Abstract
Rationale: Sarcoidosis is a multisystem disease of unknown cause. Löfgren’s syndrome (LS) is a characteristic subgroup of sarcoidosis that is associated with a good prognosis in sarcoidosis. However, little is known about its genetic architecture or its broader phenotype, non-LS sarcoidosis.
Objectives: To address the genetic architecture of sarcoidosis phenotypes, LS and non-LS.
Methods: An association study in a white Swedish cohort of 384 LS, 664 non-LS, and 2,086 control subjects, totaling 3,134 subjects using a fine-mapping genotyping platform was conducted. Replication was performed in four independent cohorts, three of white European descent (Germany, n = 4,975; the Netherlands, n = 613; and Czech Republic, n = 521), and one of black African descent (United States, n = 1,657), totaling 7,766 subjects.
Measurements and Main Results: A total of 727 LS-associated variants expanding throughout the extended major histocompatibility complex (MHC) region and 68 non-LS–associated variants located in the MHC class II region were identified and confirmed. A shared overlap between LS and non-LS defined by 17 variants located in the MHC class II region was found. Outside the MHC region, two LS-associated loci, in ADCY3 and between CSMD1 and MCPH1, were observed and replicated.
Conclusions: Comprehensive and integrative analyses of genetics, transcription, and pathway modeling on LS and non-LS indicates that these sarcoidosis phenotypes have different genetic susceptibility, genomic distributions, and cellular activities, suggesting distinct molecular mechanisms in pathways related to immune response with a common region.
Keywords: genetic epidemiology of sarcoidosis, Löfgren’s syndrome, non-Löfgren’s syndrome, genome-wide associations, Immunochip
At a Glance Commentary
Scientific Knowledge on the Subject
The genetics in subgroups of patients with sarcoidosis have not been previously investigated.
What This Study Adds to the Field
This study delineates the genetic architecture of the specific subgroup with Löfgren’s syndrome.
Sarcoidosis is a systematic inflammatory disease of unknown cause that is characterized by noncaseating granulomas. The main affected organ is the lung, which is engaged in more than 90% of patients and about a quarter develop pulmonary fibrosis with permanent dysfunction (1). Löfgren’s syndrome (LS) is an acute form of sarcoidosis, characterized by bilateral lymphadenopathy on chest radiography (in some cases with parenchymal infiltrates), erythema nodosum, and/or bilateral ankle arthritis or distinct periarticular inflammation (2). Patients with LS have a better prognosis compared with patients without LS, as evaluated 2 years after disease onset. In particular, patients with LS carrying the HLA-DRB1*03 allele run a good disease course (3).
Sarcoidosis has a peak incidence in the third and fourth decade of life, with a higher disease rate in females, and has major effects on quality of life, the ability to work, and basic activities of daily living (4, 5). In Sweden, the incidence rate of sarcoidosis has been estimated at approximately 64 per 100,000 persons (6) and is highly represented by LS (7) with an estimate of 15 per 100,000 persons yearly as observed in our respiratory unit.
In the present study, we aimed to identify genetic variants linked specifically with two sarcoidosis phenotypes, LS and non-LS, in a sample of white Swedish persons. Replication analysis of identified genetic variants was conducted in independent cohorts with similar phenotypic characterization of European ancestry from Germany, the Netherlands, and Czech Republic and of African American ancestry from the United States. A genome-wide association approach was adopted using a high-density genetic mapping technique that has been used across a range of autoimmune and inflammatory diseases (8–11) but so far not for sarcoidosis phenotypes.
Methods
Study Design and Participants
All studies had protocols approved by local institutional review boards. All participants in Sweden, Germany, the Netherlands, Czech Republic, and United States provided written informed consent and gave permission to use their DNA for research purposes. Baseline measures of clinical and demographic characteristics were measured at the time of the participants’ enrollment in each study. Details of study cohorts, genotyping, and quality control of genetic data are available in the Methods section of the online supplement.
Statistical Analysis
Association study of LS and non-LS
For both LS and non-LS, logistic regression analysis of genotyped and imputed single-nucleotide polymorphisms (SNPs) was performed with expected genotype count using the posterior probabilities. Analyses were adjusted for sex and were performed using PLINK v1.07 (12) on Immunochip data. Genome-wide significance was defined as genomic control-corrected PGC less than 5 × 10−8 and a suggestive significance threshold of PGC greater than 5 × 10−8 and less than 5 × 10−5 was established for identifying susceptible variants. The suggestive P value was chosen based on published works (9, 11, 13) using Immunochip data and proposed threshold significance PIchip less than 10−4. Moreover, conditional analysis on the lead SNP was performed as to evaluate for other potential variants. Additionally, conditional analysis on HLA-DRB1 alleles was also performed. Each HLA-DRB1 allele was analyzed as a covariate along with sex and was coded as positive (assigned a “1”) or negative (assigned a “0”) for the allele in question.
Replication of significant SNPs
Replication of associated SNPs was conducted in four independent cohorts, three of white European descent and one of black African descent, totaling 7,766 subjects. Specifically, in samples from Germany (64 LS, 413 non-LS, and 4,498 control subjects) and from African Americans (781 non-LS and 876 control subjects), SNPs identified at PGC less than 5 × 10−5 in the discovery cohort were extracted from an available dataset typed on the Immunochip. In subjects from the Netherlands (241 LS, 180 non-LS, and 192 control subjects) and Czech Republic (47 LS, 263 non-LS, and 211 control subjects), the top SNP associated with LS and non-LS, respectively, was genotyped by TaqMan assay. A nominal P value threshold for replication significance was set to P less than 0.05 with Bonferroni correction.
Meta-analysis of associated SNPs
We conducted meta-analysis by combining association results from all cohorts. An inverse variance weighting approach on regression estimates and standard errors was used as implemented in the Metafor (14) R-package for high and suggestive signals associated with LS and non-LS, respectively. We also applied genomic control to the discovery cohort (given that the inflation factor, lambda, was >1.00 [i.e., 1.16 in LS, and 1.17 in non-LS]) by multiplying standard errors by the square root of lambda. For both phenotypes, we calculated a meta-analysis log odds ratio (OR), 95% confidence interval (CI), meta-P value, along with model statistic measures, amount of heterogeneity in the true log effect size (τ2), heterogeneity P value (τ2 P), and total variability in effect size (I2). Fixed-effect and random-effect models were used for carrying out meta-analysis. We established a significance threshold of meta-P less than 5 × 10−8 as genome-wide significant.
Gene-centric analysis
To explore the genetic effects of variants located within a gene including those in introns, coding, 5′ untranslated region (5′ UTR), and 3′ UTR, we performed a genome-wide association gene-based approach for LS and non-LS, respectively, using the versatile gene-based association test as implemented in VEGAS (Versatile Gene-based Association Study) (15) software, which requires genic SNPs and individual marker P values from genome-wide association test to compute gene-based P value. From the association summary statistics of LS and non-LS, we extracted 48,740 SNPs tagging 6,840 genes across the genome and used the genomic-controlled P value (PGC) as P value entry. To account for LD structure, the HapMap phase 2 CEU population (Utah residents with ancestry from northern and western Europe) was used as reference along with the top SNP option. A Bonferroni-corrected threshold of P less than 7.3 × 10−6 (0.05/6840) was used for genome-wide significance and P less than 0.05 for nominal significance.
Integrative Analyses on Significant Findings
Bioinformatic analysis for refinement of associated variants
To explore the potential functionality of genetic variants associated with LS and non-LS and to substantiate their associations with the phenotypes in question, we evaluated genetic variant involvement in expression quantitative trait loci (eQTL), which are known to be directly linked to DNA variation. Using public/published eQTL databases (16–25), we classified a SNP as cis-eQTL SNP if it was found associated with eQTL at P less than 0.05. For SNPs with no direct cis-eQTL association, we computed the linkage disequilibrium (LD; defined by R2 > 0.8 and Dʹ = 1) using HapMap and 1,000 Genomes datasets to establish whether the SNP was near a cis-eQTL SNP. Additionally, to investigate whether a SNP had a role in machineries of gene regulation, we used available datasets from ENCODE (the Encyclopedia of DNA Elements) and the National Institutes of Health Roadmap Epigenomics Mapping Consortium. Using RegulomeDB (26) and HaploReg v2 (27) databases, we obtained measured/predicted data for all SNPs in question across various tissues and cell types. We classified a SNP as a regulatory SNP if it had at least one measured/predicted regulatory function present across collected data.
Enrichment and pathway analyses of associated variants
Enrichment and pathway analyses were conducted based on significant confirmed variants using MetaCore (Thomson Reuters, New York, NY) integrated software. Herein, we followed two different approaches: examination of biologic implication by associated variants; and examination of biologic implication by pooling together associated variants and differentially expressed genes. In the latter, three public available expression datasets from sarcoidosis (GEO accession numbers GSE18781 [28], GSE19314 [29], and GSE32887 [30]) were considered. For each Gene Expression Omnibus (GEO-GSE) dataset, we selected sarcoidosis cases and control subjects at a minimum 1:1 ratio (details are provided in Table E7 in the online supplement) and performed expression analysis using the online GEOR2 tool (http://www.ncbi.nlm.nih.gov/geo/geo2r/). Subsequently, results were downloaded and mapped to chromosome/position using the Affymetrix-Human-Genome-U133-Plus-2.0-Array annotation-data (chip_hgu133plus2) Bioconductor R-package (Affymetrix, Inc., Santa Clara, CA). After mapping, a subset of probe-IDs with false discovery rate less than 0.05 identified as differentially expressed was extracted and uploaded onto MetaCore.
Results
Association Findings
LS versus healthy control subjects
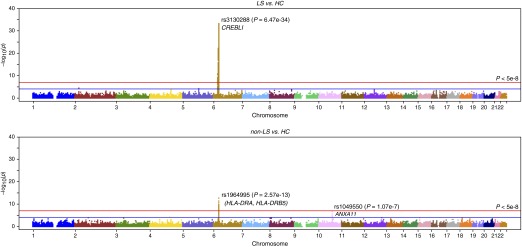
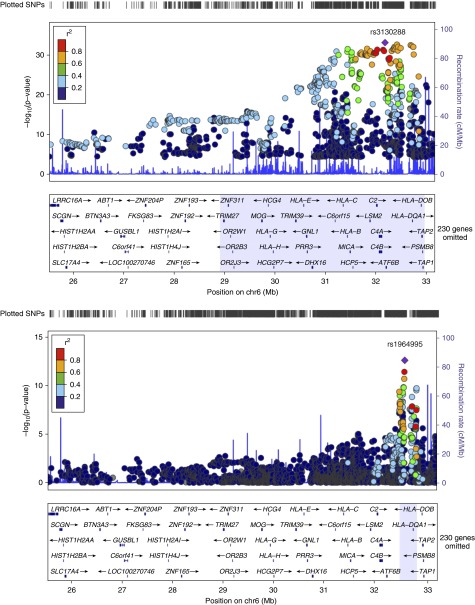
The association analysis on LS (345 cases vs. 2,025 control subjects) revealed rs3130288 (OR, 4.01; 95% CI, 3.26–4.95; PGC = 6.47 × 10−34) located in the 3′ UTR of ATF6B (also known as CREBL1), which localizes in major histocompatibility complex (MHC) class III region as the most significant finding. A large number of high signals (1,246 SNPs at PGC < 5 × 10−8) was identified spanning from the extended MHC class I to class II regions (from LRRC16, chr6:25558005 to HLA-DPB2, chr6:33191175). The top 25 significant SNPs are provided in Table 1. The entire list of LS-associated SNPs can be found as Table E1. Outside the MHC region, rs13407913 (OR, 1.48; 95% CI, 1.25–1.74; PGC = 2.31 × 10−5) located in an intron of ADCY3 on chromosome 2 was observed. Manhattan plot of association statistics is shown in Figure 1A. A close-up association regional plot is shown in Figure 2. Genomic control showed small dispersion for LS with an inflation factor lambda of 1.16. Q-Q plot for the observed versus expected P values and distribution of significant SNPs (defined by PGC < 5 × 10−8) by variant location are shown in Figures E1 and E2, respectively. Suggestive signals (692 SNPs at 5 × 10−8 < PGC < 5 × 10−5) enriching the extended MHC region were also identified and are provided in Table E1.
Table 1.
Results of Immunochip Analysis on LS versus Healthy Control Subjects Highlighting the Top 25 SNPs at Significance P Less Than 5 × 10−8
| SNP | CHR | BP | Gene (Nearby Genes) | CA/NCA | CAF | Discovery Cohort (Sweden, European Ancestry) (n = 2,470) |
Replicating Cohort (Germany, European Ancestry) (n = 4,498) |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | PGC | OR (95% CI) | P Value | ||||||
| rs3130288 | 6 | 32203979 | CREBL1 | A/C | 0.345 | 4.02 (3.26–4.95) | 3.29E-39 | 6.47E-34 | 4.13 (2.83–6.02) | 1.66E-15 |
| rs3129927 | 6 | 32441805 | C6orf10 | C/A | 0.343 | 3.99 (3.24–4.92) | 1.09E-38 | 1.82E-33 | 4.19 (2.87–6.11) | 7.79E-16 |
| rs2187668 | 6 | 32713862 | HLA-DQA1 | A/G | 0.359 | 3.97 (3.22–4.89) | 1.80E-38 | 2.79E-33 | 3.84 (2.65–5.57) | 2.93E-14 |
| rs2854275 | 6 | 32736406 | HLA-DQB1 | A/C | 0.357 | 3.96 (3.21–4.88) | 2.64E-38 | 3.87E-33 | 3.74 (2.57–5.44) | 1.42E-13 |
| rs3129950 | 6 | 32466179 | (C6orf10, BTNL2) | C/G | 0.342 | 3.98 (3.22–4.91) | 5.22E-38 | 6.96E-33 | 4.33 (2.98–6.31) | 7.89E-17 |
| rs9268219 | 6 | 32392086 | C6orf10 | C/A | 0.334 | 3.97 (3.22–4.90) | 5.59E-38 | 7.38E-33 | 4.34 (2.98–6.33) | 1.14E-16 |
| rs3129716 | 6 | 32765414 | (HLA-DQB1, HLA-DQA2) | G/A | 0.356 | 3.92 (3.19–4.83) | 5.78E-38 | 7.60E-33 | 3.73 (2.57–5.43) | 1.61E-13 |
| rs2856674 | 6 | 32767623 | (HLA-DQB1, HLA-DQA2) | G/A | 0.357 | 3.93 (3.19–4.84) | 6.08E-38 | 7.93E-33 | 3.74 (2.57–5.44) | 1.49E-13 |
| rs2395149 | 6 | 32433540 | C6orf10 | A/G | 0.343 | 3.92 (3.19–4.83) | 7.82E-38 | 9.85E-33 | 4.19 (2.87–6.11) | 7.79E-16 |
| rs9268235 | 6 | 32398186 | C6orf10 | A/G | 0.343 | 3.91 (3.18–4.82) | 1.06E-37 | 1.28E-32 | 4.18 (2.86–6.09) | 8.86E-16 |
| rs1265757 | 6 | 32410360 | C6orf10 | A/G | 0.343 | 3.91 (3.18–4.82) | 1.06E-37 | 1.28E-32 | 4.18 (2.87– 6.10) | 8.31E-16 |
| rs3135394 | 6 | 32516475 | HLA-DRA | G/A | 0.344 | 3.84 (3.13–4.72) | 1.19E-37 | 1.41E-32 | 4.21 (2.89–6.14) | 6.03E-16 |
| rs3130490 | 6 | 31847099 | C6orf27 | A/C | 0.339 | 3.83 (3.12–4.70) | 1.84E-37 | 2.05E-32 | 3.79 (2.58–5.56) | 2.97E-13 |
| rs7775397 | 6 | 32369230 | C6orf10 | C/A | 0.343 | 3.87 (3.14–4.76) | 2.45E-37 | 2.63E-32 | 4.18 (2.87–6.10) | 8.42E-16 |
| rs9268177 | 6 | 32382860 | C6orf10 | A/C | 0.343 | 3.87 (3.14–4.76) | 2.56E-37 | 2.73E-32 | 4.18 (2.87–6.10) | 8.31E-16 |
| rs389884 | 6 | 32048876 | STK19 | G/A | 0.342 | 3.82 (3.11–4.69) | 3.80E-37 | 3.84E-32 | 4.09 (2.80–5.96) | 2.76E-15 |
| rs1150753 | 6 | 32167845 | TNXB | G/A | 0.341 | 3.83 (3.11–4.71) | 4.27E-37 | 4.24E-32 | 4.12 (2.82–6.01) | 1.88E-15 |
| rs3132971 | 6 | 32338234 | (NOTCH4, C6orf10) | C/A | 0.341 | 3.86 (3.13–4.75) | 8.76E-37 | 7.86E-32 | 4.18 (2.86–6.09) | 8.97E-16 |
| rs4143332 | 6 | 31456344 | A/G | 0.337 | 3.78 (3.07–4.65) | 1.56E-36 | 1.29E-31 | 3.64 (2.50–5.31) | 6.88E-13 | |
| rs3131618 | 6 | 31542600 | (HCP5, 3.8-1) | G/A | 0.340 | 3.81 (3.09–4.69) | 2.40E-36 | 1.87E-31 | 3.72 (2.55–5.42) | 2.69E-13 |
| rs3132449 | 6 | 31733992 | (APOM, C6orf47) | A/G | 0.340 | 3.71 (3.03–4.55) | 2.46E-36 | 1.91E-31 | 3.94 (2.70–5.75) | 1.73E-14 |
| rs3117577 | 6 | 31835453 | MSH5 | G/A | 0.339 | 3.70 (3.02–4.54) | 2.85E-36 | 2.17E-31 | 3.96 (2.72–5.77) | 1.36E-14 |
| rs519417 | 6 | 31986412 | (ZBTB12, C2) | A/G | 0.340 | 3.71 (3.03–4.56) | 3.26E-36 | 2.43E-31 | 4.01 (2.75–5.85) | 6.91E-15 |
| rs497309 | 6 | 32000463 | (ZBTB12, C2) | C/A | 0.340 | 3.71 (3.02–4.55) | 3.79E-36 | 2.77E-31 | 4.03 (2.76–5.88) | 5.71E-15 |
| rs1270942 | 6 | 32026839 | CFB | G/A | 0.339 | 3.72 (3.03–4.56) | 4.06E-36 | 2.94E-31 | 4.05 (2.78–5.91) | 4.43E-15 |
Definition of abbreviations: BP = base pairs; CA = coded allele; CAF = coded allele frequency; CHR = chromosome; CI = confidence interval; LS = Löfgren’s syndrome; NCA = noncoded allele; OR = odds ratio; P = unadjusted P value; PGC = adjusted P value by genomic control (lambda = 1.16); SNP = single-nucleotide polymorphism.
Genetic effects (OR) adjusted according to sex, coded allele. OR is the risk on LS (0/1) per allele based on the additive genetic model. 95% CI is the 95% CI for the OR.
The entire list of LS-associated SNPs is provided in Table E2.
Figure 1.
Manhattan plot of all markers from the Immunochip. Each dot represents the PGC value of association analysis [−log10(PGC)] on the y-axis and position in chromosome on the x-axis (PGC is genomic-controlled P value). (Top) Löfgren’s syndrome (LS) versus control subjects and (bottom) non-LS versus control subjects. Red line indicates PGC < 5 × 10−8; blue line is PGC < 5 × 10−5. HC = healthy control subjects.
Figure 2.
Regional association plots of chr6:25527073–33191175. Löfgren’s syndrome (LS)-associated single-nucleotide polymorphisms (SNPs) (top) at PGC < 5 × 10−8 highlight region chr6:28910128–3295881. Non-LS–associated SNPs (bottom) at PGC < 5 × 10−8 highlight region chr6: 32447326–32789970. PGC = genomic-controlled P value.
Results after conditioning on the top signal rs3130288 (see Table E2) showed that residual associations in the MHC classical class II locus were predominant, as indicated by leading SNP rs4642516 (Pcond,GC = 6.69 × 10−7) located between HLA-DQB1 and HLA-DQA2. Additionally, findings at Pcond,GC less than 5 × 10−5 were also noted in non-MHC genomic loci, 55 SNPs located near ERAP1, in ERAP2 and LNPEP, and near LIX1, all on chromosome 5, and one SNP between CSMD1 and MCPH1 on chromosome 8. The Manhattan and Q-Q plots after conditioning are shown in Figure E3. Regional association plots for LS-associated SNPs on chromosome 6 at PGC less than 5 × 10−8 before and after conditioning on the top signal are illustrated in Figures E4A and E4B. Regional association plot for suggestive finding is shown in Figure E5.
Additionally, because HLA-DRB1*03 plays a strong role in the course of the disease, which is particularly seen in patients with LS, we stratified our LS dataset (cases and control subjects) by HLA-DRB1*03 carriers and noncarriers and performed Immunochip analysis on these subgroups. The association test in HLA-DRB1*03 carriers (513 healthy control subjects and 253 LS) identified rs2248462 (PGC = 1.20 × 10−4), an intergenic SNP located HCP5 and MICB on chr6:31554775 as the top signal. Similarly, in HLA-DBR1*03 noncarriers (1,573 healthy control subjects and 131 LS), the test revealed rs3104407 (PGC = 1.25 × 10−5), an intergenic SNP located between HLA-DQB1 and HLA-DQA2 on chr6:32790430 as the top signal (further details on these analyses and results are given in the online supplement Note). Notably, Immunochip results for HLA-DRB1*03 stratified groups were consistent with Immunochip results for conditioning on HLA-DRB1*03 (see the Note in the online supplement and Figure E6), namely all identified associations for LS substantially attenuate (with P > 5 × 10−8) when the effect of HLA-DRB1*03 is considered.
Non-LS versus healthy control subjects
Results from association analysis on non-LS (568 cases vs. 2,086 control subjects) showed rs1964995 (OR, 0.55; 95% CI, 0.47–0.64; PGC = 2.57 × 10−13) located between HLA-DRA and HLA-DRB5, which localizes to MHC class II locus as the most significant signal. Fifty-two high signals located between C6orf10 and HLA-DQA2 (chr6:32447326–32789970) and between PLAC9 and ANXA11 (chr10:8191705–81916682) at PGC < 5 × 10−8 were identified. The top 25 significant SNPs are provided in Table 2. The entire list of non-LS–associated SNPs can be found in Table E3. Manhattan plot for Immunochip associations is illustrated in Figure 1B. A close-up association regional plot is shown in Figure 2. Genomic control for non-LS defined by inflation factor lambda was 1.17. The Q-Q plot and distribution of significant SNPs (PGC < 5 × 10−8) by variant type are illustrated in Figures E7 and E8, respectively. Suggestive signals (122 SNPs included in Table E3) at PGC greater than 5 × 10−8 and less than 5 × 10−5 revealed additional signals in the MHC region, particularly clustering between classes III and II regions and outside the extended MHC on chromosome 10 in ANXA11 and near PLAC9, DYDC2, C10orf58, and TSPAN14.
Table 2.
Results of Immunochip Analysis on Non-LS versus Healthy Control Subjects Highlighting the Top 25 SNPs at Significance P Less Than 5 × 10−8
| SNP | CHR | BP | Gene (Nearby Genes) | CA/NCA | CAF | Discovery Cohort (Sweden, European Ancestry) (n = 2,750) |
Replicating Cohort (Germany, European Ancestry) (n = 4,911) |
Replicating Cohort (African American Ancestry) (n = 1,657) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | PGC | OR (95% CI) | P Value | OR (95% CI) | P Value | ||||||
| rs1964995 | 6 | 32557389 | (HLA-DRA, HLA-DRB5) | G/A | 0.296 | 0.55 (0.47–0.64) | 2.47E-15 | 2.57E-13 | 0.56 (0.48–0.66) | 1.19E-12 | 0.70 (0.59–0.82) | 1.95E-05 |
| rs9269110 | 6 | 32551247 | (HLA-DRA, HLA-DRB5) | A/C | 0.389 | 1.74 (1.51–2.01) | 5.78E-14 | 3.82E-12 | 1.34 (1.16–1.56) | 6.20E-05 | — | — |
| rs9269081 | 6 | 32549078 | (HLA-DRA, HLA-DRB5) | A/C | 0.376 | 1.71 (1.48–1.97) | 4.01E-13 | 2.01E-11 | 1.33 (1.14–1.54) | 1.63E-04 | — | — |
| rs6923504 | 6 | 32536164 | (HLA-DRA, HLA-DRB5) | G/C | 0.381 | 1.66 (1.44–1.91) | 4.05E-12 | 1.46E-10 | 1.35 (1.16–1.56) | 6.09E-05 | — | — |
| rs6903608 | 6 | 32536263 | (HLA-DRA, HLA-DRB5) | G/A | 0.381 | 1.66 (1.44–1.91) | 4.05E-12 | 1.46E-10 | 1.36 (1.17–1.57) | 3.55E-05 | — | — |
| rs9268882 | 6 | 32539601 | (HLA-DRA, HLA-DRB5) | A/G | 0.381 | 1.66 (1.44–1.91) | 4.54E-12 | 1.61E-10 | 1.36 (1.17–1.57) | 3.86E-05 | — | — |
| rs3998158 | 6 | 32789970 | (HLA-DQB1, HLA-DQA2) | G/A | 0.168 | 0.53 (0.45–0.64) | 5.35E-12 | 1.85E-10 | 0.60 (0.49–0.74) | 9.45E-07 | 1.19 (0.98–1.43) | 7.47E-02 |
| rs9275582 | 6 | 32788048 | (HLA-DQB1, HLA-DQA2) | A/G | 0.172 | 0.53 (0.45–0.64) | 5.56E-12 | 1.91E-10 | 0.59 (0.48–0.72) | 2.42E-07 | 1.29 (1.08–1.54) | 4.29E-03 |
| rs9275580 | 6 | 32787440 | (HLA-DQB1, HLA-DQA2) | G/A | 0.178 | 0.54 (0.46–0.65) | 9.71E-12 | 3.08E-10 | 0.60 (0.50–0.74) | 4.99E-07 | 1.42 (1.20–1.67) | 3.98E-05 |
| rs2395153 | 6 | 32453573 | (C6orf10, BTNL2) | C/G | 0.273 | 0.59 (0.51–0.69) | 1.46E-11 | 4.38E-10 | 0.68 (0.58–0.79) | 8.61E-07 | 0.63 (0.53–0.75) | 2.12E-07 |
| rs3129948 | 6 | 32462622 | (C6orf10, BTNL2) | C/A | 0.390 | 1.63 (1.41–1.88) | 4.11E-11 | 1.06E-09 | 1.35 (1.17–1.56 | 5.66E-05 | 1.35 (1.14–1.60) | 6.34E-04 |
| rs3117098 | 6 | 32466491 | (C6orf10, BTNL2) | G/A | 0.390 | 1.59 (1.38–1.84) | 2.37E-10 | 4.78E-09 | 1.35 (1.16–1.56) | 6.45E-05 | 1.36 (1.14–1.61) | 4.63E-04 |
| rs3129954 | 6 | 32473558 | BTNL2 | A/G | 0.390 | 1.59 (1.38–1.84) | 2.37E-10 | 4.78E-09 | 1.35 (1.16–1.56) | 6.10E-05 | 1.34 (1.13–1.59) | 8.91E-04 |
| rs4424066 | 6 | 32462406 | (C6orf10, BTNL2) | G/A | 0.314 | 0.62 (0.54–0.72) | 2.54E-10 | 5.06E-09 | 0.70 (0.60–0.81) | 3.09E-06 | 0.75 (0.64–0.87) | 1.97E-04 |
| rs3129961 | 6 | 32486918 | (BTNL2, HLA-DRA) | G/A | 0.390 | 1.59 (1.38–1.84) | 2.82E-10 | 5.53E-09 | 1.34 (1.16–1.55) | 7.52E-05 | 1.35 (1.14–1.60) | 4.18E-04 |
| rs3129955 | 6 | 32473818 | BTNL2 | A/G | 0.385 | 1.59 (1.38–1.84) | 3.49E-10 | 6.64E-09 | 1.35 (1.17–1.56) | 5.19E-05 | 1.34 (1.13–1.59) | 8.24E-04 |
| rs4373382 | 6 | 32458846 | (C6orf10, BTNL2) | C/A | 0.313 | 0.63 (0.54–0.73) | 4.16E-10 | 7.72E-09 | 0.69 (0.59–0.80) | 1.58E-06 | 0.75 (0.65–0.87) | 1.73E-04 |
| rs3817973 | 6 | 32469089 | (C6orf10, BTNL2) | A/G | 0.314 | 0.63 (0.54–0.73) | 4.35E-10 | 8.02E-09 | 0.69 (0.59–0.80) | 1.57E-06 | 0.75 (0.65–0.88) | 2.68E-04 |
| rs2076529 | 6 | 32471933 | BTNL2 | G/A | 0.314 | 0.63 (0.54–0.73) | 4.35E-10 | 8.02E-09 | 0.69 (0.59–0.80) | 1.48E-06 | 0.75 (0.65–0.88) | 2.56E-04 |
| rs9268472 | 6 | 32463583 | (C6orf10, BTNL2) | A/G | 0.314 | 0.63 (0.54–0.73) | 4.61E-10 | 8.43E-09 | 0.69 (0.59–0.80) | 1.57E-06 | 0.76 (0.65–0.89) | 4.44E-04 |
| rs9271588 | 6 | 32698931 | (HLA-DRB1, HLA-DQA1) | G/A | 0.351 | 0.64 (0.56–0.74) | 8.35E-10 | 1.40E-08 | 0.67 (0.58–0.78) | 2.40E-07 | — | — |
| rs2858332 | 6 | 32789139 | (HLA-DQB1, HLA-DQA2) | A/C | 0.354 | 0.65 (0.56–0.75) | 1.47E-09 | 2.28E-08 | 1.36 (1.18–1.57) | 3.07E-05 | — | — |
| rs3830135 | 6 | 32656442 | HLA-DRB1 | A/G | 0.054 | 0.42 (0.32–0.56) | 1.82E-09 | 2.74E-08 | 0.56 (0.42–0.74) | 3.16E-05 | — | — |
Definition of abbreviations: BP = base pairs; CA = coded allele; CAF = coded allele frequency; CHR = chromosome; CI = confidence interval; LS = Löfgren’s syndrome; NCA = noncoded allele; OR = odds ratio; P = unadjusted P value; PGC = adjusted P value by genomic control (lambda = 1.17); SNP = single-nucleotide polymorphism.
Genetic effects (OR) adjusted according to sex, coded allele. OR is the risk on LS (0/1) per allele based on the additive genetic model. 95% CI is the 95% CI for the OR.
The entire list of non-LS–associated SNPs is provided in Table E3.
Results after conditioning on top signal rs1964995 (see Table E4) stressed rs1049550 (Pcond,GC = 2.19 × 10−7) located in a coding region of ANXA11 as the highest peak, followed by four intergenic SNPs, adjacent to previously observed genes. Suggestive signals were also observed, of which most (intergenic SNPs) were clustered together among the HLA class II genes (chr6:32411670–32873563). Manhattan and Q-Q plots after conditioning are shown in Figure E9. Regional association plots for non-LS–associated SNPs on chromosome 6 at PGC less than 5 × 10−8 before and after conditioning on the top signal are illustrated in Figures E10A and E10B. Regional association plot for suggestive signals at PGC less than 5 × 10−5 is illustrated in Figure E11. Association results from evaluating both phenotypes, LS versus non-LS including conditioning on the top signal and on HLA-DRB1 alleles, and replication results are provided in the Note in the online supplement.
Additionally, considering that the non-LS phenotype may be confounded by phenotype heterogeneity because of the presence of extrapulmonary manifestations as often seen in patients without LS, we reran Immunochip analysis by excluding patients without LS with confirmed extrapulmonary manifestations. Herein, 451 non-LS cases without extrapulmonary manifestations and 2,086 healthy control subjects were reanalyzed. Immunochip results showed rs1964995 (OR, 0.50; 95% CI, 0.42–0.59; PGC = 4.31 × 10−13) as the top association along with similar high and suggested signals as identified in the main Immunochip for non-LS, thus implying that the same dominating gene associations are important in both non-LS groups with or without extrapulmonary manifestations. Results from Immunochip for non-LS without pulmonary manifestation are adjoined with main Immunochip findings for non-LS as given in Table E3.
Replication of Significant SNPs
Results from the association analysis on LS (64 LS vs. 4,498 control subjects) conducted in an independent German cohort confirmed the association of many signals. In the LS group, out of 1,246 high signals identified in the discovery cohort, 671 were confirmed with Bonferroni-adjusted P less than 4 × 10−5 (0.05/1,246) to account for multiple testing. The top signal rs3130288 identified in the discovery cohort was well replicated (Germany: OR, 4.13; 95% CI, 2.83–6.02; P = 1.66 × 10−15) (the Netherlands: OR, 4.54; 95% CI, 2.97–6.93; P = 2.82 × 10−12) (Czech Republic: OR, 4.94; 95% CI, 2.53–9.65; P = 2.88 × 10−6). Confirmed signals localized to the extended MHC region spanning from ZNF184 to HLA-DPB2 (chr6:27521903–33191175), as illustrated in the regional association plot (Figure 2). Replication for assessing 692 suggestive signals identified with PGC greater than 5 × 10−8 and less than 5 × 10−5 in the Swedish population resulted in the confirmation of 56 suggested signals with Bonferroni-adjusted P less than 7.22 × 10−5 (0.05/692), which localized to MHC class I and class II regions. Non-MHC suggested signals located in LRRC16A (rs6921589, P = 6.54 × 10−2 and rs9295661, P = 6.61 × 10−2) were borderline significant. Signals in ADCY3 (rs13407913, rs6752378, and rs10182181) were not confirmed in the German cohort, possibly because of few number of LS cases. Results from this analysis are given in Table E1.
In the non-LS group, out of 38 high signals identified in the discovery cohort, 27 were confirmed in the German cohort and 20 in the African American cohort with Bonferroni-adjusted P less than 1.32 × 10−3 (0.05/38). The lead signal rs1964995 was well replicated in all participating cohorts (Germany: OR, 0.56; 95% CI, 0.49–0.66; P = 1.19 × 10−12) (African American: OR, 0.70; 95% CI, 0.59–0.82; P = 1.95 × 10−5) (the Netherlands: OR, 0.71; 95% CI, 0.51–0.97; P = 3.31 × 10−2) (Czech Republic: OR, 0.55; 95% CI, 0.39–0.77; P = 4.05 × 10−4). All confirmed signals localized to MHC class II region, clustering among C6orf10, BTNL2, HLA-DRA, HLA-DRB5, HLA-DRB1, HLA-DQA1, and HLA-DQA2, as illustrated in the regional association plot (Figure 1). Replication assessment of 135 suggestive signals confirmed 41 suggestive signals in the Germany cohort and 30 suggested signals in the USA-AA cohort with Bonferroni-adjusted P less than 3.70 × 10−4 (0.05/135). These localized in the same regions as confirmed high signals, thus enriching the density of the associated region. Out of the seven signals located outside the MHC, three signals (rs1953600, rs2573346, and rs1049550; in strong LD, R2 ≥ 0.87, D′≥ 0.93) located between PLAC9 and ANXA11 (intergenic), and inside ANXA11 (intronic and coding) were replicated with nominal significance P less than 0.05. Results from this analysis are given in Table E3.
Results from evaluating the association of six SNPs located outside the MHC region and conducted in two independent cohorts from Czech Republic and the Netherlands led to the confirmation of two SNPs. For LS, rs10108612 (discovery PGC = 1.97 × 10−2) located between CSMD1 and MCPH1 on chromosome 8 was marginally significant (P = 5.81 × 10−2) in the Czech cohort with and nonsignificant in the Dutch cohort. Similarly, rs13407913 (discovery PGC = 2.31 × 10−5) located in an intron of ADCY3 on chromosome 2 was marginally significant (P = 7.8 × 10−2) in the Dutch LS group and nonsignificant in the Czech LS group. For non-LS, no associations among the tested SNPs were found in the either Czech or Dutch samples.
In summary, combining replicated high and suggestive signals, we observed that 710 confirmed variants were associated exclusively to LS compared with control subjects, 51 to non-LS compared with control subjects, and 17 were associated to both LS and non-LS. Whereas in a population of African ancestry, out of 50 signals associated with non-LS, 17 were LS-associated signals found in populations of European ancestry. In populations with European ancestry, the shared genomic loci associated to both LS and non-LS spanned between two intergenic regions between C6orf10 and BTNL2 and between HLA-DRA and HLA-DRB5 (chr6:32453573–32528157), whereas in a population with African ancestry it extended along chromosome 6 to an intergenic region located between HLA-DQB1 and HLA-DQA2 (chr6: 32453573–32778286).
Meta-analysis of Associated SNPs
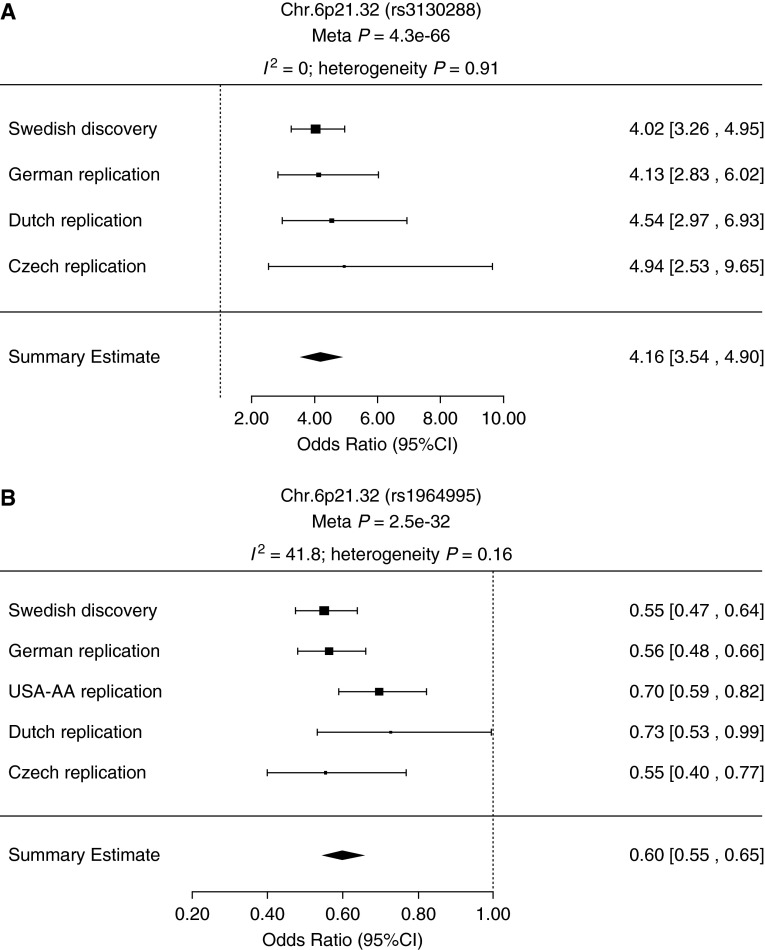
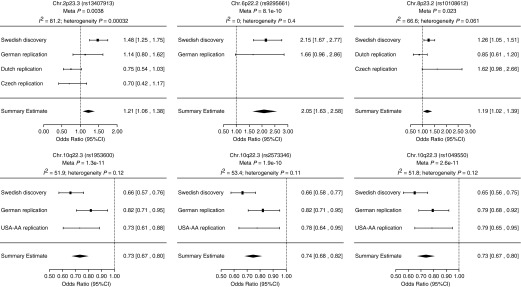
In the meta-analysis, results for estimating the overall effect size (OR) of lead SNPs associated with LS and non-LS, respectively, were highly significant (rs3130288 meta-P = 4.3 × 10−66 and rs1964995 meta-P = 2.5 × 10−32) (Figure 3). Likewise, results for SNPs located outside the MHC region showed a similar pattern: for LS, rs13407913 (meta-P = 3.8 × 10−3), rs9295661 (meta-P = 8.1 × 10−10), and rs10108612 (meta-P = 2.3 × 10−2); and for non-LS, rs1953600 (meta-P = 1.3 × 10−11), rs2573346 (meta-P = 1.9 × 10−10), and rs1049550 (meta-P = 2.6 × 10−1a). Forest plots for SNPs outside the MHC are illustrated in Figure 4.
Figure 3.
Forest plots showing Löfgren’s syndrome (LS) and non-LS lead single-nucleotide polymorphism (SNP) associations inside the major histocompatibility complex. Plots show the study-specific association estimates (odds ratio) and 95% confidence intervals (CIs) for discovery set (Swedish) and all replication studies (German, African American [USA-AA], Dutch, and Czech). Rs3130288 (A) and rs1964995 (B) are the lead SNPs associated with LS and non-LS, respectively. The summary estimate includes the association estimate and CI under fixed-effect model for the meta-analysis combining the discovery and replicating cohort shown as a diamond. I2 indicates the percentage of total variation in study estimates because of heterogeneity in the meta-analysis.
Figure 4.
Forest plots showing Löfgren’s syndrome (LS) and non-LS single-nucleotide polymorphism (SNP) associations outside the major histocompatibility complex. Plots show the study-specific association estimates (odds ratio) and 95% confidence intervals (CIs) for discovery set (Swedish) and replication studies (German, African American [USA-AA], Dutch, Czech). Results for LS-associated SNPs rs133409913, rs9295661, and rs10108612 are shown in the top. Results for non-LS–associated SNP rs1953600, rs2573346, and rs1049550 are shown in the bottom. The association estimate and CI (estimated under fixed-effect model) for the meta-analysis combining the discovery and replicating cohort are shown as a diamond. Blank spaces indicate occasions in which a particularly study was not able to provide results for a given SNP. I2 indicates the percentage of total variation in study estimates because of heterogeneity in the meta-analysis.
Combining association summary statistics from both Swedish and German cohorts, results from meta-analysis on 1,938 high and suggested LS signals revealed 1,499 signals (defining 267 loci; 22/1,499 signals had heterogeneity P < 0.05 and I2 = 0%) under the fixed-effects model and 1,320 signals (defining 241 loci; 116/1,320 had I2 = [ 0.7%, 62.4%]) under the random-effects model with strong evidence for association at meta-P less than 5 × 10−8. Table 3 provides the top 25 LS-associated findings. The entire list for LS-associated SNPs can be found in Table E5. Similarly, meta-analysis combining association summary statistics from Swedish, German, and African American cohorts for evaluating 173 high and suggested non-LS signals showed 47 signals (defining 12 loci) with strong evidence for association under the fixed-effects model and 37 signals (defining eight loci) under the random-effects model, with strong evidence for association at meta-P less than 5 × 10−8. Non-LS signals were selected with heterogeneity P greater than 0.05. Table 4 provides the 25 top non-LS–associated signals. The entire list for non-LS–associated SNPs can be found in Table E6.
Table 3.
Results from Combined Meta-analysis on LS-associated SNPs at Significance Meta-P Less Than 5 × 10−8 (Top 25 Findings)
| SNP | CHR | BP | CA/NCA | Gene (Nearby Genes) | SNP Location | Cohorts | Fixed-Effects Model |
Random-Effects Model |
||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Meta-P Value | OR (95% CI) | Meta-P Value | |||||||
| rs3130288 | 6 | 32203979 | A/C | CREBL1 | 3′ UTR | GER, SWE | 4.16 (3.54–4.90) | 4.30E-66 | 4.16 (3.54–4.90) | 4.30E-66 |
| rs3129927 | 6 | 32441805 | C/A | C6orf10 | Intron | GER, SWE | 4.04 (3.36–4.84) | 1.29E-50 | 4.04 (3.36–4.84) | 1.29E-50 |
| rs9268219 | 6 | 32392086 | C/A | C6orf10 | Intron | GER, SWE | 4.05 (3.37–4.87) | 1.51E-50 | 4.05 (3.37–4.87) | 1.51E-50 |
| rs3129950 | 6 | 32466179 | C/G | (C6orf10, BTNL2) | Intergenic | GER, SWE | 4.06 (3.38–4.88) | 1.79E-50 | 4.06 (3.38–4.88) | 1.79E-50 |
| rs2395149 | 6 | 32433540 | A/G | C6orf10 | Intron | GER, SWE | 3.98 (3.32–4.77) | 3.55E-50 | 3.98 (3.32–4.77) | 3.55E-50 |
| rs3135394 | 6 | 32516475 | G/A | HLA-DRA | Intron | GER, SWE | 3.92 (3.27–4.70) | 7.66E-50 | 3.92 (3.27–4.70) | 7.66E-50 |
| rs1265757 | 6 | 32410360 | A/G | C6orf10 | Intron | GER, SWE | 3.97 (3.31–4.77) | 8.02E-50 | 3.97 (3.31–4.77) | 8.02E-50 |
| rs9268235 | 6 | 32398186 | A/G | C6orf10 | Intron | GER, SWE | 3.97 (3.31–4.76) | 8.22E-50 | 3.97 (3.31–4.76) | 8.22E-50 |
| rs7775397 | 6 | 32369230 | C/A | C6orf10 | Coding | GER, SWE | 3.94 (3.28–4.73) | 2.94E-49 | 3.94 (3.28–4.73) | 2.94E-49 |
| rs9268177 | 6 | 32382860 | A/C | C6orf10 | Intron | GER, SWE | 3.94 (3.28–4.73) | 2.98E-49 | 3.94 (3.28–4.73) | 2.98E-49 |
| rs2187668 | 6 | 32713862 | A/G | HLA-DQA1 | Intron | GER, SWE | 3.94 (3.28–4.73) | 3.29E-49 | 3.94 (3.28–4.73) | 3.29E-49 |
| rs389884 | 6 | 32048876 | G/A | STK19 | Intron | GER, SWE | 3.88 (3.24–4.65) | 4.28E-49 | 3.88 (3.24–4.65) | 4.28E-49 |
| rs3132971 | 6 | 32338234 | C/A | (NOTCH4, C6orf10) | Intergenic | GER, SWE | 3.93 (3.28–4.72) | 6.55E-49 | 3.93 (3.28–4.72) | 6.55E-49 |
| rs1794282 | 6 | 32774504 | A/G | (HLA-DQB1, HLA-DQA2) | Intergenic | GER, SWE | 3.92 (3.26–4.70) | 9.82E-49 | 3.92 (3.26–4.70) | 9.82E-49 |
| rs1150753 | 6 | 32167845 | G/A | TNXB | Intron | GER, SWE | 3.89 (3.25–4.67) | 1.26E-48 | 3.89 (3.25–4.67) | 1.26E-48 |
| rs3129716 | 6 | 32765414 | G/A | (HLA-DQB1, HLA-DQA2) | Intergenic | GER, SWE | 3.88 (3.23–4.65) | 1.77E-48 | 3.88 (3.23–4.65) | 1.77E-48 |
| rs2854275 | 6 | 32736406 | A/C | HLA-DQB1 | Intron | GER, SWE | 3.91 (3.25–4.69) | 2.39E-48 | 3.91 (3.25–4.69) | 2.39E-48 |
| rs2856674 | 6 | 32767623 | G/A | (HLA-DQB1, HLA-DQA2) | Intergenic | GER, SWE | 3.88 (3.24–4.66) | 2.80E-48 | 3.88 (3.24–4.66) | 2.80E-48 |
| rs3130490 | 6 | 31847099 | A/C | C6orf27 | Intron | GER, SWE | 3.82 (3.19–4.58) | 6.92E-48 | 3.82 (3.19–4.58) | 6.92E-48 |
| rs1270942 | 6 | 32026839 | G/A | CFB | Intron | GER, SWE | 3.79 (3.17–4.54) | 7.12E-48 | 3.79 (3.17–4.54) | 7.12E-48 |
| rs3132449 | 6 | 31733992 | A/G | (APOM, C6orf47) | Intergenic | GER, SWE | 3.76 (3.14–4.50) | 1.11E-47 | 3.76 (3.14–4.50) | 1.11E-47 |
| rs519417 | 6 | 31986412 | A/G | (ZBTB12, C2) | Intergenic | GER, SWE | 3.78 (3.16–4.52) | 1.34E-47 | 3.78 (3.16–4.52) | 1.34E-47 |
| rs497309 | 6 | 32000463 | C/A | (ZBTB12, C2) | Intergenic | GER, SWE | 3.78 (3.16–4.53) | 1.82E-47 | 3.78 (3.16–4.53) | 1.82E-47 |
| rs3117577 | 6 | 31835453 | G/A | MSH5 | Intron | GER, SWE | 3.76 (3.14–4.49) | 1.96E-47 | 3.76 (3.14–4.49) | 1.96E-47 |
| rs3117582 | 6 | 31728499 | C/A | (BAT3, APOM) | Intergenic | GER, SWE | 3.73 (3.12–4.46) | 2.53E-47 | 3.73 (3.12–4.46) | 2.53E-47 |
Definition of abbreviations: BP = base pairs; CA = coded allele; CHR = chromosome; CI = confidence interval; GER = Germany; LS = Löfgren’s syndrome; NCA = noncoded allele; OR = odds ratio; SNP = single-nucleotide polymorphism; SWE = Sweden; UTR = untranslated region.
The entire list of LS-associated SNPs is provided in Table E5.
The overall association estimates (OR) and 95% CI for LS-associated SNPs under fixed-effect and random-effect models, respectively. Cohorts from SWE and GER are of European ancestry.
Table 4.
Results from Combined Meta-analysis on Non-LS–associated SNPs at Significance Meta-P Less Than 5 × 10−8 (Top 25 Findings)
| SNP | CHR | BP | CA/NCA | Gene (Nearby Genes) | SNP Location | Cohorts | Fixed-Effects Model |
Random-Effects Model |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Meta-P Value | OR (95% CI) | Meta-P Value | τ2P Value | I2 (%) | |||||||
| rs1964995 | 6 | 32557389 | G/A | (HLA-DRA, HLA-DRB5) | Intergenic | SWE, GER, USA-AA | 0.60 (0.55–0.65) | 2.53E-32 | 0.61 (0.54–0.68) | 5.99E-17 | 1.55E-01 | 41.79 |
| rs2213585 | 6 | 32521128 | G/A | (HLA-DRA, HLA-DRB5) | Intergenic | SWE, GER, USA-AA | 1.51 (1.39–1.63) | 8.82E-23 | 1.51 (1.34–1.70) | 3.36E-11 | 1.11E-01 | 54.69 |
| rs2227139 | 6 | 32521437 | G/A | (HLA-DRA, HLA-DRB5) | Intergenic | SWE, GER, USA-AA | 1.50 (1.39–1.63) | 8.96E-23 | 1.51 (1.33–1.70) | 6.87E-11 | 1.03E-01 | 56.15 |
| rs2213586 | 6 | 32521072 | A/G | (HLA-DRA, HLA-DRB5) | Intergenic | SWE, GER, USA-AA | 1.50 (1.38–1.63) | 1.29E-22 | 1.50 (1.33–1.70) | 4.58E-11 | 1.09E-01 | 54.98 |
| rs7192 | 6 | 32519624 | A/C | HLA-DRA | Coding | SWE, GER, USA-AA | 1.50 (1.39–1.63) | 1.31E-22 | 1.51 (1.34–1.69) | 8.78E-12 | 1.27E-01 | 51.57 |
| rs2395153 | 6 | 32453573 | C/G | (C6orf10, BTNL2) | Intergenic | SWE, GER, USA-AA | 0.63 (0.58–0.69) | 1.76E-22 | 0.63 (0.58–0.69) | 1.76E-22 | 5.17E-01 | 0.00 |
| rs7195 | 6 | 32520517 | A/G | HLA-DRA | UTR | SWE, GER, USA-AA | 1.50 (1.38–1.63) | 1.77E-22 | 1.50 (1.34–1.68) | 9.57E-13 | 1.52E-01 | 46.70 |
| rs3763327 | 6 | 32521808 | G/C | (HLA-DRA, HLA-DRB5) | Intergenic | SWE, USA-AA | 1.57 (1.42–1.73) | 7.36E-19 | 1.57 (1.34–1.85) | 2.75E-08 | 1.08E-01 | 61.20 |
| rs477515 | 6 | 32677669 | A/G | (HLA-DRB1, HLA-DQA1) | Intergenic | SWE, GER, USA-AA | 0.64 (0.58–0.71) | 3.93E-18 | 0.63 (0.54–0.75) | 5.32E-08 | 7.79E-02 | 61.89 |
| rs2395182 | 6 | 32521295 | C/A | (HLA-DRA, HLA-DRB5) | Intergenic | SWE, GER, USA-AA | 1.49 (1.36–1.62) | 7.05E-18 | 1.47 (1.22–1.78) | 4.95E-05 | 1.48E-02 | 76.86 |
| rs4373382 | 6 | 32458846 | C/A | (C6orf10, BTNL2) | Intergenic | SWE, GER, USA-AA | 0.69 (0.63–0.75) | 1.17E-17 | 0.69 (0.62–0.76) | 5.81E-13 | 2.48E-01 | 28.87 |
| rs3129952 | 6 | 32467741 | C/G | (C6orf10, BTNL2) | Intergenic | SWE, USA-AA | 1.64 (1.46–1.84) | 1.54E-17 | 1.64 (1.46–1.84) | 1.54E-17 | 4.18E-01 | 0.00 |
| rs4424066 | 6 | 32462406 | G/A | (C6orf10, BTNL2) | Intergenic | SWE, GER, USA-AA | 0.69 (0.63–0.75) | 1.73E-17 | 0.69 (0.62–0.76) | 3.44E-12 | 2.28E-01 | 32.82 |
| rs2076529 | 6 | 32471933 | G/A | BTNL2 | Coding | SWE, GER, USA-AA | 0.69 (0.63–0.75) | 1.74E-17 | 0.69 (0.62–0.76) | 1.70E-12 | 2.37E-01 | 30.92 |
| rs3817973 | 6 | 32469089 | A/G | (C6orf10, BTNL2) | Intergenic | SWE, GER, USA-AA | 0.69 (0.63–0.75) | 1.96E-17 | 0.69 (0.62–0.76) | 2.39E-12 | 2.34E-01 | 31.62 |
| rs2239802 | 6 | 32519824 | C/G | HLA-DRA | Intron | SWE, GER, USA-AA | 1.48 (1.35–1.61) | 1.97E-17 | 1.46 (1.22–1.76) | 5.85E-05 | 1.56E-02 | 76.65 |
| rs9268472 | 6 | 32463583 | A/G | (C6orf10, BTNL2) | Intergenic | SWE, GER, USA-AA | 0.69 (0.63–0.75) | 3.84E-17 | 0.69 (0.62–0.77) | 3.06E-11 | 2.04E-01 | 37.34 |
| rs3177928 | 6 | 32520413 | A/G | HLA-DRA | UTR | SWE, GER, USA-AA | 0.53 (0.46–0.62) | 8.37E-17 | 0.53 (0.46–0.62) | 8.37E-17 | 6.77E-01 | 0.00 |
| rs2076530 | 6 | 32471794 | G/A | BTNL2 | Coding | SWE, GER, USA-AA | 0.69 (0.64–0.76) | 1.21E-16 | 0.69 (0.64–0.76) | 1.21E-16 | 4.96E-01 | 0.00 |
| rs9268969 | 6 | 32542327 | A/G | (HLA-DRA, HLA-DRB5) | Intergenic | SWE, GER, USA-AA | 0.66 (0.60–0.73) | 1.29E-16 | 0.66 (0.60–0.73) | 1.29E-16 | 6.60E-01 | 0.00 |
| rs3129888 | 6 | 32519704 | G/A | HLA-DRA | Intron | SWE, GER, USA-AA | 1.48 (1.35–1.62) | 2.07E-16 | 1.47 (1.25–1.74) | 6.22E-06 | 4.00E-02 | 68.94 |
| rs3129948 | 6 | 32462622 | C/A | (C6orf10, BTNL2) | Intergenic | SWE, GER, USA-AA | 1.44 (1.32–1.58) | 2.30E-16 | 1.44 (1.27–1.63) | 1.46E-08 | 1.31E-01 | 50.83 |
| rs2294878 | 6 | 32475773 | A/C | BTNL2 | Intron | SWE, GER, USA-AA | 0.70 (0.64–0.76) | 3.66E-16 | 0.70 (0.64–0.76) | 3.66E-16 | 9.24E-01 | 0.00 |
| rs3117098 | 6 | 32466491 | G/A | (C6orf10, BTNL2) | Intergenic | SWE, GER, USA-AA | 1.44 (1.32–1.57) | 6.96E-16 | 1.43 (1.28–1.60) | 4.11E-10 | 2.01E-01 | 38.74 |
| rs3129961 | 6 | 32486918 | G/A | (BTNL2, HLA-DRA) | Intergenic | SWE, GER, USA-AA | 1.43 (1.31–1.56) | 1.02E-15 | 1.43 (1.28–1.60) | 6.67E-10 | 1.95E-01 | 39.66 |
Definition of abbreviations: BP = base pairs; CA = coded allele; CHR = chromosome; CI = confidence interval; GER = Germany; LS = Löfgren’s syndrome; NCA = noncoded allele; OR = odds ratio; SNP = single-nucleotide polymorphism; SWE = Sweden; USA-AA = US African American; UTR = untranslated region.
The entire list of non-LS–associated SNPs is provided in Table E6.
The overall association estimates (OR) and 95% CI for non-LS–associated SNPs under fixed-effect and random-effect models, respectively.
τ2 P represents the P value significance for the amount of heterogeneity in the true log odds; the I2 statistic represents how much of the total variability in the effect size estimate can be attributed to heterogeneity among the true effects. Cohorts from SWE and GER are of European ancestry; the USA-AA cohort is of African ancestry.
Gene-centric Analysis
A gene-based approach identified five genomic loci (BTNL2, HLA-DRA, and HLA-DRB1 on chr6, and PLAC9 and ANXA11 on chr10) associated with non-LS and 179 genomic loci (from SCGN to HLA-DPB1 localizing to extended MHC on chr6) associated with LS as exceeding a Bonferroni-corrected threshold of P less than 7.3 × 10−6. Remarkably, significant gene associations for both LS and non-LS identified by this method were similar to those identified by associated SNPs from Immunochip analyses, and thus strengthened our findings. Table 5 lists the 15 most significant LS- and non-LS–associated genes obtained from VEGAS. Results for LS and non-LS at nominal P less than 0.05 are provided in Tables E7 and E8, respectively.
Table 5.
VEGAS Results for the 15 Most Significant Genes from Immunochip Analysis on LS and non-LS (48,740 SNPs and 6,840 Genes)
| CHR | Gene | nSNPs | Start Position | Stop Position | Test Statistic | P Value | Best SNP | SNP P Value |
|---|---|---|---|---|---|---|---|---|
| LS vs. HC | ||||||||
| 6 | TNXB | 50 | 32116910 | 32185129 | 1206.77 | 1.00E-06 | rs3130288 | 6.47E-34 |
| 6 | CREBL1 | 51 | 32191022 | 32203995 | 1409.21 | 1.00E-06 | rs3130288 | 6.47E-34 |
| 6 | FKBPL | 49 | 32204461 | 32206045 | 1464.31 | 1.00E-06 | rs3130288 | 6.47E-34 |
| 6 | PRRT1 | 61 | 32224117 | 32227698 | 2290.62 | 1.00E-06 | rs3130288 | 6.47E-34 |
| 6 | PPT2 | 68 | 32229278 | 32239430 | 2779.84 | 1.00E-06 | rs3130288 | 6.47E-34 |
| 6 | EGFL8 | 69 | 32240382 | 32244040 | 2728.04 | 1.00E-06 | rs3130288 | 6.47E-34 |
| 6 | AGPAT1 | 84 | 32243966 | 32253820 | 3085.42 | 1.00E-06 | rs3130288 | 6.47E-34 |
| 6 | C6orf10 | 149 | 32368452 | 32447634 | 5678.59 | 1.00E-06 | rs3129927 | 1.82E-33 |
| 6 | BTNL2 | 109 | 32470490 | 32482878 | 3946.51 | 1.00E-06 | rs3129927 | 1.82E-33 |
| 6 | HLA-DRB1 | 2 | 32654524 | 32665540 | 144.87 | 1.00E-06 | rs2187668 | 2.79E-33 |
| 6 | HLA-DQA1 | 2 | 32713160 | 32719407 | 288.31 | 1.00E-06 | rs2187668 | 2.79E-33 |
| 6 | HLA-DQB1 | 2 | 32735634 | 32742444 | 288.31 | 1.00E-06 | rs2187668 | 2.79E-33 |
| 6 | HLA-DRA | 61 | 32515624 | 32520802 | 2278.93 | 1.00E-06 | rs3135394 | 1.41E-32 |
| 6 | C2 | 50 | 32003472 | 32021427 | 1027.18 | 1.00E-06 | rs389884 | 3.84E-32 |
| 6 | CFB | 45 | 32021699 | 32027840 | 831.90 | 1.00E-06 | rs389884 | 3.84E-32 |
| Non-LS vs. HC | ||||||||
| 6 | HLA-DRA | 61 | 32515624 | 32520802 | 671.61 | 1.00E-06 | rs3129882 | 5.33E-10 |
| 6 | BTNL2 | 109 | 32470490 | 32482878 | 935.64 | 1.00E-06 | rs3129882 | 5.33E-10 |
| 6 | HLA-DRB1 | 2 | 32654524 | 32665540 | 36.76 | 1.00E-06 | rs3830135 | 2.74E-08 |
| 10 | ANXA11 | 6 | 81904859 | 81955308 | 82.55 | 1.00E-06 | rs1049550 | 1.07E-07 |
| 10 | PLAC9 | 3 | 81882237 | 81894764 | 60.30 | 1.00E-06 | rs1049550 | 1.07E-07 |
| 6 | HLA-DPB1 | 58 | 33151737 | 33162954 | 363.18 | 9.40E-05 | rs9277357 | 1.61E-05 |
| 18 | PTPN2 | 54 | 12775479 | 12874334 | 317.12 | 9.70E-05 | rs2852151 | 1.55E-04 |
| 6 | HLA-DPA1 | 46 | 33140771 | 33149356 | 326.14 | 1.28E-04 | rs9277357 | 1.61E-05 |
| 12 | PAH | 2 | 1.02E+08 | 1.02E+08 | 18.94 | 1.85E-04 | rs2242381 | 1.10E-04 |
| 6 | LST1 | 61 | 31661949 | 31664665 | 249.78 | 2.11E-04 | rs2239704 | 1.53E-05 |
| 6 | TNF | 60 | 31651328 | 31654091 | 297.74 | 2.92E-04 | rs2239704 | 1.53E-05 |
| 6 | LTB | 61 | 31656314 | 31658181 | 269.43 | 3.19E-04 | rs2239704 | 1.53E-05 |
| 6 | NCR3 | 60 | 31664650 | 31668741 | 225.25 | 3.71E-04 | rs2239704 | 1.53E-05 |
| 6 | PSMB9 | 87 | 32929915 | 32935606 | 357.37 | 4.05E-04 | rs3819717 | 2.43E-06 |
| 6 | PSMB8 | 87 | 32916471 | 32920690 | 357.37 | 4.25E-04 | rs3819717 | 2.43E-06 |
Definition of abbreviations: CHR = chromosome; HC = healthy control subjects; LS = Löfgren’s syndrome; nSNPs = number of SNPs included within a gene; SNP = single-nucleotide polymorphism; VEGAS = Versatile Gene-based Association Study.
The entire lists of LS and non-LS genic SNPs at nominal P < 0.05 are provided in Tables E7 and E8, respectively.
Integrative Analyses on Significant Findings
Bioinformatic analysis for refinement of associated variants
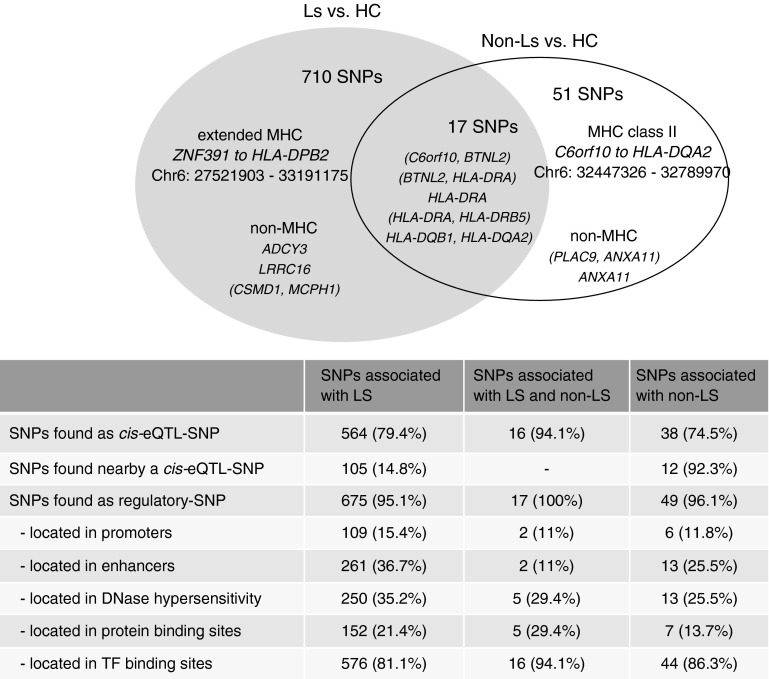
Results from studying the potential influence of confirmed variants on gene expression levels in various tissues and cell types revealed 564 of 710 LS-associated SNPs, 51 of 68 non-LS–associated SNPs, and 16 of 17 mutually associated SNPs as cis-eQTL SNPs (Figure 5).
Figure 5.
Distribution of cis-expression quantitative trait loci (eQTL) and regulatory single-nucleotide polymorphisms (SNPs) among associated confirmed variants of Löfgren’s syndrome (LS), non-LS, and both. Percentages are derived from the total number in each group, respectively. Genes enclosed in parentheses denote intergenic regions. HC = healthy control subjects; MHC = major histocompatibility complex; TF = transcription factor.
Advanced query on unclassified SNPs using extended LD information uncovered 105 LS-associated SNPs and 12 non-LS–associated SNPs positioned near a cis-eQTL SNP located on the same haplotype and in high LD (R2 > 0.8). Moreover, 41 LS-associated SNPs and three non-LS–associated SNPs were found with no cis-eQTL mapping.
Results from assessing confirmed variants in elements of transcription regulation using datasets from ENCODE and the National Institutes of Health Roadmap Epigenomics Mapping Consortium identified 675 LS-associated SNPs, 49 non-LS–associated SNPs, and all 17 mutually SNPs as regulatory SNPs. A distribution diagram of confirmed variants by transcription-element classification is illustrated in Figure 5.
Enrichment and pathway analyses of associated variants
Results from enrichment analysis based on 727 LS-associated confirmed variants at false discovery rate less than 0.05 identified pathway maps related to immune response of T-cell regulation and processes, and cytoskeleton remodeling. Pooling together 727 LS-associated SNPs and differentially expressed genes (from each GSE dataset tested) yielded similar pathway maps (see Table E9). In non-LS, assessment based on the 68 confirmed variants yielded no pathway maps. However, when combined with differentially expressed genes, identification of pathway maps related to immune response, differentiation, and regulation of T cells (see Table E10) was detected.
Noteworthy, many of the pathway maps that were identified in LS and non-LS, respectively, were overlapping by sharing genes with other pathways, and in some cases were subsets of each other. For example, in LS, the pathway maps labeled as a, b, c, d, f, g, i, and k (see Table E9) shared three common components: MHC class II, nuclear factor-κB, and CD86. Likewise, in non-LS, pathway maps labeled 1, 2, 4, 6, 8, 11, 13, and 14 (see Table E10) shared one common component: PCL-gamma 1. Further analysis on enrichment results from both phenotypes pinpointed the differences and shared pathway maps among these. Explicitly, pathway maps b-c-e-g-h (see Table E9) were unique to LS; pathway maps 1-2-3-4-6-7-8-9-11-12-13-14 (see Table E10) were unique to non-LS; and pathway maps a-10, c-17, d-5, i-16, k-15 were common to both. In the latter, the nuclear factor-κB was unanimously present across the shared pathway maps.
Discussion
In this study, we report for the first time results of Immunochip analysis of LS and non-LS segregated as two different sarcoidosis entities in well-phenotyped cohorts with replicated and meta-analysis results. In this work, we aimed to investigate the genetic contributions to LS and non-LS by using the custom Illumina Infinium array that leveraged the remarkable genetic overlap of 186 loci associated with autoimmunity. Results from various approaches in LS (using populations of European ancestry) and non-LS (using populations of European and African ancestry) show that genetic susceptibility of these phenotypes is different and that only a small number of loci are shared. Thus, we were able to capture genetic differences between LS and non-LS.
LS-associated variants expand through the extended MHC gene map (31), ranging from ZNF184 to HLA-DPB2. Outside the extended MHC region, variants in ADCY3, LRR16A, and between CSMD1 and MCPH1 shall be further studied to explain their association, which may be linked to eQTL, as elucidated by cis-eQTL SNPs in gene expression of immunity cells (18, 21, 26, 32). The susceptibility of non-LS localized to a smaller region of the MHC class II, ranging from C6orf10 to HLA-DQA2. Noteworthy, outside this region, confirmed variants highlighted exclusive associations with PLAC9 and ANXA11 (a previously discovered sarcoidosis-associated locus) on chromosome 10. An observed overlap between LS and non-LS defined by 17 variants that localized to MHC class II region is worth further exploration. Our efforts to gain insights on the role of HLA-DRB1 alleles in non-LS and LS phenotypes reinforced these findings (results and further discussion are available in the Note in the online supplement). Likewise, results from a genome-wide gene-centric approach underlined the significance of LS and non-LS identified gene regions, which may well establish the groundwork for investigating these phenotypes independently.
Individual assessment of each gene variant based on bioinformatic exploration provides an extensive knowledge for supporting variant association by highlighting potential regulatory and epigenetic mechanisms ensuing within the associated regions.
As reported from many genome-wide association studies (33), common genetic variants can only explain a relatively small portion of the phenotype variability (explained variance in LS, 4.1–4.7%; non-LS, 1.28–2.13%) and heritability thus stressing the plausibility for involvement of other genetic and regulatory elements to account for the portion of phenotypic variance. In non-LS, the results showed a presence of non-HLA genes, thus raising the possibility for gene x factor (a factor is defined as a gene, regulatory/epigenetic element, or protein) interactions across chromosomes, resembling a polygenic phenotype. Thus if so, the phenotype variability may be more likely explained by genes than rather by phenotype heterogeneity within non-LS (i.e., ocular, skin, heart, neurologic sarcoidosis, and many more), as we have demonstrated. Whereas in LS, the results show a higher presence of HLA-genes, suggesting that the phenotype variability may be explained by HLA-genes and gene x factor interactions located within one chromosome.
The presence of eQTL SNPs, regulatory elements, and epigenetic factors in the genomic regions associated with LS and non-LS, prompt new research questions that urge to be undertaken. eQTL genes have been proposed to act as master cis- and trans-regulators (34), whereas epigenetic elements alter chromatin structure and influence gene expression imposing specific heritable patterns on progeny cells (35–39). Based on this knowledge, it is not surprising to observe a predominant presence of cis-eQTL SNPs among the genetics of LS and non-LS. Analogously, the presence of epigenetic factors in the associated regions is expected given the numerous essential interactions between HLA and non-HLA genes. Hence, the observed implications and differences in eQTLs and epigenetics events among LS and non-LS support our hypothesis that mechanisms underlying the genetic structure and pathogenesis of these phenotypes are distinct. The limitation of this bioinformatic analysis is the lack of expression and epigenetic data available from our datasets, which could facilitate an advanced understanding of biologic implications for the genomic-associated regions.
With the present collective information comprising associated variants and differently expressed genes in sarcoidosis, we are able to present a better understanding of the functional mechanisms that may shed light on the pathogenesis of LS and non-LS, and on the overlap between these phenotypes. The molecular similarities between both phenotypes may be elucidated by potential sharing mechanisms that were captured on pathway maps that identified several overlapping pathways underlining immune responses. For example, pathways maps highlighting immune response to naive CD4+ T, Th1, and Th2 cells shall be further studied to understand their correlation with these phenotypes. Particularly, in LS, pathway maps involving Th9, Th17, and Th22 cells and dendritic cells shall be favorably considered for investigation. Given that, significant involvement of Th17 cells determined by higher levels of IL-17 in bronchoalveolar lavage fluid of patients with LS has been reported (40). Naturally, to facilitate such investigations additional omics studies, such as transcriptomics and epigenomics, are necessary before functional studies take place.
In summary, we present a comprehensive genetic study of LS and non-LS phenotypes of sarcoidosis in populations of European and African ancestry, thus focusing mainly on the genetic structure of each phenotype. For the first time, the genetic architecture of LS and non-LS is analyzed and presented, independently, identifying 175 loci associated with LS and 11 loci associated with non-LS by replication in independent cohorts. Moreover, out of the total number of loci identified, five are common to both phenotypes. We provide evidence to sustain our claim, that LS and non-LS are two different phenotypes with a shared genetic overlap and a reasonable number of specific factors. Through various analyses, we demonstrated that LS and non-LS have different genetic susceptibility and thus different genomic distributions that comprise distinctive gene regulatory elements associated with various relevant pathway maps of immune responses.
Acknowledgments
Acknowledgment
The authors thank the EIRA group for information regarding control subjects in the Swedish cohort. They thank Dr. F. Mrazek (University Hospital Olomouc) for assistance in providing samples of control subjects in the Czech Republic cohort. They thank Professor Wolfgang Lieb (PopGen), Professor Carsten Büning, and Professor Stephan Brand for providing German control samples.
Footnotes
The Swedish cohort was funded by Swedish Heart-Lung Foundation, by the Swedish Medical Research Council, and through the regional agreement on medical training and clinical research between Stockholm County Council and the Karolinska Institutet. The Czech Republic cohort was funded by grant IGA_LF_2015_020 (M.P.). The German study was supported by the Federal Ministry for Education and Research in Germany through the National Genome Research Network, within the framework of the e:Med (sysINFLAME) research and funding concept (01ZX1306A), the Genome Research Network “MooDS,” and the PopGen 2.0 network (01EY1103); by the German Research Foundation through grants FI 1935/1-1, MU 692/8-1, and BR 1912/6-1; through the Clusters of Excellence “Inflammation at Interfaces” and “ImmunoSensation”; by the Heinz Nixdorf Foundation; and by the Else-Kröner-Fresenius-Stiftung (Else Kröner-Exzellenzstipendium 2010_EKES.32). The African American study was supported by National Institutes of Health grant P30GM110766.
Author Contributions: Substantial contribution to design of work and clinical/genetic data for replication and meta-analysis: K.S., A. Franke, M.M.N., J.M.-Q., S.S., A. Fischer, C.H.M.v.M., B.K., J.C.G., Z.N., V.K., M.P., I.A., B.A.R., M.C.I., and C.M. Drafting the work and/or revising it critically for intellectual content: N.V.R., K.S., M.R., A.E., L.P., J.G., A. Fischer, M.P., and C.H.M.v.M. Accountability for all aspects of the work in ensuring that questions related to the accuracy/integrity of any part of the work are appropriately investigated and resolved: N.V.R., L.P., and J.G. All authors have given final approval of the version submitted for publication.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201507-1372OC on December 10, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007;357:2153–2165. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- 2.Bauer HJ, Löfgren S. International study of pulmonary sarcoidosis in mass chest radiography. Acta Med Scand Suppl. 1964;425:103–105. doi: 10.1111/j.0954-6820.1964.tb05712.x. [DOI] [PubMed] [Google Scholar]
- 3.Grunewald J, Eklund A. Löfgren’s syndrome: human leukocyte antigen strongly influences the disease course. Am J Respir Crit Care Med. 2009;179:307–312. doi: 10.1164/rccm.200807-1082OC. [DOI] [PubMed] [Google Scholar]
- 4.De Vries J, Van Heck GL, Drent M. Gender differences in sarcoidosis: symptoms, quality of life, and medical consumption. Women Health. 1999;30:99–114. doi: 10.1300/j013v30n02_07. [DOI] [PubMed] [Google Scholar]
- 5.Drent M, Marcellis R, Lenssen A, De Vries J. Association between physical functions and quality of life in sarcoidosis. Sarcoidosis. 2014;31:117–128. [PubMed] [Google Scholar]
- 6.Belfer MH, Stevens RW. Sarcoidosis: a primary care review. Am Fam Physician. 1998;58:2041–2050, 55–6. [PubMed] [Google Scholar]
- 7.Bauer HJ, Löfgren S. International study of pulmonary sarcoidosis in mass chest radiography. Acta Med Scand Suppl. 1964;425:103–105. doi: 10.1111/j.0954-6820.1964.tb05712.x. [DOI] [PubMed] [Google Scholar]
- 8.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, Bakker SF, Bardella MT, Bhaw-Rosun L, Castillejo G, et al. Spanish Consortium on the Genetics of Coeliac Disease (CEGEC); PreventCD Study Group; Wellcome Trust Case Control Consortium (WTCCC) Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, Zhernakova A, Stahl E, Viatte S, McAllister K, et al. Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate; Wellcome Trust Case Control Consortium. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, et al. Collaborative Association Study of Psoriasis (CASP); Genetic Analysis of Psoriasis Consortium; Psoriasis Association Genetics Extension; Wellcome Trust Case Control Consortium 2. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, Spencer C, Booth D, Goris A, et al. International Multiple Sclerosis Genetics Consortium (IMSGC); Wellcome Trust Case Control Consortium 2 (WTCCC2); International IBD Genetics Consortium (IIBDGC) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellinghaus D, Baurecht H, Esparza-Gordillo J, Rodríguez E, Matanovic A, Marenholz I, Hübner N, Schaarschmidt H, Novak N, Michel S, et al. High-density genotyping study identifies four new susceptibility loci for atopic dermatitis. Nat Genet. 2013;45:808–812. doi: 10.1038/ng.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viechtbauer W.Conducting meta-analyses in R with the Metafor package J Stat Softw 2010363:1–48 [Google Scholar]
- 15.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, et al. AMFS Investigators. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Gutierrez Arcelus M, Sekowska M, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 19.Gaffney DJ, Veyrieras JB, Degner JF, Pique-Regi R, Pai AA, Crawford GE, Stephens M, Gilad Y, Pritchard JK. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 2012;13:R7. doi: 10.1186/gb-2012-13-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nica AC, Parts L, Glass D, Nisbet J, Barrett A, Sekowska M, Travers M, Potter S, Grundberg E, Small K, et al. MuTHER Consortium. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, et al. Genetics and beyond: the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle APHE, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma SM, Choi D, Planck SR, Harrington CA, Austin CR, Lewis JA, Diebel TN, Martin TM, Smith JR, Rosenbaum JT. Insights in to the pathogenesis of axial spondyloarthropathy based on gene expression profiles. Arthritis Res Ther. 2009;11:R168. doi: 10.1186/ar2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koth LL, Solberg OD, Peng JC, Bhakta NR, Nguyen CP, Woodruff PG. Sarcoidosis blood transcriptome reflects lung inflammation and overlaps with tuberculosis. Am J Respir Crit Care Med. 2011;184:1153–1163. doi: 10.1164/rccm.201106-1143OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Judson MA, Marchell RM, Mascelli M, Piantone A, Barnathan ES, Petty KJ, Chen D, Fan H, Grund H, Ma K, et al. Molecular profiling and gene expression analysis in cutaneous sarcoidosis: the role of interleukin-12, interleukin-23, and the T-helper 17 pathway. J Am Acad Dermatol. 2012;66:901–910. doi: 10.1016/j.jaad.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 31.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 32.Ferreira MA, Mangino M, Brumme CJ, Zhao ZZ, Medland SE, Wright MJ, Nyholt DR, Gordon S, Campbell M, McEvoy BP, et al. International HIV Controllers Study. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am J Hum Genet. 2010;86:88–92. doi: 10.1016/j.ajhg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, Thorsteindottir U, Shin SY, Richards HB, Soranzo N, et al. GIANT Consortium; MAGIC Investigators; DIAGRAM Consortium; MuTHER Consortium. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 37.Merkenschlager M, Wilson CB. RNAi and chromatin in T cell development and function. Curr Opin Immunol. 2008;20:131–138. doi: 10.1016/j.coi.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 40.Ostadkarampour M, Eklund A, Moller D, Glader P, Olgart Höglund C, Lindén A, Grunewald J, Wahlström J. Higher levels of interleukin IL-17 and antigen-specific IL-17 responses in pulmonary sarcoidosis patients with Löfgren’s syndrome. Clin Exp Immunol. 2014;178:342–352. doi: 10.1111/cei.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]