Abstract
Nitrogen-15 Carr-Purcell-Meiboom-Gill (CPMG) transverse relaxation experiment are widely used to characterize protein backbone dynamics and chemical exchange parameters. Although an accurate value of the transverse relaxation rate, R2, is needed for accurate characterization of dynamics, the uncertainty in the R2 value depends on the experimental settings and the details of the data analysis itself. Here, we present an analysis of the impact of CPMG pulse phase alternation on the accuracy of the 15N CPMG R2. Our simulations show that R2 can be obtained accurately for a relatively wide spectral width, either using the conventional phase cycle or using phase alternation when the r.f. pulse power is accurately calibrated. However, when the r.f. pulse is miscalibrated, the conventional CPMG experiment exhibits more significant uncertainties in R2 caused by the off-resonance effect than does the phase alternation experiment. Our experiments show that this effect becomes manifest under the circumstance that the systematic error exceeds that arising from experimental noise. Furthermore, our results provide the means to estimate practical parameter settings that yield accurate values of 15N transverse relaxation rates in the both CPMG experiments.
Keywords: relaxation, Monte Carlo, dynamics, protein, NMR, CPMG
Introduction
NMR relaxation is one of powerful methods to characterize internal motion of individual sites of proteins (Bruschweiler 2003; Dayie et al. 1996; Farrow et al. 1995; Fushman & Cowburn 2001; Igumenova et al. 2006; Ishima & Torchia 2000; Jarymowycz & Stone 2006; Kay 2005; Palmer 2001; Peng & Wagner 1995; Redfield 2004). Several relaxation rates, including the transverse relaxation rate (R2), are typically used to characterize the degree of internal motion in biomolecules (Kay et al. 1989; Lipari & Szabo 1982a; Lipari & Szabo 1982b; Nirmala & Wagner 1988). R2 is also used to detect slow conformational changes in biomolecules (Carver & Richards 1972; Davis et al. 1994; Ishima et al. 1998; Loria et al. 1999; Mulder et al. 1999; Orekhov et al. 1994). In biomolecular relaxation experiments, accurate error estimation is important in order to compare with NMR-derived dynamics parameters with those obtained by other methods, such as Gibbs free energy estimated from chemical exchange (Farrow et al. 1995; Huang & Oas 1995; Szyperski et al. 1993), molecular dynamics simulation data (Chandrasekhar et al. 1992; Eriksson et al. 1993; Horita et al. 2000; Smith et al. 1995; Wrabl et al. 2000; Yamasaki et al. 1995), and conformational entropy estimated from generalized order parameters (Akke & Palmer 1996; Li et al. 1996; Yang & Kay 1996). The accuracy and precision of R2 measurements, have been carefully analyzed (Bain et al. 2010; Bain et al. 2011; Bretthorst et al. 2005; Czisch et al. 1997; Ishima & Torchia 2005; Istratov & Vyvenko 1999; Korzhnev et al. 2000; Long et al. 2008; Myint et al. 2009; Palmer et al. 1991; Ross et al. 1997; Skelton et al. 1993; Viles et al. 2001; Yip & Zuiderweg 2004). However, some of the R2 measurements remain unclear, yet.
One approach to improve the accuracy of R2 measurements, by reducing cumulative pulse error, is an alternative phase scheme that applies pairs of orthogonal 180° pulses (XXYȲ) rather than the conventional CPMG (XXXX) (Bain et al. 2010; Bain et al. 2011; Gullion et al. 1990; Long et al. 2008; Yip & Zuiderweg 2004). The alternative phase scheme with a correction factor given to account for longitudinal relaxation during the pulsing was found to provide more accurate transverse relaxation rates covering a wider off-resonance frequency range than the conventional scheme (Bain et al. 2011; Yip & Zuiderweg 2004). This conclusion was supported by simulation results as well as experimental results. However, the studies assumed relatively low power radio-frequency (r.f.) pulses (~2.5 kHz), which is far smaller than in typical applications (Bain et al. 2011; Yip & Zuiderweg 2004). Although we and others have studied off-resonance errors (Bain et al. 2011; Czisch et al. 1997; Ishima & Torchia 2005; Korzhnev et al. 2000; Myint et al. 2009; Ross et al. 1997), quantitative analysis of the uncertainty of R2 determine using the alternative CPMG phase scheme has not been studied. Most 15N CPMG relaxation experiments applied to protein backbone dynamics studies have been performed using the conventional CPMG sequence (Meiboom & Gill 1958), and the practical advantage of the alternative phase scheme method has not yet been established.
The purpose of this article is to establish the condition under which the alternative phase scheme provides a practical advantage to the conventional CPMG scheme in measuring accurate R2 values. In particular, we aim to identify whether it is advantageous or not when a relatively strong r.f. is employed for CPMG pulse train. In order to achieve this goal, we simulated 15N CPMG relaxation data using three different phase schemes, and determined R2 and its uncertainty for each as a function of off-resonance frequencies. The R2 uncertainty was estimated by Monte Carlo error estimations in which a Gaussian distribution was assumed to generate synthetic datasets for the samplings. We generated the synthetic dataset by employing two different standard deviations (a) given by a rmsd deviation of the noise of the NMR spectra and (b) by a rmsd deviation of the residuals (Ii − Ifit) in the fit. Both have been used for 15N relaxation data analysis (Nicholson et al. 1992; Palmer et al. 1991; Skelton et al. 1993). The former sampling reflects only the statistical noise uncertainties of R2 but not any additional error. The later reflects overall fit-uncertainties that include both experimental statistical noise and systematic errors. By comparing the R2 uncertainties, magnitudes of the uncertainties other than those due to the experimental noise are estimated (Ishima & Torchia 2005). Our results show that in CPMG experiments performed by employing a radio-frequency of B1 > 5 kHz, R2 obtained using the conventional CPMG (XXXX) phase scheme is close to the one obtained using the alternative phase scheme (XXYȲ) at the off-resonance frequency, ωoff/2π, up to 1400 Hz. However, the conventional CPMG is expected to introduce higher systematic error even with ωoff/2π < 1400 Hz presumably due to non-exponential behavior of magnetization decay. We conducted 15N CPMG relaxation experiments to verify the results of the calculations. Experimental results further showed that the differences in R2 uncertainties in the two schemes are manifest only when the noise-to-signal ratio is significantly smaller than the fractional systematic error on R2.
Methods
NMR Experiments
15N transverse relaxation experiments were conducted using a 0.8 mM 15N labeled ubiquitin at pH 4.5 on a Bruker Avance 900 NMR instrument. Ubiquitin sample was purchased and prepared as described previously (Myint et al. 2009). Three transverse relaxation experiments were performed with a conventional CPMG phase scheme (XX – XX, here noted 00-00), an alternative phase scheme (XX –YȲ, here noted as 00-13), and a long alternative phase scheme (XXYȲ – XXYȲ, here noted as 0013-0013). Here, X and Y indicate pulse phases, and a hyphen indicates the timing applied to the 1H 180° pulses to suppress cross-correlation by 1H-15N dipolar interaction and 15N chemical shift anisotropy (CSA) (Kay et al. 1992; Palmer et al. 1992). We used the same CPMG pulse sequence that was applied previously ((Freedberg et al. 2002), except for an additional semi-constant time for the t1 evolution, not in the original sequence (Kay et al. 1992). The total phase cycle is 8 with a phase inversion for the CPMG period. All the 15N pulses are applied even number of times at each pulse scheme. Delays for the 00-00 and 00-13 experiments were varied: 0, 8, 16, 24, 32, 40, and 48 ms for the 00-00 and 00-13. Delays for the 0013-0013 experiments were varied: 0, 16, 32, and 48 ms. The maximum delay is set shorter than the expected 1/R2 because of the high sensitivity of an instrument equipped with a cryogenic probe and to avoid significant sample heating (Myint et al. 2009). Radio-frequency power for the 15N pulses was 5.56 kHz (90 μs as a 180° pulse), and a half duration between 15N CPMG pulses, τCP, was 0.5 ms. Here, 2τCP is the time between the centers of two adjacent CPMG pulses. 15N pulses were applied at 130 ppm as a carrier frequency to investigate off-resonance effects. The number of scans was 8. Using the same pulse powers, 15N longitudinal relaxation was recorded for the analysis of the 00-13 type relaxation data. The delays applied for the longitudinal relaxation measurements were: 0, 0.05, 0.1, 0.25, and 0.5 s.
15N transverse relaxation experiments were also performed using a 250 μM Human Immunodeficiency virus-1 (HIV-1) protease at pH 5.8 on a Bruker Avance 600 MHz NMR instrument. The protease was over-expressed and purified mainly using the previous protocol (Nalam et al. 2007). Data was recorded using a conventional CPMG phase scheme (XX – XX) with delays of 0, 8, 16, 24, 32, 48, and 64 ms. Data were recorded by employing the same r.f. field strength for the 15N pulses, 5.56 kHz, as the same that of 900 MHz. However, the carrier frequency of the pulse was placed at 117 ppm that is approximately the center of the amide 15N chemical shifts of the protease sample. 64 scans were accumulated for each free induction decay.
Data analysis
Free induction decay signals were zero-filled four times, apodized with a sine window function with 40% offset, and Fourier transformed in both dimensions using NMRpipe software (Delaglio et al. 1995). Signal intensities at individual positions were taken instead of peak volumes using the NMRdraw software (Delaglio et al. 1995). Line-shape fitting algorithm was not used to estimate peak heights. Experimental noise was obtained for each two-dimensional spectrum, but assumed to be the same in each CPMG experiments.
For each time-course of magnetization decay, a transverse relaxation rate, R2, was optimized assuming a mono-exponential decay function, I(t) = I0exp(−t·R2). In theory, if the initial intensity is correct, there is only one unknown parameter in this equation. However, since there is small loss of intensity at time zero, both I0 and R2 are typically optimized. Note that the same equation is used for R1 determination as well as R2 in protein 15N NMR relaxation since the Freeman-Hill method is used (Freeman & Hill 1971).
Once R2 values were optimized, Monte Carlo error estimation was performed to estimate uncertainty of R2. In this method, first unknown parameters, R2 and the initial intensity, were optimized using the experimental data, and the set of the fit “ideal” intensities were back-calculated using the optimized parameters. Second, a hundred number of sets of synthetic intensity data were generated to allow a Gaussian distribution with the ideal intensities as the mean. Finally, optimization was repeated for these synthesized data sets to calculate the standard deviation of R2 and the initial intensity. The Gaussian distribution to generate synthetic intensities was defined in two ways:
a root-mean-square deviation (rmsd) of the NMR spectral noise (Palmer et al. 1991; Skelton et al. 1993)
a standard deviation of the residual (Ii − Ifit) of the fit (Nicholson et al. 1992). In this article, R2 errors that were calculated using the experimental noise are denoted R2noise_err, and those were calculated using the residual of the fit are denoted R2fit_err. The former reflects only experimental noise error and the later reflects overall fit-uncertainties that include both experimental statistic noise and systematic noise. By comparing the two types of R2 uncertainties the uncertainties other than those due the experimental noise are estimated (Ishima & Torchia 2005).
The error in the R2 value, measured using the 00-13 and 0013-0013 sequences, caused by R1 relaxation during the r.f. pulses is recommended to be corrected using the equation introduced by Zuiderweg’s group (Yip & Zuiderweg 2004):
| (1) |
Here, d indicates a duty cycle of the r.f. pulse: total duration of the r.f. pulses divided by the entire CPMG delay. However, in the following analysis, no correction was made because the correction factor is calculated to be small. When pulse power γNB1/2π = 5.6 kHz is applied with a reasonable delay (τCP = 0.45 ms and 0.5 ms for simulation and experiments, respectively), the observed R2 will be only 2.5% and 2.25% smaller than the correct R2, respectively when R1 = 0 (equation (1)). When 0 < R1 < R2, the difference becomes less than 2.5% and 2.25%, respectively. This is in contrast to the condition that was used by Yip and Zuiderweg in which ca. 8% correction of R2 was needed (Yip & Zuiderweg 2004). Note that since this equation does not contain any off-resonance frequency, some of the error caused by the combination of the effects of pulse imperfection and off-resonance is not corrected by this equation. We did not use the 00130031 sequence (Yip & Zuiderweg 2004) because the minimum cycle, each CPMG loop (n=1) of (00130031-00130031)2, needed to suppress cross correlation between 15N CSA and 1H-15N dipolar interactions is long, i.e., 32 ms when τCP = 0.5 ms.
Simulation
The time evolution of bulk nuclear magnetization of a scalar-coupled 15N-1H spin system was calculated to determine transverse relaxation rates. For this, we used a relaxation matrix that contains 16 Cartesian product operators as the base set, as described previously (Allard et al. 1997; Allard et al. 1998; Myint et al. 2009; Myint & Ishima 2009).
| E/2 | NX | NY | NZ | HX | HY | HZ | 2NXHX |
| 2NYHX | 2NZHX | 2NXHY | 2NYHY | 2NZHY | 2NXHZ | 2NYHZ | 2NZHZ |
Time dependence of the relaxation was calculated step by step for each time increment, t1, by solving the M(t0+ t1)=exp(R2*t1)M(t0). τCP is 0.45 ms, and each CPMG loop of (τ′CP −180N − τ′CP − τ′CP −180N − τ″CP −180H − τ″CP −180N − τ′CP − τ′CP −180N − τ′CP)2 was set to be a 7.2 ms, and incremented to a total 72 ms. Here, τ′CP and τ″CP satisfy τCP = τ′CP + 90N and τCP = τ″CP + 90N + 90H. Simulation was done twice with the starting 15N transverse magnetization at X and −X and detection at X and −X, respectively, for the phase cycle. The output is the average time course of the two phase cycles. In the relaxation matrix, 1H and 15N chemical shift and the 90-Hz 1H-15N J-coupling evolution, r.f. pulse effects, auto and cross relaxation terms, and cross-correlation terms were included (Allard et al. 1997; Allard et al. 1998; Myint et al. 2009; Myint & Ishima 2009). In the calculation, 15N and 1H r.f. pulses were applied at field strengths of 5.56 kHz and 25 kHz, respectively, and 15N signal off-resonance frequency, ωoff/2π, was varied from −3000 to 3000 Hz in 200 Hz steps. Off-resonance frequency of the 1H signal was set at 2250 Hz, which corresponds to 7.5 ppm at 900 MHz NMR. Relaxation terms in the matrix were calculated assuming a simple Lorentzen spectral density function with a 5 ns rotational correlation time, 170 ppm 15N CSA, and 1.02 Å N-H distance. Additional 1H longitudinal and transverse relaxation rates of 10 s−1 and 20 s−1 were added, in addition to the 1H-15N dipolar term, to the 1H relaxation (Myint et al. 2009).
Once the time dependence was calculated, R2 and its uncertainty, R2fit_err, were determined by Monte-Carlo error estimation using the standard deviation of the residual (Ii − Ifit) of the fit as the uncertainty in Ii (Nicholson et al. 1992). However, R2noise_err was not calculated because experimental noise itself was not assumed in the simulation. The effect of the inhomogeneity of the B1 field was simulated by assuming that relative magnetization intensities of 0.375, 0.5, and 0.125 experienced respective r,f, field strengths of 1.0B1, 1.05B1 and 1.1B1, respectively, where B1 is the correctly calibrated field strength. Then, R2 and R2fit_err were determined from simulations of the weighted time course of the average magnetization. The simulation analysis for B1 inhomogeneity was performed for the [00-00] and the [00-13] schemes individually. Calculations were performed using MATLAB software (The Mathworks Inc., Natick, MA).
Results and Discussions
The purposes of this study are to further identify systematic errors, that persist even when experiments are repeated (Bevington & Robinson 1992; Heinrich & Lyons 2007), in 15N transverse relaxation rates, which are used to characterize protein backbone dynamics. In particular, uncertainties in CPMG R2 values caused by systematic errors present when using phase schemes, [00-00], [00-13], and [0013-0013] are compared. For this purpose, we will firstly compare values of R2 and R2fit_err derived from simulations of measurments obtained using phase-schemes, [00-00] and [00-13]. Next, we will compare experimental results obtained using the [00-00], [00-13], and [0013-0013] schemes. In the evaluation of the experimental results, R2 errors estimated from two sets of Monte-Carlo methods, R2fit_err and R2noise_err, are compared as well as the R2 values to idenity systematic errors.
Simulated off-resonance frequency dependence of [00-00] and [00-13] at a practical r.f. power level
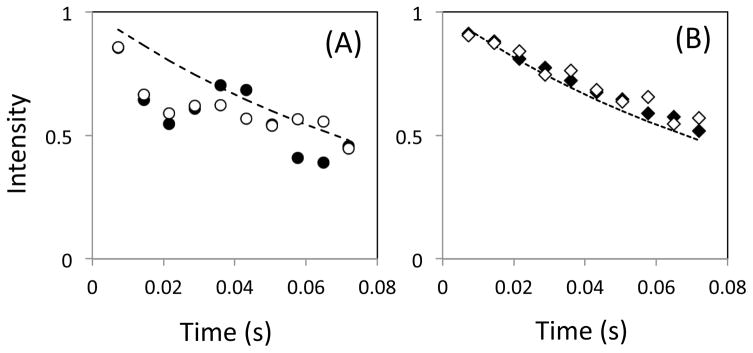
The time course of magnetization in a scalar-coupled two-spin system was calculated for the conventional phase [00-00] and the alternative phase [00-13], using r.f. pulse power of γNB1/2π = 5.6 kHz (i.e., 90 μs, 180° pulse) at varying off-resonance frequency, ωoff/2π. Time courses of magnetization obtained by employing the [00-00] scheme at ωoff/2π = ±2400 Hz exhibit non single-exponential decay profiles (Figure 1A). The profiles are basically consistent to the previous results (For example, Fig. 4 of the reference, (Yip & Zuiderweg 2004)). In contrast, the time course of magnetization obtained by employing the [00-13] scheme under the same conditions exhibited profiles that are closer to a single-exponential decay (Figure 1B). This result is consistent with previous observations (Bain et al. 2011; Yip & Zuiderweg 2004). Note that our simulation shows slightly different profiles at positive and negative ωoff/2π. Such a different is not observed in the simulation for a single spin system, or when 1H signal is located at the r.f. carrier frequency in the two-spin system (data not shown). Thus, although an even number of 1H pulses is applied in each CPMG cycle, the difference in profiles is most likely due to an asymmetry caused by an 1H off-resonance effect. The effect of the coupled spin was not taken into account in the previous CPMG simulations for the alternative phase schemes (Bain et al. 2011; Yip & Zuiderweg 2004).
Figure 1.
Simulated time course of transverse magnetization simulated using (A) the [00-00] sequence (circles) and (B) the [00-13] sequences (diamonds). In each figure, calculations were performed with ωoff/2π set equal to +2400 Hz (open circles) and −2400 Hz (closed circles).
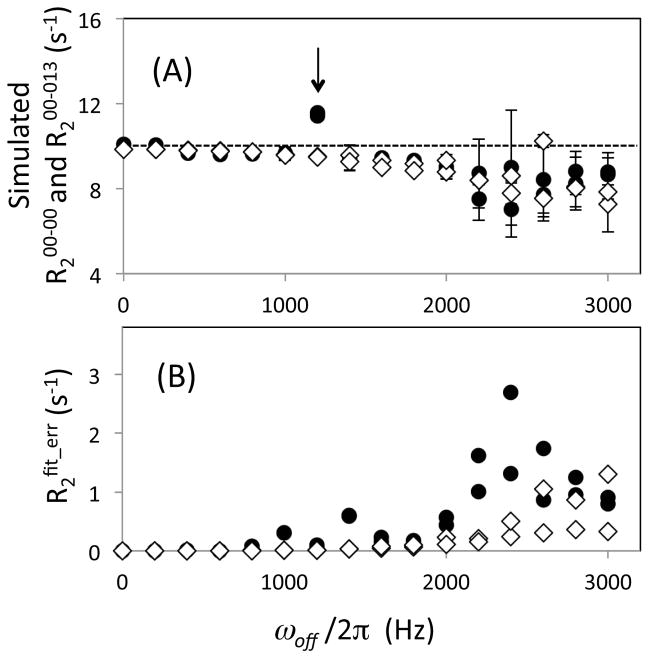
Once the time course of magnetization was simulated, R2 was determined by assuming that each time course decayed as a single-exponential function. The resultant R2 values are shown as a function of the absolute value of ωoff/2π in Figure 2A. R2 values were determined using both [00-00] and [00-13] schemes and are denoted as R200-00 and R200-13, respectively. Both R2 values were quite similar up to an off-resonance frequency, ωoff/2π = 1400 Hz (i.e., ωoff/(γNB1) = 0.25), with average difference in R2 1.2% and with the maximum difference in R2 of 2.2% (Figure 2A). Within each individual scheme, the fractional root-mean square deviations of R2 values in the range from ωoff/2π 0 to 1400 Hz were 0.20% and 0.17% for R200-00 and R200-13, respectively. These results were obtained including all data except for the R200-00 point at 1200 Hz. As shown by an arrow in Figure 2A, the R200-00 at 1200 Hz was significantly higher than others. When the magnetization along X-axis starts CPMG duration and the pulses are applied from the X-axis, the Y-component of the transverse magnetization vector rotates through the Y-Z plane during the pulsing whereas the X-component stays mostly in the transverse plane. Thus, pulse imperfection effect caused by chemical shift precession during the pulsing becomes largest when a magnetization vector undergoes almost 360° precession between the adjacent CPMG pulses, as has been shown in other simulation and experimental results (Bain et al. 2011; Yip & Zuiderweg 2004).
Figure 2.
(A) Transverse relaxation rate, R2, and (B) its error, R2fit_err, determined by simulation using the [00-00] (filled circles) and the [00-13] (open diamonds) sequences, plotted as a function of the absolute off-resonance frequency, ωoff/2π. Simulation of the time dependence of magnetization was performed varying ωoff/2π from −3000 to 3000 Hz at a 200 Hz step. Note that there are two data points determined using each pulse scheme at each ωoff/2π point, i.e., at the positive and the negative frequencies (Here, the two data points are shown by the same symbols). The arrow in (A) indicates a data point at a frequency that is close to the inverse of the CPMG pulse duration, a condition in which magnetization undergoes almost 360° precession during the period between the two adjacent CPMG pulses (see text). In (A), the dashed line indicates the original R2 determined only by the auto relaxation. R2 values are all plotted with R2fit_err as the error bars, while some of the error bars may not be notably large. R2fit_errs were estimated from the residual of the fits (see Methods section).
At ωoff/2π > 1400 Hz, deviations of R2 by the off-resonance effects are observable in both R200-00 and R200-13 (Figure 2). In previous results, R200-13 profiles were reported to exhibit a smaller and smoother change than those of R200-00 (Bain et al. 2011; Yip & Zuiderweg 2004). This discrepancy between these results and ours is mostly caused by differences in experimental and simulation parameter: their applied B1 field strength (γNB1/2π ≤ 2.5 kHz) was smaller than that of ours (γNB1/2π = 5.6 kHz), and their τCP (0.35 ms) was shorter than ours (0.45 ms and 0.5 ms). The latter set of the parameters make the [00-00] profile unfavorable. In addition, even in the previous experimental results, reduced but clear deviations of R2 caused by off-resonance effects were shown even in R200-13 (Bain et al. 2011; Yip & Zuiderweg 2004). Errors in R200-13 found at a large ωoff/2π values are not corrected by equation (1) because equation (1) does not contain off-resonance frequency as one of calibration parameters. Overall, using our practical conditions for 15N relaxation measurements, R200-00 and R200-13 exhibit similar profiles almost within the entire experimentally required ωoff/2π range.
Uncertainty of each R2 value, R2fit_err, was estimated from the Monte-Carlo method by assuming an average residual of the simulated intensities from the fit-intensities as a standard deviation for the normal distribution (Figure 2B). This R2fit_err solely reflects a discrepancy from a single-exponential decay model. R2fit_err for R200-13 exhibits very small uncertainties (<0.1 s−1 that correspond to <1% of R200-13) in the entire ωoff/2π range. In contrast, R2fit_err obtained for R200-00 exhibited large uncertainties at ωoff/2π > 1800 Hz, reflecting non-exponential behavior of magnetization simulated using the [00-00] scheme (Figure 2B). Nevertheless, R2fit_err values for R200-00 (< 2%) remained small for ωoff/2π < 1800 Hz. Again this frequency range covers nearly all 15N signals of backbone amides in diamagnetic proteins up to 20 ppm at a 91 MHz 15N resonance frequency (Figure 2B). Overall, application of the practical B1 field strength (γNB1/2π = 5.6 kHz) provides similar R2 values in both R200-00 and R200-13 schemes, within the spectral range of ±1400 Hz.
Effects of pulse strength miscalibration on R2 error, determined by simulation
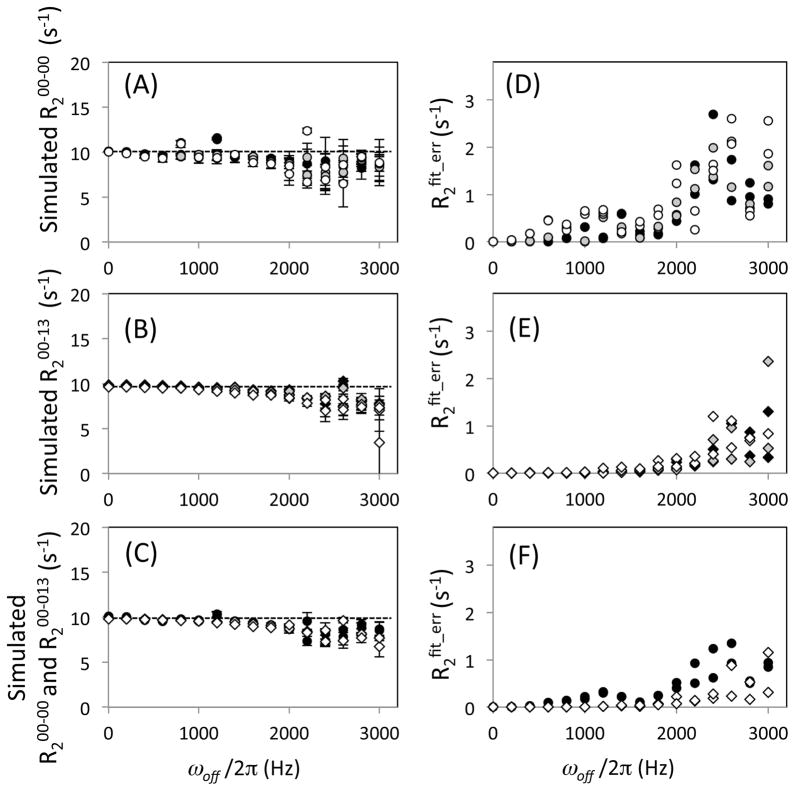
Similar simulations of magnetization and the determinations of R2 were performed varying the B1 field strength keeping the pulse width constant (i.e., power miscalibration). At a 5% or 10% increase (miscalibration) of the B1 field strength, R200-00 exhibits more variation of R2 values at ωoff/2π < 1400 Hz (Figure 3A), which is larger than the variation of R200-13 (Figure 3B). Similar to these observations, but more significantly, R2fit_err for R200-00 (Figure 3D) became larger than that of R200-13 at the miscalibrated B1 field strength (Figure 3E). In particular, the R2fit_err for R200-00 increased in proportion to the power miscalibration (Figure 3D). Such differences between R200-00 and R200-13, and between R2fit_err for R200-00 and R2fit_err for R200-13 are reduced when B1 inhomogeneity is taken into account in the simulations (Figures 3C and F). Overall, the simulations demonstrate that the difference between R2 obtained using R200-00 and R200-13 is insignificant for a relatively wide spectral width, provided that r.f. pulses are properly calibrated (Figure 2). However, the difference becames significant when r.f. pulses are miscalibrated (Figure 3). In particular, the [00-13] scheme is more tolerant to miscalibration errors than the [00-00] scheme.
Figure 3.
(A, B, C) Transverse relaxation rate, R2, and (D, E, F) its error, R2fit_err, determined by simulation, plotted as a function of the off-resonance frequency, ωoff/2π. Simulation was performed (A, D) using the [00-00] scheme, (B, E) using the [00-13] scheme, and (C, F) assuming the B1 inhomogeneity for both. In all figures, circle and diamond symbols indicate for R2 obtained using the [00-00] and the [00-13] schemes, respectively. In (A–E), simulation of the time dependence of magnetization was done at a correctly calibrated B1 field strength of 5.56 kHz CPMG 180° pulse of 90 μs (filled symbols), at a 5% stronger B1 field strength (gray symbols), and at a 10% stronger B1 field strength (open symbols). In (C) and (D), R2 and R2fit_err determination assuming B1 inhomogeneity was performed for both of the [00-00] and the [00-13] schemes (see Methods section). In the calculation, ωoff/2π was varied from −3000 to 3000 Hz at a 200 Hz step. There are two data points determined using each condition at each ωoff/2π, i.e., at the positive and the negative frequencies. In (A–C), R2 values are all plotted with R2fit_err as the error bars while some of the error bars may not be notably large.
Experimental R2 determined using [00-00], [00-13], and [0013-0013] sequences
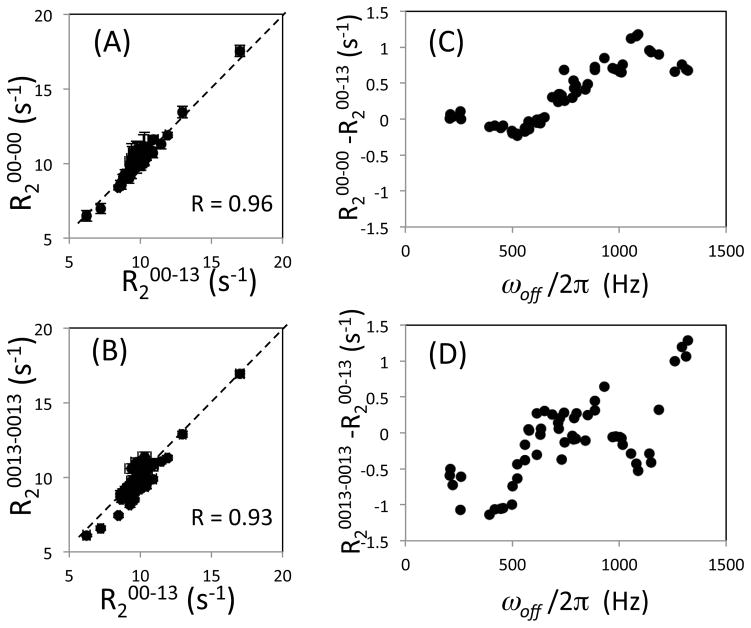
To verify the predictions of these simulations, we conducted 15N R2 experiments for ubiquitin using a 900 MHz NMR instrument, and evaluated experimental results only for signals that were fit by an single-exponential decay function with R2err < 1 s−1 (Figure 4). Most of the R200-00 values were very similar to R200-13 values (Figure 4A). As shown in Figure 4C, R200-00 and R200-13 did not exhibit significant difference for signals at ωoff/2π < 500 Hz, but the R2 values increaslingly differ as ωoff/2π (off-resonance) increases. In contrast, although the effect of resonance off-set for R20013-0013 is basically similar to that of R200-13 (Figure 4B), R20013-0013 exhibits differences from R200-13 even when, ωoff/2π < 500 Hz (Figure 4D). Since the [0013-0013] scheme is more symmetric with respect to the 1H 180° pulses than the [00-13] scheme, a possible explanation of the poor performance of the [0013-0013] scheme when ωoff/2π is small may be incomplete suppression of CSA-dipolar cross correlation. In the [0013-0013] scheme the period between 1H 180° pulses is twice that of [00-13] scheme. As shown previously (Kay et al. 1992; Palmer et al. 1992), the cross correlation is suppressed in a two-spin system. However, artifacts in R2 measurements may become significant because of the interactions with external protons that have been neglected in our simulations.
Figure 4.
Transverse relaxation rate, R2, for ubiquitin experimentally determined using the [00-13] sequence was compared with (A) those obtained using the conventional [00-00] sequence and (B) those obtained using the [0013-0013] sequence using a 900 MHz NMR instrument. Difference of (C) the rates determined between the [00-00] and the [00-13] schemes, and (D) between the [0013-0013] and the [00-13] schemes are shown as a function of the off-resonance frequency, ωoff/2π. Note that the number of relaxation data points corrected for the [0013-0013] sequence were smaller than those of [00-00] and [00-13] due to the longer cycle length. For this reason, a [00130031-00130031] sequence (Yip & Zuiderweg 2004) was not applied. Only the data points at ωoff/2π < 1500 Hz were include because large error bars at larger resonance off-sets make it difficult to view the correlation plots. In (A) and (B), R2 values are all plotted with R2fit_err error bars but may not be notable when the errors are small.
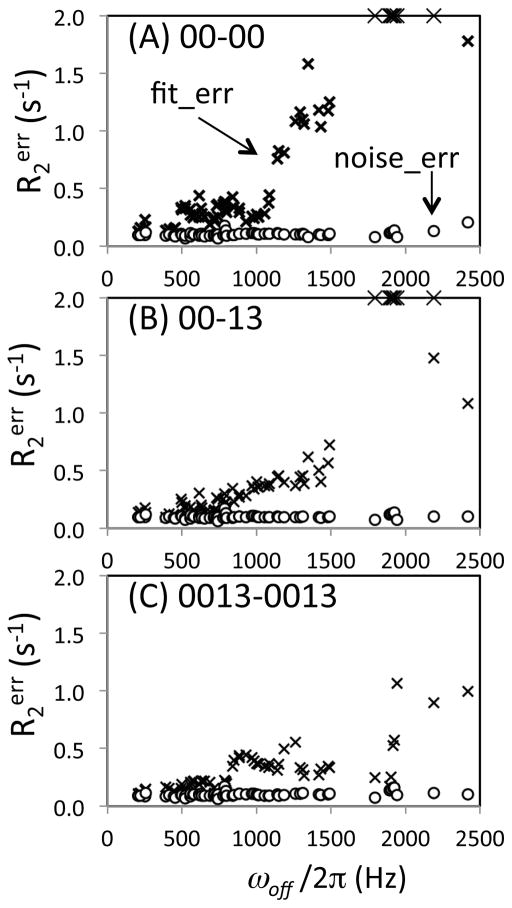
Monte-Carlo error estimation was calculated for ubiquitin R2 values in two ways: using the spectral noise (R2noise_err) and using the residual of the fits (R2fit_err), to generate the synthetic data. Since ubiquitin is a rigid, small protein, amide signal heights were quite uniform (except for E24 that undergoes chemical exchange and exhibits severe line broadening at 900 MHz) as is evident in the R2 correlations plotted in Figure 4. Therefore, R2noise_err was approximately the same for all three datasets (Figure 5, circles). In contrast, R2fit_err exhibits quite different profiles for R200-00 (Figure 5A, ×-symbols) compared with R200-13 and R20013-0013 (Figures 5B and 5C, ×-symbols). R2fit_err for R200-00 begins to increase at ωoff/2π ca. 1000 Hz, and becomes over 2 s−1 at ωoff/2π > 1800 Hz (Figure 5A, ×-symbols). This profile is consistent with the simulations that incorporate the effect of pulse miscalibration, Figure 3F, demonstrating the excellent performance of the calculations. R2fit_err for R200-13 and R20013-0013 exhibits a more gradual increases as a function of ωoff/2π, and stays smaller than R2fit_err for R200-00 even at ωoff/2π ~1500 Hz. This small gradual increase of R2err for the phase alternation experiments is also consistent with the simulation results, Figure 3F. Overall, in both R200-00 and R200-13, R2fit_err almost equals R2noise_err when ωoff/2π is small, and increases as ωoff/2π increases. This increasing discrepancy between the R2fit_err and R2noise_err as ωoff/2π increases is clear evidence of the off-resonance related systematic error.
Figure 5.
Uncertainties, R2err, of the transverse relaxation rates for ubiquitin experimentally determined using (A) [00-00], (B) [00-13], and (C) [0013-0013] sequences on a 900 MHz NMR instrument, shown as a function of the off-resonance frequency, ωoff/2π. R2 values that are shown in Figure 4 are used. An additional 16 sets of data that exhibited R2err > 1 s−1 were also included in the graphs in order to show how the uncertainty increases as a function of ωoff/2π. R2err values were determined by two different Monte-Carlo error estimations in which the standard deviations of a Gaussian distribution function that generates synthetic data sets were given by rmsd of spectral noise, R2noise_err (circle), and by rmsd of residuals of the initial fit intensities, R2fit_err (×-symbols). Difference of the two uncertainties indicates errors other than experimental noise. R2err values above 2 s−1 are shown as the ceiling values.
Off-resonance systematic error is not observed in large proteins
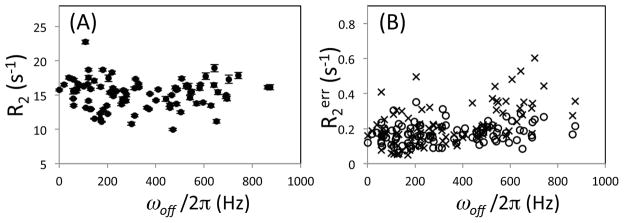
To test if off-resonance effects are observable in the case of a larger protein with less spectra sensitivity than ubiquition, the CPMG experiments were also performed for HIV-1 protease using a 600 MHz instrument. In this experiment, the 15N pulse carrier was placed at 117 ppm which is almost the center of the 15N chemical shifts of the protease backbone amides. R200-00 for the 92 residues are on average 15.22 s−1 (±2.1 s−1), which is approximately 1.5 times larger than that of ubiquitin and consistent with previous protease measurements (Freedberg et al. 2002). R200-00 values are relatively uniform in this ωoff/2π range (Figure 6A). R2noise_err is 0.17 s−1 whereas R2fit_err is 0.22 s−1 (Figure 6B). The R2noise_err values are approximately 2% of R200-00. Although some R2fit_err are higher at ωoff/2π > 500 Hz, the differences between R2noise_err and R2fit_err are not significant. Thus, differences in R2err estimated by two methods are not observed because the noise error is larger than the off-resonance error. This result is in contrast to that obtained for ubiquitin.
Figure 6.
(A) Transverse relaxation rate, R2, and (B) its uncertainty, R2err, for HIV-1 protease experimentally determined using the [00-00] sequence on a 600 MHz NMR instrument with 117 ppm as 15N pulse carrier frequency. In (A), R2 values are all plotted with R2fit_err error bars but may not be notable when the errors are small. In (B), R2err values were determined by two different Monte-Carlo error estimations in which the standard deviations of a Gaussian distribution function that generates synthetic data sets were given by rmsd of spectral noise, R2noise_err (circle), and by rmsd of residuals of the initial fit intensities, R2fit_err (×-symbols).
Summary
In this study, 15N CPMG experiments with and without the phase alternation were investigated at the practical high pulse power (γNB1/2π = 5.6 kHz) and also with 1H 180° pulses that are applied to suppress dipolar/CSA cross correlation. Since CPMG performance is determined by the field strength of the r.f. pulse (γNB1/2π), the delay between the CPMG pulses (τCP), the off-resonance frequency (ωoff/2π), and relaxation rates and scalar couplings, it is important to investigate the performance at optimal parameter settings. First, our results showed that, in the high power condition, both pulse phase schemes yield similar results unless resonance off-sets are very large. Our simulations reproduced the reduction of the measurement accuracy as ωoff/2π increases even with the phase alternation method, which is consistent with the experimental results. Second, when the pulse power is miscalibrated, the fit error becomes larger in the conventional CPMG than the phase alternation approach, even when the resonance off-set is small. Third, although the full (longer) phase alternation cycle has a better performance in theory, our experimental results show that the actual performance of the longer cycle was not improved in the 15N experiments with 1H 180° pulses.
Acknowledgments
We thank Dennis Torchia for useful discussions and critical reading of the manuscript. This study was financially supported by the American Heart Association (Great Rivers affiliate) new investigator grant 0765348U, National Science Foundation research grant MCB 0814905, and subcontract of National Institutes of Health program project 5P01GM066524-08.
Abbreviations
- CPMG
Carr-Purcell-Meiboom-Gill
- R2
transverse relaxation rate
- R2err
uncertainty of transverse relaxation rate
- ωoff/2π
off-resonance frequency
- r.f
radio frequency
References
- Akke M, Palmer AG., III Monitoring Macromolecular Motions on Microsecond to Millisecond Time Scales by R1ϱ–R1 Constant Relaxation Time NMR Spectroscopy. J Am Chem Soc. 1996;118:911–912. [Google Scholar]
- Allard P, Helgstrand M, Hard T. A Method for Simulation of NOESY, ROESY, and Off-Resonance ROESY Spectra. J Magn Reson. 1997;129:19–29. doi: 10.1006/jmre.1997.1252. [DOI] [PubMed] [Google Scholar]
- Allard P, Helgstrand M, Hard T. The complete homogeneous master equation for a heteronuclear two-spin system in the basis of Cartesian product operators. J Magn Reson. 1998;134:7–16. doi: 10.1006/jmre.1998.1509. [DOI] [PubMed] [Google Scholar]
- Bain AD, Anand CK, Nie Z. Exact solution to the Bloch equations and application to the Hahn echo. J Magn Reson. 2010;206:227–40. doi: 10.1016/j.jmr.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Bain AD, Anand CK, Nie Z. Exact solution of the CPMG pulse sequence with phase variation down the echo train: application to R2 measurements. J Magn Reson. 2011;209:183–194. doi: 10.1016/j.jmr.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill; Neew York: 1992. [Google Scholar]
- Bretthorst GL, Hutton WC, Garbow JR, Ackerman JJH. Exponential parameter estimation (in NMR) using Bayesian probability theory. Concepts Magn Reson. 2005;27A:55–63. [Google Scholar]
- Bruschweiler R. New approaches to the dynamic interpretation and prediction of NMR relaxation data from proteins. Curr Opin Structl Biol. 2003;13:175–183. doi: 10.1016/s0959-440x(03)00036-8. [DOI] [PubMed] [Google Scholar]
- Carver JP, Richards RE. General 2-Site Solution for Chemical Exchange Produced Dependence of T2 Upon Carr-Purcell Pulse Separation. J Magn Reson. 1972;6:89–105. [Google Scholar]
- Chandrasekhar I, Clore GM, Szabo A, Gronenborn AM, Brooks BR. A 500 ps molecular dynamics simulation study of interleukin-1 beta in water. Correlation with nuclear magnetic resonance spectroscopy and crystallography. J Mol Biol. 1992;226:239–250. doi: 10.1016/0022-2836(92)90136-8. [DOI] [PubMed] [Google Scholar]
- Czisch M, King GC, Ross A. Removal of systematic errors associated with off-resonance oscillations in T2 measurements. J Magn Reson. 1997;126:154–157. doi: 10.1006/jmre.1997.1168. [DOI] [PubMed] [Google Scholar]
- Davis DG, Perlman ME, London RE. Direct Measurements of the Dissociation-Rate Constant for Inhibitor-Enzyme Complexes Via the T-1-Rho and T-2 (Cpmg) Methods. J Magn Reson, Ser B. 1994;104:266–275. doi: 10.1006/jmrb.1994.1084. [DOI] [PubMed] [Google Scholar]
- Dayie KT, Wagner G, Lefevre JF. Theory and practice of nuclear spin relaxation in proteins. Annual Review of Physical Chemistry. 1996;47:243–282. doi: 10.1146/annurev.physchem.47.1.243. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Eriksson MA, Berglund H, Härd T, Nilsson L. A comparison of 15N NMR relaxation measurements with a molecular dynamics simulation: backbone dynamics of the glucocorticoid receptor DNA-binding domain. Proteins. 1993;17:375–90. doi: 10.1002/prot.340170406. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Zhang O, Forman-Kay JD, Kay LE. Comparison of the backbone dynamics of a folded and an unfolded SH3 domain existing in equilibrium in aqueous buffer. Biochemistry. 1995;34:868–878. doi: 10.1021/bi00003a021. [DOI] [PubMed] [Google Scholar]
- Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich I, Louis JM, Torchia DA. Rapid structural fluctuations of the free HIV protease flaps in solution. Protein Sci. 2002;11:221–32. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman R, Hill HDW. Fourier Transform Study of NMR Spin-Lattic Relaxation by “Progressive Saturation”. J Chem Phys. 1971;54:3367–3377. [Google Scholar]
- Fushman D, Cowburn D. Nuclear magnetic resonance relaxation in determination of residue-specific N-15 chemical shift tensors in proteins in solution: Protein dynamics, structure, and applications of transverse relaxation optimized spectroscopy. In. Methods in Enzymology. Nuclear Magnetic Resonance of Biological Macromolecules. 2001;339(Pt B):109–126. doi: 10.1016/s0076-6879(01)39312-6. [DOI] [PubMed] [Google Scholar]
- Gullion T, Baker DB, Conradi MS. New, Compensated Carr-Purcell Sequences. J Magn Reson. 1990;89:479–484. [Google Scholar]
- Heinrich J, Lyons L. Systematic errors. Ann Rev Nucl Part Sci. 2007;57:145–169. [Google Scholar]
- Horita DA, Zhang W, Smithgall TE, Gmeiner WH, Byrd RA. Dynamics of the Hck-SH3 domain: comparison of experiment with multiple molecular dynamics simulations. Protein Sci. 2000;9:95–103. doi: 10.1110/ps.9.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GS, Oas TG. Submillisecond folding of monomeric lambda repressor. Proc Natl Acad Sci U S A. 1995;92:6878–82. doi: 10.1073/pnas.92.15.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igumenova TI, Frederick KK, Wand AJ. Characterization of the fast dynamics of protein amino acid side chains using NMR relaxation in solution. Chem Rev. 2006;106:1672–1699. doi: 10.1021/cr040422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishima R, Torchia DA. Protein dynamics from NMR. Nat Struct Biol. 2000;7:740–3. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- Ishima R, Torchia DA. Error estimation and global fitting in transverse-relaxation dispersion. J Biomol NMR. 2005;32:41–54. doi: 10.1007/s10858-005-3593-z. [DOI] [PubMed] [Google Scholar]
- Ishima R, Wingfield PT, Stahl SJ, Kaufman JD, Torchia DA. Using amide H-1 and N-15 transverse relaxation to detect millisecond time-scale motions in perdeuterated proteins: Application to HIV-1 protease. J Am Chem Soc. 1998;120:10534–10542. [Google Scholar]
- Istratov AA, Vyvenko OF. Exponential analysis in physical phenomena. Rev Sci Instrum. 1999;70:1233–1257. [Google Scholar]
- Jarymowycz VA, Stone MJ. Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem Rev. 2006;106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- Kay LE. NMR studies of protein structure and dynamics. J Magn Reson. 2005;173:193–207. doi: 10.1016/j.jmr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Kay LE, Nicholson LK, Delaglio F, Bax A, Torchia DA. Pulse Sequences for Removal of the Effects of Cross Correlation between Dipolar and Chemical-Shift Anisotropy Relaxation Mechanisms on the Measurement of Heteronuclear T1 and T2 Values in Proteins. J Magn Reson. 1992;97:359–375. [Google Scholar]
- Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Tischenko EV, Arseniev AS. Off-resonance effects in N-15 T-2 CPMG measurements. J Biomol NMR. 2000;17:231–237. doi: 10.1023/a:1008348827208. [DOI] [PubMed] [Google Scholar]
- Li Z, Raychaudhuri S, Wand AJ. Insights into the local residual entropy of proteins provided by NMR relaxation. Protein Sci. 1996;5:2647–2650. doi: 10.1002/pro.5560051228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982a;104:4546–4559. [Google Scholar]
- Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc. 1982b;104:4559–4570. [Google Scholar]
- Long D, Liu M, Yang D. Accurately probing slow motions on millisecond timescales with a robust NMR relaxation experiment. J Am Chem Soc. 2008;130:2432–2433. doi: 10.1021/ja710477h. [DOI] [PubMed] [Google Scholar]
- Loria JP, Rance M, Palmer AG., III A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. Journal of the American Chemical Society. 1999;121:2331–2332. [Google Scholar]
- Meiboom S, Gill D. Modified Spin-Echo Method for Measuring Nuclear Relaxation Times. Rev Sci Instrm. 1958;29:688–691. [Google Scholar]
- Mulder FA, van Tilborg PJ, Kaptein R, Boelens R. Microsecond time scale dynamics in the RXR DNA-binding domain from a combination of spin-echo and off-resonance rotating frame relaxation measurements. J Biomol NMR. 1999;13:275–288. doi: 10.1023/a:1008354232281. [DOI] [PubMed] [Google Scholar]
- Myint W, Gong Q, Ishima R. Practical aspects of 15N CPMG transverse relaxation experiments for proteins in solution. Concepts Magn Reson. 2009;34A:63–75. [Google Scholar]
- Myint W, Ishima R. Chemical exchange effects during refocusing pulses in constant-time CPMG relaxation dispersion experiments. J Biomol NMR. 2009;45:207–16. doi: 10.1007/s10858-009-9344-9. [DOI] [PubMed] [Google Scholar]
- Nalam MN, Peeters A, Jonckers TH, Dierynck I, Schiffer CA. Crystal structure of lysine sulfonamide inhibitor reveals the displacement of the conserved flap water molecule in human immunodeficiency virus type 1 protease. J Virol. 2007;81:9512–8. doi: 10.1128/JVI.00799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson LK, Kay LE, Baldisseri DM, Arango J, Young PR, Bax A, Torchia DA. Dynamics of methyl groups in proteins as studied by proton-detected 13C NMR spectroscopy. Application to the leucine residues of Staphylococcal nuclease. Biochemistry. 1992;31:5253–63. doi: 10.1021/bi00138a003. [DOI] [PubMed] [Google Scholar]
- Nirmala NR, Wagner G. Measurement of C-13 relaxation-times in proteins by two-dimensional heteronuclear H1-C-13 correlation spectroscopy. J Am Chem Soc. 1988;110:7557–7558. [Google Scholar]
- Orekhov VY, Pervushin KV, Arseniev AS. Backbone dynamics of (1-71)bacterioopsin studied by two-dimensional 1H-15N NMR spectroscopy. Eur J Biochem. 1994;219:887–896. doi: 10.1111/j.1432-1033.1994.tb18570.x. [DOI] [PubMed] [Google Scholar]
- Palmer AG., III Nmr probes of molecular dynamics: overview and comparison with other. Annu Rev Biophys Biomol Struct. 2001;30:129–55. doi: 10.1146/annurev.biophys.30.1.129. [DOI] [PubMed] [Google Scholar]
- Palmer AG, III, Rance M, Wright PE. Intramolecular motions of a zinc finger DNA-binding domain from Xfin characterized by proton-detected natural abundance 13C Heteronuclear NMR spectroscopy. J Am Chem Soc. 1991;113:4371–4380. [Google Scholar]
- Palmer AG, III, Skelton NJ, Chazin WJ, Wright PE, Rance M. Suppression of the Effects of Cross-Correlation between Dipolar and Anisotropic Chemical-Shift Relaxation Mechanisms in the Measurement of Spin Spin Relaxation Rates. Mol Phys. 1992;75:699–711. [Google Scholar]
- Peng JW, Wagner G. Frequency spectrum of NH bonds in eglin c from spectral density mapping at multiple fields. Biochemistry. 1995;34:16733–16752. doi: 10.1021/bi00051a023. [DOI] [PubMed] [Google Scholar]
- Redfield C. Using nuclear magnetic resonance spectroscopy to study molten globule states of proteins. Methods Mol Biol. 2004;34:121–132. doi: 10.1016/j.ymeth.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Ross A, Czisch M, King GC. Systematic errors associated with the CPMG pulse sequence and their effect on motional analysis of biomolecules. J Magn Reson. 1997;124:355–365. [Google Scholar]
- Skelton NJ, Palmer AG, III, Akke M, Kordel J, Rance M, Chazin WJ. Practical Aspects of 2-Dimensional Proton-Detected N-15 Spin Relaxation Measurements. J Magn Reson Ser B. 1993;102:253–264. [Google Scholar]
- Smith LJ, Mark AE, Dobson CM, van Gunsteren WF. Comparison of MD simulations and NMR experiments for hen lysozyme. Analysis of local fluctuations, cooperative motions, and global changes. Biochemistry. 1995;34:10918–31. doi: 10.1021/bi00034a026. [DOI] [PubMed] [Google Scholar]
- Szyperski S, Luginbühl P, Otting G, Güntert P, Wüthrich K. Protein dynamics studied by rotating frame. J Biomol NMR. 1993;3:151–164. doi: 10.1007/BF00178259. [DOI] [PubMed] [Google Scholar]
- Viles JH, Duggan BM, Zaborowski E, Schwarzinger S, Huntley JJ, Kroon GJ, Dyson HJ, Wright PE. Potential bias in NMR relaxation data introduced by peak intensity analysis and curve fitting methods. J Biomol NMR. 2001;21:1–9. doi: 10.1023/a:1011966718826. [DOI] [PubMed] [Google Scholar]
- Wrabl JO, Shortle D, Woolf TB. Correlation between changes in nuclear magnetic resonance order parameters and conformational entropy: molecular dynamics simulations of native and denatured staphylococcal nuclease. Proteins. 2000;38:123–33. doi: 10.1002/(sici)1097-0134(20000201)38:2<123::aid-prot2>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Saito M, Oobatake M, Kanaya S. Characterization of the internal motions of Escherichia coli ribonuclease HI by a combination of 15N-NMR relaxation analysis and molecular dynamics simulation: examination of dynamic models. Biochemistry. 1995;34:6587–601. doi: 10.1021/bi00020a003. [DOI] [PubMed] [Google Scholar]
- Yang DW, Kay LE. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: Application to protein folding. J Mol Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- Yip GN, Zuiderweg ER. A phase cycle scheme that significantly suppresses offset-dependent artifacts in the R2-CPMG 15N relaxation experiment. J Magn Reson. 2004;171:25–36. doi: 10.1016/j.jmr.2004.06.021. [DOI] [PubMed] [Google Scholar]