01450.x. [DOI] [PubMed] [Google Scholar]
- 87.Alderton JM, Steinhardt RA. Calcium influx through calcium leak channels is responsible for the elevated levels of calcium-dependent proteolysis in dystrophic myotubes. J Biol Chem. 2000;275:9452–60. doi: 10.1074/jbc.275.13.9452. [DOI] [PubMed] [Google Scholar]
- 88.Mallouk N, Jacquemond V, Allard B. Elevated subsarcolemmal Ca2+ in mdx mouse skeletal muscle fibers detected with Ca2+-activated K + channels. Proc Natl Acad Sci USA. 2000;97:4950–5. doi: 10.1073/pnas.97.9.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turner PR, Westwood T, Regen CM, Steinhardt RA. Increased protein degradation results from elevated free calcium levels found in muscle from mdx mice. Nature. 1988;335:735–8. doi: 10.1038/335735a0. [DOI] [PubMed] [Google Scholar]
- 90.Gissel H. The role of Ca2 + in muscle cell damage. Ann N Y Acad Sci. 2005;1066:166–80. doi: 10.1196/annals.1363.013. [DOI] [PubMed] [Google Scholar]
- 91.Iwata Y, Katanosaka Y, Arai Y, Shigekawa M, Wakabayashi S. Dominant-negative inhibition of Ca2+ influx via TRPV2 ameliorates muscular dystrophy in animal models. Hum Mol Genet. 2009;18:824–34. doi: 10.1093/hmg/ddn408. [DOI] [PubMed] [Google Scholar]
- 92.Millay DP, Goonasekera SA, Sargent MA, Maillet M, Aronow BJ, Molkentin JD. Calcium influx is sufficient to induce muscular dystrophy through a TRPC-dependent mechanism. Proc Natl Acad Sci USA. 2009;106:19023–8. doi: 10.1073/pnas.0906591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klinge L, Laval S, Keers S, Haldane F, Straub V, Barresi R, et al. From T-tubule to sarcolemma: damage-induced dysferlin translocation in early myogenesis. FASEB J. 2007;21:1768–76. doi: 10.1096/fj.06-7659com. [DOI] [PubMed] [Google Scholar]
- 94.Lennon NJ, Kho A, Bacskai BJ, Perlmutter SL, Hyman BT, Brown RH., Jr Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J Biol Chem. 2003;278:50466–73. doi: 10.1074/jbc.M307247200. [DOI] [PubMed] [Google Scholar]
- 95.Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, et al. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat Genet. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- 96.Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, et al. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat Genet. 1998;20:31–6. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- 97.Ho M, Post CM, Donahue LR, Lidov HG, Bronson RT, Goolsby H, et al. Disruption of muscle membrane and phenotype divergence in two novel mouse models of dysferlin deficiency. Hum Mol Genet. 2004;13:1999–2010. doi: 10.1093/hmg/ddh212. [DOI] [PubMed] [Google Scholar]
- 98.Klinge L, Harris J, Sewry C, Charlton R, Anderson L, Laval S, et al. Dysferlin associates with the developing T-tubule system in rodent and human skeletal muscle. Muscle Nerve. 2010;41:166–73. doi: 10.1002/mus.21166. [DOI] [PubMed] [Google Scholar]
- 99.Beckmann JS, Spencer M. Calpain 3, the “gatekeeper” of proper sarcomere assembly, turnover and maintenance. Neuromuscul Disord. 2008;18:913–21. doi: 10.1016/j.nmd.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duguez S, Bartoli M, Richard I. Calpain 3: a key regulator of the sarcomere? FEBS J. 2006;273:3427–36. doi: 10.1111/j.1742-4658.2006.05351.x. [DOI] [PubMed] [Google Scholar]
- 101.Sorimachi H, Hata S, Ono Y. Calpain chronicle—an enzyme family under multidisciplinary characterization. Proc Jpn Acad Ser B Phys Biol Sci. 2011;87:287–327. doi: 10.2183/pjab.87.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kramerova I, Kudryashova E, Venkatraman G, Spencer MJ. Calpain 3 participates in sarcomere remodeling by acting upstream of the ubiquitin-proteasome pathway. Hum Mol Genet. 2005;14:2125–34. doi: 10.1093/hmg/ddi217. [DOI] [PubMed] [Google Scholar]
- 103.Kramerova I, Kudryashova E, Wu B, Ottenheijm C, Granzier H, Spencer MJ. Novel role of calpain-3 in the triad-associated protein complex regulating calcium release in skeletal muscle. Hum Mol Genet. 2008;17:3271–80. doi: 10.1093/hmg/ddn223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang Y, de Morree A, van Remoortere A, Bushby K, Frants RR, Dunnen JT, et al. Calpain 3 is a modulator of the dysferlin protein complex in skeletal muscle. Hum Mol Genet. 2008;17:1855–66. doi: 10.1093/hmg/ddn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Linke WA, Kruger M. The giant protein titin as an integrator of myocyte signaling pathways. Physiology. 2010;25:186–98. doi: 10.1152/physiol.00005.2010. [DOI] [PubMed] [Google Scholar]
- 106.Kruger M, Linke WA. The giant protein titin: a regulatory node that integrates myocyte signaling pathways. J Biol Chem. 2011;286:9905–12. doi: 10.1074/jbc.R110.173260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gautel M. The sarcomeric cytoskeleton: who picks up the strain? Curr Opin Cell Biol. 2011;23:39–46. doi: 10.1016/j.ceb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 108.Haravuori H, Vihola A, Straub V, Auranen M, Richard I, Marchand S, et al. Secondary calpain3 deficiency in 2q-linked muscular dystrophy: titin is the candidate gene. Neurology. 2001;56:869–77. doi: 10.1212/wnl.56.7.869. [DOI] [PubMed] [Google Scholar]
- 109.Richard I, Roudaut C, Marchand S, Baghdiguian S, Herasse M, Stockholm D, et al. Loss of calpain 3 proteolytic activity leads to muscular dystrophy and to apoptosis-associated IkappaBalpha/nuclear factor kappaB pathway perturbation in mice. J Cell Biol. 2000;151:1583–90. doi: 10.1083/jcb.151.7.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kramerova I, Kudryashova E, Tidball JG, Spencer MJ. Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum Mol Genet. 2004;13:1373–88. doi: 10.1093/hmg/ddh153. [DOI] [PubMed] [Google Scholar]
- 111.Fanin M, Nascimbeni AC, Fulizio L, Trevisan CP, Meznaric-Petrusa M, Angelini C. Loss of calpain-3 autocatalytic activity in LGMD2A patients with normal protein expression. Am J Pathol. 2003;163:1929–36. doi: 10.1016/S0002-9440(10)63551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vainzof M, de Paula F, Tsanaclis AM, Zatz M. The effect of calpain 3 deficiency on the pattern of muscle degeneration in the earliest stages of LGMD2A. J Clin Pathol. 2003;56:624–6. doi: 10.1136/jcp.56.8.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tagawa K, Taya C, Hayashi Y, Nakagawa M, Ono Y, Fukuda R, et al. Myopathy phenotype of transgenic mice expressing active site-mutated inactive p94 skeletal muscle-specific calpain, the gene product responsible for limb girdle muscular dystrophy type 2A. Hum Mol Genet. 2000;9:1393–402. doi: 10.1093/hmg/9.9.1393. [DOI] [PubMed] [Google Scholar]
- 114.Ojima K, Kawabata Y, Nakao H, Nakao K, Doi N, Kitamura F, et al. Dynamic distribution of muscle-specific calpain in mice has a key role in physical-stress adaptation and is impaired in muscular dystrophy. J Clin Invest. 2010;120:2672–83. doi: 10.1172/JCI40658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ojima K, Ono Y, Ottenheijm C, Hata S, Suzuki H, Granzier H, et al. Non-proteolytic functions of calpain-3 in sarcoplasmic reticulum in skeletal muscles. J Mol Biol. 2011;407:439–49. doi: 10.1016/j.jmb.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ermolova N, Kudryashova E, Difranco M, Vergara J, Kramerova I, Spencer MJ. Pathogenity of some limb girdle muscular dystrophy mutations can result from reduced anchorage to myofibrils and altered stability of calpain 3. Hum Mol Genet. 2011;20:3331–45. doi: 10.1093/hmg/ddr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Charton K, Daniele N, Vihola A, Roudaut C, Gicquel E, Monjaret F, et al. Removal of the calpain 3 protease reverses the myopathology in a mouse model for titinopathies. Hum Mol Genet. 2010;19:4608–24. doi: 10.1093/hmg/ddq388. [DOI] [PubMed] [Google Scholar]
- 118.Huebsch KA, Kudryashova E, Wooley CM, Sher RB, Seburn KL, Spencer MJ, et al. Mdm muscular dystrophy: interactions with calpain 3 and a novel functional role for titin's N2A domain. Hum Mol Genet. 2005;14:2801–11. doi: 10.1093/hmg/ddi313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Garvey SM, Rajan C, Lerner AP, Frankel WN, Cox GA. The muscular dystrophy with myositis (mdm) mouse mutation disrupts a skeletal muscle-specific domain of titin. Genomics. 2002;79:146–9. doi: 10.1006/geno.2002.6685. [DOI] [PubMed] [Google Scholar]
- 120.Hance JE, Fu SY, Watkins SC, Beggs AH, Michalak M. Alpha-actinin-2 is a new component of the dystrophin-glycoprotein complex. Arch Biochem Biophys. 1999;365:216–22. doi: 10.1006/abbi.1999.1172. [DOI] [PubMed] [Google Scholar]
- 121.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton. 2004;58:104–11. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 122.Chiu C, Bagnall RD, Ingles J, Yeates L, Kennerson M, Donald JA, et al. Mutations in alpha-actinin-2 cause hypertrophic cardiomyopathy: a genome-wide analysis. J Am Coll Cardiol. 2010;55:1127–35. doi: 10.1016/j.jacc.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 123.North KN, Yang N, Wattanasirichaigoon D, Mills M, Easteal S, Beggs AH. A common nonsense mutation results in alpha-actinin-3 deficiency in the general population. Nat Genet. 1999;21:353–4. doi: 10.1038/7675. [DOI] [PubMed] [Google Scholar]
- 124.Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, et al. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet. 2003;73:627–31. doi: 10.1086/377590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Niemi AK, Majamaa K. Mitochondrial DNA and ACTN3 genotypes in Finnish elite endurance and sprint athletes. Eur J Hum Genet. 2005;13:965–9. doi: 10.1038/sj.ejhg.5201438. [DOI] [PubMed] [Google Scholar]
- 126.Papadimitriou ID, Papadopoulos C, Kouvatsi A, Triantaphyllidis C. The ACTN3 gene in elite Greek track and field athletes. Int J Sports Med. 2008;29:352–5. doi: 10.1055/s-2007-965339. [DOI] [PubMed] [Google Scholar]
- 127.Roth SM, Walsh S, Liu D, Metter EJ, Ferrucci L, Hurley BF. The ACTN3 R577X nonsense allele is under-represented in elite-level strength athletes. Eur J Hum Genet. 2008;16:391–4. doi: 10.1038/sj.ejhg.5201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Clarkson PM, Devaney JM, Gordish-Dressman H, Thompson PD, Hubal MJ, Urso M, et al. ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. J Appl Physiol. 2005;99:154–63. doi: 10.1152/japplphysiol.01139.2004. [DOI] [PubMed] [Google Scholar]
- 129.Moran CN, Yang N, Bailey ME, Tsiokanos A, Jamurtas A, MacArthur DG, et al. Association analysis of the ACTN3 R577X polymorphism and complex quantitative body composition and performance phenotypes in adolescent Greeks. Eur J Hum Genet. 2007;15:88–93. doi: 10.1038/sj.ejhg.5201724. [DOI] [PubMed] [Google Scholar]
- 130.MacArthur DG, Seto JT, Chan S, Quinlan KG, Raftery JM, Turner N, et al. An Actn3 knockout mouse provides mechanistic insights into the association between alpha-actinin-3 deficiency and human athletic performance. Hum Mol Genet. 2008;17:1076–86. doi: 10.1093/hmg/ddm380. [DOI] [PubMed] [Google Scholar]
- 131.Seto JT, Lek M, Quinlan KG, Houweling PJ, Zheng XF, Garton F, et al. Deficiency of {alpha}-actinin-3 is associated with increased susceptibility to contraction-induced damage and skeletal muscle remodeling. Hum Mol Genet. 2011;20:2914–27. doi: 10.1093/hmg/ddr196. [DOI] [PubMed] [Google Scholar]
- 132.Beltran-Valero de Bernabe D, Voit T, Longman C, Steinbrecher A, Straub V, Yuva Y, et al. Mutations in the FKRP gene can cause muscle-eye-brain disease and Walker-Warburg syndrome. J Med Genet. 2004;41:e61. doi: 10.1136/jmg.2003.013870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brockington M, Blake DJ, Prandini P, Brown SC, Torelli S, Benson MA, et al. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin alpha2 deficiency and abnormal glycosylation of alpha-dystroglycan. Am J Hum Genet. 2001;69:1198–209. doi: 10.1086/324412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brown SC, Torelli S, Brockington M, Yuva Y, Jimenez C, Feng L, et al. Abnormalities in alpha-dystroglycan expression in MDC1C and LGMD2I muscular dystrophies. Am J Pathol. 2004;164:727–37. doi: 10.1016/s0002-9440(10)63160-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Muntoni F, Torelli S, Brockington M. Muscular dystrophies due to glycosylation defects. Neurotherapeutics. 2008;5:627–32. doi: 10.1016/j.nurt.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kawahara G, Guyon JR, Nakamura Y, Kunkel LM. Zebrafish models for human FKRP muscular dystrophies. Hum Mol Genet. 2010;19:623–33. doi: 10.1093/hmg/ddp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang CH, Chan YM, Tang RH, Xiao B, Lu P, Keramaris-Vrantsis E, et al. Post-natal knockdown of fukutin-related protein expression in muscle by long-term RNA interference induces dystrophic pathology [corrected] Am J Pathol. 2011;178:261–72. doi: 10.1016/j.ajpath.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ackroyd MR, Skordis L, Kaluarachchi M, Godwin J, Prior S, Fidanboylu M, et al. Reduced expression of fukutin related protein in mice results in a model for fukutin related protein associated muscular dystrophies. Brain. 2009;132:439–51. doi: 10.1093/brain/awn335. [DOI] [PubMed] [Google Scholar]
- 139.Chan YM, Keramaris-Vrantsis E, Lidov HG, Norton JH, Zinchenko N, Gruber HE, et al. Fukutin-related protein is essential for mouse muscle, brain and eye development and mutation recapitulates the wide clinical spectrums of dystroglycanopathies. Hum Mol Genet. 2010;19:3995–4006. doi: 10.1093/hmg/ddq314. [DOI] [PubMed] [Google Scholar]
- 140.Bortolanza S, Nonis A, Sanvito F, Maciotta S, Sitia G, Wei J, et al. AAV6-mediated systemic shRNA delivery reverses disease in a mouse model of facioscapulohumeral muscular dystrophy. Mol Ther. 2011 Aug 9; doi: 10.1038/mt.2011.153. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van Spaendonck-Zwarts K, van Hessem L, Jongbloed JD, de Walle HE, Capetanaki Y, van der Kooi AJ, et al. Desmin-related myopathy: a review and meta-analysis. Clin Genet. 2011;80:354–66. doi: 10.1111/j.1399-0004.2010.01512.x. [DOI] [PubMed] [Google Scholar]
- 142.Goldfarb LG, Dalakas MC. Tragedy in a heartbeat: malfunctioning desmin causes skeletal and cardiac muscle disease. J Clin Invest. 2009;119:1806–13. doi: 10.1172/JCI38027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dalakas MC, Park KY, Semino-Mora C, Lee HS, Sivakumar K, Goldfarb LG. Desmin myopathy, a skeletal myopathy with cardiomyopathy caused by mutations in the desmin gene. N Engl J Med. 2000;342:770–80. doi: 10.1056/NEJM200003163421104. [DOI] [PubMed] [Google Scholar]
- 144.Milner DJ, Weitzer G, Tran D, Bradley A, Capetanaki Y. Disruption of muscle architecture and myocardial degeneration in mice lacking desmin. J Cell Biol. 1996;134:1255–70. doi: 10.1083/jcb.134.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li Z, Colucci-Guyon E, Pincon-Raymond M, Mericskay M, Pournin S, Paulin D, et al. Cardiovascular lesions and skeletal myopathy in mice lacking desmin. Dev Biol. 1996;175:362–6. doi: 10.1006/dbio.1996.0122. [DOI] [PubMed] [Google Scholar]
- 146.Thornell L, Carlsson L, Li Z, Mericskay M, Paulin D. Null mutation in the desmin gene gives rise to a cardiomyopathy. J Mol Cell Cardiol. 1997;29:2107–24. doi: 10.1006/jmcc.1997.0446. [DOI] [PubMed] [Google Scholar]
- 147.Milner DJ, Taffet GE, Wang X, Pham T, Tamura T, Hartley C, et al. The absence of desmin leads to cardiomyocyte hypertrophy and cardiac dilation with compromised systolic function. J Mol Cell Cardiol. 1999;31:2063–76. doi: 10.1006/jmcc.1999.1037. [DOI] [PubMed] [Google Scholar]
- 148.Milner DJ, Mavroidis M, Weisleder N, Capetanaki Y. Desmin cytoskeleton linked to muscle mitochondrial distribution and respiratory function. J Cell Biol. 2000;150:1283–98. doi: 10.1083/jcb.150.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Weisleder N, Soumaka E, Abbasi S, Taegtmeyer H, Capetanaki Y. Cardiomyocyte-specific desmin rescue of desmin null cardiomyopathy excludes vascular involvement. J Mol Cell Cardiol. 2004;36:121–8. doi: 10.1016/j.yjmcc.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 150.Maloyan A, Robbins J. Autophagy in desmin-related cardiomyopathy: thoughts at the halfway point. Autophagy. 2010;6:665–66. doi: 10.1161/CIRCRESAHA.109.212639. [DOI] [PubMed] [Google Scholar]
- 151.Schrickel JW, Stockigt F, Krzyzak W, Paulin D, Li Z, Lubkemeier I, et al. Cardiac conduction disturbances and differential effects on atrial and ventricular electrophysiological properties in desmin deficient mice. J Interv Card Electrophysiol. 2010;28:71–80. doi: 10.1007/s10840-010-9482-8. [DOI] [PubMed] [Google Scholar]
- 152.Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, et al. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–7. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 153.Kostareva A, Gudkova A, Sjoberg G, Kiselev I, Moiseeva O, Karelkina E, et al. Desmin mutations in a St. Petersburg cohort of cardiomyopathies. Acta Myol. 2006;25:109–15. [PubMed] [Google Scholar]
- 154.Xu H, Wu XR, Wewer UM, Engvall E. Murine muscular dystrophy caused by a mutation in the laminin α2 (Lama2) gene. Nat Genet. 1994;8:297–302. doi: 10.1038/ng1194-297. [DOI] [PubMed] [Google Scholar]
- 155.Helbling-Leclerc A, Zhang X, Topaloglu H, Cruaud C, Tesson F, Weissenbach J, et al. Mutations in the laminin alpha 2-chain gene (LAMA2) cause merosin-deficient congenital muscular dystrophy. Nat Genet. 1995;11:216–8. doi: 10.1038/ng1095-216. [DOI] [PubMed] [Google Scholar]
- 156.Guo LT, Zhang XU, Kuang W, Xu H, Liu LA, Vilquin JT, et al. Laminin alpha2 deficiency and muscular dystrophy; genotype-phenotype correlation in mutant mice. Neuromuscul Disord. 2003;13:207–15. doi: 10.1016/s0960-8966(02)00266-3. [DOI] [PubMed] [Google Scholar]
- 157.Geranmayeh F, Clement E, Feng LH, Sewry C, Pagan J, Mein R, et al. Genotype-phenotype correlation in a large population of muscular dystrophy patients with LAMA2 mutations. Neuromuscul Disord. 2010;20:241–50. doi: 10.1016/j.nmd.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 158.Gawlik KI, Durbeej M. Skeletal muscle laminin and MDC1A: pathogenesis and treatment strategies. Skelet Muscle. 2011;1:9. doi: 10.1186/2044-5040-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu H, Christmas P, Wu XR, Wewer UM, Engvall E. Defective muscle basement membrane and lack of M-laminin in the dystrophic dy/dy mouse. Proc Natl Acad Sci USA. 1994;91:5572–6. doi: 10.1073/pnas.91.12.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sunada Y, Bernier SM, Utani A, Yamada Y, Campbell KP. Identification of a novel mutant transcript of laminin α2 chain gene responsible for muscular dystrophy and dysmyelination in dy2J mice. Hum Mol Genet. 1995;4:1055–61. doi: 10.1093/hmg/4.6.1055. [DOI] [PubMed] [Google Scholar]
- 161.Kuang W, Xu H, Vachon PH, Liu L, Loechel F, Wewer UM, et al. Merosin-deficient congenital muscular dystrophy. Partial genetic correction in two mouse models. J Clin Invest. 1998;102:844–52. doi: 10.1172/JCI3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Miyagoe Y, Hanaoka K, Nonaka I, Hayasaka M, Nabeshima Y, Arahata K, et al. Laminin α2 chain-null mutant mice by targeted disruption of the Lama2 gene: a new model of merosin (laminin 2)-deficient congenital muscular dystrophy. FEBS Lett. 1997;415:33–9. doi: 10.1016/s0014-5793(97)01007-7. [DOI] [PubMed] [Google Scholar]
- 163.Shelton GD, Liu LA, Guo LT, Smith GK, Christiansen JS, Thomas WB, et al. Muscular dystrophy in female dogs. J Vet Intern Med. 2001;15:240–4. doi: 10.1892/0891-6640(2001)015<0240:mdifd>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 164.Bonaldo P, Russo V, Bucciotti F, Doliana R, Colombatti A. Structural and functional features of the alpha 3 chain indicate a bridging role for chicken collagen VI in connective tissues. Biochemistry. 1990;29:1245–54. doi: 10.1021/bi00457a021. [DOI] [PubMed] [Google Scholar]
- 165.Kuo HJ, Maslen CL, Keene DR, Glanville RW. Type VI collagen anchors endothelial basement membranes by interacting with type IV collagen. J Biol Chem. 1997;272:26522–9. doi: 10.1074/jbc.272.42.26522. [DOI] [PubMed] [Google Scholar]
- 166.Schessl J, Zou Y, Bönnemann CG. Congenital muscular dystrophies and the extracellular matrix. Semin Pediatr Neurol. 2006;13:80–9. doi: 10.1016/j.spen.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 167.Bernardi P, Bonaldo P. Dysfunction of mitochondria and sarcoplasmic reticulum in the pathogenesis of collagen VI muscular dystrophies. Ann N Y Acad Sci. 2008;1147:303–11. doi: 10.1196/annals.1427.009. [DOI] [PubMed] [Google Scholar]
- 168.Specks U, Mayer U, Nischt R, Spissinger T, Mann K, Timpl R, et al. Structure of recombinant N-terminal globule of type VI collagen alpha 3 chain and its binding to heparin and hyaluronan. EMBO J. 1992;11:4281–90. doi: 10.1002/j.1460-2075.1992.tb05527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bethlem J, van Wijngaarden GK. Benign myopathy, with autosomal dominant inheritance: a report on three pedigrees. Brain. 1976;99:91–100. doi: 10.1093/brain/99.1.91. [DOI] [PubMed] [Google Scholar]
- 170.Jöbsis GJ, Boers JM, Barth PG, de Visser M. Genetic localization of Bethlem myopathy. Neurology. 1996;46:779–82. doi: 10.1212/wnl.46.3.779. [DOI] [PubMed] [Google Scholar]
- 171.Jöbsis GJ, Keizers H, Vreijling JP, de Visser M, Speer MC, Wolterman RA, et al. Type VI collagen mutations in Bethlem myopathy an autosomal dominant myopathy with contractures. Nat Genet. 1996;14:113–5. doi: 10.1038/ng0996-113. [DOI] [PubMed] [Google Scholar]
- 172.Speer MC, Tandan R, Rao PN, Fries T, Stajich JM, Bolhuis PA, et al. Evidence for locus heterogeneity in the Bethlem myopathy and linkage to 2q37. Hum Mol Genet. 1996;5:1043–6. doi: 10.1093/hmg/5.7.1043. [DOI] [PubMed] [Google Scholar]
- 173.Camacho Vanegas O, Bertini E, Zhang RZ, Petrini S, Minosse C, Sabatelli P, et al. Ullrich scleroatonic muscular dystrophy is caused by recessive mutations in collagen type VI. Proc Natl Acad Sci USA. 2001;98:7516–21. doi: 10.1073/pnas.121027598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Demir E, Sabatelli P, Allamand V, Ferreiro A, Moghadaszadeh B, Makrelouf M, et al. Mutations in COL6A3 cause severe and mild phenotypes of Ullrich congenital muscular dystrophy. Am J Hum Genet. 2002;70:1446–58. doi: 10.1086/340608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bonaldo P, Braghetta P, Zanetti M, Piccolo S, Volpin D, Bressan GM. Collagen VI deficiency induces early onset myopathy in the mouse: an animal model for Bethlem myopathy. Hum Mol Genet. 1998;7:2135–40. doi: 10.1093/hmg/7.13.2135. [DOI] [PubMed] [Google Scholar]
- 176.Irwin WA, Bergamin N, Sabatelli P, Reggiani C, Megighian A, Merlini L, et al. Mitochondrial dysfunction and apoptosis in myopathic mice with collagen VI deficiency. Nat Genet. 2003;35:367–71. doi: 10.1038/ng1270. [DOI] [PubMed] [Google Scholar]
- 177.Bertini E, Guglielmina P. Collagen type VI and related disorders: Bethlem myopathy and Ullrich scleroatonic muscular dystrophy. Eur J Paediatr Neurol. 2002;6:193–8. doi: 10.1053/ejpn.2002.0593. [DOI] [PubMed] [Google Scholar]
- 178.Tiepolo T, Angelin A, Palma E, Sabatelli P, Merlini L, Nicolosi L, et al. The cyclophilin inhibitor Debio 025 normalizes mitochondrial function, muscle apoptosis and ultrastructural defects in Col6a1–/– myopathic mice. Br J Pharmacol. 2009;157:1045–52. doi: 10.1111/j.1476-5381.2009.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Telfer WR, Busta AS, Bönnemann CG, Feldman EL, Dowling JJ. Zebrafish models of collagen VI-related myopathies. Hum Mol Genet. 2010;19:2433–44. doi: 10.1093/hmg/ddq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.de Greef JC, Lemmers RJ, van Engelen BG, Sacconi S, Venance SL, Frants RR, et al. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat. 2009;30:1449–59. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- 181.Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Matteotti C, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611–23. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 182.Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–3. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.van der Maarel SM, Tawil R, Tapscott SJ. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol Med. 2011;17:252–8. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gabellini D, D'Antona G, Moggio M, Prelle A, Zecca C, Adami R, et al. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 2006;439:973–7. doi: 10.1038/nature04422. [DOI] [PubMed] [Google Scholar]
- 185.Wallace LM, Garwick-Coppens SE, Tupler R, Harper SQ. RNA interference improves myopathic phenotypes in mice over-expressing FSHD region gene 1 (FRG1) Mol Ther. 2011 doi: 10.1038/mt.2011.118. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Harper PS, Johnson K. Myotonic dystrophy. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited disease. III. New York: McGraw-Hill; 2001. pp. 5525–50. [Google Scholar]
- 187.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–48. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, et al. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–14. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, et al. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–14. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 191a.Gomes-Pereira M, Cooper TA, Gourdon G. Myotonic dystrophy mouse models: towards rational therapy development. Trends Mol Med. 2011;17:506–17. doi: 10.1016/j.molmed.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191b.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–73. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 192.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–9. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Seznec H, Agbulut O, Sergeant N, Savouret C, Ghestem A, Tabti N, et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum Mol Genet. 2001;10:2717–26. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- 194.Orengo JP, Chambon P, Metzger D, Mosier DR, Snipes GJ, Cooper TA. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc Natl Acad Sci USA. 2008;105:2646–51. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–80. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 196.Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:3614–22. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Muir LA, Chamberlain JS. Emerging strategies for cell and gene therapy of the muscular dystrophies. Expert Rev Mol Med. 2009;11:e18. doi: 10.1017/S1462399409001100. [DOI] [PMC free article] [PubMed] [Google Scholar]