Abstract
Antifreeze proteins (AFPs) found in many organisms can noncolligatively lower the freezing point of water without altering the melting point. The difference between the depressed freezing point and the melting point, termed thermal hysteresis (TH), is usually a measure of the antifreeze activity of AFPs. Certain low molecular mass molecules and proteins can further enhance the antifreeze activity of AFPs. Interaction between an enhancer and arginine is known to play an important role in enhancing the antifreeze activity of an AFP from the beetle Dendroides canadensis (DAFP-1). Here, we examined the enhancement effects of several prevalent phosphate-containing coenzymes on the antifreeze activity of DAFP-1. β-Nicotinamide adenine dinucleotide (reduced) (NADH) is identified as the most efficient enhancer of DAFP-1, which increases the antifreeze activity of DAFP-1 by around 10 times. Examination of the enhancement abilities of a series of NADH analogs and various molecular fragments of NADH reveals that the modifications of nicotinamide generate a series of highly efficient enhancers, though none as effective as NADH itself, and the whole molecular structure of NADH is necessary for its highly efficient enhancement effect. We also demonstrated a 1:1 binding between DAFP-1 and NADH. The binding was characterized by high-performance liquid chromatography (HPLC) using the gel filtration method of Hummel and Dreyer. The data analysis suggests binding between DAFP-1 and NADH with a dissociation constant in the micromolar range. Interactions between DAFP-1 and NADH are discussed along with molecular mechanisms of enhancer action.
Keywords: arginine, phosphate, nicotinamide adenine dinucleotide, antifreeze protein, antifreeze activity enhancement
INTRODUCTION
Antifreeze proteins (AFPs) have been found in a variety of organisms, such as fish, plants, and insects, which allow them to survive at sub-freezing temperatures (DeVries, 1971; DeVries and Wohlschlag, 1969; Duman, 1977; Tomchaney et al., 1982; Urrutia et al., 1992). Although AFPs from different species have a great diversity of structures, they are all characterized by their ability to depress the freezing point of aqueous solutions in a noncolligative manner. The widely accepted adsorption–inhibition mechanism for AFP function proposes that the specific adsorption of AFPs onto ice crystal surfaces results in the inhibition of ice growth because of the Kelvin effect (Jia and Davies, 2002; Knight and Devries, 1994; Raymond and DeVries, 1977). The resulting difference between the nonequilibrium freezing and melting points is termed thermal hysteresis (TH), which can be used as a measure of the antifreeze activity (or TH activity) of AFPs. Insect AFPs are usually much more active than fish and plant AFPs (DeVries and Cheng, 1992; Duman and Olsen, 1993; Duman and Serianni, 2002).
The purified AFPs from Dendroides canadensis hemolymph (DAFPs), however, produce much less TH activity than the hemolymph itself. The high antifreeze activity in the hemolymph may in fact result from the presence of enhancers (Wang and Duman, 2005; Wang and Duman, 2006). Known enhancers of DAFPs include some proteins (e.g., antibodies and other DAFP isoforms) and low-mass molecules (e.g., polycarboxylates and polyols) (Amornwittawat et al., 2009; Amornwittawat et al., 2008; Li et al., 1998a; Wang and Duman, 2005; Wang and Duman, 2006; Wu et al., 1991). The detailed molecular mechanism of AFP enhancer action is still not well understood. It was hypothesized that the protein enhancers may increase the antifreeze activities of DAFPs by interacting with the DAFPs that bind to the surface of the ice crystal, increasing the surface of the potential seed ice crystal blocked by the DAFPs (Wang et al., 2009b; Wu et al., 1991), and DAFPs need to bind to both the protein enhancers and the ice simultaneously (Duman and Serianni, 2002). The low molecular mass enhancers do not have antifreeze activity themselves. Different mechanisms are probably involved in the antifreeze activity enhancement by the low molecular mass enhancers (Duman and Serianni, 2002). Some low molecular mass enhancers may optimize the higher-order structure of DAFPs for binding to ice by providing subtle stabilization and/or by promoting interactions between DAFPs and other protein enhancers (Li et al., 1998a). For some common monovalent Hofmeister salts (their TH enhancement effects are very small), the salting-out effect may be the only mechanism for their TH enhancement (Wang et al., 2009a). Recently, the importance of arginine has been demonstrated for the enhancement of antifreeze activity in DAFP-1, and a 1:1 binding between DAFP-1 and citrate (a relatively strong, carboxylate-containing enhancer for DAFP-1) has been demonstrated (Wang et al., 2009b).
In proteins, arginine often serves as the positively charged center for the recognition and binding of anionic substrates, such as carboxylate and phosphate anions (Hannon and Anslyn, 1993; Schmuck, 2006). The phosphate-containing coenzymes, such as reduced nicotinamide adenine dinucleotide (NADH) and its oxidized form (NAD), play a central role in the metabolic activities of living cells. The role of arginines in the recognition of phosphate-containing coenzymes has been extensively studied (Fujioka and Takata, 1981; Jairajpuri et al., 1998; Lange et al., 1974; Riordan et al., 1977; Sabri et al., 2009; Sem and Kasper, 1993). Can phosphate-containing coenzymes bind to DAFP-1 and act as efficient enhancers for the antifreeze activity of DAFP-1? In the present study, we addressed the above question and provided insights into the molecular basis of AFP enhancers. We investigated the enhancement effects of a series of phosphate-containing coenzymes (e.g., NAD(H), flavin adenine dinucleotide (FAD), and coenzyme A (CoA)) on the antifreeze activity of DAFP-1 and demonstrated that NADH is the most efficient enhancer for the antifreeze activity of DAFP-1 identified so far. Furthermore, the binding between NADH and DAFP-1 has been characterized. In order to further investigate the governing interaction between NADH and DAFP-1, we characterized the effects of molecular fragments of NADH on the antifreeze activity of DAFP-1. Considering the nonphysiological high concentrations of the coenzyme enhancers used in this study as well as the compartmentation of the coenzymes and DAFP-1, the probabilities that these coenzymes act as physiological enhancers are much less compared to earlier identified polyhydroxy enhancers (Amornwittawat et al., 2009). Nevertheless, the discovery of highly efficient coenzyme enhancers for DAFP-1 is significant with respect to the many potential biotechnological applications of AFPs, such as in cold preservation and frozen foods (Barrett, 2001).
MATERIALS AND METHODS
Materials
The chemicals were ACS grade or better, and the high-performance liquid chromatography (HPLC)–grade solvents and chemicals were purchased from Sigma-Aldrich (St. Louis, MO). NADH, NAD, thionicotinamide adenine dinucleotide (TNAD), 3-pyridine aldehyde adenine dinucleotide (PAAD), FAD, CoA, ribose, D-ribose-5-phosphate (R5P), pyrophosphate, adenosine, (—)-adenosine 5′-monophosphate (AMP), adenosine 5′-diphosphate (ADP), and adenosine 5′-diphosphoribose (ADPR) (Sigma) were used without additional purification. Milli-Q water produced from a Synergy water system (Millipore) with a minimum resistivity of 18MΩ·cm was used to prepare all the solutions.
DAFP-1 sample preparation
DAFP-1 was prepared as previously described (Amornwittawat et al., 2009). Briefly, DAFP-1 was expressed as a fusion protein containing a His tag in Escherichia coli Origami B cells. The cells were harvested by centrifugation at 4°C and lysed by French press (Thermo Fisher). The crude protein was purified using Ni–NTA agarose (Qiagen). The tags were cleaved using enterokinase (New England Biolabs) and removed using Ni–NTA agarose. The cleaved protein was finally purified using ÄKTA Purifier 10 (GE Healthcare) with a Sephacryl S-100 gel filtration column (GE Healthcare). The identity of purified DAFP-1 was confirmed by SDS-PAGE, HPLC, and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. The protein concentration was determined by UV-Vis spectroscopy (Varian) using the absorption at 280nm (Gill and von Hippel, 1989).
Antifreeze activity measurements
The antifreeze (or TH) activities of the samples were assessed using differential scanning calorimetry (DSC) following the known procedure (Amornwittawat et al., 2008). The measurements were performed on a DSC 823e (Mettler Toledo, OH) with an HSS7 high sensitivity sensor and a Julabo FT900 intracooler chiller (Julabo Company, Allentown, PA). The enhancement effects of the following compounds on the TH activity of DAFP-1 were assessed: NADH and the NADH analogs, including NAD, TNAD, PAAD, and two other coenzymes, FAD and CoA; the molecule fragments of NADH, including ribose, R5P, pyrophosphate, adenosine, AMP, ADP, and ADPR; and previously identified enhancers, triethylenetetramine-N,N,N′,N″,N″′,N″′-hexaacetate (TTHA) and trehalose, for comparison. The concentrations of these potential enhancers were varied from 0.00M to 0.80M, depending on their solubilities. The concentration of DAFP-1 was 453μM in all the samples, and the TH activity of the protein sample alone was 0.69±0.01°C. The TH activity of DAFP-1 is unchanged from pH 2 to pH 12. To eliminate possible enhancement effects of buffer compounds, the pHs of the samples were carefully adjusted using 0.30M HCl and 0.10M NaOH when necessary. The pH of the final samples was 7.5. Each combination of DAFP-1 and enhancer was prepared twice, and the TH of a given sample was measured at least twice. The TH values were presented as means, and the standard deviations were within 2% of the mean values.
MALDI mass spectrometry
The experiments were performed on a PerSeptive Biosystems/Voyager-DE MALDI-TOF mass spectrometer at Stanford PAN facility. The mass spectrometer was calibrated using external mass standards before the sample analysis. A saturated solution of the matrix, sinapinic acid, was prepared in 1:1 (v/v) water:acetonitrile with 0.1% trifluoroacetic acid (TFA). The sample solution was made by mixing 2.0μl of 150μM DAFP-1 and 2.0μl of 500μM NADH (sodium salt), pH 7.5. The sample was then incubated for 30min at 4°C. After desalting by Ziptip C18 resin (Millipore), the sample was dissolved in double-distilled H2O with 0.1% TFA, and 0.5μl of the sample solution was mixed with an equal volume of the saturated matrix solution. The prepared sample mixture was then dropped in a MALDI sample plate and air dried before analysis.
Size-exclusion HPLC analysis of NADH binding to DAFP-1
The binding of NADH to DAFP-1 was characterized using HPLC by an improved gel filtration method of Hummel and Dreyer (Hummel and Dreyer, 1962; Parsons, 1980). The Dionex HPLC system (Sunnyvale, CA) consisting of an Ultimate 3000 pump and an Ultimate 3000 Photodiode Array Detector was used in this study. A BioSuite 125, 4-μm UHR SEC HPLC column, 4.6 × 300mm (Waters, Milford, MA) was equilibrated with buffer containing 0.10M Na2SO4, 0.05% NaN3, 0.10M NaPi, and NADH at a desired concentration, pH 7.00. The DAFP-1 samples were prepared by adding a known amount of DAFP-1 in the above buffer. After equilibration for at least 30min, the DAFP-1 sample was injected into the column. The injection volume was 10μl in all experiments. Elution was performed at a flow rate of 0.5μl/min at ambient temperature. The eluates were monitored at 220nm. The experiments were performed at various concentrations of DAFP-1 (1.5–20μM) and NADH (0–270μM). The initial concentrations of NADH were determined using the molar extinction coefficient (ε340=6.22 × 103M−1cm−1) (Dawson et al., 1985). The binding of NADH to DAFP-1 resulted in a trough at the retention time of NADH. The area of the trough depends directly on the amount of bound NADH that is depleted by binding to DAFP-1. The amount of bound NADH was determined by the internal calibration method (Parsons 1980; Sun and Hsiao, 1993). The amount of free NADH is obtained from the difference of total NADH and calculated bound NADH. The Klotz plot (or semilog plot) was used to analyze the binding data (Klotz, 1982). The dissociation constant (Kd) and stoichiometry for the binding of NADH to DAFP-1 were estimated by curve fitting using nonlinear regression analysis.
RESULTS
Effects of NADH, its analogs, and other coenzymes on DAFP-1 activity
The TH activity of DAFP-1 can be enhanced by around 10 times in the presence of NADH (Figure 1). The enhancement ability of NADH is much higher than any of the known enhancers of DAFP-1 activity (Amornwittawat et al., 2009; Amornwittawat et al., 2008; Li et al., 1998a; Wang and Duman, 2005; Wang and Duman, 2006; Wang et al., 2009a; Wu et al., 1991). The previously identified best carboxylate containing enhancer, TTHA, and hydroxyl containing enhancer, trehalose, are shown in Figure 1 for comparison. To examine the effects of changes in the structure of NADH on the TH activity of DAFP-1, a series of NADH analogs and other coenzymes were assessed for their enhancement abilities (Figure 2). The molar enhancement effectiveness order, NADH>NAD>TNAD>PAAD, was observed for the NADH analogs (Figure 3). The enhancement ability of NAD is very similar to, although a little lower than, that of its reduced form, NADH. TNAD, in which the oxygen of the carboxamide is substituted by sulfur, and PAAD, in which the carboxamide is substituted by aldehyde, also show a very efficient enhancement ability of the TH activity of DAFP-1. The three analogs, NAD, TNAD, and PAAD, are more efficient than any previously identified enhancers for DAFP-1. Both of the other two coenzymes, FAD and CoA, can enhance the TH activity of DAFP-1; however, their enhancement efficiencies are less than those of NADH and NAD analogs (Figure 3). The data acquired for FAD is limited by its solubility in the test system.
Figure 1.
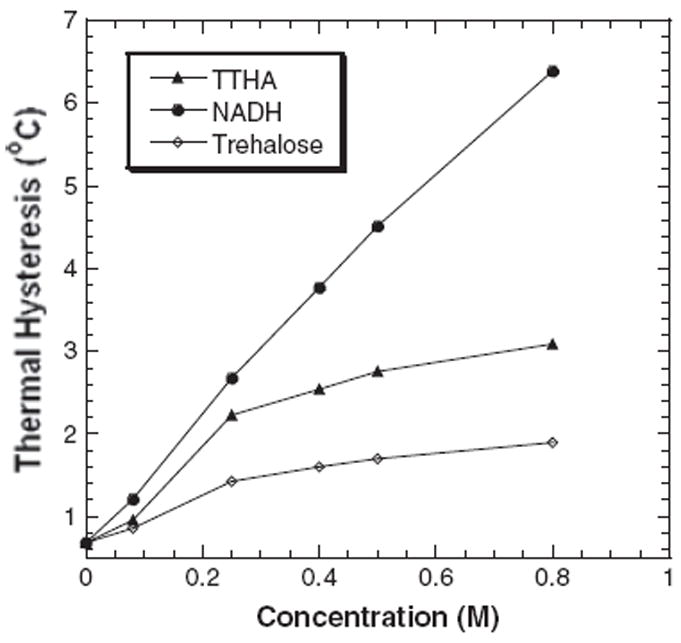
The antifreeze activities of DAFP-1 (453μM) in the presence of NADH compared with those in the presence of trehalose (the previously identified most efficient naturally occurring enhancer) and TTHA (the previously identified most efficient enhancer). The TH values were presented as means, and the standard deviations were with 2% of the mean values. The concentrations of the enhancers were in the range of 0–0.80M. The pH of the samples was 7.5.
Figure 2.
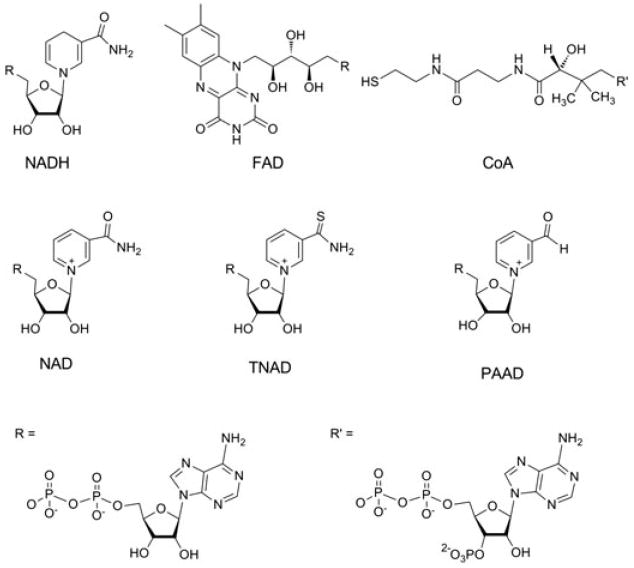
Structures of the central coenzymes in metabolism, NADH, FAD, and CoA, and various NAD(H) analogs (NAD, TNAD, and PAAD) examined as potential enhancers of DAFP-1 in this study.
Figure 3.
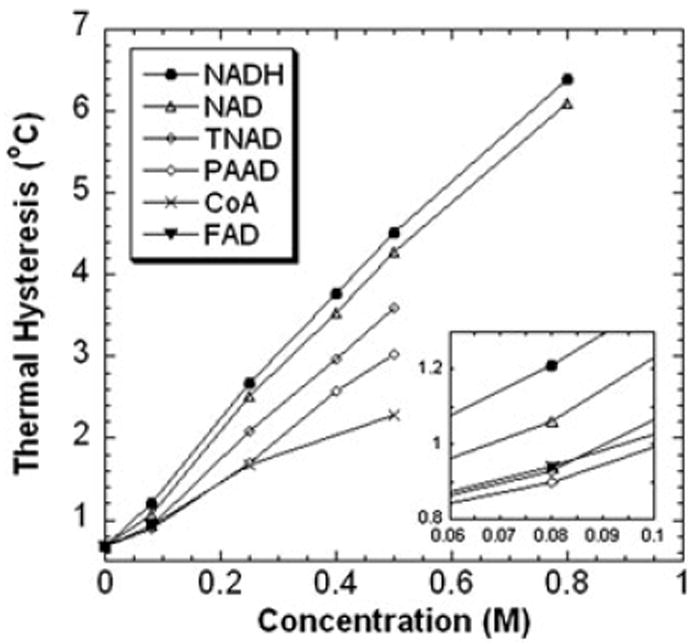
The antifreeze activities of DAFP-1 (453μM) in the presence of NADH, FAD, CoA, and the NAD(H) analogs (NAD, TNAD, and PAAD). The TH values were presented as means, and the standard deviations were with 2% of the mean values. The concentrations of the enhancers were in the range of 0–0.80M. The pH of the samples was 7.5. The inset shows the expansion of the low concentration region of the enhancers (0.06–0.10M).
Effects of molecular fragments of NADH on DAFP-1 activity
To examine the effects of structural moieties of NADH on the TH activity of DAFP-1, the enhancement effects of various molecular fragments of NADH were assessed (Figures 4 and 5). All the molecular fragments of NADH are much less efficient enhancers for DAFP-1 compared to the entire molecule of NADH (Figure 5). The following molar enhancement effectiveness order was observed for the selected fragments: ADPR>AMP>ADP>adenosine> R5P>pyrophosphate>ribose. ADPR, which only lacks the nicotinamide, is the fragment containing the most structural moieties of NADH (Figure 4). ADPR has the most efficient enhancement ability among these fragments and is very close to that of TTHA (Figures 1 and 5). The TH enhancement abilities of all the other fragments are lower than that of TTHA. ADP lacks a ribose moiety compared to ADPR, and the TH enhancement ability of ADP is less than that of ADPR (Figure 5). Adenosine lacks a phosphate group compared to AMP. At 0.080M, the enhancement ability of adenosine is lower than that of ADP. Data at higher concentrations, however, are not available for adenosine due to its low solubility. Ribose is the smallest structural component of NADH among the fragments; it also has the least enhancement of the TH activity of DAFP-1. This trend was also observed for the other fragments. Generally, it seems that the fragment containing more structural moieties of NADH has better enhancement efficiency for the TH activity of DAFP-1. The notable exception to this trend is the transposition of AMP and ADP in this series. The sum of the TH enhancements of the fragments that collectively constitute a complete NADH moiety is not equal to that of NADH alone. This indicates that the entire molecular structure of NADH is important for its highly efficient enhancement of the TH activity of DAFP-1.
Figure 4.
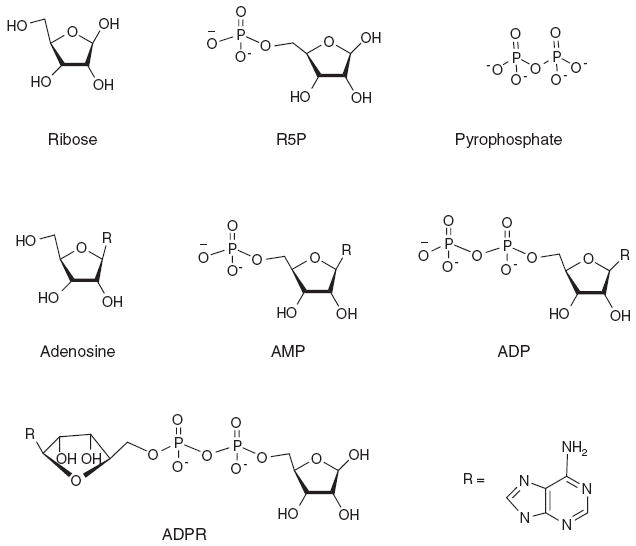
The structures of the molecular fragments of NADH examined as potential enhancers in this study. These molecular fragments include ribose, R5P, pyrophosphate, adenosine, AMP, ADP, and ADPR.
Figure 5.
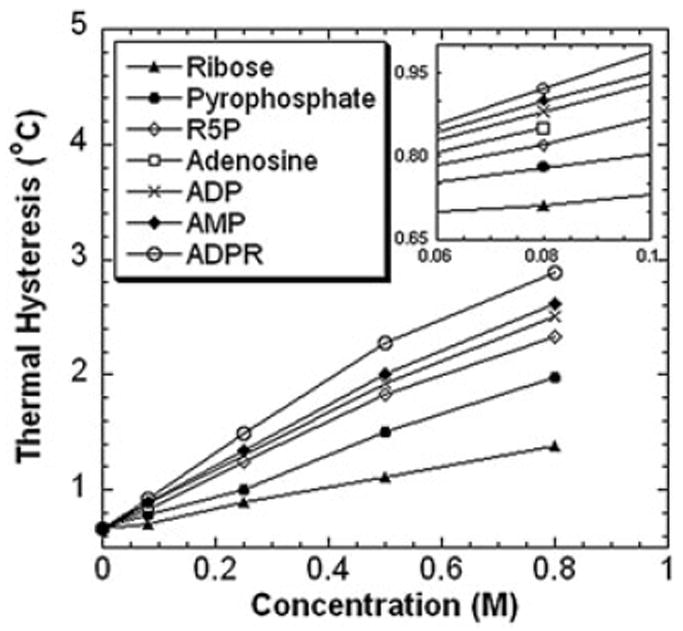
The antifreeze activities of DAFP-1 (453μM) in the presence of the molecular fragments of NADH (i.e., ribose, R5P, pyrophosphate, adenosine, AMP, ADP, and ADPR). The TH values were presented as means, and the standard deviations were with 2% of the mean values. The concentrations of the enhancers were in the range of 0–0.80M. The pH of the samples was 7.5. The inset shows the expansion of the low concentration region of the enhancers (0.06–0.10M).
Binding of NADH to DAFP-1
A 1:1 binding of NADH to DAFP-1 can be detected by MALDI-TOF mass spectrometry, indicating that the intact complex exists under the experimental conditions. The calculated molecular mass of DAFP-1 is 8968.8Da. In Figure 6, the Na+ adduct ion of DAFP-1 was observed as a major peak at m/z=8990.9. The peak observed at m/z=9656.9 corresponds to the Na+ adduct ion of the 1:1 complex of DAFP-1 and NADH.
Figure 6.
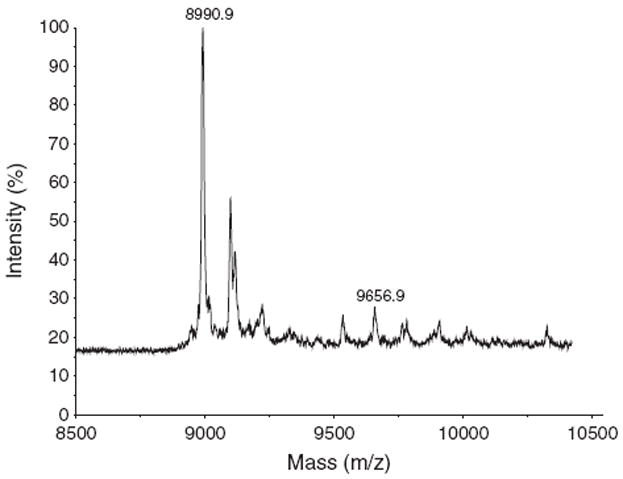
The MALDI-TOF mass spectrum of DAFP-1 (0.3nmol) incubated with NADH (1.0nmol). Formations of sodium adduct ions of DAFP-1 (m/z=8990.9) and the 1:1 complex of DAFP-1 and NADH (m/z=9656.9) are indicated.
The formation of 1:1 complex of DAFP-1 and NADH was further confirmed by HPLC analysis. With the gel filtration method of Hummel and Dreyer, the size exclusion column is equilibrated with a buffer containing the ligand at a known concentration. The protein sample prepared in the buffer (containing the ligand at the same concentration) is injected into the equilibrated column. If binding occurs, the concentrations of the ligand in the elution buffer and in the elution are usually different. As a result, a positive peak corresponding to the protein (the ligated protein and the free protein if there is any) appears at the retention time of the macromolecule, followed by a trough at the retention time of the ligand, indicating the decrease of the ligand concentration due to the binding. The size exclusion column would not be able to discriminate between the free protein and the ligated protein if the size difference between the protein and the protein–ligand complex is beyond the limit of the column. The trough area depends directly on the amount of the bound ligand that is depleted by binding to the protein. The bound ligand can be determined more precisely using the internal calibration method (Parsons, 1980; Sun and Hsiao, 1993).
Figure 7 shows a representative HPLC chromatogram of NADH binding to DAFP-1 using the Hummel and Dreyer method. DAFP-1 was eluted at 10.0min. The amount of NADH in the column depleted by binding to DAFP-1 was observed as a trough at 11.5min. The resulting semilog plot is shown in Figure 8. The curve fitting results suggest that the binding of NADH to DAFP-1 is characterized by a Kd of 0.28 ± 0.02μM and a stoichiometry of 1:1.
Figure 7.
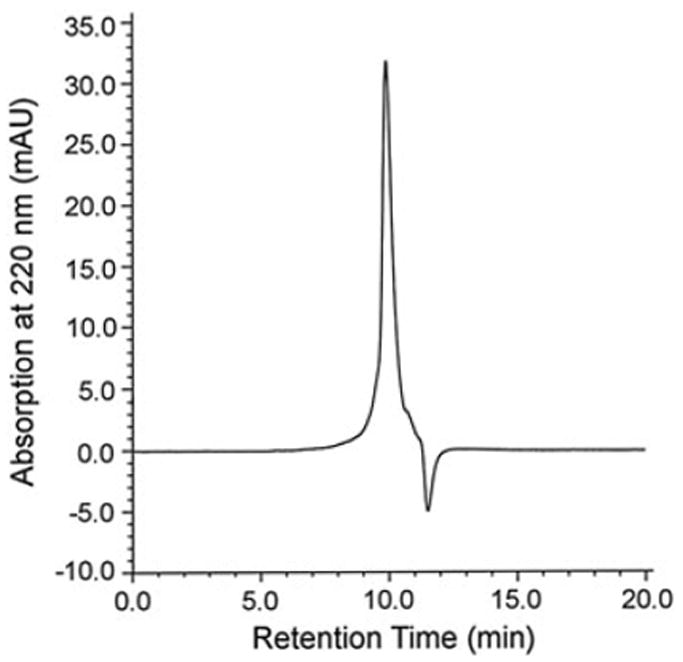
A typical HPLC chromatogram of the binding of NADH to DAFP-1 using the Hummel and Dreyer method. Flow rate was 0.35ml/min. The buffer contained 0.10M Na2SO4, 0.05% NaN3, 0.10M NaPi, and NADH at 0.50μM, pH 7.00. 1.51μM DAFP-1 in the above buffer was injected into the equilibrated column. The positive peak was observed at the retention time of DAFP-1, 10.0min. The trough was observed at 11.5min, the retention time of NADH.
Figure 8.
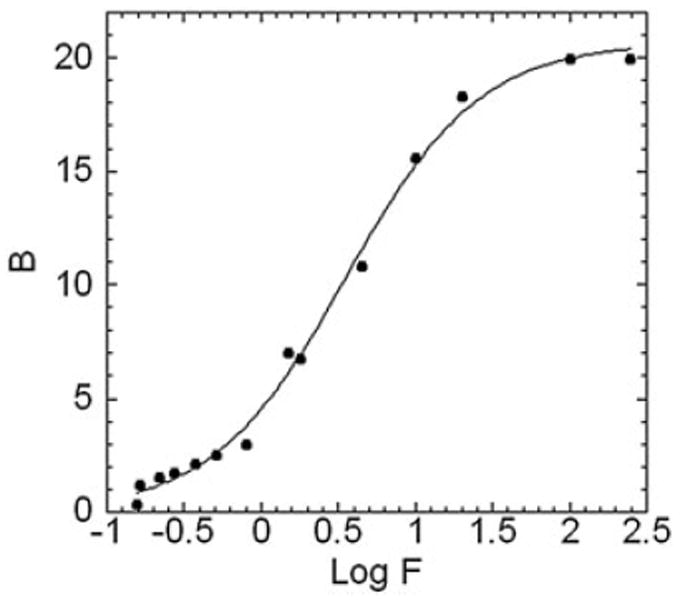
The HPLC data are displayed as a Klotz plot. B and F are the concentrations of bound and free NADH in micromolars. The solid curve represents the nonlinear regression fitting of the data points to a single-site binding model, B=Bmax × [Kd × F / (1+Kd × F)], where Bmax is the binding capacity of DAFP-1, which is equal to the ratio of the moles of bound NADH to the moles of total DAFP-1 in the column. The Kd was estimated to be 0.28 ± 0.02μM.
DISCUSSION
By correlating the enhancement abilities of the enhancers with their physicochemical properties, two series of efficient low molecular mass enhancers, polycarboxylates and polyols, were identified for the TH activity of DAFP-1 (Amornwittawat et al., 2009; Amornwittawat et al., 2008). Among these enhancers, TTHA, which contains six carboxylate groups, was reported as the most efficient enhancer of DAFP-1. Some polyhydroxy enhancers, such as trehalose, glycerol, and glucose, are present at high concentrations in insect, and trehalose is the most efficient polyhydroxy enhancer identified so far. Considering that almost all the known enhancers can interact with arginine, it was identified as the key residue involved in the enhancement of antifreeze activity of DAFP-1 (Wang et al., 2009b). In addition, the binding between DAFP-1 and citrate was demonstrated, and arginine was proposed to be involved in the binding of this enhancer (Wang et al., 2009b).
The discovery of the importance of arginine in DAFP-1 for its enhancement by low molecular mass enhancers provides a new clue to the identification of highly efficient enhancers of DAFP-1. NADH and other coenzymes were selected to be examined in this work based on the speculation that arginines are often involved in the binding of phosphate-containing coenzymes. We found that the TH activity of DAFP-1 can be enhanced approximately 10 times in the presence of NADH. The enhancement efficiency of NADH is much greater than that of trehalose, the previously identified naturally occurring enhancer, TTHA, the most efficient enhancer of DAFP-1, and the other coenzymes tested in this work. Furthermore, the binding between DAFP-1 and NADH has been demonstrated. Under our experimental conditions, the intact 1:1 complex of DAFP-1 and NADH has been detected by MALDI-TOF mass spectrometry. Usually, only tight binding noncovalent protein–ligand complex can be observed under the MALDI conditions (Glocker et al., 1996). The formation of a 1:1 complex of DAFP-1 and NADH was further confirmed by HPLC analysis, using the gel filtration method of Hummel and Dreyer (Hummel and Dreyer, 1962; Parsons, 1980). The Klotz treatment of the data analysis suggests a 1:1 binding between DAFP-1 and NADH, with a Kd of 0.28 ± 0.02μM, but we cannot rule out the possibility of nonspecific binding of two or multiple NADH molecules to one DAFP-1.
Examination of the enhancement abilities of the various NADH analogs and their structures indicated that the modifications of the nicotinamide ring generate a series of highly efficient enhancers whose enhancement abilities are stronger than that of any previously identified enhancers but weaker than that of NADH. NAD has the most similar structure, and its enhancement effect is comparable to that of NADH. Compared to NAD, the sulfur replaces oxygen in the carboxamide moiety of TNAD, and the carboxamide group is replaced by an aldehyde in PAAD. TNAD and PAAD are less efficient enhancers than NAD, and PAAD is the least effective enhancer among the analogs. The results suggest that the carboxamide moiety is essential for the greatest enhancement.
The investigation of the enhancement of the various molecular fragments of NADH indicated that all the molecular fragments of NADH can enhance the TH activity but are much less effective than that of NADH. Thus, the whole molecular structure of NADH is necessary for its highly efficient enhancement. Among the fragments, ADPR containing the most structural moieties of NADH (i.e., only lacks the nicotinamide) has the most efficient enhancement ability among the fragments; ribose, the smallest structural component of NADH, has the least enhancement effect. Generally, the fragment containing more structural moieties of NADH appears to have better enhancement efficiency for the TH activity. The relative enhancement efficiencies, AMP>ADP, however, cannot be explained by their molecular sizes. Compared to ADP, AMP lacks one phosphate group and is less negatively charged. The mechanism of the enhancer action has been suggested to involve the binding to an arginine in DAFP-1. Considering the free energy change of the ionic hydration, the binding of ADP to arginine is less favorable than the binding of AMP to arginine (Fraústo da Silva and Williams, 1976). The balance between ionic interaction and hydration in aqueous solution may be the explanation for our observation that the enhancement ability of ADP for the TH activity of DAFP-1 is lower than that of AMP.
We have hypothesized that extensive interactions, involving ionic binding, hydrogen bonding, and hydrophobic interaction, between an enhancer and DAFP-1 are required for the enhancer to be a highly efficient one (Wang et al., 2009a; Wang et al., 2009b). Ionic interactions are critical in the binding of the pyrophosphate moiety of NADH to the guanidinium group of arginine in DAFP-1. However, the ionic interaction between NADH and DAFP-1 alone cannot be very stable since the ion pair stability is greatly decreased by the hydration of the ions (i.e., the guanidinium in arginine and the phosphate in NADH) in aqueous solutions. To form a tight complex, the binding between DAFP-1 and NADH must involve various interactions, including ionic interaction, hydrogen bonding, and hydrophobic interaction, with the involvement of other important amino acids in DAFP-1. For example, hydrophobic interactions may occur in the binding of the adenine moiety and the nicotinamide moiety of NADH to some nonpolar residues of DAFP-1, and hydrogen bonding may be essential for orientating the two ribose moieties of NADH in the binding. The identification of the series of highly efficient enhancers and the results on the enhancement effects of the molecular fragments of NADH support our previous hypothesis.
An interesting and important question is how the interactions between DAFP-1 and NADH affect the TH activity of DAFP-1. In aqueous solutions, DAFP-1 and enhancer molecules are solvated by water molecules. Therefore, effects on the surrounding water molecules must be taken into account. The water molecules bound to the interacting surfaces of DAFP-1 and NADH are expected to be released upon formation of the complex (Figure 9). The released water into the bulk solution may result in a more disordered system (i.e., entropically favorable) counteracting the entropically unfavorable freezing process. Furthermore, guanidinium–phosphate interactions are generally enthalpically favorable. According to the adsorption–inhibition mechanism, by a simple estimation shown in Supporting Information, the energy released upon the interactions may contribute to the inhibition of the growth of the ice fraction. The Kd of the binding of NADH to DAFP-1 is in the micromolar range, which is in fast exchange regime on the NMR time scale in solution (Fielding, 2007). The high concentration used for the enhancer may help shift the chemical equilibrium (Figure 9) to the formation of the AFP–enhancer complex, and the free enhancers in the solution rapidly exchange with the AFP-bound enhancers, which allows NADH to continue to act as an enhancer as long as the enhancer and the AFP molecules are stable in the solution. This dynamic process in solution may also provide a barrier for water to join the ice surface. Besides promoting the formation of AFP–enhancer complexes, the high concentration of enhancer may salt out AFP (i.e., the salting-out effect of the enhancers), which has been reported to contribute to the enhancement mechanism by small molecules (Li et al., 1998a; Wang et al., 2009b). Thus, in contrast to the protein enhancers that help block a larger ice growth surface, different mechanisms seem to be involved in these low molecular mass enhancers.
Figure 9.
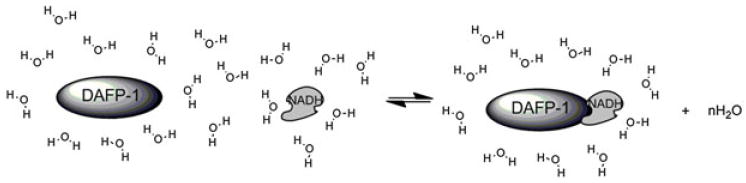
Schematic representation of the release of water molecules upon NADH binding to DAFP-1.
The highly efficient enhancers identified in this work have particular interest. Some of them, such as NAD(H), FAD, and CoA, are prevalent coenzymes that play a central role in the metabolic activities of plants, animals, and microorganisms. It should be noted that the concentrations of NADH used in this study are much higher than those found in cytosol (Yamada et al., 2006; Yang et al., 2007). DAFPs have been shown to be present in the hemolymph, gut fluid, and epidermis of D. canadensis larvae (Duman et al., 2002). The amounts of NADH in the above sites are too limited to serve as an effective physiological enhancer for the TH activity of DAFPs despite its potent enhancement ability. In contrast, the previously identified polyol enhancers, such as glycerol and trehalose, may be present in all compartments of the insect and thus be available for enhancer function in vivo (Amornwittawat et al., 2009). Although the physiological relevance of this finding is not yet appreciated, it has its significance in the potential biomedical and biotechnological applications of AFPs (e.g., cold preservation of tissues and organs) by providing a new and efficient means for increasing the potency of AFPs.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health Grant GM086249 and Research Corporation Cottrell College Science Award CC10492 to X.W. We thank Dr. John Duman at the University of Notre Dame for providing the cDNAs of DAFP-1.
Abbreviations
- AFP
antifreeze protein
- TH
thermal hysteresis
- DSC
differential scanning calorimetry
- DAFP
Dendroides canadensis antifreeze protein
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- HPLC
high-performance liquid chromatography
- NADH
β-nicotinamide adenine dinucleotide, reduced
- TTHA
triethylenetetramine-N,N,N′,N″,N″′,N″′-hexaacetate
- NAD
β-nicotinamide adenine dinucleotide
- TNAD
thionicotinamide adenine dinucleotide
- PAAD
3-pyridine aldehyde adenine dinucleotide
- FAD
flavin adenine dinucleotide
- CoA
coenzyme A
- R5P
d-ribose-5-phosphate
- ribose
D-(—)-ribose
- AMP
(—)-adenosine 5′-monophosphate
- ADP
adenosine 5′-diphosphate
- ADPR
adenosine 5′-diphosphoribose
- For simplicity, the positive charge is omitted from NAD, PAAD, and TNAD
Footnotes
Supporting information may be found in the online version of this paper.
References
- Amornwittawat N, Wang S, Banatlao J, Chung M, Velasco E, Duman JG, Wen X. Effects of Polyhydroxy compounds on beetle antifreeze protein activity. Biochim Biophys Acta. 2009;1794:341–346. doi: 10.1016/j.bbapap.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amornwittawat N, Wang S, Duman JG, Wen X. Polycarboxylates enhance beetle antifreeze protein activity. Biochim Biophys Acta. 2008;1784:1942–1948. doi: 10.1016/j.bbapap.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. Thermal hysteresis proteins. Int J Biochem Cell Biol. 2001;33:105–117. doi: 10.1016/S1357-2725(00)00083-2.l. [DOI] [PubMed] [Google Scholar]
- Dawson RMC, Elliott DC, Elliott WH, Jones KM. Data for biochemical research. 3. University Press; Oxford: 1985. p. 122. [Google Scholar]
- DeVries AL. Glycoproteins as biological antifreeze agents in Antarctic fishes. Science. 1971;172:1152–1155. doi: 10.1126/science.172.3988.1152. [DOI] [PubMed] [Google Scholar]
- DeVries AL, Cheng C-HC. The role of antifreeze glycopeptides and peptides in the survival of cold-water fishes. In: Osmond CB, editor. Water and Life: Comparative analysis of water relationships at the organismic, cellular and molecular level. Springer-Verlag; Berlin, Heidelberg: 1992. pp. 301–315. [Google Scholar]
- DeVries AL, Wohlschlag DE. Freezing resistance in some Antarctic fishes. Science. 1969;163:1073–1075. doi: 10.1126/science.163.3871.1073. [DOI] [PubMed] [Google Scholar]
- Duman JG. The role of macromolecular antifreeze in the darkling beetle, Meracantha contracta. J Comp Physiol B. 1977;115:279–286. doi: 10.1017/CBO9780511675997. [DOI] [PubMed] [Google Scholar]
- Duman JG, Olsen TM. Thermal hysteresis protein activity in bacteria, fungi, and phylogenetically diverse plants. Cryobiology. 1993;30:322–328. doi: 10.1006/cryo.1993.1031. [DOI] [Google Scholar]
- Duman JG, Serianni AS. The role of endogenous antifreeze protein enhancers in the hemolymph thermal hysteresis activity of the beetle Dendroides canadensis. J Insect Physiol. 2002;48:103–111. doi: 10.1016/S0022-1910(01)00150-0. [DOI] [PubMed] [Google Scholar]
- Duman JG, Verley D, Li N. Site specific forms of antifreeze proteins in the beetle Dendroides canadensis. J Comp Physiol B. 2002;172:547–552. doi: 10.1007/s00360-002-0284-x. [DOI] [PubMed] [Google Scholar]
- Fielding L. NMR methods for the determination of protein-ligand dissociation constants. Prog NMR Spec. 2007;51:219–242. doi: 10.2174/1568026033392705. [DOI] [PubMed] [Google Scholar]
- Fraústo da Silva JJR, Williams RJP. The uptake of elements by Biological Systems. Struct Bonding. 1976;29:67–121. doi: 10.1007/BFb0116519. [DOI] [Google Scholar]
- Fujioka M, Takata Y. Role of arginine residue in saccharopine dehydrogenase (L-lysine-forming) from bakers’ yeast. Biochemistry. 1981;20:468–472. doi: 10.1021/bi00506a004. [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Glocker MO, Bauer SHJ, Kast J, Volz J, Przybylski M. Characterization of specific noncovalent protein complexes by UV matrix-assisted laser desorption ionization mass spectrometry. J Mass Spectrom. 1996;31:1221–1227. doi: 10.1002/(SICI)1096-9888(199611). [DOI] [PubMed] [Google Scholar]
- Hannon CL, Anslyn EV. Bioorganic Chemistry Frontiers. Springer Verlag; Berlin: 1993. p. 193. [Google Scholar]
- Hummel JP, Dreyer WJ. Measurement of protein-binding phenomena by gel filtration. Biochim Biophys Acta. 1962;63:530–532. doi: 10.1016/0006-3002(62)90124-5. [DOI] [PubMed] [Google Scholar]
- Jairajpuri MA, Azam N, Baburaj K, Bulliraju E, Durani S. Charge and solvation effects in anion recognition centers: an inquiry exploiting reactive arginines. Biochemistry. 1998;37:10780–10791. doi: 10.1021/bi980058e. [DOI] [PubMed] [Google Scholar]
- Jia ZC, Davies PL. Antifreeze proteins: an unusual receptor-ligand interaction. Trends Biochem Sci. 2002;27:101–106. doi: 10.1016/S0968-0004(01)02028-X. [DOI] [PubMed] [Google Scholar]
- Klotz IM. Numbers of receptor sites from Scatchard graphs: facts and fantasies. Science. 1982;217:1247–1249. doi: 10.1126/science.6287580. [DOI] [PubMed] [Google Scholar]
- Knight CA, Devries AL. Effects of a polymeric, nonequilibrium ’antifreeze’ upon ice growth from water. J Cryst Growth. 1994;143:301–310. doi: 10.1016/0022-0248(94)90071-X. [DOI] [Google Scholar]
- Lange LG, Riordan JF, Vallee BL. Functional arginyl residues as NADH binding sites of alcohol dehydrogenases. Biochemistry. 1974;13:4361–4370. doi: 10.1021/bi00718a019. [DOI] [PubMed] [Google Scholar]
- Li N, Andorfer C, Duman J. Enhancement of insect antifreeze protein activity by solutes of low molecular mass. J Exp Biol. 1998a;201:2243–2251. doi: 10.1242/jeb.201.15.2243. [DOI] [PubMed] [Google Scholar]
- Parsons DL. Determination of ligand-macromolecule binding parameters and the method of Hummel and Dreyer. J Chromatogr A. 1980;193:520–521. [Google Scholar]
- Raymond JA, DeVries AL. Adsorption inhibition as a mechanism of freezing resistance in polar fishes. Proc Natl Acad Sci U S A. 1977;74:2589–2593. doi: 10.1073/pnas.74.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JF, McElvany KD, Borders CL., Jr Arginyl residues: anion recognition sites in enzymes. Science. 1977;195:884–886. doi: 10.1126/science.190679. [DOI] [PubMed] [Google Scholar]
- Sabri M, Dunford AJ, McLean KJ, Neeli R, Scrutton NS, Leys D, Munro AW. Characterization of coenzyme binding and selectivity determinants in Mycobacterium tuberculosis flavoprotein reductase A: analysis of Arg199 and Arg200 mutants at the NADP(H) 2′-phosphate binding site. Biochem J. 2009;417:103–112. doi: 10.1042/BJ20080466. [DOI] [PubMed] [Google Scholar]
- Schmuck C. How to improve guanidinium cations for oxoanion binding in aqueous solution?: The design of artificial peptide receptors. Coord Chem Rev. 2006;250:3053–3067. doi: 10.1016/j.ccr.2006.04.001. [DOI] [Google Scholar]
- Sem DS, Kasper CB. Interaction with arginine 597 of NADPH-cytochrome P-450 oxidoreductase is a primary source of the uniform binding energy used to discriminate between NADPH and NADH. Biochemistry. 1993;32:11548–11558. doi: 10.1021/bi00094a011. [DOI] [PubMed] [Google Scholar]
- Sun SF, Hsiao CL. Hummel-Dreyer method in high-performance liquid chromatography for the determination of drug-protein binding parameters. J Chromatogr A. 1993;648:325–331. doi: 10.1016/0021-9673(93)80414-4. [DOI] [Google Scholar]
- Tomchaney AP, Morris JP, Kang SH, Duman JG. Purification, composition, and physical properties of a thermal hysteresis “antifreeze” protein from larvae of the beetle Tenebrio molitor. Biochemistry. 1982;21:716–721. doi: 10.1021/bi00533a020. [DOI] [PubMed] [Google Scholar]
- Urrutia ME, Duman JG, Knight CA. Plant thermal hysteresis proteins. Biochim Biophys Acta. 1992;1121:199–206. doi: 10.1016/0167-4838(92)90355-H. [DOI] [PubMed] [Google Scholar]
- Wang L, Duman JG. Antifreeze proteins of the beetle Dendroides canadensis enhance one another’s activities. Biochemistry. 2005;44:10305–10312. doi: 10.1021/bi050728y. [DOI] [PubMed] [Google Scholar]
- Wang L, Duman JG. A thaumatin-like protein from larvae of the beetle Dendroides canadensis enhances the activity of antifreeze proteins. Biochemistry. 2006;45:1278–1284. doi: 10.1021/bi051680r. [DOI] [PubMed] [Google Scholar]
- Wang S, Amornwittawat N, Banatlao J, Chung M, Kao Y, Wen X. Hofmeister Effects of Common Monovalent Salts on the Beetle Antifreeze Protein Activity. J Phys Chem B. 2009a;113:13891–13894. doi: 10.1021/jp907762u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Amornwittawat N, Juwita V, Kao Y, Duman JG, Pascal TA, Goddard WA, Wen X. Arginine, a key residue for the enhancing ability of an antifreeze protein of the beetle Dendroides canadensis. Biochemistry. 2009b;48:9696–9703. doi: 10.1021/bi901283p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DW, Duman JG, Xu L. Enhancement of insect antifreeze protein activity by antibodies. Biochim Biophys Acta. 1991;1076:416–420. doi: 10.1016/0167-4838(91)90485-I. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hara N, Shibata T, Osago H, Tsuchiya M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal Biochem. 2006;352:282–285. doi: 10.1016/j.ab.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


