Abstract
Human 15-lipoxygenase-1 (h-15-LOX-1) is an important mammalian lipoxygenase and plays an important role in several inflammatory lung diseases such as asthma, COPD and chronic bronchitis. Novel potent inhibitors of h-15-LOX-1 are required to explore the role of this enzyme further and to enable drug discovery efforts. In this study, we applied an approach in which we screened a fragment collection that is focused on a diverse substitution pattern of nitrogen containing heterocycles such as indoles, quinolones, pyrazoles etc. We denoted this approach Substitution Oriented fragment Screening (SOS), because it is focuses on identification of novel substitution patterns rather than on novel scaffolds. This approach enabled the identification of hits with good potency and clear structure activity relationships (SAR) for h-1-5-LOX-1 inhibition. A molecular modeling enabled the rationalization of the observed SAR and supported structure-based design for further optimization to obtain inhibitor 14d that binds with a Ki of 36 nM to the enzyme. In vitro and ex vivo biological evaluations of our best inhibitor demonstrate significant increase of interleukin-10 (IL-10) gene expression, which indicates anti-inflammatory properties.
Keywords: 15-lipoxygenase, indole-based inhibitor, fragment screening, anti-inflammatory, interleukin-10, ex vivo studies, precision-cut lung slices
Introduction
Inflammatory lung diseases like asthma and chronic obstructive pulmonary disease (COPD) can have detrimental effects on patients’ health.1 Fortunately, nowadays these diseases can be alleviated by various therapeutic agents. Nevertheless, expansion of the therapeutic possibilities is needed, because for some patients the currently available medicines are ineffective or cause severe side effects.2 Therefore, the development of novel compounds targeting enzymes that are involved in inflammation is highly important.
The regulatory function of macrophages is gaining increasing attention in drug discovery, because they play key regulatory roles in the different disease stages of asthma and COPD as they polarize into different subclasses according to the cytokines they encounter in their environment.3 Based upon the signals they receive, their role, and cytokine profile, macrophages are categorized in to three subpopulations: M1 (induced by LPS/IFNγ), M2 (induced by IL-4/IL-13) and M2-like subsets (combination of Toll-like receptor stimulation). M1 macrophages play a role in inflammatory responses to intracellular pathogens and M2 are involved in scavenging debris, promoting angiogenesis, help in tissue remodeling/repair, and are therefore considered as wound-healing macrophages. The third class are the M2-like macrophages; as the name implies these are macrophages which resemble M2. M2-like macrophages are able to produce TGF-β and IL-10 implying an anti-inflammatory role.4–6
An enzyme class highly expressed in macrophages and other immune cells are the lipoxygenases (LOXs). These enzymes are non-heme iron containing enzymes that metabolize polyunsaturated fatty acids (PUFAs) such as arachidonic acid and linoleic acid into lipid signaling molecules such as leukotrienes and lipoxins.7–9 Human 15-lipoxygenase-1 (h-15-LOX-1, also denoted 12/15-LOX) is an important mammalian lipoxygenase, responsible for the biosynthesis of antiinflammatory and pro-inflammatory mediators (signaling molecules) such as lipoxins and eoxins.10,11 This enzyme is highly expressed in monocytes, broncho-alveolar epithelial cells, and in eosinophils and macrophages of asthmatic patients. 12–14 Growing evidence suggests that h-15-LOX-1 may modulate inflammatory responses. It was demonstrated that h-15-LOX-1 regulates the expression of IL-12 in a cell-type and stimuli-restricted manner.15 In addition, in lungs, it has been demonstrated that signaling products of h-15-LOX-1 can stimulate inflammation and mucus secretion.16 The crucial regulatory role of h-15-LOX-1 in several respiratory diseases such as asthma, COPD and chronic bronchitis14,17–20 and their role in modulating the inflammatory response calls for development of novel potent and selective inhibitors.
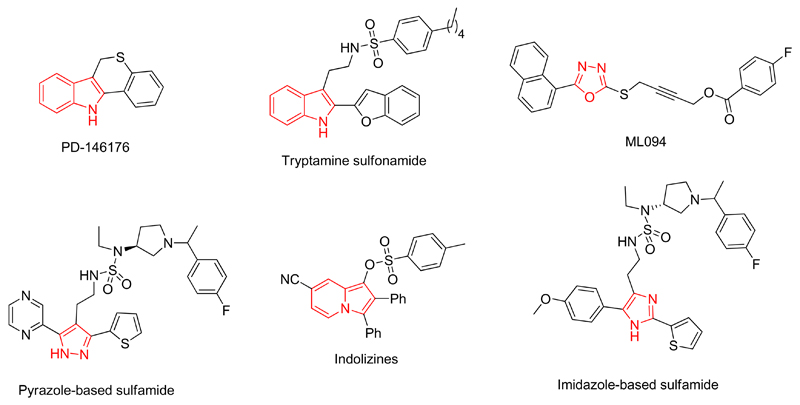
Despite the fact that the key role of h-15-LOX-1 was exemplified as target in drug discovery for several inflammatory diseases, the discovery of very potent h-15-LOX-1 inhibitors and their role as a novel therapeutic strategy is still in an early phase (Figure 1). Indole-based inhibitors, such as PD-146176 by Pfizer21 and tryptamine sulfonamides by Bristol-Myers Squibb (BMS)22 exhibited inhibitory potency against r12/15-LOX with IC50 value of 3.81 μM and 21 nM respectively. The inhibitor PD-146176 also showed downregulation of interleukin-12 (IL-12) after stimulation with LPS.15 However, the inhibitory potency of the PD-146176 is relatively low. Furthermore, five-membered heterocycles like pyrazole-based sulfonamide and sulfamides (IC50 = 1.4 nM, r12/15-LOX),23 oxadiazole or oxazole derivatives as ML094 (IC50 = 10 nM, h-15-LOX-1)24 and ML351 (IC50 = 200 nM, h-15-LOX-1)25 but also imidazole-based compounds (IC50 = 75 nM, r12/15-LOX)26 are reported as 15-LOX inhibitors. In addition, indolizine (IC50 = 25 μM, r12/15-LOX),27 thiourea-based (IC50 = 1.8 μM, soybean 15-LOX)28 and thiadiazine (IC50 = 9 μM, soybean 15-LOX)29 derivatives were developed as 15-LOX inhibitors, although they demonstrate a relatively low inhibitory potency. Recently, anacardic acid derived salicylates were described by our research group as LOX inhibitors.30–32 Although the potency of these inhibitors is moderate or good they often suffer from unfavourable physicochemical properties33 and limited ligand efficiency values (LE), which limits their utility as drug candidates. Taking into account all the above, new chemotypes are needed in order to explore the structure activity relationships (SAR) and the drugability of this enzyme further.
Figure 1.
Examples of previously reported 15-LOX inhibitors
The currently available inhibitors are frequently built around nitrogen containing heterocycles such as indoles, imidazoles or pyrazoles. We anticipate that generally the identification of the right substitution pattern is more demanding than finding a scaffold that provides inhibition. We therefore developed an approach for screening of fragments that is focused on specific nitrogen containing heterocycles, such as indoles, quinolones, pyrazoles, etc. that have a very diverse substitution pattern. We denoted this approach Substitution Oriented fragment Screening (SOS). After having identified fragment hits with the right substitution pattern to give inhibition in the micromolar range we assume that the active hits are sufficiently potent to provide initial SAR. This will enable docking studies and structure-based design to optimize the substitution pattern further.
Here we set out to test the SOS approach for fragment-based screening to find potent substituted scaffolds that inhibit the enzyme h-15-LOX-1. The hits were used to establish an initial SAR and docking studies were performed to enable the proposal of a plausible binding mode for the new hits in line with the SAR. The hits were further optimized using structure-based design to yield inhibitors with potency in the nanomolar range. The effects of the most potent inhibitor on pro-and anti-inflammatory gene expression were investigated in RAW 264.7 macrophages and precision-cut lung slices (PCLS) of mouse lung tissue.
Results And Discussion
Initial screening and discovery of potent inhibitors
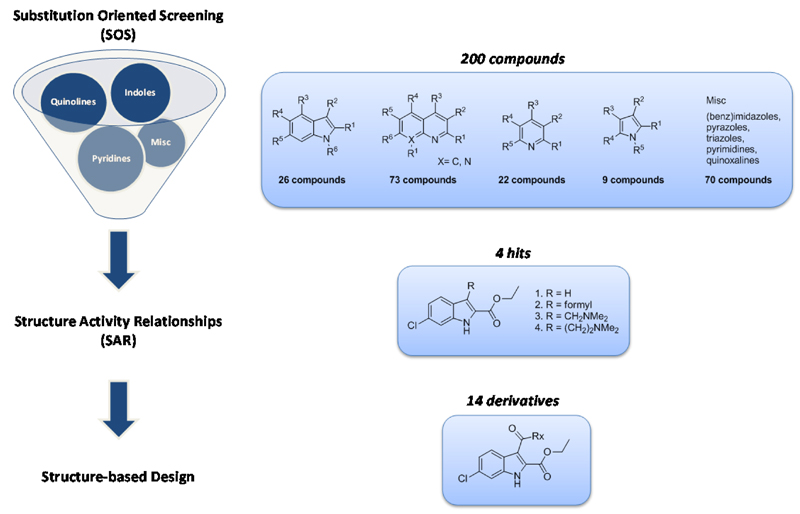
For our SOS approach we employed a library of 200 fragments containing 26 indoles, 73 quinolones, 22 pyrroles and 79 other heterocycles with a diverse substitution pattern (Figure 2). This library consists mostly of di- or tri-substituted scaffolds with substituents varying from EWG, EDG, aliphatic and aromatic moieties.
Figure 2.
Substitution Oriented fragment Screening (SOS) enables the identification of unique substitution patterns around known scaffolds, which can be further optimized by identification of Structure Activity Relationships (SAR) to support structure-based design.
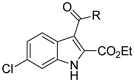
For the h-15-LOX-1 activity studies an enzyme assay was used that employs the UV absorption of the h-15-LOX-1 product (λmax 234 nm) that is formed upon enzymatic conversion from linoleic acid (Figure S1). This assay was done in a 96-well format, which is suited for medium-throughput screening, measurement of inhibition concentration 50% (IC50), and enzyme kinetic studies. Using this assay the SOS library was screened and four hits were identified that provide more than 80% inhibition of the enzyme activity at 50 μM, which indicates an inhibitory potency in the low micromolar range. The potency was confirmed by determination of the IC50 values for compounds 1and 2. The hits contain an indole scaffold as previously found in h-15-LOX-1 inhibitors such as for example PD-146176 or tryptamine sulfonamides (Figure 1). Interestingly, the hits 6-chloro-1H-indole-2-ethyl acetate (1) and 6-chloro-3-formyl-1H-indole-2-ethyl acetate (2) have both a unique substitution pattern around the indole scaffold, which has not been identified previously. This stresses the utility of the SOS approach to identify novel substitution patterns around known scaffolds.
Hits 1 and 2 have beneficial properties for further development, because halogen substitution in the 5 or 6 position could prevent hydroxylation of the indole thereby increasing its metabolic stability.34,35 The ethyl acetate substitution in the 2-position of the indole ring is beneficial because the 2-position constitutes an active position that is receptive to nucleophilic substitutions especially with an EWG in the 3-position. Combining all the above facts, an indole derivative which is substituted at 2nd, 3rd and 5th or 6th position would be a good starting point for our further studies.
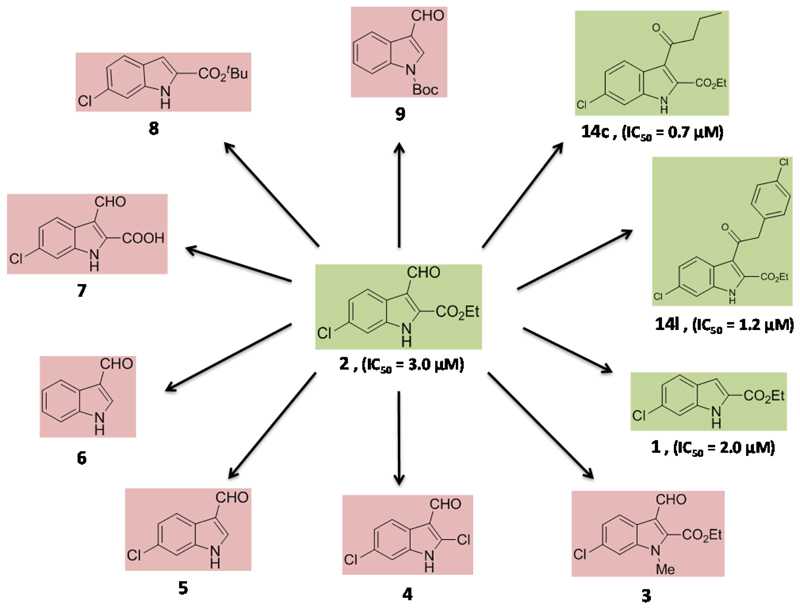
An initial SAR was derived by comparison of 1 and 2 with a selection of compounds with a closely related structure (Figure 3). We investigated the importance of the indole NH as potential hydrogen bond donor by methylation using MeI/K2CO3 to give compound 3, which inhibits the enzyme activity less than 50% at 50 μM. This indicates that the free NH is important for binding, possibly due to hydrogen bond formation. Importantly, also compounds 4, 5, 7 and 8 are inactive, which indicates that the ethyl acetate substitution in 2-position is important for binding to the enzyme.
Figure 3.
A study of structure-activity relations of compound 2. The compounds with red background do not inhibit h-15-LOX-1 in concentration below 50 μM, whereas the compounds with green background are active in the low micromolar range.
Comparison of 1 and 2 indicates that the 3-formyl substitution does not greatly change the inhibitory potency thus indicating that larger substituents could be linked to this position. This suggestion is in line with literature on related indole based inhibitors where substitution of the indole heterocycle at the 3-position markedly improved the h-15-LOX-1 inhibitory potency as observed for inhibitors like PD-146176 or tryptamine sulfonamides (Figure 1). This triggered us to synthesize compounds 14c and 14l that are elongated on 3-position. Compounds 14c and 14l were synthesized after acylation of the 6-chloro-1H-indole-2-ethyl acetate (1) with butyric anhydride and 4-chlorophenylacetyl chloride respectively via a Friedel-Crafts type reaction.36 The IC50 values were calculated to be 0.7 ± 0.3 μM for the compound 14c and 1.2 ± 0.5 μM for the compound 14l which means 2 to 2.5 folds better inhibitory potency, probably due to lipophilic interactions. The fact that these inhibitors are more potent then compound 1 and 2 indicates that substitution in this position is possible and could improve the potency of this type of inhibitors. To conclude, we confirmed that both the –NH and -ethyl acetate groups are playing a vital role in the inhibition of h-15-LOX-1 while attachment of extended substitutions in 3-position is possible with retention or even improvement of affinity. Having established these qualitative SAR we performed molecular modeling studies and structure-based design in order to provide a rationale for our findings and optimize the substitution in the 3-position.
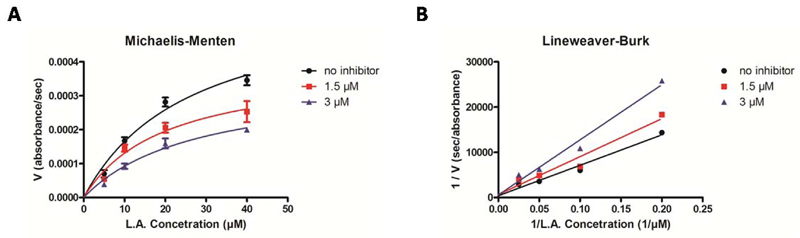
In order to establish the mechanism of h-15-LOX-1 inhibition, Michaelis-Menten enzyme kinetics analysis in presence of inhibitor 14l was performed (Figure 4A,B). The Lineweaver-Burk double reciprocal plot shows that inhibitor 14l causes an increase in the Km values, whereas the Vmax values remain more constant (Table S3), indicating competitive inhibition. Using the Cheng-Prusoff equation the binding affinities (Ki) of all the inhibitors were calculated and presented in Table 1.
Figure 4.
Steady-State kinetic characterization of h-15-LOX-1 in the presence of different concentrations of compound 14l: A) Michaelis-Menten representation and B) Lineweaver-Burk representation.
Table 1.
Compound collection of the synthesized inhibitor with IC50 and Ki values.
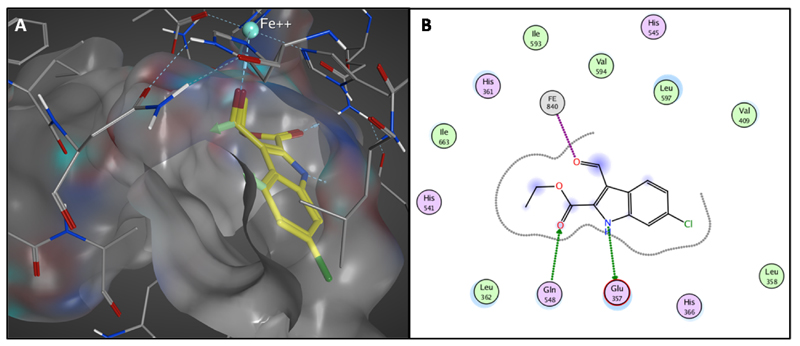
In order to link the observed SAR to structural information, compound 2 was docked in the active site of the enzyme due to the identified competitive inhibition. Due to the fact that there is no crystal structure of human 15-LOX-1 available, the docking study was performed with the crystal structure of rabbit 15-LOX (PDB: 1LOX) as determined by Gillmor et al.37 This is justified by the high sequence similarity (87%) between rabbit 15-LOX and human 15-LOX-1 as described before.32 In three of the top five highest scoring poses, inhibitor 2 binds in the same orientation in the active site of the enzyme, forming a hydrogen bond with Glu357 (Figure S4). Subsequent energy minimization in the active site provided the final binding model for inhibitor 2 (Figure 5). The modeling shows the presence of two hydrogen bonds, the first between Gln548 and the -ethyl acetate group and the second between the –NH group and the Glu357 (Figure 5B). The formyl group in 3-position gives an interaction with the iron of the enzyme and there is space for extension of the substitution in this position due to presence of free space in the substrate binding pocket (Figure 5A). These features in this model are consistent with the observed SAR thus justifying the application of this model as a basis for structure-based design to optimize the substituent in the 3-position.
Figure 5.
A) Binding orientation of inhibitor 2 in the active site of h-15-LOX-1 proposed by molecular modeling studies. The surface of the active site of the enzyme is presented with grey while the light green vectors in the compound shows the direction for possible extension of the molecule. B) 2D representation of the interactions of compound 2 with the active site of the enzyme.
Structure-based design
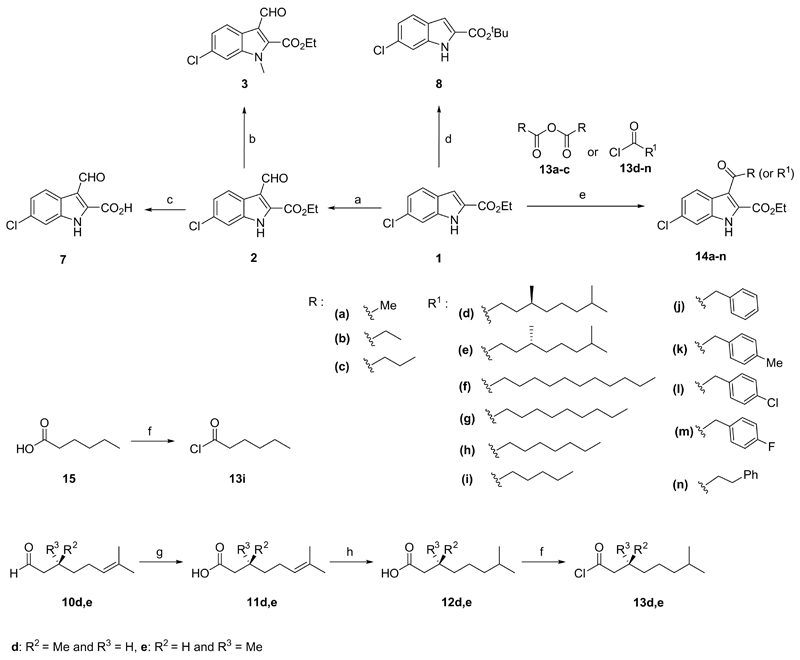
Our SAR based model indicates that optimization of the inhibitor by acylations in the indole 3-position is feasible. The free space in the enzyme binding pocket that should be addressed this way has mainly a hydrophobic character in line with the character required for binding to its natural substrate linoleic acid. Taking this into account, we synthesized a library of 14 derivatives to explore the SAR for substitution in this position. Exclusive acylation on the 3-position of inhibitor 1 as starting material was achieved using a Friedel-Craft type acylation with different acid chlorides or anhydrides. The reactions were performed in the presence of SnCl4 and CH3NO2 in DCM, yielding the corresponding compounds 14a-n in good to excellent yields (Scheme 1). All the compounds were analytically pure after simple washing steps, underscoring the facility of their synthetic preparation. The acid chlorides, which are not commercially available, were synthesized from the corresponding acids using oxalyl chloride in DCM in quantitative yields and used without further purification. The enantiomerically pure products 14d,e were prepared in a four step reaction sequence starting from the commercially available (R)-(+)- or (S)-(-)-Citronellal. Initially, the aldehydes 10d,e were oxidized to carboxylic acids 11d,e using Tollens' reagent and the double bonds were reduced with Pd/C under H2 atmosphere to give 12d,e. Next, the acid chlorides 13d,e were synthesized and coupled with the 6-chloro-1H-indole-2-ethyl acetate (1) resulting in the desired products 14d,e (Scheme 1).
Scheme 1.
General synthesis. Reagents and conditions: a) POCl3, DMF, 60 °C, overnight; b) K2CO3, CH3I, acetone, rt, 2 h; c) LiOH, EtOH/H2O, rt, 24 h; d) H2SO4, MgSO4, DCM, tBuOH, rt, overnight; e) SnCl4, CH3NO2, DCM, rt, overnight; f) (COCl)2, DCM, 0 °C for 30 min then rt for 3 h; g) Ag2O, H2O, rt, overnight; h) H2, Pd/C, EtOH, 40 °C, overnight.
All the newly synthesized compounds were subjected for IC50 determination in a h-15-LOX-1 inhibition assay and the values are shown in Table 1. We first explored inhibitors with a phenyl group in the 3-substituent in which we varied both the position of the phenyl group as well as the electronic and steric properties (14j-n). However, changing the phenyl substitution in 14j to 14m, 14l and 14k did not improve the inhibition. Also changing the position of the phenyl-group in 14j to 14n did not greatly affect the inhibition although the potency with 0.56 μM is reasonably good. We secondly explored another route of optimization with compounds 14a-i, in which the length of the lipophilic tale is varied from one to eleven carbon atoms. This series of inhibitors showed a clear structure activity relationship with optimal binding for 14g with 9 aliphatic carbons in the lipophilic tail. The clear activity dependence on the length of the aliphatic tale, from 2.3 μM to 0.4 μM (Table 1), indicates the role for lipophilic interactions in the binding, which is according to the molecular modeling study. The successful optimization of the inhibitors with an aliphatic acylation in the 3-position triggered our interest to include an aliphatic tail that was identified in a previous study32 as reflected in inhibitors 14d,e. Previously, we found that an aliphatic tail that is branching at 3,7-position improves the inhibitory activity for h-15-LOX-1.32 We included these structural features in our inhibitors as a further optimization of the aliphatic substituent in the 3-position. The resulting inhibitors 14e and 14d contain a stereogenic center and the stereochemistry being either R or S affected the affinity by a 3-fold difference in potency. The S enantiomer (14d) provided the highest potency thus reaching an IC50 value of 0.09 ± 0.03 μM, which reflects a Ki value of 0.03 ± 0.01 μM.
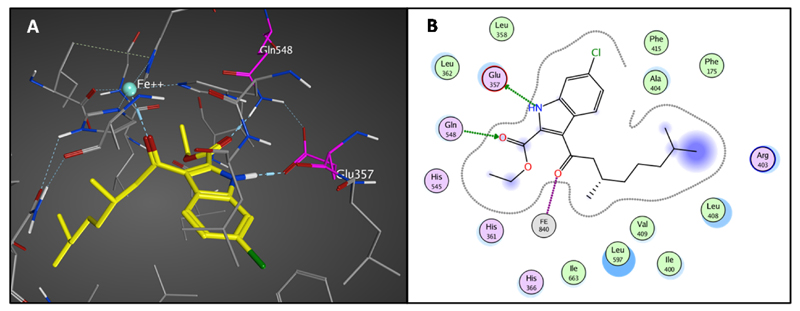
Molecular modeling of the inhibitor 14d in the active site of the enzyme using procedures applied for inhibitor 2 provided a binding configuration in which the 6-chloro-1H-indole-2-ethyl acetate moiety binds similar to the docking of inhibitor 2. The extension of the substituent in the 3-position fills the lipophilic binding pocket available in this position. The docking shows again the presence of the two hydrogen bonds with Gln548 and Glu357 and the interaction with the iron, while the branched aliphatic tail occupies the lipophilic binding pocket completely (Figure 6, S5). This SAR is in total agreement with the observed activity data in which 14g is the most potent inhibitor with a linear aliphatic tail and 14d reaches the highest affinity with a branched aliphatic tail.
Figure 6.
A) Binding of compound 14d in the active site of 15-LOX-1 after molecular modeling studies. B) 2D illustration of the interactions of compound 14d with the active site of the enzyme.
Ligand efficiency metrics
The SOS based development strategy provided 14d as an inhibitor with nanomolar potency that can be further investigated in biochemical studies. To support development of inhibitors from this stage towards successful application in drug discovery projects they need to show acceptable physicochemical properties to achieve an acceptable ADME-Tox (absorption, distribution, metabolism, excretion and toxicity) profile in vivo. In this perspective, ligand efficiency metrics rendered into a generally accepted tool to estimate the value of lead compounds in this perspective.38 Therefore, ligand efficiency metrics and indices such as ligand efficiency (LE), binding efficiency index (BEI) and the surface efficiency index (SEI) were calculated for 14d in comparison to previously described inhibitors (table 2). Ligand efficiency is defined as the binding affinity of a ligand in relation to the number of non-hydrogen atoms (HA) according the Equation LE = (1.37/HA) x pKi.39 Inhibitor 14d has 26 heavy atoms (non-hydrogen) and the Ki value is 0.03 μM, so the LE is equal to 0.40 kcal per mol per heavy atom. LE values > 0.3 kcal per mol are considered to be acceptable values for LE of drug candidates. 40 Inhibitor 14d satisfies the requirements and exceeds the values calculated for previously described inhibitors, except the inhibitor PD-146176. Another parameter is BEI, which is the binding efficiency index relating potency to molecular weight on a per kDa scale and SEI is the surface efficiency index monitoring the potency gains as related to the increase in polar surface area (PSA) referred to 100 Å2.41 The BEI and the SEI for 14d and the other inhibitors were calculated according to the Equations BEI = pKi/MW and SEI = pKi/(PSA/100 Å2 ).41 The MW of inhibitor 14d is 377 Da or 0.377 KDa, so the BEI is 19.90. The polar surface area (PSA) was estimated from MarvinSketch software to 59.16 and according to that the SEI for the compound 14d is equal to 12.71. Reference values for these parameters were obtained from mapping of the surface-binding and binding efficiency indices (SEI–BEI values) for 92 examples of marketed drugs, which provided average values for SEI of 17.9 ± 15.7 and BEI of 28.0 ± 11.5.38 The parameters for inhibitor 14d lay well with this range (Table 2). In conclusion, the ligand efficiency metrics for 14d demonstrate a very favourable binding efficiency compared to previously described inhibitors, which justifies further investigation of this molecule in biochemical studies.
Table 2.
Binding affinity and ligand efficiency metrics and indices of the previously published 15-LOX-1 inhibitors as well as the newly synthesized inhibitor 14d
| 15-LOX inhibitors | Ki (µM) | LE* | BEI | SEI |
|---|---|---|---|---|
| 14d | 0.036 | 0.40 | 19.90 | 12.71 |
| PD-146176 | 3.81 | 0.43 | 22.79 | 34.31 |
| Tryptamine sulfonamide | 0.021 | 0.30 | 15.71 | 10.78 |
| Pyrazole-based sulfamide | 0.001 | 0.32 | 15.54 | 8.26 |
| Imidazole-based sulfamide | 0.073 | 0.24 | 11.93 | 7.88 |
| ML094 | 0.010 | 0.36 | 19.12 | 12.26 |
| Indolizine | 25.0 | 0.19 | 10.47 | 9.63 |
The units for LE are (kcal/mol·HA)
Gene expression profiling
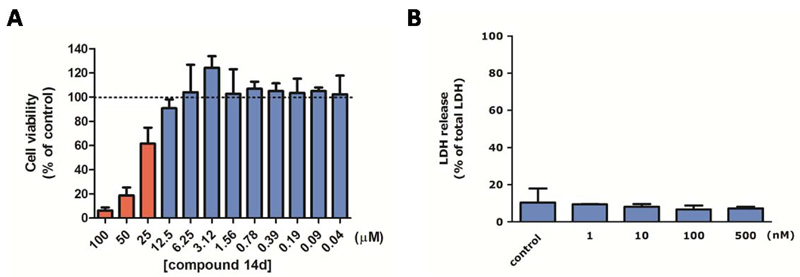
Having identified 14d as a potent inhibitor for h-15-LOX-1 we moved on to study its effects in model systems for inflammatory lung diseases using in vitro studies with RAW 264.7 macrophages and ex vivo studies with precision-cut lung slices (PCLS). As a first step the cytotoxicity of compound 14d was investigated in RAW 264.7 cells as well as in PCLS in order to identify the concentration range in which this inhibitor does not affect cell viability. The inhibitor 14d did not inhibit cell viability at concentration below 12.5 μM in RAW 264.7 cells (Figure 7A) as determined using an MTS assay. The viability of PCLS was determined by measuring the release of lactate dehydrogenase (LDH) into the medium upon incubation with different concentrations of inhibitor 14d. The LDH release demonstrated that inhibitor 14d is not affecting the PCLS viability at concentrations of 500 nM and lower (Figure 7B).
Figure 7.
A) LC50 graph after MTS assay. The compound 14d was incubated with RAW 264.7 cells at concentrations from 0.05 μM to 100 μM. Red bars are statistically different compared to non-treated control cells. B) Viability of precision-cut lung slices (PCLS) measured by release of lactate dehydrogenase (LDH) into the incubation medium. PCLS were treated with various concentrations of 14d (1, 10, 100 and 500 nM) and 10 μM linoleic acid for 20 h. Maximal LDH content of the PCLS was determined by lysis with 1% Triton X-100. LDH release was plotted relative to maximal LDH. Data are presented as mean values ± SD of 3 independent experiments.
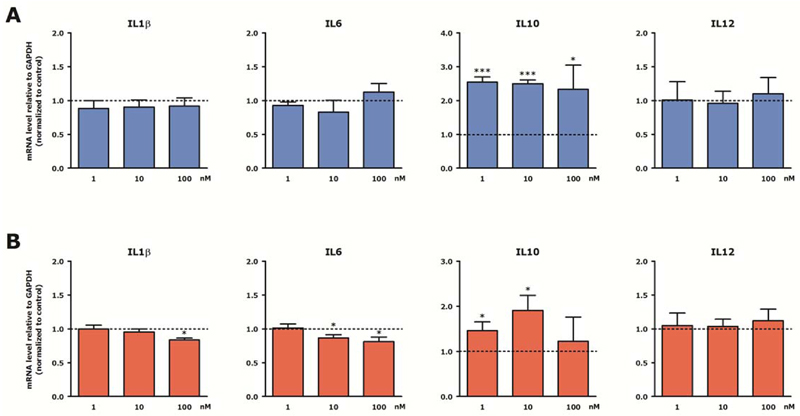
Having identified the non-toxic concentration for inhibitor 14d its influence on gene expression was investigated. In these models LPS/INFγ was applied as inflammatory stimulus and the influence of 14d on the expression of pro-inflammatory genes IL-1β, IL-6 or IL-12 and the antiinflammatory gene IL-10 was investigated. Since the cellular concentration of free fatty acids (preferred substrate of mammalian LOXs) is rather low, we supplemented the culture medium with linoleic acid (C18:Δ2, n – 6) which is the most abundant polyenoic fatty acid in mammalian cells and serves as a natural substrate for 15-LOX-1. Linoleic acid itself did not alter the expression of IL-1β, IL-6, IL-10 and IL-12 in RAW 264.7 cells and PCLS (data not shown). In LPS/INFγ-stimulated RAW 264.7 macrophages incubation with the h-15-LOX-1 inhibitor 14d did not change the expression of the genes IL-1β, IL-6, and IL-12 but provided a more then 2-fold increase of the IL-10 expression at concentrations as low as 1 nM (Figure 8A). Since IL-10 has anti-inflammatory properties, and is connected with the anti-inflammatory M2 macrophage phenotype, the observed changes in gene expression are indicative for a potential anti-inflammatory effect. Next, the effect of 14d on the expression of LPS/INFγ-induced cytokines in PCLS was assessed. In line with the results obtained in macrophages the expression of IL-10 is upregulated in PCLS. In addition, there is a small but significant downregulation of the pro-inflammatory genes IL-1β and IL-6 (Figure 8B). Remarkably, the expression of IL-12 remained unchanged while others have reported that h-15-LOX-1 regulates the production of IL-12 in macrophages.15 The effect on IL-10 expression is remarkable because the important role of IL-10 in inflammatory lung diseases has been previously highlighted in a study that showed diminished IL-10 production in lung tissue of COPD patients after LPS stimulation as compared to lung tissue of patients with normal lung function.42 In addition, it has been demonstrated that the level of IL-10 and IL-10-positive macrophages in sputum of COPD patients and healthy smokers was decreased as compared to healthy non-smokers.43 This suggests that macrophages in COPD have lost their ability to produce anti-inflammatory cytokines like IL-10 and are therefore unable to effectively dampen inflammation. Our newly developed inhibitor 14b could, among others, alleviate the lack of IL10 production in these diseases. Taken together, we demonstrate that the effects observed for treatment with inhibitor 14d are in agreement with an anti-inflammatory effect that could be beneficial in inflammatory lung diseases. This sets the stage for further investigation and optimization of inhibitor 14d for in vivo applications towards the development of novel therapeutics for inflammatory lung diseases.
Figure 8.
Effect of h-15-LOX-1 inhibition on interleukin expression in RAW 264.7 cells and precision-cut lung slices (PCLS). Stimulated RAW 264.7 cells (A) and PCLS (B) were subjected to 14d in combination with 10 μM linoleic acid for 20 h and stimulated with LPS/IFNγ for the last 4 h. Subsequently, RAW 264.7 cells and PCLS were lysed and gene expression was assessed by RT-qPCR and expressed as fold change compared to control (LPS/IFNγ/linoleic acid-treated) group. Data are presented as mean values ± SEM of 4-6 independent experiments. * p < 0.05; *** p < 0.001 compared to control.
Conclusions
In this study, we employed an efficient strategy to identify structurally new inhibitors for the enzyme h-15-LOX-1, which is an emerging drug target in various inflammatory diseases. Our approach started with a substitution oriented fragment screening (SOS) of a focused fragment library containing diversely substituted heterocycles. After the discovery of four hits, two inhibitors were selected to derive structure activity relationships and subjected to enzyme kinetic analysis, which indicated competitive inhibition. These observations were applied to support a molecular modeling study proposing a binding configuration in the active site of the enzyme h-15-LOX-1. Based on this model the substitution of the inhibitor was further optimized using structure-based design to provide inhibitor 14d with a Ki of 36 nM and good ligand efficiency metrics. This inhibitor was evaluated in cell-based studies using RAW 264.7 macrophages and ex vivo studies in mouse precision-cut lung slices. The inhibitor provided an increase in the expression of IL-10 both in macrophages and mouse lung tissue. In the future, these inhibitors could be used as lead compounds for further optimization or as a starting point of drug discovery efforts targeting h-15-LOX-1.
Experimental Section
Chemistry
General
The solvent and reagents were purchased from Sigma-Aldrich and Acros chemicals and were used without further purification unless otherwise noted. Reactions were monitored by thin layer chromatography (TLC). Merck silica gel 60 F254 plates were used and spots were detected under UV light or after staining with potassium permanganate for the non UV-active compounds. MP Ecochrom silica 32-63, 60Å was used for flash column chromatography. 1H NMR (500 MHz) and 13C NMR (126 MHz) spectra were recorded with a Bruker Avance 4-channel NMR Spectrometer with TXI probe. Chemical shifts were referenced to the residual proton and carbon signal of the deuterated solvent CDCl3: δ = 7.26 ppm (1H) and 77.05 ppm (13C). The following abbreviations were used for spin multiplicity: s = singlet, br s = broad singlet, d = doublet, t = triplet, q = quartet, quin = quintet, dd = double of doublets, ddd = double of doublet of doublets, m = multiplet. Fourier Transform Mass Spectrometry (FTMS) and electrospray ionization (ESI) were recorded on an Applied Biosystems/SCIEX API3000-triple quadrupole mass spectrometer.
Synthetic procedure 1: indole 3-acylation
Indole (1.0 mmol) was dissolved in 4.0 mL DCM under nitrogen atmosphere and the solution was cooled down to 0°C. To the stirring solution, SnCl4 (1.2 mmol) was added in a single portion via syringe. After the ice bath was removed, the mixture was stirred at room temperature for 30 min and then the acyl chloride or anhydride (1.0 mmol) was added in small portions, followed by the addition of nitromethane (3.0 mL). The reaction mixture was stirred at room temperature overnight. After being quenched with cold water (10 mL), the organic material extracted with EtOAc (2 x 15 mL), dried over MgSO4, filtered and concentrated under reduced pressure.
Ethyl 3-acetyl-6-chloro-1H-indole-2-carboxylate (14a, DN397)
The final product was obtained after 3-acylation of indole 1 using the synthetic procedure 1. Pink solid, yield 75%. 1H NMR (500 MHz, CDCl3) δ 9.28 (br s, 1H), 8.00 (d, J = 10.0 Hz, 1H), 7.41 (d, J = 0.5 Hz, 1H), 7.22 (dd, J = 8.7, 1.7 Hz, 1H), 4.48 (q, J = 7.2 Hz, 2H), 2.75 (s, 3H), 1.45 (t, J = 7.2 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 198.3, 160.8, 135.4, 132.3, 127.3, 125.4, 124.0, 123.9, 121.7, 111.8, 62.5, 32.1, 14.3. MS (ESI): m/z 264.0 [M-H]+.
Ethyl 6-chloro-3-propionyl-1H-indole-2-carboxylate (14b, N239)
The final product was obtained after 3-acylation of indole 1 using the synthetic procedure 1. Orange solid, yield 82%. 1H NMR (500 MHz, CDCl3) δ 9.17 (s, 1H), 7.86 (d, J = 8.7 Hz, 1H), 7.41 (d, J = 1.6 Hz, 1H), 7.20 (dd, J = 8.7, 1.6 Hz, 1H), 4.45 (q, J = 7.1 Hz, 2H), 3.08 (q, J = 7.3 Hz, 2H), 1.43 (t, J = 7.1 Hz, 3H), 1.23 (t, J = 7.3 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 201.7, 160.6, 135.3, 132.3, 126.1, 125.2, 123.7, 123.6, 121.9, 111.5, 62.1, 37.3, 14.2, 8.6. MS (ESI): m/z 302.1 [M+Na]+.
Ethyl 3-butyryl-6-chloro-1H-indole-2-carboxylate (14c, N238)
The final product was obtained after 3-acylation of indole 1 using the synthetic procedure 1. Orange solid, yield 83%. 1H NMR (500 MHz, CDCl3) δ 9.16 (s, 1H), 7.85 (d, J = 8.7 Hz, 1H), 7.41 (d, J = 1.8 Hz, 1H), 7.20 (dd, J = 8.7, 1.8 Hz, 1H), 4.46 (q, J = 7.1 Hz, 2H), 3.04 (t, J = 7.4 Hz, 2H), 1.76 (m, 2H), 1.43 (t, J = 7.1 Hz, 3H), 0.98 (t, J = 7.4 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 201.3, 160.6, 135.2, 132.2, 126.1, 125.2, 123.6, 123.5, 122.2, 111.5, 62.1, 46.1, 18.2, 14.3, 13.9. MS (ESI): m/z 316.1 [M+Na]+.
Synthetic procedure 2: oxidation with Tollens' reagent
Fresh silver oxide was prepared by dropwise addition of a solution of AgNO3 (2.3 mmol) in water (4.0 mL) to a stirred solution of NaOH (4.9 mmol) in water (4.0 mL) at 0 °C and further stirring for 30 min in the dark while a brown precipitate formed. Under continuous stirring, the aldehyde (1.0 mmol) was added dropwise and the reaction was stirred overnight. The resulting suspension was filtered, washed with hot water (2 × 10 mL) and the clear solution was acidified with aqueous HCl (4N) to pH<1. The solution was extracted with Et2O (3 × 20 mL), dried over MgSO4, filtered and concentrated under reduced pressure.
Synthetic procedure 3: reduction of the alkene
The alkene (1.0 mmol) and Pd/C (10%) (0.10 mmol) were suspended in EtOH (10 mL). The suspension was stirred under H2 atmosphere at 40 °C overnight. Subsequently, the mixture was filtered through Celite and after that, the solvent was evaporated under reduced pressure.
Synthetic procedure 4: conversion of carboxylic acids to acid chlorides
The acid was dissolved in dry DCM (5.0 mL) under nitrogen atmosphere and the solution was cooled down to 0 °C. A solution of oxalyl chloride (1.2 mmol) in dry DCM (4.0 mL) was added dropwise and the solution was stirred under nitrogen atmosphere at room temperature for an additional 3 h. The solvent and the excess of oxalyl chloride were evaporated under reduced pressure yielding the product as yellow oil.
(S)-3,7-dimethyloct-6-enoic acid (11d, N240)
The final product was obtained after oxidation of (S)-3,7-dimethyloct-6-enal with Tollens' reagent, using the synthetic procedure 2. Yellow oil, yield 94%. 1H NMR (500 MHz, CDCl3) δ 10.40 (s, 1H), 5.08 (t, J = 7.1 Hz, 1H), 2.35 (dd, J = 15.0, 5.8 Hz, 1H), 2.14 (dd, J = 15.0, 8.3 Hz, 1H), 2.07 – 1.92 (m, 3H), 1.67 (s, 3H), 1.59 (s, 3H), 1.37 (m, 1H), 1.24 (m, 1H), 0.97 (d, J = 6.7 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 179.9, 131.6, 124.2, 41.6, 36.7, 29.8, 25.7, 25.4, 19.6, 17.6.
(S)-3,7-dimethyloctanoic acid (12d, N242)
The final product was obtained after reduction of the alkene of (S)-3,7-dimethyloct-6-enoic acid using synthetic procedure 3. Yellow oil, quantitative yield. 1H NMR (500 MHz, CDCl3) δ 10.68 (s, 1H), 2.33 (dd, J = 15.0, 5.9 Hz, 1H), 2.13 (dd, J = 15.0, 8.2 Hz, 1H), 1.95 (m, 1H), 1.51 (m, 1H), 1.34 – 1.12 (m, 6H), 0.95 (d, J = 6.8 Hz, 3H), 0.85 (d, J = 7.0 Hz, 6H). 13C NMR (126 MHz, CDCl3) δ 180.0, 41.7, 39.0, 36.9, 30.2, 27.9, 24.7, 22.7, 22.6, 19.7.
(R) or (S)-ethyl 6-chloro-3-(3,7-dimethyloctanoyl)-1H-indole-2-carboxylate (14e (N246), 14d (N247))
The final product was obtained after 3-acylation of indole 1 using the synthetic procedure 1. White solid, yield 89%. 1H NMR (500 MHz, CDCl3) δ 9.20 (s, 1H), 7.84 (d, J = 8.7 Hz, 1H), 7.41 (s, 1H), 7.20 (d, J = 8.7 Hz, 1H), 4.55 (q, J = 7.1 Hz, 2H), 3.09 (dd, J = 15.8, 5.7 Hz, 1H), 2.88 (dd, J = 15.8, 8.2 Hz, 1H), 2.11 (m, 1H), 1.43 (t, J = 7.1 Hz, 3H), 1.35 – 1.06 (m, 7H), 0.94 (d, J = 6.7 Hz, 3H), 0.83 (d, J = 6.7 Hz, 6H). 13C NMR (126 MHz, CDCl3) δ 201.3, 160.6, 135.3, 132.2, 126.0, 125.2, 123.6, 123.5, 122.5, 111.6, 62.1, 51.6, 39.1, 37.3, 30.4, 27.9, 24.8, 22.7, 22.6, 20.0, 14.3. MS (ESI): m/z 400.2 [M+Na]+.
Ethyl 6-chloro-3-dodecanoyl-1H-indole-2-carboxylate (14f, DN433)
The final product was obtained after 3-acylation of indole 1 using the synthetic procedure 1. Red solid, yield 91%. 1H NMR (500 MHz, CDCl3) δ 9.28 (br s, 1H), 7.84 (d, J = 8.7 Hz, 1H), 7.41 (d, J = 1.8 Hz, 1H), 7.20 (dd, J = 8.7, 1.8 Hz, 1H), 4.46 (q, J = 7.1 Hz, 2H), 3.06 (t, J = 7.6 Hz, 2H), 1.75-1.71 (m, 2H), 1.36 (t, J = 7.1 Hz, 3H), 1.33-1.24 (m, 16H), 0.89-0.86 (m, 3H). 13C NMR (126 MHz, CDCl3) δ 201.7, 160.9, 135.5, 132.4, 126.2, 125.4, (2x) 123.7, 122.4, 111.7, 62.3, 44.5, 32.1, 29.8, 29.7, 29.5, 24.9, 22.9, 14.4, 14.3. MS (ESI): m/z 428.2 [M+Na]+.
Ethyl 6-chloro-3-decanoyl-1H-indole-2-carboxylate (14g, DN432)
The final product was obtained after 3-acylation of indole 1 using synthetic procedure 1. Red solid, yield 91%. 1H NMR (500 MHz, CDCl3) δ 9.25 (br s, 1H), 7.84 (d, J = 8.7 Hz, 1H), 7.41 (d, J = 1.8 Hz, 1H), 7.21 (dd, J = 8.7, 1.8 Hz, 1H), 4.45 (q, J = 7.2 Hz, 2H), 3.05 (t, J = 7.5 Hz, 2H), 1.74-1.71 (m, 2H), 1.43 (t, J = 7.2 Hz, 3H), 1.33-1.24 (m, 14H), 0.87 (t, J = 7.1 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 201.7, 160.8, 135.5, 132.4, 126.2, 125.4, 123.8, 123.7, 122.4, 111.7, 62.3, 44.5, 32.1, 29.6, 29.6, 29.5, 29.4, 24.9, 22.9, 14.4, 14.3. MS (ESI): m/z 400.2 [M+Na]+.
Ethyl 6-chloro-3-octanoyl-1H-indole-2-carboxylate (14h, N214)
The final product was obtained after 3-acylation of indole 1 using synthetic procedure 1. Orange solid, yield 93%. 1H NMR (500 MHz, CDCl3) δ 9.25 (s, 1H), 7.84 (d, J = 8.7 Hz, 1H), 7.40 (d, J = 1.8 Hz, 1H), 7.19 (dd, J = 8.7, 1.8 Hz, 1H), 4.45 (q, J = 7.2 Hz, 2H), 3.05 (t, J = 7.5 Hz, 2H), 1.74 – 1.68 (m, 2H), 1.43 (t, J = 7.2 Hz, 3H), 1.37 – 1.23 (m, 8H), 0.86 (t, J = 6.7 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 201.4, 160.6, 135.3, 132.2, 126.0, 125.2, 123.6, 123.5, 122.2, 111.5, 62.1, 44.2, 31.7, 29.3, 29.2, 24.7, 22.6, 14.2, 14.1. MS (ESI): m/z 372.2 [M+Na]+.
Ethyl 6-chloro-3-hexanoyl-1H-indole-2-carboxylate (14i, DN441)
The final product was obtained after 3-acylation of indole 1 using the synthetic procedure 1. Yellow solid, yield 95%. 1H NMR (500 MHz, CDCl3) δ 9.17 (br s, 1H), 7.85 (d, J = 8.7 Hz, 1H), 7.41 (d, J = 1.8 Hz, 1H), 7.20 (dd, J = 8.7, 1.8 Hz, 1H), 4.45 (q, J = 7.1 Hz, 2H), 3.06 (t, J = 7.4 Hz, 2H), 1.76-1.70 (m, 2H), 1.37 (t, J = 7.1 Hz, 3H), 1.33-1.26 (m, 4H), 0.90-0.88 (m, 3H). 13C NMR (126 MHz, CDCl3) δ 201.4, 160.8, 135.4, 132.4, 126.2, 125.5, 123.9, 123.7, 122.4, 111.7, 62.3, 44.4, 31.7, 24.6, 24.6, 22.8, 14.4. MS (ESI): m/z 322.1 [M+H]+.
Ethyl 6-chloro-3-(2-phenylacetyl)-1H-indole-2-carboxylate (14j, DN309)
The final product was obtained after 3-acylation of indole 1 using synthetic procedure 1. Yellow solid, yield 99%. 1H NMR (500 MHz, CDCl3) δ 9.49 (s, 1H), 7.78 (d, J = 8.7 Hz, 1H), 7.36 (s, 1H), 7.28 (d, J = 7.2 Hz, 2H), 7.27-7.20 (m, 3H), 7.15 (d, J = 8.7, 1.7 Hz, 1H), 4.48 (q, J = 7.1 Hz, 2H), 4.45 (s, 2H), 1.43 (t, J = 7.1 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 198.3, 160.5, 135.2, 134.7, 133.2, 129.6, 128.5, 126.8, 126.4, 125.4, (2x)123.6, 123.5, 121.5, 111.6, 62.2, 50.5, 14.3. MS (ESI): m/z 340.2 [M-H]+.
Ethyl 6-chloro-3-(2-(p-tolyl)acetyl)-1H-indole-2-carboxylate (14k, N225)
The final product was obtained after 3-acylation of indole 1 using synthetic procedure 1. White solid, yield 94%. 1H NMR (500 MHz, CDCl3) δ 9.18 (s, 1H), 7.60 (d, J = 8.6 Hz, 1H), 7.42 (s, 1H), 7.21 – 7.11 (m, 5H), 4.42 (q, J = 7.1 Hz, 2H), 3.62 (s, 2H), 2.33 (s, 3H), 1.42 (t, J = 7.1 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 177.4, 162.2, 137.2, 137.0, 131.3, 130.3, 129.4, 129.2, 128.1, 126.0, 123.5, 121.8, 111.8, 108.7, 61.4, 40.6, 21.1, 14.4. MS (ESI): m/z 378.1 [M+Na]+.
Ethyl 6-chloro-3-(2-(4-chlorophenyl)acetyl)-1H-indole-2-carboxylate (14l, DN522)
The final product was obtained after 3-acylation of indole 1 using synthetic procedure 1. Yellow solid, yield 95%. 1H NMR (500 MHz, DMSO-d6) δ 12.5 (br s, 1H), 7.79 (d, J = 8.4 Hz, 1H), 7.53 (s, 1H), 7.35 (d, J = 7.8 Hz, 2H), 7.25 (d, J = 7.8 Hz, 2H), 7.21 (d, J = 8.4 Hz, 1H), 4.44 (q, J = 6.6 Hz, 2H), 4.37 (s, 2H), 1.36 (t, J = 6.6 Hz, 3H). 13C NMR (126 MHz, DMSO-d6) δ 197.1, 161.1, 136.2, 134.8, 132.0, 131.7, 130.4, 128.8, 128.6, 125.1, 123.7, 123.2, 120.1, 112.7, 62.3, 48.9, 14.5. MS (ESI): m/z 374.0 [M-H]+.
Ethyl 6-chloro-3-(2-(4-fluorophenyl)acetyl)-1H-indole-2-carboxylate (14m, DN312)
The final product was obtained after 3-acylation of indole 1 using the synthetic procedure 1. Yellow solid, yield 95%. 1H NMR (500 MHz, CDCl3) δ 9.20 (br s, 1H), 7.81 (d, J = 8.0 Hz, 1H), 7.40 (s, 1H), 7.23-7.18 (m, 3H), 6.98-7.01 (m, 2H), 4.50 (q, J = 7.2 Hz, 2H), 4.43 (s, 2H), 1.46 (t, J = 7.2 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 198.3, 160.6, 135.4, 132.7, (2x) 131.4, 126.6, 125.7, 124.0, 123.9, 115.6, 115.5, 111.7, 62.5, 49.6, 40.1, 14.5. MS (ESI): m/z 358.0 [M-H]+.
Ethyl 6-chloro-3-(3-phenylpropanoyl)-1H-indole-2-carboxylate (14n, DN427)
The final product was obtained after 3-acylation of indole 1 using the synthetic procedure 1. Red solid, yield 96%. 1H NMR (500 MHz, CDCl3) δ 9.20 (br s, 1H), 7.83 (d, J = 8.8 Hz, 1H), 7.39 (s, 1H), 7.28-7.17 (m, 6H), 4.41 (q, J = 7.1 Hz, 2H), 3.42 (t, J = 7.7 Hz, 2H), 3.09 (t, J = 7.7 Hz, 2H), 1.38 (t, J = 7.1 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 200.1, 160.7, 141.4, 135.4, 134.8, 132.5, (2x) 128.60, 125.5, 124.0, (2x) 123.8, 111.7, 62.4, 45.7, 30.8, 14.4. MS (ESI): m/z 378.1 [M+Na]+.
Ethyl 6-chloro-1H-indole-2-carboxylate (1).44,45
White solid, yield 75%. 1H NMR (500 MHz, CDCl3) δ 8.89 (s, 1H), 7.60 (d, J = 8.7 Hz, 1H), 7.42 (s, 1H), 7.20 (d, J = 8.7 Hz, 1H), 7.13 (dd, J = 8.7, 0.5 Hz, 1H), 4.41 (q, J = 7.1 Hz, 2H), 1.43 (t, J = 7.1 Hz, 3H).
Ethyl 6-chloro-3-formyl-1H-indole-2-carboxylate (2).46,47
Yellow solid, yield 85%. 1H NMR (500 MHz, CDCl3) δ 10.7 (s, 1H), 9.34 (br s, 1H), 8.38 (d, J = 8.7 Hz, 1H), 7.45 (s, 1H), 7.30 (d, J = 8.7 Hz, 1H), 4.52 (q, J = 7.1 Hz, 2H), 1.46 (t, J = 7.1 Hz, 3H).
Ethyl 6-chloro-3-formyl-1-methyl-1H-indole-2-carboxylate (3).46
Yellow solid, yield 95%. 1H NMR (500 MHz, CDCl3) δ 10.6 (s, 1H), 8.43 (d, J = 8.7 Hz, 1H), 7.43 (s, 1H), 7.32 (d, J = 8.7 Hz, 1H), 4.53 (q, J = 7.1 Hz, 2H), 1.48 (t, J = 7.1 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 188.4, 161.0, 134.8, 132.6, (2x) 125.1, 123.2, 120.0, (2x) 110.6, 62.5, 32.8, 14.5.
6-chloro-3-formyl-1H-indole-2-carboxylic acid (7).46
Pink solid, yield 75%. 1H NMR (500 MHz, CDCl3) δ 10.7 (s, 1H), 8.27 (d, J = 8.7 Hz, 1H), 7.57 (d, J = 0.5 Hz, 1H), 7.23 (dd, J = 8.77, 0.5 Hz, 1H).
tert-butyl 6-chloro-1H-indole-2-carboxylate (8).48
Pink solid, yield 75%. 1H NMR (500 MHz, CDCl3) δ 7.57 (d, J = 10 Hz, 1H), 7.41 (s, 1H), 7.11-7.09 (m, 2H), 1.62 (s, 9H).
Enzyme inhibition studies
Human 15-LOX-1 screening UV assay
The h-15-LOX-1 enzyme was expressed and purified as described before.32 The h-15-LOX-1 activity was determined by the conversion of linoleic acid to 13S-hydroperoxy-9Z,11E-octadecadienoic acid (13(S)-HpODE) (λmax of 234 nm). The conversion rate was followed by UV-absorbance at 234 nm over time. The linear increase in absorbance was used to determine the enzyme activity. That conversion rate was evaluated at the linear part of the plot before the substrate depletion. The linear part usually covers the first 10 to 16 minutes depending on the enzyme concentration. The optimum concentration of h-15-LOX-1 was determined by an enzyme activity assay (x 640 times dilution). The raw data of the conversion of linoleic acid in present (Positive Control) or absent of the enzyme (Blank) following with UV-absorbance assay at 234 nm over time are presenting in the Figure S1.
The assay buffer consists of 25 mM HEPES titrated to pH 7.5 using a concentrated aqueous solution of NaOH. The substrate, linoleic acid (LA) (Sigma Aldrich, L1376) was diluted in ethanol to 500 µM. The inhibitor (100 mM in DMSO) was diluted with the assay buffer to 125 µM. The inhibitor solution of 80 µL was mixed with 60 µL assay buffer, 50 µL of 1:160 enzyme solution and incubated for 10 min at rt. After incubation, 10 µL of 500 µM LA was added to the mixture which provided a mixture with a final dilution of the enzyme of 1:640 and 25 µM LA. The linear absorbance increase in absence of the inhibitor was set as 100%, whereas the absorbance increase in absence of the enzyme was set to 0%. All experiments were performed in triplicate and the average triplicate values and their standard deviations are plotted. The half maximal inhibitor concentration (IC50) of the inhibitors for h-15-LOX-1 was determined using the same assay. The inhibitor was diluted with assay buffer. Using a serial dilution, the desired concentrations of the inhibitor were obtained ranging from 0.09-50 µM or 0.009-20 µM depending of the inhibitory potency. The data analysis was performed using Microsoft Excel Professional Plus 2013 and GraphPad Prism 5.01.
Michaelis-Menten Enzyme Kinetics
In this kinetic study, 25 mM HEPES buffer, pH 7.5, was used as an assay buffer. The enzyme was diluted 1:160 with assay buffer while linoleic acid (Sigma Aldrich, L1376-1G) was diluted with EtOH to 4 mM. LA concentrations ranging from 0.05 to 3 mM were made using the 4 mM LA stock solution. The enzyme activity was measured in the absence or presence of fixed concentrations of the inhibitor (0 µM, 1.5 µM and 3.0 µM). 50 µL of the enzyme solution was mixed with 80 µL of inhibitor (25 µM or 50 µM), 60 µL assay buffer and incubated for 10 min at rt. In absence of the inhibitor, the amount of inhibitor is substituted with assay buffer. Subsequently, 10 µL of LA solution ranging from 0.05 to 4 mM was added to the mixture to provide a mixture with a final dilution of the enzyme of 1:640. The mixtures were immediately measured after 5 sec of mixing the enzyme with the inhibitor and substrate for 16 min. These experiments were performed in triplicate. The reaction velocities (v) were plotted against the substrate concentrations in a Michaelis-Menten plot and the Km and Vmax in the presence of the inhibitor were derived. The reciprocal of the velocities were taken and plotted against the reciprocal of the LA concentrations in a Lineweaver-Burk plot. Substrate inhibition was obtained in concentration above 60 µM. All experiments were performed in triplicate and the average triplicate values and their standard deviations are plotted. The data analysis was performed using Microsoft Excel Professional Plus 2013 and GraphPad Prism 5.01.
Gene expression profiling
Animals
C57bl/6 male mice (weight 20-25 g; age 8-10 weeks) were purchased from Harlan (Zeist, the Netherlands). Animals were maintained on mouse chow and tap water ad libitum in a humidity- and temperature-controlled room at 24°C with a 12 h light/dark cycle. All experiments were performed according to national guidelines and upon approval of the experimental procedures by the local Animal Care and Use committee of Groningen University, DEC number 6962A. Mice were randomly assigned to the experiments.
Precision-cut lung slices
Mouse precision-cut lung slices (PCLS) were prepared as previously described for guinea pig with the following modifications.49 Male mice were anesthetized by subcutaneous injection of ketamin (75 mg/kg, Alfasan, Woerden, the Netherlands) and dexdomitor (0.5 mg/kg, Orion Pharma, Mechelen, Belgium). Subsequently, the trachea was cannulated and the animal was exsanguinated by cutting the jugular vein, after which the lungs were filled trough the cannula with 1.5 ml low melting-point agarose solution (1.5% final concentration (Gerbu Biotechnik GmbH, Wieblingen, Germany) in CaCl2 (0.9 mM), MgSO4 (0.4 mM), KCl (2.7 mM), NaCl (58.2 mM), NaH2PO4 (0.6 mM), glucose (8.4 mM), NaHCO3 (13 mM), Hepes (12.6 mM, Gibco® by Life Technologies, Bleiswijk, the Netherlands), sodium pyruvate (0.5 mM, GE Healthcare Life Sciences, Eindhoven, the Netherlands), glutamine (1 mM, Gibco® by Life Technologies), MEM-amino acid mixture (1:50, Gibco® by Life Technologies) and MEM-vitamins mixture (1:100, Gibco® by Life Technologies), pH 7.2). The lungs were placed on ice for 15 min to solidify the agarose for slicing. The lobes were separated and tissue cores were prepared of the individual lobes, after which the lobes were sliced at a thickness of 250 μm in medium composed of CaCl2 (1.8 mM), MgSO4 (0.8 mM), KCl (5.4 mM), NaCl (116.4 mM), NaH2PO4 (1.2 mM), glucose (16.7 mM), NaHCO3 (26.1 mM), Hepes (25.2 mM), pH 7.2, using a tissue slicer (Compresstome™ VF-300 microtome, Precisionary Instruments, San Jose, CA, USA). Tissue slices were incubated at 37 °C in a humid atmosphere under 5% CO2/95% air. In order to remove the agarose and cell debris from the tissue, slices were washed every 30 min (four times in total) in medium composed of CaCl2 (1.8 mM), MgSO4 (0.8 mM), KCl (5.4 mM), NaCl (116.4 mM), NaH2PO4 (1.2 mM), glucose (16.7 mM), NaHCO3 (26.1 mM), Hepes (25.2 mM), sodium pyruvate (1 mM), glutamine (2 mM), MEM-amino acid mixture (1:50), MEM-vitamins mixture (1:100), penicillin (100 U/ml, Gibco® by Life Technologies) and streptomycin (100 μg/ml, Gibco® by Life Technologies), pH 7.2. Chemicals to prepare the media described above were obtained from Sigma-Aldrich (Zwijndrecht, the Netherlands) unless stated otherwise, and were of analytical grade.
Treatment of lung slices
PCLS were incubated in Dulbecco’s Modification of Eagle’s Medium (DMEM, Gibco® by Life Technologies) supplemented with sodium pyruvate (1 mM), MEM non-essential amino acid mixture (1:100, Gibco® by Life Technologies), gentamycin (45 μg/ml, Gibco® by Life Technologies), penicillin (100 U/ml), streptomycin (100 μg/ml) and amphotericin B (1.5 μg/ml, Gibco® by Life Technologies). Slices were cultured at 37 °C in a humidified atmosphere under 5% CO2/95% air in 12-well tissue culture plates (Costar Europe, Badhoevedorp, the Netherlands), using 3 slices per well. Slices were treated with the LOX inhibitor (LOXi) 14d at a final concentration of 1-500 nM for 20 h, and were co-incubated with 10 μM Linoleic acid (L1376; Sigma Aldrich). The last 4 h of the experiments tissue slices were stimulated with 10 ng/ml lipopolysaccharide (LPS, Escherichia coli, serotype 0111:B4; Sigma-Aldrich) and 10 ng/ml interferon gamma (IFNγ, cat.#315-05; PeproTech, Hamburg, Germany).
Assessment of tissue viability using lactate dehydrogenase
To assess the viability of the PCLS subjected to increasing concentrations 14d, the amount of lactate dehydrogenase (LDH) released from the tissue slices into the incubation medium was analyzed. Maximal LDH release was determined by lysing 3 slices with 1% Triton X-100 for 30 min at 37 °C at the start of the experiments. Supernatants were stored at -80 °C. LDH release was determined using an assay form Roche Diagnostics (Mannheim, Germany), and was measured using a Hitachi automatic analyzer (Modular Analytics, Roche Diagnostics). LDH release from the PCLS into the incubation medium was plotted relative to maximal LDH release.
Cell culture and LOXi treatment
RAW 264.7 murine macrophage cells were obtained from ATCC (Wesel, Germany) and cultured in plastic tissue culture plates or flasks at 37 °C under 5% CO2/95% air in DMEM + GlutaMAX™ (Gibco® by Life Technologies) supplemented with 10% (v/v) heat inactivated fetal bovine serum (FBS; Invitrogen, Breda, the Netherlands), 2 mM additional GlutaMAX™ (Gibco® by Life Technologies), 100 U/ml penicillin and 100 µg/ml streptomycin. RAW 264.7 cells were used between passage 7 and 16.
To obtain identical cell density at the start of the experiment, RAW 264.7 cells were seeded at 20,000 cells/cm2 in 12-well tissue culture plates one day prior to the experiment. Cells were treated with the LOXi 14d(final concentration 1-500 nM) for 20 h, and were co-incubated with 10 μM Linoleic acid. Cells were stimulated with 10 ng/ml LPS and IFNγ for the last 4 h of the experiment.
Gene expression analysis by RT-qPCR
RAW 264.7 cells and PCLS were washed twice with ice-cold DPBS (Gibco® by Life Technologies) and total RNA was isolated from RAW 264.7 cells using the SV Total RNA Isolation System (Promega, Leiden, the Netherlands) and from PCLS using the Maxwell® 16 LEV simplyRNA Tissue Kit (Promega), both according to the protocol of the manufacturer. RNA integrity was determined by 28S/18S ratio detection on an agarose gel, which was consistently found intact. For gene expression analysis, RNA was reverse transcribed using a reverse transcription kit (Promega). Subsequently, 10 ng of cDNA was applied for each real-time PCR, which was performed on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Nieuwerkerk a/d IJssel, the Netherlands). The primers for IL-1β (Mm00434228_m1), IL-6 (Mm00446190_m1) IL-10 (Mm00439614_m1), IL-12 (Mm00434174_m1) and GAPDH (Mm99999915_g1) were purchased as Assay-on-Demand (Applied Biosystems). For each sample, the real-time PCR reactions were performed in duplicate or triplicate and the averages of the obtained Ct values were used for further calculations. Gene expression levels were normalized to the expression of the reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was not influenced by the experimental conditions resulting in the ΔCt value. Gene expression levels were calculated by the comparative Ct method (2-ΔΔCt). 50
Statistical analysis
Statistical analysis of the results was performed by a two-tailed unpaired Student's t-test, assuming equal variances to compare two replicate groups. Analysis of differences between multiple replicate groups was analyzed with one-way ANOVA followed by Tukey post hoc analysis. p values <0.05 were considered to be significant. Data were analyzed with GraphPad Prism (GraphPad software 5.00, San Diego, CA, USA).
Supplementary Material
Supporting Information.1H NMR and 13C NMR spectra data, dataset of focused library (Table S1), screening results (Table S2), IC50 graphs (Figure S2), enzyme kinetic parameters (Table S3), molecular modeling docking poses (Figure S4,5) and positive and negative control of enzymatic conversion (Figure S1) were reported. The Supporting Information is available free of charge on the ACS Publications website at DOI: …
Acknowledgments
We acknowledge the European Research Counsel for providing an ERC starting grant (309782) and the Netherlands Organisation for Scientific Research (NWO) for providing a VIDI grant (016.122.302) to F.J.D. The work (C.N and A.D) was financially supported from the NIH (1R01GM097082-01) and by Innovative Medicines Initiative (grant agreement n° 115489). We acknowledge Prof. dr. Reinoud Gosens (Department of Molecular Pharmacology, University of Groningen) for his support with ex vivo experiments. We also acknowledge T. Holman (University of California, Santa Cruz) for providing the h-15-LOX-1 plasmid.
Abbreviations
- BOC
tert-butyl carbonate
- DCM
dichloromethane
- DMF
dimethylformamide
- EtOAc
ethyl acetate
- EtOH
ethanol
- Et2O
diethyl ether
- rt
room temperature
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Barnes PJ. Immunology of Asthma and Chronic Obstructive Pulmonary Disease. Nat Rev Immunol. 2008;8(3):183–192. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- 2.Corey EJ, Barbara Czakó LK. Molecules and Medicine. WILEY-VCH Verlag; 2007. [Google Scholar]
- 3.Boorsma CE, Draijer C, Melgert BN. Macrophage Heterogeneity in Respiratory Diseases. Mediators Inflamm. 2013;2013 doi: 10.1155/2013/769214. 769214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon S, Taylor PR. Monocyte and Macrophage Heterogeneity. Nat Rev Immunol. 2005;5(12):953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 5.Gordon S, Plüddemann A, Martinez Estrada F. Macrophage Heterogeneity in Tissues: Phenotypic Diversity and Functions. Immunol Rev. 2014;262(1):36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneberger D, Aharonson-Raz K, Singh B. Monocyte and Macrophage Heterogeneity and Toll-like Receptors in the Lung. Cell and Tissue Research. 2011:97–106. doi: 10.1007/s00441-010-1032-2. [DOI] [PubMed] [Google Scholar]
- 7.Brash AR. Lipoxygenases: Occurrence, Functions, Catalysis, and Acquisition of Substrate. J Biol Chem. 1999;274(34):23679–23682. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni A. Lipoxygenase-a Versatile Biocatalyst for Biotransformation of Endobiotics and Xenobiotics. Cell Mol Life Sci C. 2001;58(12-13):1805–1825. doi: 10.1007/PL00000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haeggström JZ, Funk CD. Lipoxygenase and Leukotriene Pathways: Biochemistry, Biology, and Roles in Disease. Chem Rev. 2011;111(10):5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- 10.Joo Y-C, Oh D-K. Lipoxygenases: Potential Starting Biocatalysts for the Synthesis of Signaling Compounds. Biotechnol Adv. 2012;30(6):1524–1532. doi: 10.1016/j.biotechadv.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Samuelsson B, Dahlen S, Lindgren J. Leukotrienes and Lipoxins: Structures, Biosynthesis, and Biological Effects. Science (80-. ) 1987;237(4819):1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 12.Sigal E, Dicharry S, Highland E, Finkbeiner WE. Cloning of Human Airway 15-Lipoxygenase: Identity to the Reticulocyte Enzyme and Expression in Epithelium. Am J Physiol. 1992;262(4 Pt 1):L392–L398. doi: 10.1152/ajplung.1992.262.4.L392. [DOI] [PubMed] [Google Scholar]
- 13.Conrad DJ, Kuhn H, Mulkins M, Highland E, Sigal E. Specific Inflammatory Cytokines Regulate the Expression of Human Monocyte 15-Lipoxygenase. Proc Natl Acad Sci USA. 1992;89(1):217–221. doi: 10.1073/pnas.89.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mabalirajan U, Rehman R, Ahmad T, Kumar S, Leishangthem GD, Singh S, Dinda AK, Biswal S, Agrawal A, Ghosh B. 12/15-Lipoxygenase Expressed in Non-Epithelial Cells Causes Airway Epithelial Injury in Asthma. Sci Rep. 2013;3:1540. doi: 10.1038/srep01540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Middleton MK, Rubinstein T, Pure E. Cellular and Molecular Mechanisms of the Selective Regulation of IL-12 Production by 12/15-Lipoxygenase. J Immunol. 2006;176(1):265–274. doi: 10.4049/jimmunol.176.1.265. [DOI] [PubMed] [Google Scholar]
- 16.Levy BD, Serhan CN. Resolution of Acute Inflammation in the Lung. Annu Rev Physiol. 2014;76:467–492. doi: 10.1146/annurev-physiol-021113-170408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okunishi K, Peters-Golden M. Leukotrienes and Airway Inflammation. Biochim Biophys Acta - Gen Subj. 2011;1810(11):1096–1102. doi: 10.1016/j.bbagen.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, O’Donnell VB, Balzar S, Croix CM, Trudeau JB, Wenzel SE. 15-Lipoxygenase 1 Interacts with Phosphatidylethanolamine-Binding Protein to Regulate MAPK Signaling in Human Airway Epithelial Cells. Proc Natl Acad Sci USA. 2011;108(34):14246–14251. doi: 10.1073/pnas.1018075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mabalirajan U, Rehman R, Ahmad T, Kumar S, Singh S, Leishangthem GD, Aich J, Kumar M, Khanna K, Singh VP, Dinda AK, et al. Linoleic Acid Metabolite Drives Severe Asthma by Causing Airway Epithelial Injury. Sci Rep. 2013;3:1349. doi: 10.1038/srep01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindley AR, Crapster-Pregont M, Liu Y, Kuperman DA. 12/15-Lipoxygenase Is an Interleukin-13 and Interferon-Γ Counterregulated-Mediator of Allergic Airway Inflammation. Mediators Inflamm. 2010;2010:727305. doi: 10.1155/2010/727305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sendobry SM, Cornicelli JA, Welch K, Bocan T, Tait B, Trivedi BK, Colbry N, Dyer RD, Feinmark SJ, Daugherty A. Attenuation of Diet-Induced Atherosclerosis in Rabbits with a Highly Selective 15-Lipoxygenase Inhibitor Lacking Significant Antioxidant Properties. Br J Pharmacol. 1997;120(7):1199–1206. doi: 10.1038/sj.bjp.0701007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinstein DS, Liu W, Gu Z, Langevine C, Ngu K, Fadnis L, Combs DW, Sitkoff D, Ahmad S, Zhuang S, Chen X, et al. Tryptamine and Homotryptamine-Based Sulfonamides as Potent and Selective Inhibitors of 15-Lipoxygenase. Bioorg Med Chem Lett. 2005;15(5):1435–1440. doi: 10.1016/j.bmcl.2004.12.081. [DOI] [PubMed] [Google Scholar]
- 23.Ngu K, Weinstein DS, Liu W, Langevine C, Combs DW, Zhuang S, Chen X, Madsen CS, Harper TW, Ahmad S, Robl Ja. Pyrazole-Based Sulfonamide and Sulfamides as Potent Inhibitors of Mammalian 15-Lipoxygenase. Bioorg Med Chem Lett. 2011;21(14):4141–4145. doi: 10.1016/j.bmcl.2011.05.107. [DOI] [PubMed] [Google Scholar]
- 24.Rai G, Kenyon V, Jadhav A, Schultz L, Armstrong M, Jameson JB, Hoobler E, Leister W, Simeonov A, Holman TR, Maloney DJ. Discovery of Potent and Selective Inhibitors of Human Reticulocyte 15-Lipoxygenase-1. J Med Chem. 2010;53(20):7392–7404. doi: 10.1021/jm1008852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rai G, Joshi N, Jung JE, Liu Y, Schultz L, Yasgar A, Perry S, Diaz G, Zhang Q, Kenyon V, Jadhav A, et al. Potent and Selective Inhibitors of Human Reticulocyte 12/15-Lipoxygenase as Anti-Stroke Therapies. J Med Chem. 2014;57(10):4035–4048. doi: 10.1021/jm401915r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinstein DS, Liu W, Ngu K, Langevine C, Combs DW, Zhuang S, Chen C, Madsen CS, Harper TW, Robl JA. Discovery of Selective Imidazole-Based Inhibitors of Mammalian 15-Lipoxygenase: Highly Potent against Human Enzyme within a Cellular Environment. Bioorg Med Chem Lett. 2007;17(18):5115–5120. doi: 10.1016/j.bmcl.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Gundersen L-L, Malterud KE, Negussie AH, Rise F, Teklu S, Østby OB. Indolizines as Novel Potent Inhibitors of 15-Lipoxygenase. Bioorg Med Chem. 2003;11(24):5409–5415. doi: 10.1016/j.bmc.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 28.Mahdavi M, Shirazi MS, Taherkhani R, Saeedi M, Alipour E, Moghadam FH, Moradi A, Nadri H, Emami S, Firoozpour L, Shafiee A, et al. Synthesis, Biological Evaluation and Docking Study of 3-Aroyl-1-(4-Sulfamoylphenyl)thiourea Derivatives as 15-Lipoxygenase Inhibitors. Eur J Med Chem. 2014;82:308–313. doi: 10.1016/j.ejmech.2014.05.054. [DOI] [PubMed] [Google Scholar]
- 29.Asghari T, Bakavoli M, Rahimizadeh M, Eshghi H, Saberi S, Karimian A, Hadizadeh F, Ghandadi M. Synthesis and Evaluation of a New Series of 3,5-bis((5-Bromo-6-Methyl-2-T-Aminopyrimidin-4-Yl)thio)-4H-1,2,4-Triazol-4-Amines and Their Cyclized Products “Pyrimidinylthio Pyrimidotriazolothiadiazines” as 15- Lipo-Oxygenase Inhibitors. Chem Biol Drug Des. 2015;85(2):216–224. doi: 10.1111/cbdd.12375. [DOI] [PubMed] [Google Scholar]
- 30.Wisastra R, Ghizzoni M, Boltjes A, Haisma HJ, Dekker FJ. Anacardic Acid Derived Salicylates Are Inhibitors or Activators of Lipoxygenases. Bioorg Med Chem. 2012;20(16):5027–5032. doi: 10.1016/j.bmc.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 31.Wisastra R, Kok PAM, Eleftheriadis N, Baumgartner MP, Camacho CJ, Haisma HJ, Dekker FJ. Discovery of a Novel Activator of 5-Lipoxygenase from an Anacardic Acid Derived Compound Collection. Bioorg Med Chem. 2013;21(24):7763–7778. doi: 10.1016/j.bmc.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eleftheriadis N, Thee S, Te Biesebeek J, van der Wouden P, Baas B-J, Dekker FJ. Identification of 6-Benzyloxysalicylates as a Novel Class of Inhibitors of 15-Lipoxygenase-1. Eur J Med Chem. 2015;94:265–275. doi: 10.1016/j.ejmech.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Rai G, Joshi N, Jung JE, Liu Y, Schultz L, Yasgar A, Perry S, Diaz G, Zhang Q, Kenyon V, Jadhav A, et al. Potent and Selective Inhibitors of Human Reticulocyte 12/15-Lipoxygenase as Anti-Stroke Therapies. J Med Chem. 2014;57(10):4035–4048. doi: 10.1021/jm401915r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A WBP, H YE. The Synthesis and Pharmacological Activity of Some Chloro-A-Alkyltryptamines. J Med Chem. 1963;6:378–380. doi: 10.1021/jm00340a009. [DOI] [PubMed] [Google Scholar]
- 35.Taylor P, Krishna C, Al L. Synthesis and CNS Activity of Some Fluorine Containing 3-Indolylglyoxamides and Tryptamines. Agric Biol Chem. 1978;(42):1723–1726. [Google Scholar]
- 36.Ottoni O, Neder AV, Dias AK, Cruz RP, Aquino LB. Acylation of Indole under Friedel-Crafts Conditions-an Improved Method to Obtain 3-Acylindoles Regioselectively. Org Lett. 2001;3(7):1005–1007. [PubMed] [Google Scholar]
- 37.Gillmor SA, Villaseñor A, Fletterick R, Sigal E, Browner MF. The Structure of Mammalian 15-Lipoxygenase Reveals Similarity to the Lipases and the Determinants of Substrate Specificity. Nat Struct Biol. 1997;4(12):1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 38.Abad-Zapatero C, Metz J. Ligand Efficiency Indices as Guideposts for Drug Discovery. Drug Discov Today. 2005;10(7) doi: 10.1016/S1359-6446(05)03386-6. [DOI] [PubMed] [Google Scholar]
- 39.Kuntz ID, Chen K, Sharp KA, Kollman PA. The Maximal Affinity of Ligands. Proc Natl Acad Sci USA. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hopkins AL, Keserü GM, Leeson PD, Rees DC, Reynolds CH. The Role of Ligand Efficiency Metrics in Drug Discovery. Nat Rev Drug Discov. 2014;13(2):105–121. doi: 10.1038/nrd4163. [DOI] [PubMed] [Google Scholar]
- 41.Abad-Zapatero C. Ligand Efficiency Indices for Effective Drug Discovery. Expert Opin Drug Discov. 2007;2(4):469–488. doi: 10.1517/17460441.2.4.469. [DOI] [PubMed] [Google Scholar]
- 42.Hackett TL, Holloway R, Holgate ST, Warner JA. Dynamics of pro-Inflammatory and Anti-Inflammatory Cytokine Release during Acute Inflammation in Chronic Obstructive Pulmonary Disease: An Ex Vivo Study. Respir Res. 2008;9:47. doi: 10.1186/1465-9921-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takanashi S, Hasegawa Y, Kanehira Y, Yamamoto K, Fujimoto K, Satoh K, Okamura K. Interleukin-10 Level in Sputum Is Reduced in Bronchial Asthma, COPD and in Smokers. Eur Respir J. 1999;14(2):309–314. doi: 10.1034/j.1399-3003.1999.14b12.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang R, Shi H-F, Zhao J-F, He Y-P, Zhang H-B, Liu J-P. Design, Synthesis and Aromatase Inhibitory Activities of Novel Indole-Imidazole Derivatives. Bioorg Med Chem Lett. 2013;23(6):1760–1762. doi: 10.1016/j.bmcl.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 45.Louvet P, Lallement G, Pernot-Marino I, Luu-Duc C, Blanchet G. Novel Benzimidazoles as Ligands for the Strychnine-Insensitive N-Methyl-D-Aspartate-Linked Glycine Receptor. Eur J Med Chem. 1993;28(1):71–75. [Google Scholar]
- 46.Doemling A, Holak T. Novel p53-mdm2/p53-mdm4 Antagonists to Treat Proliferative Disease. 2011 Sep 1; [Google Scholar]
- 47.Boettcher A, Buschmann N, Furet P, Groell JM, Kallen J, Hergovich LJ, Masuya K, Mayr L, Vaupel A. 3-Imidazolyl-Indoles for the Treatment of Proliferative Diseases. 2008 Oct 9; [Google Scholar]
- 48.Wright SW, Hageman DL, Wright AS, McClure LD. Convenient Preparations of T-Butyl Esters and Ethers from T-Butanol. Tetrahedron Lett. 1997;38(42):7345–7348. [Google Scholar]
- 49.Oenema TA, Maarsingh H, Smit M, Groothuis GMM, Meurs H, Gosens R. Bronchoconstriction Induces TGF-B Release and Airway Remodelling in Guinea Pig Lung Slices. PLoS One. 2013;8(6):e65580. doi: 10.1371/journal.pone.0065580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.