Abstract
Studies of transcriptomes are critical for understanding gene expression. Release of RNA molecules from cells is typically the first step for transcriptomic analysis. Effective cell lysis approaches that completely release intracellular materials are in high demand especially for cells that are structurally robust. In this report, we demonstrate a microfluidic electric lysis device that is effective for mRNA extraction from mycobacteria that have hydrophobic and waxy cell walls. We used a packed bed of microscale silica beads to filter M. smegmatis out of the suspension. 4000–8000 V/cm field intensity was used to lyse M. smegmatis with long pulses (i.e., up to 30 pulses that were 5 s long each). Our quantitative reverse transcription (qRT)-PCR results showed that our method yielded a factor of 10–20 higher extraction efficiency than the current state-of-the-art method (bead beating). We conclude that our electric lysis technique is an effective approach for mRNA release from hard-to-lyse cells and highly compatible with microfluidic molecular assays.
Profiling transcriptomes (the set of all RNA molecules) is critical for understanding the functional elements of the genome and disease processes.1 Various technologies have been developed in recent years, such as real time PCR,2 microarrays,3 and RNA sequencing (RNA-seq),1,4,5 to detect and quantify mRNA for understanding physiological events. The purity and integrity of input RNA are critical for the success of these RNA-based analysis. Compromise in RNA quality leads to variable results.6,7 There is a growing demand for mRNA extraction methods that allow transcriptomic profiling of all species.
Mycobacteria are nonmotile, aerobic, and acid-fast bacteria, including highly pathogenic species that cause tuberculosis and leprosy.8 Compared to other bacteria, mycobacteria have a thick cell wall that is hydrophobic, waxy, and rich in mycolic acids/mycolates. Analysis of intracellular contents from mycobacteria is challenging due to this structural characteristic.
Several methods have been developed for RNA isolation from bacteria. Chemical disruption, including the TRIzol-based method9,10 and hot-phenol-based method,11 is traditionally used for bacteria RNA extraction. However, the procedures are usually tedious and time-consuming, taking several hours to a few days.12,13 The chemicals (SDS and phenol) involved in these procedures often cause RNA fragmentation and result in variability in RNA quality and analysis results.13 Bead beating is considered to be the state of the art for RNA extraction from lysis-resistant bacterial cells including mycobacteria.14 TRIzol is often added in bead beating to improve RNA stability and facilitate cell wall disruption (via denaturing proteins and inhibiting RNases).15 Bacterial cells and beads are contained in a closed tube and then subject to high-frequency oscillation. The high shear stress generated by periodic vertical flow leads to mechanical lysis. This procedure is typically applied to a large number of cells (>108 cells). This creates difficulty for studying slow-growing mycobacteria and probing a low number of cells.16 Such release tends to be incomplete. The mechanical mechanism is also hard to replicate on a microfluidic device.
Electric lysis is a rapid physical method for cell membrane disruption and intracellular content release.17,18 Electric lysis typically refers to irreversible cell electroporation that occurs under application of electrical pulses with defined intensity and duration. When the field intensity and duration of these electrical pulses exceed certain threshold (that is specific to the cell type), cells are irreversibly lysed and intracellular molecules are released into the surrounding solution. Electric lysis can be completed rapidly (within seconds to minutes) and does not involve the use of chemical/biological reagents that may potentially interfere with downstream assays. Although electroporation has been extensively utilized for releasing intracellular proteins,17−19 there has been very little work on using it to release nucleic acids in general.20−22 None of the previous works examined the effectiveness of electric lysis on mycobacteria that are generally considered highly resistant to most lysis methods.
Here, we describe a rapid mRNA extraction from Mycobacterium smegmatis under ultrahigh-intensity (up to 8000 V/cm) electric lysis on a microfluidic device. We formed a packed bed of microscale silica beads in the device to trap the mycobacterial cells. Electric pulses were then applied to electrically lyse trapped M. smegmatis within 3 min. Cell lysate was directly used for quantitative reverse transcription (qRT)-PCR analysis without further treatment. We show that our mRNA extraction efficiency was 10–20 times higher than bead beating.
Material and Methods
Microfluidic Chip Fabrication and Operation
A two-layered PDMS chip was fabricated by multilayer soft lithography.23,24 Photomasks were designed by Freehand MX (Macromedia, San Francisco, CA) and printed on transparency film at 4000 dpi resolution. The fluidic layer master was fabricated in SU-8 2002 (Microchem, Newton, MA) and AZ 9260 (Clariant, Charlotte, NC) with the thickness being 2 and 10 μm, respectively. Micropillars were placed in fluidic chambers to avoid collapse. The master was heated at 130 °C for 30 s to form a rounded cross-sectional profile for the features in AZ 9260. The control layer master was fabricated in SU-2025 with 24 μm thickness. The control layer was made by spinning PDMS prepolymers (RTV615A/RTV615B = 20:1, R. S. Hughes, Sunnyvale, CA) at 500 rpm for 10 s and then at 1700 rpm for 30 s. This resulted in a thickness of 67 μm for the control layer PDMS. The fluidic layer had a composition of RTV615A/RTV615B = 5:1 and a thickness of ∼0.4 cm. Both layers of PDMS were cured at 80 °C for 15 min. The two layers were then aligned, brought into contact, and baked for 1 h at 80 °C. The two-layered PDMS structure was then peeled off from the control layer master, and access holes were punched. Glass slides (VWR, Radnor, PA) were cleaned in a basic solution (H2O/27% NH4OH/30% H2O2 = 5:1:1, volumetric ratio) at 80 °C for 1 h, rinsed with ultrapure water, and blown dry using nitrogen. Finally, the PDMS structure was bonded to the precleaned glass slide after plasma oxidation of both surfaces (Harrick Plasma, Ithaca, NY) and baked at 80 °C for 1 h for further strengthening of the device.
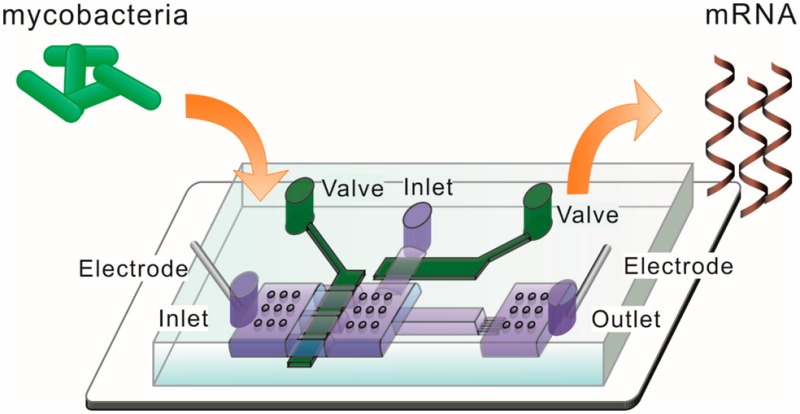
We wrote two LabVIEW (National Instruments, Austin, TX) programs to operate on-chip pneumatic valves and generate electric pulses with designated duration/direction, all via a DAQ card (NI SCB-68 from National Instruments).20,24,25 One program controlled actuation of on-chip pneumatic valves (under a pressure of 25 psi) via operation of solenoid valves (18801003-12VDC, ASCO Scientific, Florham Park, NJ). The other defined the pulse duration and direction. Two platinum electrodes were inserted into the reservoirs at inlet 2 and the outlet with direct contact to the solution (Figure 1). The two electrodes were connected to DC voltage provided by a high voltage power supply (PS350/5000V-25W, Stanford Research System, Sunnyvale, CA) via an H-bridge circuit. The H-bridge circuit consisted of 4 high voltage reed relays (5501-05-1, Coto Technology, Kingstown, RI). Two of the relays worked together (by switching closed and then open at the same time) to generate a pulse of defined duration in one direction (while the other two relays remained open). On the other hand, when the other two relays were actuated in the same fashion, a pulse in the opposite direction was generated. To eliminate potential pH changes in the electroporation solution due to electrolysis of water under long pulses, we applied a series of pulses by changing the current direction after each pulse. In this study, each individual pulse was 5 s in duration and there was a 2 s interval between pulses.
Figure 1.
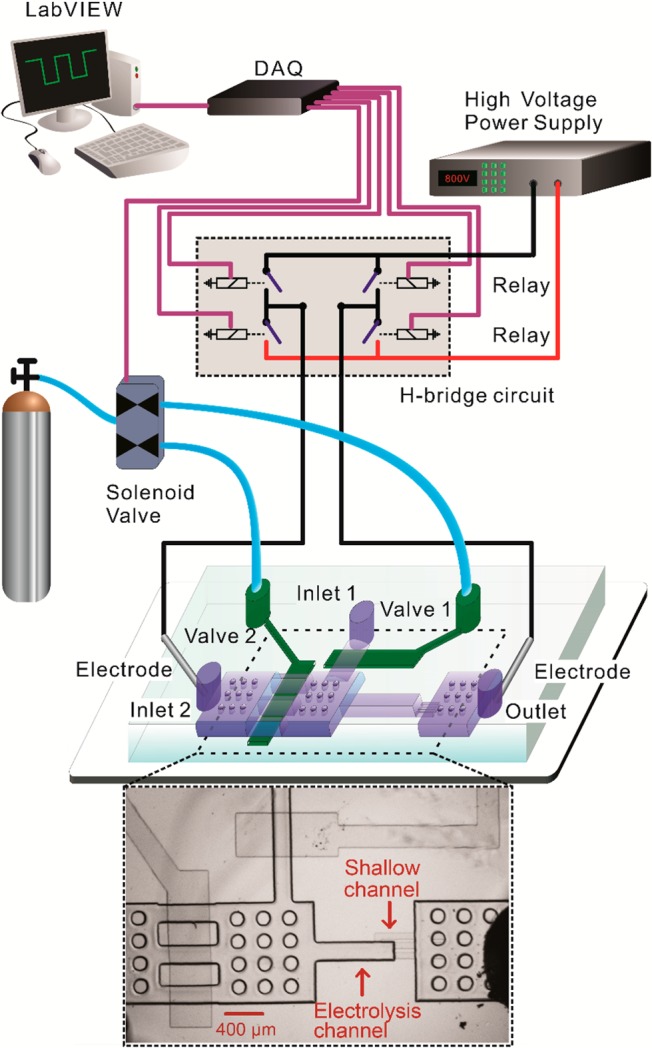
Experimental setup for conducting ultrahigh-intensity electrolysis of M. smegmatis. Electric pulses were applied across a PDMS microfluidic channel via two platinum electrodes. Pulses were generated by cutting off constant voltage generated by a power supply. The pulse duration and direction were set in a LabVIEW program that operated a H-bridge circuit (consisting of 4 high-voltage relays) via a data acquisition (DAQ) card. The two-layered microfluidic device contained two pneumatic valves. The dimensions of the electrolysis channel were 800 μm × 200 μm × 10 μm. There were circular supporting pillars in the large chambers to prevent collapsing.
Cell Culture
Mycobacterium smegmatis MC2 155 expressing green fluorescent protein (GFP) was cultured in lysogeny broth (LB) with 0.05% Tween 80 and 100 μg/μL hygromycin at 37 °C under continuous shaking at 200 rpm. The cell culture was centrifuged at 3000 rpm for 10 min and resuspended in H2O. The concentration of the cell culture was estimated by McFarland standards.
RNA Extraction by Bead Beating
The cell culture was centrifuged at 4000 rpm for 10 min; 5 ×109 to 1010 cells (with the actual value calculated on the basis of optical density reading taken with a spectrophotometer) were then resuspended in 1 mL of TRIzol (Life Technologies). The mixture was then transferred into a lysing matrix B bead beating tube (MP Biochemicals, Santa Ana, CA) on ice. Chloroform (300 μL) was added, and the bead beating tube was shaken vigorously. Bead beating using maximum power was conducted twice (45 s the first time and 30 s the second time), with a 5 min rest time in between. The mixture was centrifuged at 10 000 rpm for 15 min at 4 °C, and the top aqueous layer was collected. The sample was mixed with 600 μL of isopropanol and stored at −20 °C overnight. The RNA sample was centrifuged at 10 000 rpm for 10 min at 4 °C, washed with 75% ethanol, and resuspended in H2O.
qRT-PCR Assay
Cell lysate was eluted from the microfluidic device by 20 μL of RNase-free water and then mixed with 1× RNasecure Reagent (Life Technologies) to deactivate RNases and protect the integrity of RNA. The mixture was incubated at 60 °C for 10 min and then cooled to 4 °C. The samples were directly used in qRT-PCR (four pairs of primers are listed in Table S1) or stored at −20 °C until use. qPCR assays were performed in 20 μL aliquots in a Real-Time PCR Detection System (CFX Connect, BIO-RAD, Hercules, CA). Power SYBR Green RNA-to-CT 1-Step Kit (Life Technologies) was used to measure mRNA copy numbers following the manufacturer’s protocol. A pair of primers (100 nM each) was added into each aliquot. The mixture was incubated at 48 °C for 30 min for reverse transcription. The mixture was then denatured at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s for DNA amplification. No-template controls (NTCs) were included to monitor nonspecific amplification.
RNA Standard Synthesis and Quantification
We established a standard curve to link the copy number of mRNA with qPCR signal (Ct value). In vitro transcribed RNA was used as the template to generate the standard curve. Each of the four pairs of primers in Table S1 with added T7 promoter sequence (5′-TAA TAC GAC TCA CTA TAG GG-3′) was used to amplify a specific mRNA (from total RNA extracted by bead beating) by RT-PCR for 40 cycles. The size of RT-PCR products was verified by gel electrophoresis (Figure S1). The amplification product (DNA) contained the T7 promotor sequence on both ends that could be recognized by T7 phage RNA polymerases. About 75 ng of amplification product was mixed with nucleotides (0.5 mM in final concentration), T7 polymerase, and 1× transcription buffer polymerases (MAXIscript T7 in vitro transcription kit, Life Technologies) and incubated at 37 °C for 1 h. TURBO DNase 1 was added and incubated at 37 °C for 15 min to digest template DNA. Twenty-five mM EDTA was used to stop digestion and block heat-induced RNA degradation. The produced RNA was purified twice by precipitating with ammonium acetate/ethanol to remove unincorporated NTPs and DNase. The copy number of the RNA was measured by a Qubit 2.0 Fluorometer with Qubit RNA HS Assay Kit (Life Technologies). The RNA was serially diluted to generate samples for the standard curve using qRT-PCR.
COMSOL Modeling
COMSOL Multiphysics 4.3 (Burlington, MA) was used to simulate the electric field intensity in the microfluidic chamber. The “Electric Current (ec)” module was used to study the steady state of the model. The 3-D model was based on the actual geometry of the microfluidic structures with the exception of the packed bed. Because the resistivity of silica beads (1 × 1013 Ωm) is much higher than that of water (2 × 105 Ωm), we treated silica beads as electrically insulating. Thus, the segment that contained the packed bed was treated as having a cross-sectional area smaller than that of the channel (with the equivalent cross-sectional area occupied by the insulating beads deducted). Water was used in all other domains with electrical conductivity of 5.5 × 10–6 S/m. To simplify the modeling, we did not consider gas bubbles and treated the solution as static.
The modeling is based on the governing equation:
| 1 |
where D is electric flux density and J is the electric current density.
The electric current density is expressed by
| 2 |
where σ is the electrical conductivity and E is electric field. The electric flux density (D) is also expressed by
| 3 |
where ϵ0 is the vacuum permeability and ϵr is relative permeability. The system was electrically insulated on all boundaries. 786 V across the two reservoirs was applied.
Results and Discussion
We designed a two-layered microfluidic electric lysis chip (Figure 1). There were two inlets and one outlet. The depth of channels and chambers in the fluidic layer was 10 μm with the exception of the shallow channels (having a depth of 2 μm). Prior to the experiment, the device was primed by flowing water in via both inlets, removing debris and bubbles. Microscale silica beads (d ∼ 4.8 μm, SS05N, Bangs Laboratories, Fishers, IN) and mycobacterial cells (M. smegmatis) then flowed into the device via inlet 1, while keeping valve 2 closed. A 20 μL mixture of beads and cells (∼2 × 107 cell/mL) was packed against the downstream shallow channel to form a packed bed in the electrolysis channel with mycobacterial cells trapped inside. In our experiment, roughly 2 × 105 cells were captured in the electrolysis channel each time and loading of cells and beads was stopped by closing valve 1. Two platinum (Pt) electrodes were inserted in the inlet 2 and the outlet for application of electric pulses. Valve 1 was closed and there was a flow of 1 μL/min coming in from inlet 2 during the application of the electric pulses.
Compared to electric lysis of bacterial cells such as E. coli,18,25 electric lysis of M. smegmatis required significantly longer pulse duration and higher field intensity. Several features of our device and operation were designed to accommodate these requirements. First, we found that even low-conductivity buffer (e.g., 1 mM MgSO4, 8 mM Na2HPO4, 2 mM KH2PO4, and 250 mM sucrose) used in our previous works26−30 generated too many bubbles when the field intensity was >2000 V/cm). Thus, we flowed ultrapure water (RNase-free) during the application of electric pulses. Second, we applied a flow of 1 μL/min (coming from inlet 2) during application of electric pulses. The flow carried away generated heat and bubbles during the operation. Finally, we programmed the LabVIEW code and designed an H-bridge circuit (including four relays) so that the direction of the electric field was alternated after each pulse (of 5s duration). This was to prevent significant local pH change due to electrolysis of water associated with long pulses.
Other than the channel dimensions and the voltage applied, the field intensity of the pulses was affected by the presence of nonconductive beads. There was a substantially higher resistance associated with the packed bed of beads than that of the same channel segment without the beads. We used COMSOL Multiphysics to model the field intensity within the packed bed (Figure S2). We show that the presence of the packed bed significantly increased the local field intensity (by ∼3×) (compared to the channel segment without beads).
We used a M. smegmatis strain that expressed green fluorescent protein (GFP). In Figure 2, we show the process of bead/cell packing and the electrolysis-based release of intracellular materials. The initial stopping of beads at the interface of the shallow channel and the electrolysis channel (Figure 2a) led to the formation of a packed bed with mycobacterial cells trapped in the gaps among beads (Figure 2b). The fluorescence emitted from the cells was observed in Figure 2c before electrolysis and disappeared after electrolysis by 20, 5 s pulses with a field intensity of 6000 V/cm (Figure 2d), suggesting release of intracellular molecules (also see Video S1).
Figure 2.
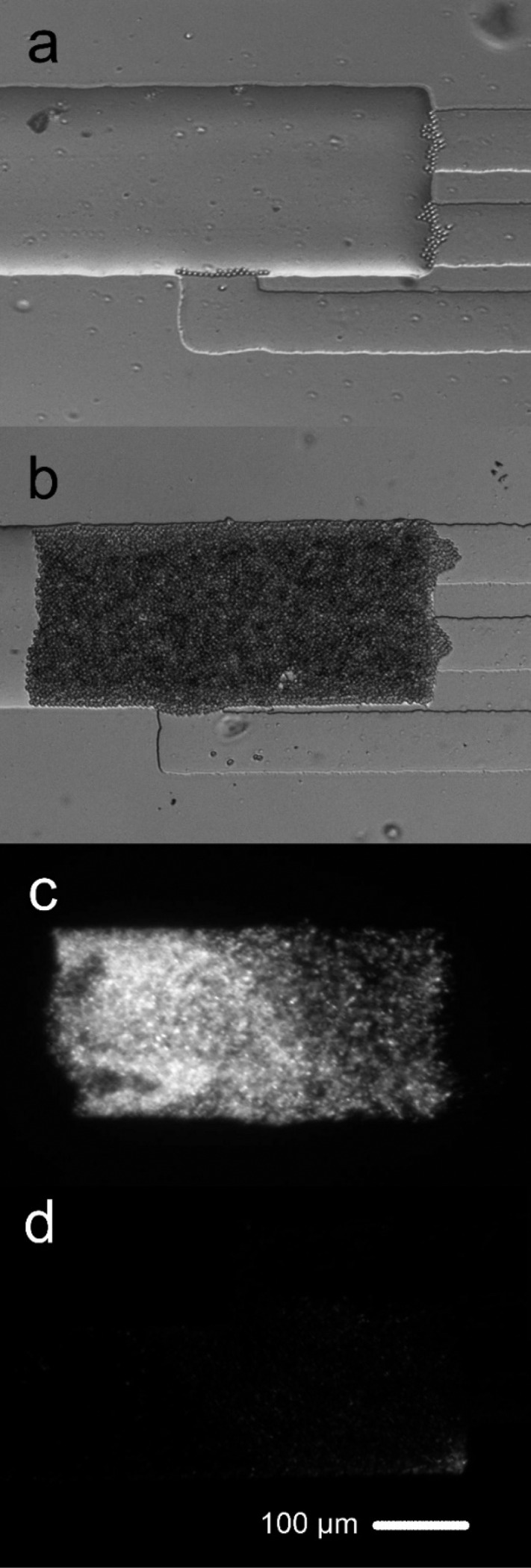
Procedure for microfluidic electrolysis of M. smegmatis. (a) Bright field image shows that beads were stopped by the shallow channel and started to accumulate in the electrolysis channel. (b) Bright field image shows the formation of packed bed of beads/M. smegmatis cells. (c) Fluorescence image shows the GFP expressing M. smegmatis cells before electrolysis. The packed bed roughly contained 2 × 105 cells. (d) Fluorescence image after electrolysis (30 pulses of 6000 V/cm and 5 s each).
We quantified the amount of RNA release by electrolysis using quantitative reverse transcription polymerase chain reaction (qRT-PCR). The RNA extraction was quantified by
| 4 |
where 1581.1 and 3.0 × 10–7 were parameters extracted from the working curve in Figure S3. Intercept and slope were calculated for each mRNA in Figure S4. Ct was the RT-qPCR signal. The number of GFP-expressing mycobacterial cells trapped in the packed bed was estimated on the basis of the amount of emitted fluorescence. The fluorescence intensity (FI) was the average of 10 z-stacking images that covered the entire depth of the packed bed (taken after the GFP-expressing mycobacterial cells occupied the packed bed). DNA% was the fraction of a mRNA copy per cell value contributed by DNA (Figure S5). Packed bed volume (V) was calculated by
| 5 |
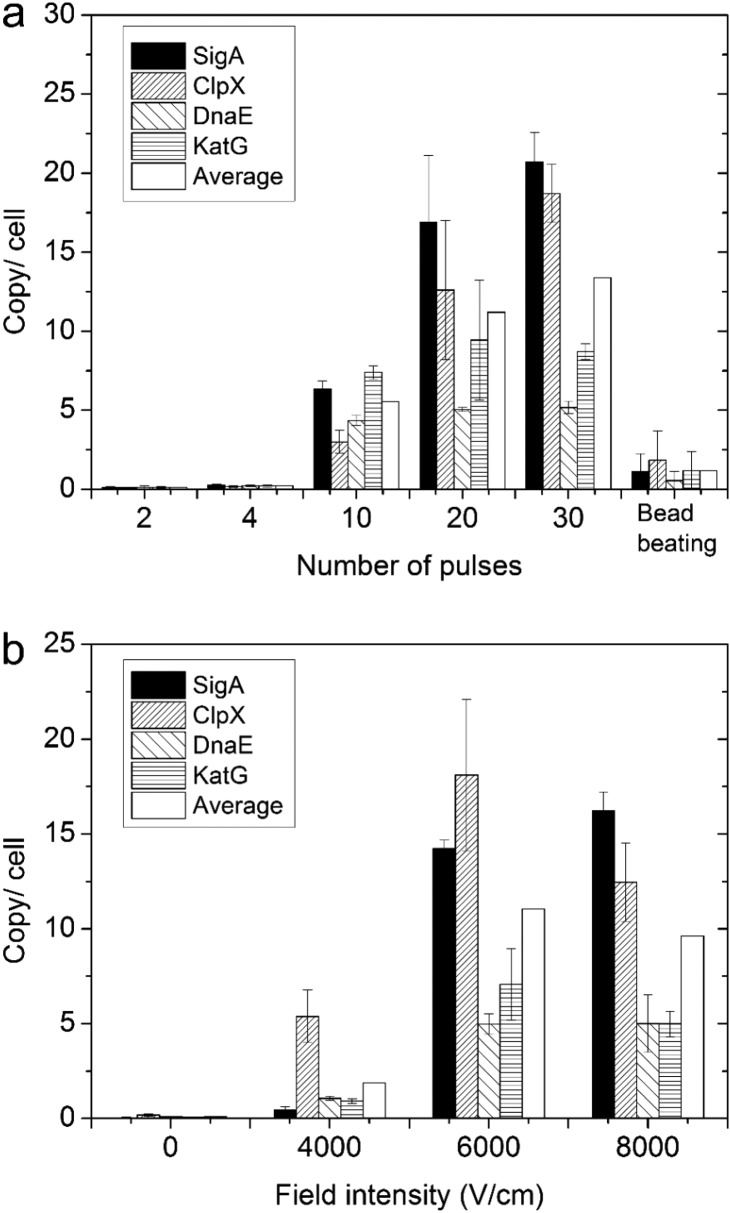
We characterized RNA extraction efficiency by the electrolysis protocol under various pulse numbers (2 to 30 pulses, all at 6000 V/cm) and field intensities (4000–8000 V/cm, all with 30 pulses) (Figure 3). We quantified the copy number per cell for 4 representative mRNAs in the cell lysate under various electric lysis conditions and compared them with those with bead beating (i.e., the conventional lysis method). The number of cells in the electrolysis channel was quantified by fluorescence, as shown in Figure S3. RNA quantification was based on standard curves established using in vitro transcribed RNAs, which offered more accuracy than using bulk RNA, cDNA, or DNA-based quantification.31Figure 3a shows that there was no substantial release of mRNA (under 2 or 4 pulses at 6000 V/cm) until the number of pulses increased to 10. The amount of mRNA release increased with higher number of pulses (from 10 to 20 and 30). Some genes experienced a drastic increase with the number of pulses (e.g., SigA from 6.3 copies/cell at 10 pulses to 16.9 and 20.7 copies/cell at 20 and 30 pulses, respectively, and ClpX from 3.0 copies/cell at 10 pulses to 12.6 and 18.7 copies/cell at 20 and 30 pulses). The results suggest that a significant number of pulses (or total electrolysis duration) was required to create sufficient cell wall damage for M. smegmatis and complete release of intracellular RNAs. Overall, electrolysis provided significantly more complete release of intracellular mRNAs than the current state of the art (i.e., bead beating). We release a factor of 18 and 10 more RNA molecules with 30 pulses than bead beating, for SigA and ClpX, respectively. Furthermore, we also examined the effect of the pulse intensity. As expected, increased field intensity of the pulses from 4000 to 6000 V/cm increased the mRNA release significantly for all genes examined (when the number of pulses was kept at 30). However, a further increase from 6000 to 8000 V/cm did not produce significant additional increase. This indicates that, with 30 pulses at 6000 V/cm, the release of mRNA was fairly complete. Further increase in the field intensity or duration did not produce better results and could potentially lead to loss of integrity and function for mRNA due to heating.
Figure 3.
Release of mRNA under various conditions for M. smegmatis. The released copy number per cell for 4 mRNAs was quantified using qRT-PCR (n = 3). The copy per cell values for microfluidics-produced samples were calibrated by deducting the fraction generated by DNA templates in the cell lysate (Figure S5). (a) The effect of pulse numbers and comparison to bead beating. Various numbers of pulses (5 s duration for each pulse with 6000 V/cm intensity) were applied. (b) The effect of pulse intensity. In each case, 30 pulses (5 s each) were applied.
Our results demonstrate that electrolysis of hard-to-lyse mycobacterial cells can be conducted with high efficiency under ultrahigh field intensity in a microfluidic device. Our microfluidic lysis protocol takes less than 30 min for each sample (including cell capture, lysis, and elution) and may potentially be run in parallel for high throughput of samples. Our microfluidic platform offers advantages for handling small quantities of cells and integration with downstream assays.
Acknowledgments
We acknowledge support from National Institutes of Health grants CA174577, EB017235, and EB019123 and funding from the Ragon Institute and the Burroughs Wellcome Foundation.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.6b00381.
The authors declare no competing financial interest.
Supplementary Material
References
- Wang Z.; Gerstein M.; Snyder M. Nat. Rev. Genet. 2009, 10, 57–63. 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D.; Livak K. J. Nat. Protoc. 2008, 3, 1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sealfon S. C.; Chu T. T. Methods Mol. Biol. 2011, 671, 3–34. 10.1007/978-1-59745-551-0_1. [DOI] [PubMed] [Google Scholar]
- Mortazavi A.; Williams B. A.; McCue K.; Schaeffer L.; Wold B. Nat. Methods 2008, 5, 621–628. 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Croucher N. J.; Thomson N. R. Curr. Opin. Microbiol. 2010, 13, 619–624. 10.1016/j.mib.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige S.; Pfaffl M. W. Mol. Aspects Med. 2006, 27, 126–139. 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Imbeaud S.; Graudens E.; Boulanger V.; Barlet X.; Zaborski P.; Eveno E.; Mueller O.; Schroeder A.; Auffray C. Nucleic Acids Res. 2005, 33, e56 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley S. S.; Splitter G.; Welch R. J. Clin. Microbiol. 1987, 25, 2227–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad T.; Roberts D.; Liao R.; Sherman D. In Mycobacteria Protocols; Parish T., Brown A. C., Eds.; Humana Press: Totowa, NJ, 2009; pp 13–22. [Google Scholar]
- Rio D. C.; Ares M.; Hannon G. J.; Nilsen T. W. Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. 10.1101/pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- Mangan J. A.; Sole K. M.; Mitchison D. A.; Butcher P. D. Nucleic Acids Res. 1997, 25, 675–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldous W. K.; Pounder J. I.; Cloud J. L.; Woods G. L. J. Clin. Microbiol. 2005, 43, 2471–2473. 10.1128/JCM.43.5.2471-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn C. E.; Charkowski A. O.; Willis D. K. J. Microbiol. Methods 2008, 75, 318–324. 10.1016/j.mimet.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Vandeventer P. E.; Weigel K. M.; Salazar J.; Erwin B.; Irvine B.; Doebler R.; Nadim A.; Cangelosi G. A.; Niemz A. J. Clin. Microbiol. 2011, 49, 2533–2539. 10.1128/JCM.02171-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite G. M.; Magan N.; Medina A. J. Microbiol. Methods 2012, 88, 413–418. 10.1016/j.mimet.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Stahl D. A.; Urbance J. W. J. Bacteriol. 1990, 172, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng T.; Lu C. Lab Chip 2013, 13, 3803–3821. 10.1039/C3LC50566A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-Y.; Bhunia A. K.; Lu C. Biosens. Bioelectron. 2006, 22, 582–588. 10.1016/j.bios.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Bao N.; Lu C. Appl. Phys. Lett. 2008, 92, 214103. 10.1063/1.2937088. [DOI] [Google Scholar]
- Vulto P.; Dame G.; Maier U.; Makohliso S.; Podszun S.; Zahn P.; Urban G. A. Lab Chip 2010, 10, 610–616. 10.1039/B913481F. [DOI] [PubMed] [Google Scholar]
- Geng T.; Bao N.; Sriranganathanw N.; Li L.; Lu C. Anal. Chem. 2012, 84, 9632–9639. 10.1021/ac3026064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.; Johnson M.; Hill P.; Gale B. K. Integr. Biol. 2009, 1, 574–586. 10.1039/b905844c. [DOI] [PubMed] [Google Scholar]
- Unger M. A.; Chou H.-P.; Thorsen T.; Scherer A.; Quake S. R. Science 2000, 288, 113–116. 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- Ma S.; Loufakis D. N.; Cao Z.; Chang Y.; Achenie L. E. K.; Lu C. Lab Chip 2014, 14, 2905–2909. 10.1039/C4LC00498A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S.; Schroeder B.; Sun C.; Loufakis D. N.; Cao Z.; Sriranganathan N.; Lu C. Integr. Biol. 2014, 6, 973–978. 10.1039/C4IB00172A. [DOI] [PubMed] [Google Scholar]
- Sun C.; Ouyang M.; Cao Z.; Ma S.; Alqublan H.; Sriranganathan N.; Wang Y.; Lu C. Chem. Commun. 2014, 50, 11536–11539. 10.1039/C4CC04730C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.; Cao Z.; Wu M.; Lu C. Anal. Chem. 2014, 86, 11403–11409. 10.1021/ac503363m. [DOI] [PubMed] [Google Scholar]
- Loufakis D. N.; Cao Z.; Ma S.; Mittelman D.; Lu C. Chem. Sci. 2014, 5, 3331–3337. 10.1039/C4SC00319E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao N.; Wang J.; Lu C. Electrophoresis 2008, 29, 2939–2944. 10.1002/elps.200700856. [DOI] [PubMed] [Google Scholar]
- Geng T.; Zhan Y.; Wang J.; Lu C. Nat. Protoc. 2011, 6, 1192–1208. 10.1038/nprot.2011.360. [DOI] [PubMed] [Google Scholar]
- Wong M. L.; Medrano J. F. BioTechniques 2005, 39, 75–85. 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.