Abstract
Atopic dermatitis is a common pruritic skin disease in which barrier dysfunction and cutaneous inflammation play a role in pathogenesis. Mechanisms underlying the associated inflammation are not fully understood, and while CD1a-expressing Langerhans cells are known to be enriched within lesions, their role in clinical disease pathogenesis has not been studied. Here we observed that house dust mite (HDM) generates neolipid antigens for presentation by CD1a to T cells in the blood and skin lesions of affected individuals. HDM-responsive CD1a-reactive T cells increased in frequency after birth and showed rapid effector function, consistent with antigen-driven maturation. To define the underlying mechanisms, we analyzed HDM-challenged human skin and observed allergen-derived phospholipase (PLA2) activity in vivo. CD1a-reactive T cell activation was dependent on HDM-derived PLA2 and such cells infiltrated the skin after allergen challenge. Filaggrin insufficiency is associated with atopic dermatitis, and we observed that filaggrin inhibits PLA2 activity and inhibits CD1a-reactive PLA2-generated neolipid-specific T cell activity from skin and blood. The most widely used classification schemes of hypersensitivity, such as Gell and Coombs are predicated on the idea that non-peptide stimulants of T cells act as haptens that modify peptides or proteins. However our results point to a broader model that does not posit haptenation, but instead shows that HDM proteins generate neolipid antigens which directly activate T cells. Specifically, the data identify a pathway of atopic skin inflammation, in which house dust mite-derived phospholipase A2 generates antigenic neolipids for presentation to CD1a-reactive T cells, and define PLA2 inhibition as a function of filaggrin, supporting PLA2 inhibition as a therapeutic approach.
Introduction
Atopic and allergic diseases affect up to 20-30% of the population and have considerable associated morbidity, mortality and health economic burden(1). Atopic dermatitis (AD) is a disease with complex genetic and environmental susceptibility factors. Whilst it is known that many loci are involved (2), null mutations in the gene encoding filaggrin are reproducibly associated with moderate to severe clinical disease (3, 4). Filaggrin is expressed in keratinocytes and functions in skin barrier, cutaneous pH regulation, hydration and antimicrobial protection. As keratinocytes proceed through cornification, profilaggrin is cleaved into filaggrin monomers which can be incorporated into the lipid envelope and exposed to the intercellular space. Thus, a general “outside-in” hypothesis predicts that filaggrin disruption acts to allow exogenous immune stimuli to enter the skin and activate immune responses (3).
Up to 50% of individuals with homozygous filaggrin null mutations, and up to 5-10% of healthy European filaggrin null carriers do not have associated atopic dermatitis, suggesting that other modifying genetic and environmental factors are important, including those that are directly involved in immune responses, such as FcεR1 and IL-4R genes (5). Indeed many cytokines modify filaggrin and anti-microbial peptide expression (6–10), and anti-IL-4Rα therapy has shown significant efficacy in atopic dermatitis treatment (11). Separately, there is convincing evidence that antigen-specific reactivity to environmental challenge has a role in the pathogenesis of atopic dermatitis. For example, the pathology of atopic dermatitis shares many features with classic delayed-type hypersensitivity, including epidermal oedema and a dominant T cell inflammatory infiltrate. Approximately 80% of individuals with atopic dermatitis have elevated serum IgE which recognize proteins derived from one or more ubiquitous environmental allergens, including house dust mite (HDM), animal dander, pollens and fungal allergens (eg Aspergillus spp) (12). Allergen peptide-specific type 2 cytokine-producing T cells have been documented in many studies to be present in the peripheral blood of affected individuals (13–17). Recently, type 2 innate lymphoid cells (ILC2) have been shown to be enriched in lesional atopic dermatitis skin and to infiltrate after HDM challenge (18). The production of type 2 cytokines by ILC2 is enhanced by PGD2, IL-33, IL-25 and TSLP and increases in the setting of reduced E-cadherin, believed to be mediated through loss of KLRG1 inhibitory signals (18, 19). Overall these data suggest that type 2 cytokine production by a number of cell types including T cells and ILC2 can compound barrier insufficiency and contribute to inflammation, supporting the alternate “inside-out” hypothesis. However there remains considerable debate about the relative roles of barrier function and cutaneous inflammation in atopic dermatitis pathogenesis. This is an important question in order to define how best to target future therapeutic strategies.
CD1a is an MHC-like antigen presenting molecule and is highly expressed by Langerhans cells of the epidermis and by a subset of dermal dendritic cells, as well as subsets of dendritic cell populations at other sites, including lung, gut and genital mucosa (20–24). CD1a presents self (25) and foreign (26) lipids to T cells, so the abundant expression of CD1a on epidermal Langerhans cells would be compatible with the detection of skin barrier compromise through binding endogenous or exogenous lipids for presentation to CD1a-reactive T cells (27). Indeed, lesional cutaneous atopic tissue carries an altered lipid profile (28–32) that is a candidate for influencing CD1a-mediated T cell activation, and CD1a+ cells are enriched within atopic dermatitis lesions (33). Recently studies show that CD1a-reactive T cells circulate at far higher frequencies than previously considered and can infiltrate normal human skin (27, 34, 35). CD1a autoreactive T cells produce cytokines that contribute to skin disease, like IL-22 and interferon-γ(27), and whereas other CD1 proteins use complex intracellular processing pathways, CD1a directly captures and displays extracellular lipids with few specialized loading requirements (36). (37–40)(41)(36)(27)(35)(40)(42, 43)Together, the natural accumulation of autoantigens, CD1a proteins and CD1a autoreactive T cells point to a natural organ-specific function in skin, but insights into clinical diseases are limited. Recently, we have identified that fatty acids generated by phospholipase activity in wasp and bee venom can be recognized by CD1a-reactive T cells (44). Further, pollen-derived phospholipids have been implicated as targets for lipid-specific T cells, including CD1d and CD1a reactive T cell clones (45). Based on these findings, we considered that CD1a might play a role in atopic dermatitis. Specifically, given the enrichment of CD1a-expressing cells and altered lipids in lesional atopic dermatitis skin, we sought to test the hypothesis that CD1a-reactive lipid-specific T cells contribute to the human response to dust mites and atopic dermatitis.
Results
House dust mite is recognized by CD1a-reactive T cells
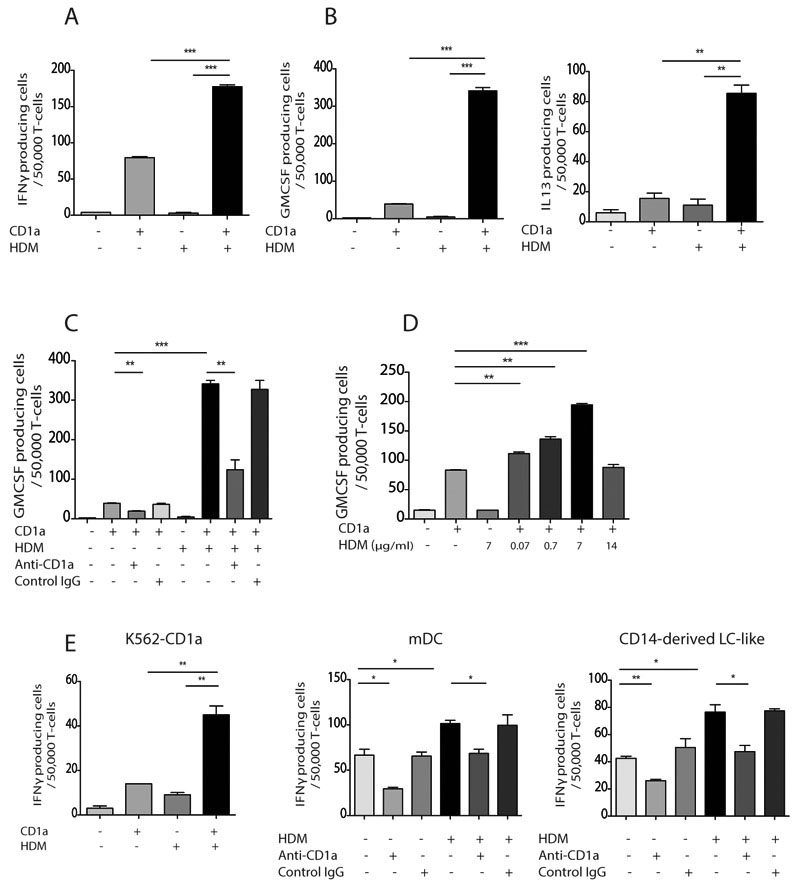
To determine whether HDM could be recognized by CD1a-reactive T cells isolated ex vivo from individuals with atopic dermatitis, we incubated polyclonal T cells with K562 target cells transfected with CD1a (K562-CD1a) in the presence or absence of whole HDM extract. K562 cells are HLAlow and thereby largely bypass alloreactivity and allow parallel testing with T cells from many unrelated donors under equivalent conditions. We measured both type 1 and type 2 cytokine production, as both have been implicated in disease pathogenesis (46–49). As seen previously (27, 35, 44), we observed a trace response to K562 cells at a rate of ˜1 in 10,000. Above this trace background rate, we detected large increments in IFNγ response to K562-CD1a cells, which likely reflects the presence and activation of autoreactive T cells recognising CD1a and endogenous K562-derived lipids (Figure 1A, donor R9500). Further, we noted significantly (P<0.0001) increased activation by K562-CD1a target cells when pulsed overnight with house dust mite extract. Importantly, no response above background was seen in response to control APCs transfected with vector alone, demonstrating that the response to dust mite extract is CD1a-mediated. Polyclonal T cells from the same donor also produced GM-CSF and IL-13 in response to dust mite products with a similar pattern, except that the rate of cellular response was particularly high for GM-CSF, reaching to more than 0.5 percent of all T cells in some cases (Figure 1B). The response was inhibited by an anti-CD1a antibody, but not by isotype control (Figure 1C). Responses were in clinically and physiologically relevant dose ranges, as titration to 0.07μg/ml HDM extract (Figure 1D), which is approximately 0.3% of doses administered in vivo during maintenance immunotherapy (up to 21μg) (50), caused detectable T cell responses.
Figure 1. Circulating CD1a-reactive house dust mite-responsive T cells produce IFNγ, GM-CSF and IL-13.
T cells were isolated by CD3 MACS beads from donor PBMC (R9500) and incubated overnight with CD1a-transfected K562 (CD1a) or untransfected K562 (EV) cells pulsed with HDM extract. IFN-γ (A), GM-CSF (B, left) and IL-13 (B, right) production were measured by ELISpot in the absence or presence of anti-CD1a antibody (C) and at different HDM concentrations (D). CD1a-expressing K562 (E, left), in vitro derived mDC (E, middle) or LC-like cells (E, right) were pulsed with HDM extract overnight and incubated with autologous peripheral blood T cells from donor R2. IFNγ production was measured by ELISpot in the presence or absence of anti-CD1a antibody. Data representative of at least three donors for each experiment are shown. Bars represent standard error. * P<0.05; ** P<0.01; ***P<0.001;****P<0.0001, t test.
K562-CD1a represent an engineered APC that has the advantage of use with any donor to test the inter-donor reproducibility of the response in a defined system, but these transformed cells might not mimic the antigens or co-stimulatory processes of the two native CD1a+ APCs: myeloid dendritic cells and Langerhans cells (LC). We differentiated monocytes with GM-CSF and IL-4 to produce monocyte-derived myeloid dendritic cells (mDCs) and also activated cells in vitro with cytokines to mimic LCs (in vitro LCs, supplementary figure 1) (51–54). Similar to prior studies that compared K562 cells, mDCs and LC-like cells side by side, we found that both mDCs and LC-like cells mediated the HDM response of polyclonal autologous T cells in a CD1a-reactive manner (Figure 1E). Overall, our data show house dust mite-responsive CD1a-reactive T cells exist in the peripheral blood of healthy individuals at high frequencies and produce IFNγ, GM-CSF and IL-13 in response to HDM challenge at relevant doses.
HDM-responsive CD1a-reactive T cells are enriched in blood and skin of atopic dermatitis patients
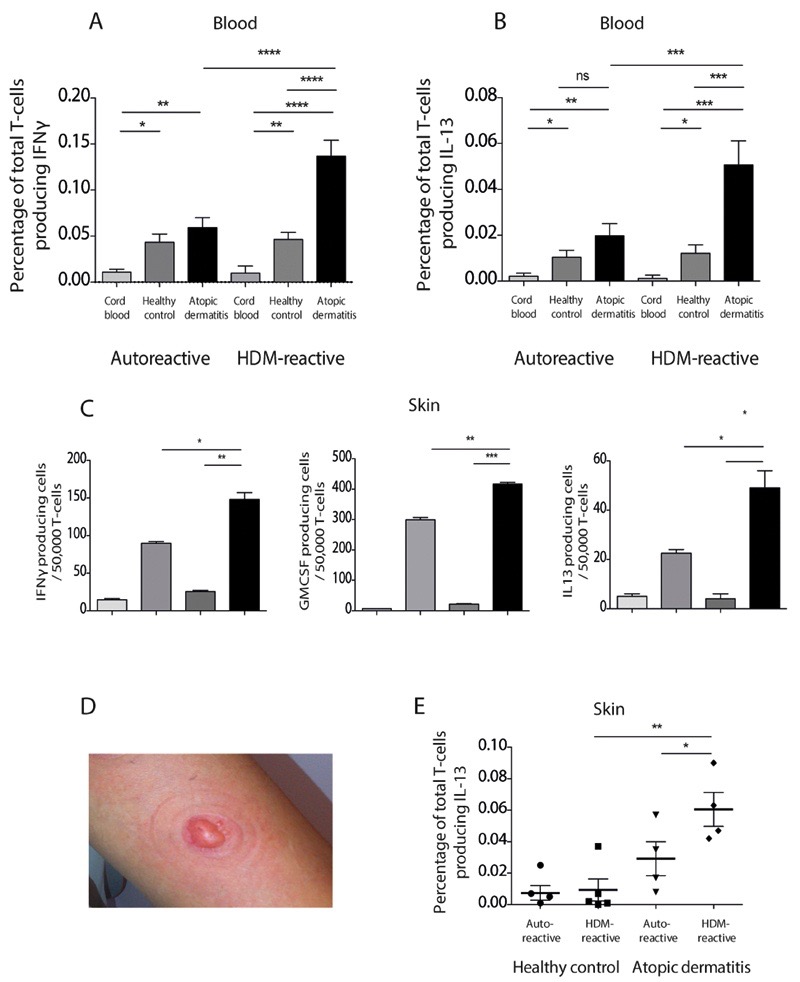
Next we examined responses to house dust mites in a larger cohort of healthy adult donors and individuals with atopic dermatitis, and compared these to responses in cord blood of neonates, which represent a control for naïve polyclonal T cells. We observed significantly higher CD1a-reactive ex vivo T cell IFNγ responses to HDM in individuals with atopic dermatitis compared to healthy non-atopic controls (Figure 2A). The frequencies of IFNγ producing cells were significantly (P<0.0001) higher in adult donors than in cord blood samples, consistent with an acquired antigen-driven T cell expansion (Figure 2A). In addition, the production of IL-13 by CD1a-reactive T cells was also significantly (P<0.001) elevated ex vivo, confirming type 1 and type 2 cytokine induction (Figure 2B). Using dual-color ELISpot, we showed while the majority of the type 1 and type 2 cytokine-producing cells are unique subsets, a mean of 7% of CD1a-reactive HDM-responsive T cells could produce both IFNγ and IL-13 (Supplementary Figure 2A). However we do not observe an increase in the IFNγ or IL-13 producing HDM-responsive CD1a-reactive T cells in patients with psoriasis (Supplementary Figure 2B). Given the association between filaggrin null mutations and moderate-severe atopic dermatitis, we genotyped the patients and controls for the two most common mutations found in Europeans (2282del4 and R501X). Frequencies of the HDM-responsive CD1a-reactive T cells were significantly increased in those individuals with filaggrin null mutations compared to those with wild-type filaggrin (contingency chi-squared 4.38, P<0.05, Supplementary Figure 3A). Furthermore, the percentage of IL-13 producing CD1a-reactive HDM-responsive T cells significantly correlated with disease severity (r2=0.445, P<0.001) and there was a significant correlation between fold increase in HDM-responsive T cells and total IgE (r2=0.588, P<0.05) and HDM-specific IgE (r2=0.502, P<0.05). Overall, these data showed that HDM-responsive, CD1a-reactive T cells increase in frequency in adults, show effector function, and are enriched in the circulation of patients with atopic dermatitis.
Figure 2. HDM-responsive CD1a-reactive T cells are enriched in blood and skin of atopic dermatitis patients.
(A,B) T cells derived from the peripheral blood of healthy controls (HC), patients with atopic dermatitis (AD) or from cord blood were incubated with CD1a-transfected K562 or untransfected K562 cells in the presence or absence of HDM extract. IFNγ (A) and IL-13 (B) production were measured by ELISpot and expressed as percentage of responding T cells (A n=30 HC, 24 AD, 10 cord blood; B n=20 HC, 17 AD, 10 cord blood). Auto-reactive is the response to CD1a-K562 in the absence of HDM. (C) Skin blister T cells from donor R229 were incubated with CD1a-transfected K562 or untransfected K562 cells in the presence or absence of HDM extract. IFNγ (C, left), GM-CSF (C, middle) and IL-13 (C, right) production were measured by ELISpot. Data are representative of at least three separate donors for each experiment. (D) Example of skin suction blister raised after 60 minutes of 200mmHg negative pressure. (E) Skin blister T cells were isolated from unchallenged skin of healthy controls (n=5) or patients with atopic dermatitis (n=4) and incubated with CD1a-transfected or untransfected K562 cells in the presence or absence of HDM extract. IFNγ production was measured by ELISpot and expressed as percentage of CD1a-reactive T cells. Bars represent standard error. * P<0.05; ** P<0.01; ***P<0.001;****P<0.0001, t test .
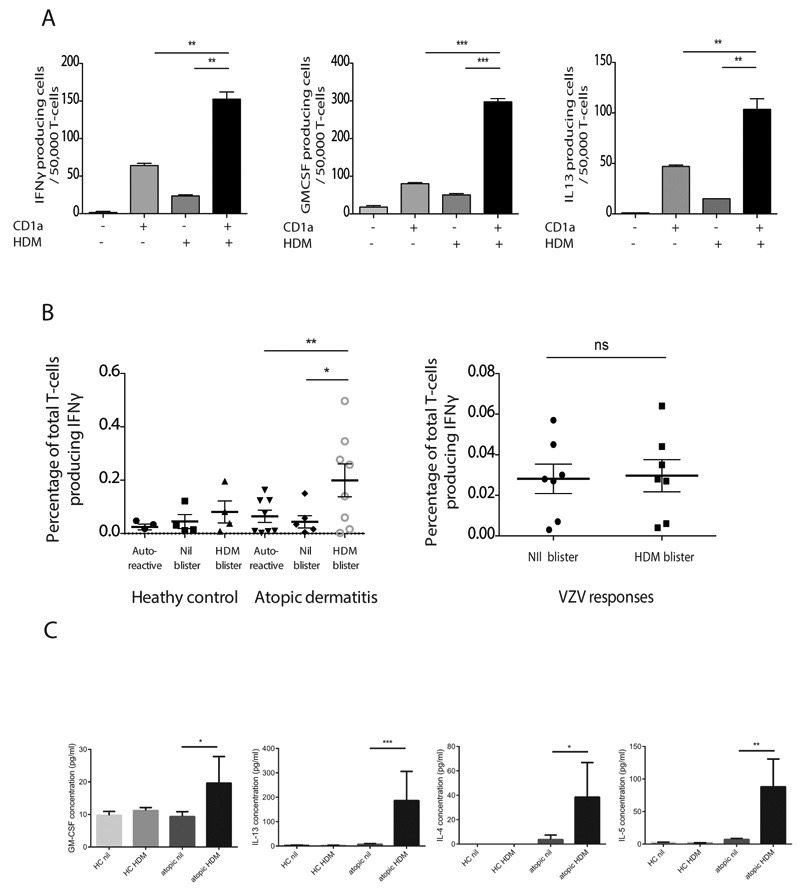
To examine whether HDM-responsive cells were also present within skin from patients with atopic dermatitis and healthy controls, we used skin suction blisters to isolate T cells from skin. This method uses low pressure, sustained suction for 60 minutes to produce extracellular blister fluids that are captured for immunological and biochemical analysis, and can be performed before or after antigen challenge (18). The approach has the advantage over conventional skin biopsies in that cells and fluid can be aspirated from the skin and used in functional assays, without the need for prolonged processing with potentially confounding treatments, such as dispase and collagenase. We observed that HDM-responsive CD1a-reactive T cells are present in skin and produce IFNγ, GM-CSF and IL-13 (Figure 2C and 2D). Furthermore the IL-13 producing T cells are enriched in the skin of patients with atopic dermatitis compared to skin of healthy controls (Figure 2E) and the frequency correlated with SCORAD disease severity (r2=0.67, P<0.01), showing that T cell infiltration into atopic dermatitis lesions correlates both with disease outcome and type 2 cytokine production in humans ex vivo. To investigate the CD1a-reactive immune response in vivo, we challenged the skin intra-epidermally with 0.7μg HDM extract and assessed skin infiltration of HDM-responsive T cells at 24 hrs. We chose a dose at <5% of the amount administered during the maintenance phase of subcutaneous immunotherapeutic approaches (typically up to 21μg) to assure safety and attempt to mimic low dose exposures (50). We noted skin infiltration of IFNγ, GM-CSF and IL-13-producing HDM-responsive T cells at 24 hours suggesting that the cells infiltrate early and may therefore contribute to the ensuing inflammation (Figure 3A and Supplementary Figure 3B). Furthermore in those with atopic dermatitis, we observed significantly greater infiltration of IFNγ-producing HDM-responsive CD1a-reactive T cells after HDM challenge than in healthy controls (Figure 3B, left panel). In contrast, there was no enrichment for varicella zoster virus-specific T cells in the skin after HDM challenge, suggesting that the enrichment is specific to HDM-responsive T cells (Figure 3B, right panel).
Figure 3. HDM-responsive CD1a-reactive T cells infiltrate skin after HDM challenge.
Skin blister T cells were isolated from donor R4 24 hours after HDM skin challenge, expanded and incubated with CD1a-transfected or untransfected K562 cells in the presence or absence of HDM extract. IFNγ (A, left), GM-CSF (A, middle) and IL-13 (A, right) production by donor R4 cells were measured by ELISpot. The data are representative of at least three separate donors for each experiment. (B, left panel) Overall frequencies of HDM-responsive CD1a-reactive IFNγ-producing T cells infiltrating human skin after saline or HDM skin challenge were compared between healthy controls (n=4) and atopic dermatitis patients (n=8). Auto-reactive refers to responses to unpulsed K562-CD1a cells. (B, right panel) Skin blister cells derived from HDM-challenged or unchallenged skin were incubated with live attenuated varicella zoster virus and IFNγ production was measured by ELISpot. (C) Concentrations of type 2 cytokines were measured in skin blister fluid by multiplex bead array after saline (nil) or HDM skin challenge in healthy controls (HC, n=8) or atopic (n=16) individuals. Bars represent standard error. * P<0.05; ** P<0.01; ***P<0.001;****P<0.0001, t test.
As described above, type 2 cytokines can influence filaggrin and anti-microbial peptide expression in the skin. We investigated the type 2 cytokine content of skin blisters derived from patients with atopic dermatitis and healthy controls with sampling at non-lesional skin, and after HDM challenge. These studies showed that after HDM challenge of non-lesional skin in atopic dermatitis patients, the concentrations of type 2 cytokines IL-4, IL-5, IL-13 and GM-CSF within skin blister fluid were significantly increased compared to healthy donors (Figure 3C).
Overall these data demonstrate that HDM-responsive CD1a-reactive T cells are present in the blood and skin of healthy donors, but are enriched in the blood and skin of patients with atopic dermatitis. Furthermore, after intra-epidermal HDM allergen challenge, the T cells infiltrate rapidly and associate with the production of type 2 cytokines in vivo.
HDM-responsive CD1a-reactive T cell responses are explained by PLA2 activity
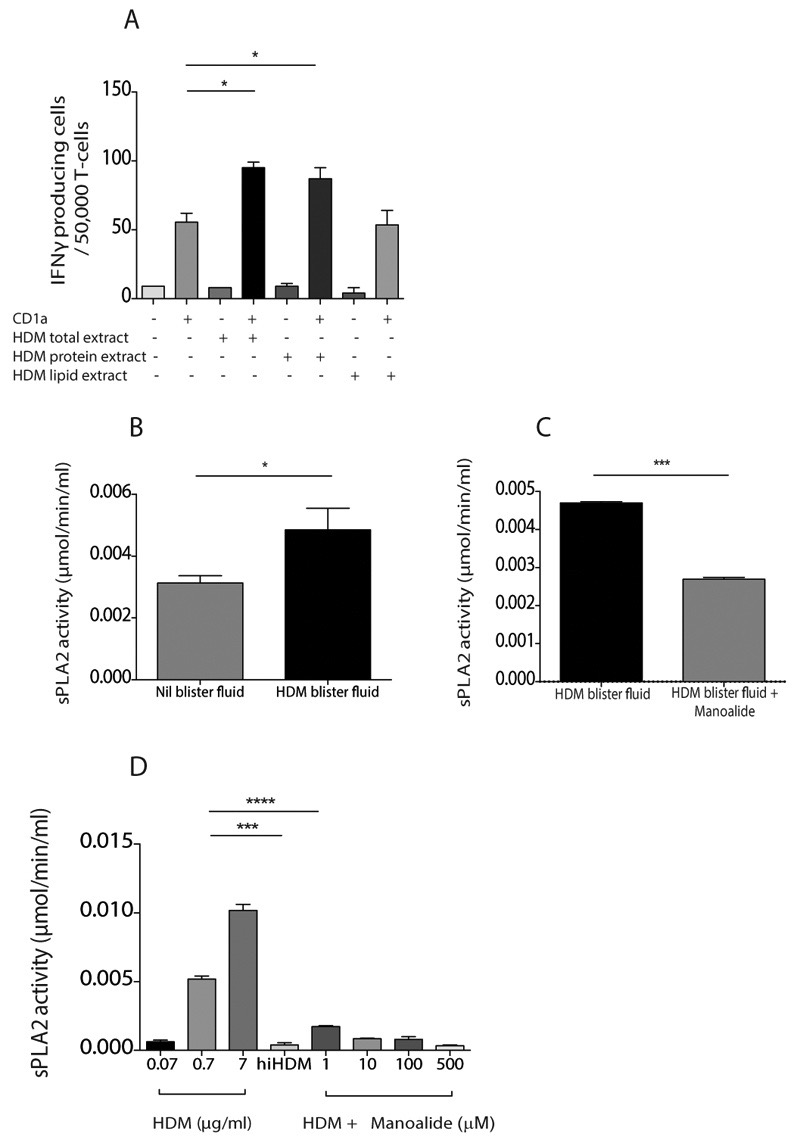
To identify the antigenic substance in house dust mites, we first treated HDM extract with chloroform and methanol to recover protein-enriched aqueous and lipid-enriched organic fractions before testing the differential capacity to sensitise CD1a-expressing K562 cells for recognition by T cells and unexpectedly observed responses to the protein-enriched fractions rather than the lipid-enriched fractions (Figure 4A). This counterintuitive result could be explained by other experiments that were recently reported (44), which showed that the origin of CD1a mediated responses to bee venom also derived from proteinaceous fractions of venom, and were identified as phospholipases. Phospholipases generated antigenic free fatty acids or lysolipids from non-antigenic phosphodiacylglycerols. We posited that a parallel mechanism existed here and tested it by examining PLA2 biochemical activity in vivo and in vitro using colorimetric thiol detection after release from diheptanoyl thio-PC substrate. Interestingly there was PLA2 activity measured in all skin blister samples, which significantly increased after HDM challenge (Figure 4B). Furthermore, HDM-induced PLA2 activity within skin blister fluid was significantly inhibited in the presence of the known PLA2 inhibitor manoalide (Figure 4C). We next demonstrated that HDM extract contains PLA2 activity in vitro, which could be inhibited by manoalide and heat inactivation (Figure 4D).
Figure 4. HDM extract contains PLA2 activity in vivo and in vitro.
(A) T cells were isolated by CD3 MACS beads from healthy donor PBMC and incubated overnight with CD1a-transfected K562 (CD1a) or untransfected K562 (EV) cells pulsed with HDM total extract, aqueous protein phase or lipid phase. IFN-γ production was measured by ELISpot (A). PLA2 activity in saline or HDM challenged skin blister fluid was detected by measuring free thiol release in the presence of the diheptanoyl thio-PC substrate in the absence (B) or presence (C) of 10μM of the PLA2 inhibitor manoalide. (D) PLA2 activity in HDM extract in vitro was detected by measuring free thiol release in the presence of the diheptanoyl thio-PC substrate and manoalide and after heat inactivation (HDMhi). Data are representative of at least three separate experiments. Bars represent standard error. * P<0.05; ** P<0.01; ***P<0.001;****P<0.0001, t test.
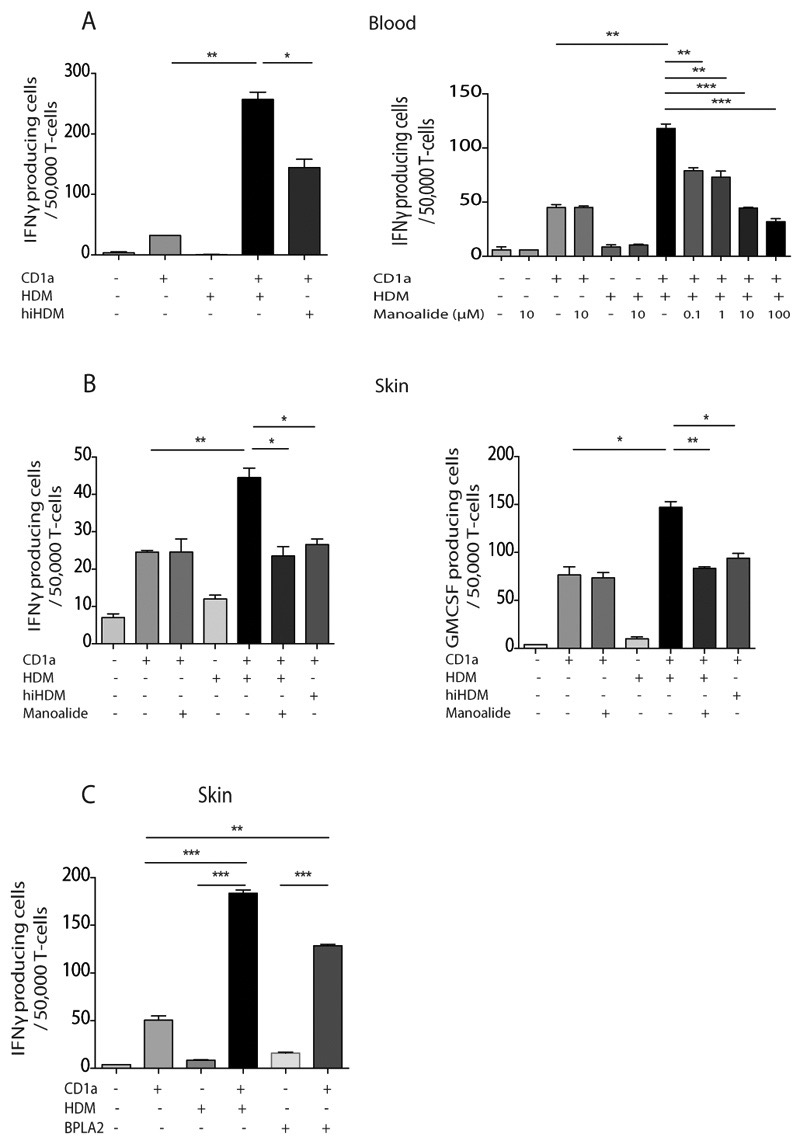
Heat inactivation of house dust mite extract or treatment with the PLA2 inhibitor manoalide abrogated the CD1a-dependent recognition of HDM-pulsed K562-CD1a cells, but did not affect the autoreactive T cell response (Figure 5A) or viral-specific T cell responses (Supplementary figure 4). Together these suggest that the CD1a-reactive HDM responses are secondary to PLA2, most likely through generation of neolipid antigens. We next sought to determine if skin-derived HDM-responsive CD1a-reactive T cells were also dependent on PLA2 activity. Figure 5B shows that IFNγ and GM-CSF production by skin T cells derived ex vivo following intra-epidermal HDM challenge to human skin are also inhibited by heat inactivation of the HDM or by treatment with manoalide. Furthermore, we show that skin HDM-responsive CD1a-reactive T cells are also able to respond to purified bee venom PLA2 (44), another recognized PLA2-containing allergen (Figure 5C). This result points to shared pathways of skin inflammation despite different depths of natural antigen delivery by venom and dust mites. Overall, HDM-derived PLA2 has the capacity to generate neolipid antigens for recognition by CD1a-reactive T cells. The lack of response with the lipid fraction of HDM extract suggests that skin derived lipid sources represent the principle substrates of HDM PLA2.
Figure 5. HDM-derived PLA2 generates neolipid antigens for presentation by CD1a to blood and skin T cells.
T cells were isolated by CD3 MACS beads from healthy donor PBMC and incubated overnight with CD1a-transfected K562 (CD1a) or untransfected K562 (EV) cells pulsed with HDM total extract. IFN-γ production was measured by ELISpot after HDM heat inactivation (hiHDM) (A, left) or overnight incubation with a dose titration of the PLA2 inhibitor manoalide (A, right). Skin blister T cells were isolated 24 hours after HDM skin challenge, expanded and incubated with CD1a-transfected or untransfected K562 cells in the presence or absence of HDM extract that had been heat inactivated or incubated with manoalide. IFNγ (B, left) and GM-CSF (B, right) production were measured by ELISpot. (C) Skin blister T cells were isolated 24 hours after HDM skin challenge, expanded and incubated with CD1a-transfected or untransfected K562 cells in the presence or absence of HDM extract or 1μg/ml purified bee venom PLA2. IFNγ production was measured by ELISpot. Data are representative of at least three separate experiments. Bars represent standard error. * P<0.05; ** P<0.01; ***P<0.001;****P<0.0001, t test.
Filaggrin inhibits HDM PLA2 and CD1a-reactive T cell responses
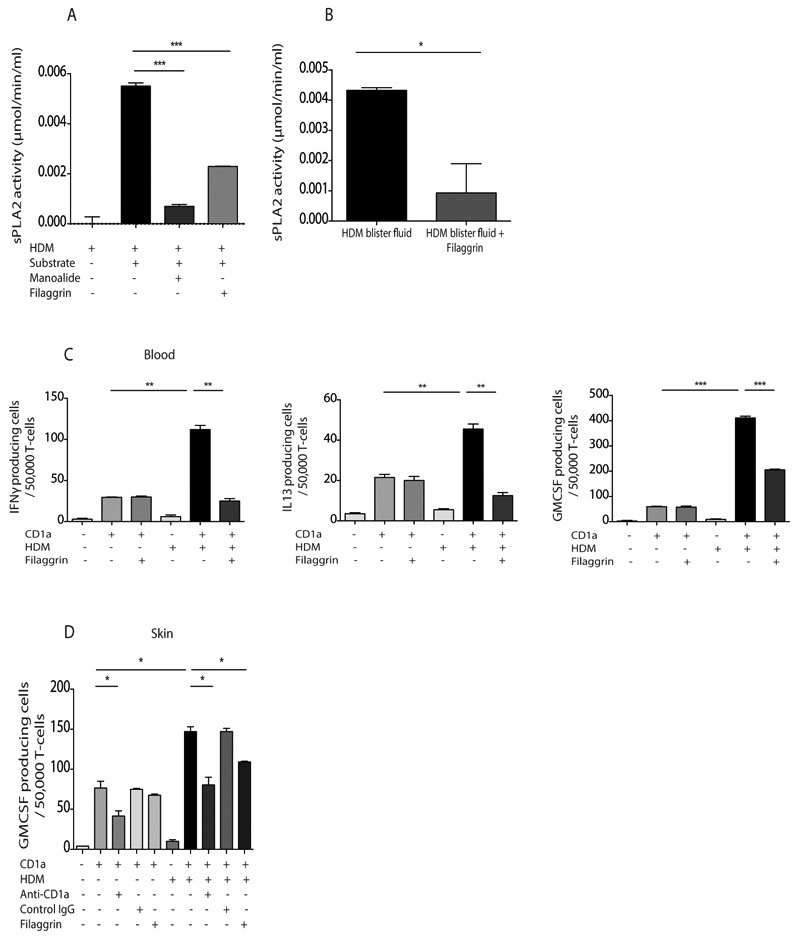
Given that filaggrin insufficiency is associated with moderate-severe atopic dermatitis and that we have shown an association between CD1a-reactive T cell responses to HDM and filaggrin null mutations, we designed experiments to test the hypothesis that filaggrin directly contributes to the CD1a-reactive T cell response. We investigated the possibility that filaggrin itself can directly inhibit PLA2 activity, and observed that filaggrin monomers efficiently inhibited HDM PLA2 biochemical activity at levels equivalent to those present in the stratum corneum in vivo (55) (Figure 6A). Furthermore recombinant filaggrin monomers inhibited the PLA2 activity observed within skin blister fluid after intra-epidermal challenge with HDM (Figure 6B). We next investigated whether filaggrin could inhibit cytokine production by HDM-responsive CD1a-reactive T cells and showed that recombinant filaggrin monomers significantly inhibited IFNγ, IL13 and GM-CSF production by T cells derived from blood ex vivo (Figure 6C). However we did not show filaggrin-inhibition of autoreactive T cells or viral specific T cells (Supplementary Figure 4), ruling out a non-specific toxic effect of the filaggrin on T cells. Lastly we demonstrated that the filaggrin monomers inhibited GM-CSF production by T cells derived ex vivo from skin after HDM skin challenge (Figure 6D). These data suggest that filaggrin may provide barrier function status signals to the innate and adaptive immune responses through effects on PLA2.
Figure 6. Filaggrin inhibits HDM PLA2 activity and inhibits responses of HDM-responsive CD1a-reactive T cells isolated from blood and skin.
(A) PLA2 activity in HDM extract in vitro was detected by measuring free thiol release in the presence of the diheptanoyl thio-PC substrate, 10μM of the PLA2 inhibitor manoalide and 1μg/ml human recombinant filaggrin. (B) PLA2 activity in HDM challenged skin blister fluid was detected by measuring free thiol release diheptanoyl thio-PC substrate in the absence or presence of filaggrin. (C) T cells were isolated by CD3 MACS beads from healthy donor PBMC and incubated overnight with CD1a-transfected K562 or untransfected K562 cells pulsed with HDM total extract. GM-CSF, IL-13 and IFNγ production were measured by ELISpot in the presence or absence of filaggrin. (D) Skin blister T cells were isolated 24 hours after HDM skin challenge and incubated with CD1a-transfected or untransfected K562 cells in the presence or absence of HDM extract, filaggrin or anti-CD1a antibody. Data are representative of at least three separate experiments. Bars represent standard error. * P<0.05; ** P<0.01; ***P<0.001;****P<0.0001, t test.
Discussion
Although CD1a protein is expressed at constitutively and at extraordinarily high density on Langerhans cells that form a broad network in the epidermis, and CD1a autoreactive T cells normally enter the skin in large numbers, there are no studies addressing the specific role of CD1a-reactive T cells in human skin disease. Here we have shown that HDM generates CD1a ligands for recognition by T cells. Such HDM-responsive CD1a-reactive T cells are enriched in the blood and skin of individuals with atopic dermatitis and correlate with IgE and disease severity, and are significantly elevated in the presence of filaggrin null mutations. Furthermore the CD1a-reactive T cells infiltrate the skin 24 hours after HDM challenge, which associates with type 2 cytokine production and phospholipase (PLA2) activity in vivo. We showed that the HDM responsiveness of CD1a-reactive T cells was explained by the presence of PLA2 activity within HDM, and that the PLA2 activity could be inhibited by recombinant filaggrin. These studies identify a pathway of atopic skin inflammation, in which neolipid antigens are generated from the skin by HDM-derived PLA2 for CD1a-mediated presentation to T cells. Loss of filaggrin inhibition of HDM PLA2 may provide neolipid signals to CD1a-reactive T cells that barrier compromise has occurred, with potential inflammatory sequelae.
All previous studies investigating the potential role of antigen-specific T cells in the pathogenesis of atopic dermatitis have focussed on peptide-specific responses that are restricted through HLA class I and class II (15, 16). The current study implicates HDM PLA2 processed skin lipids as a broad antigen class that should be added as potential candidate antigens recognized by T cells and relevant to clinical atopic disease. Langerhans cells are enriched in atopic dermatitis lesions (33), where there is a dysregulated lipid profile (28–32) consistent with a potential role of such cells in presenting lipid to T cells. It is of interest that many known allergens are lipid-binding proteins which may become targets of peptide-specific T cell and IgE responses when conjugated to their lipid cargo. Langerin is a C-type lectin expressed by subsets of dendritic cells including Langerhans cells, and is thought to contribute to lipid-loading of CD1a; it is of interest that langerin has recently been identified in a Genome-Wide Association Study of atopic dermatitis (56). The widely taught Gell and Coombs classification system summarizes a large literature that T cells play a functional role in Type IV hypersensitivity responses in humans, even though the immunogens are often not typical peptide antigens for T cells (57). The mechanisms by which non-peptide antigens lead to T cell-mediated response can involve haptenization of proteins (58), but it is unknown if this is generally true. In most cases the mechanisms for T cell stimulation by small molecules and other non-peptide antigens are unclear. The CD1 system is a particularly attractive candidate to mediate response to non-peptide immunogens, as it evolved to present lipids and small molecules to T cells (59, 60). Through its expression on Langerhans cells and a subset of dermal dendritic cells, CD1a protein is well placed to sample non-peptide antigens that are exposed to the skin. The results presented here suggest a broader model of Gell and Coombs hypersensitivity where neolipid recognition by CD1a-reactive T cells is associated with clinical atopic disease. There may therefore exist a multi-hit process where a CD1a-reactive response may be part of a barrier sensing system that if activated, can induce other adaptive immune responses and cutaneous inflammation. This is germane to animal models, in which cutaneous stress responses enhance sensitization (61). The data may also provide insights to why only certain environmental challenges are particularly allergenic. Such pro-allergenic sources, including house dust mite, may preferentially activate a barrier distress system through generation of inflammatory lipids, leading to immune responses to co-existent proteins and subsequent clinical disease. These processes may be compounded by genetically-determined or acquired filaggrin insufficiency.
CD1a protein is a member of the group 1 CD1 family, along with CD1b, CD1c and CD1e. The group 1 family is not present in mice, yet each member has a distinct cytoplasmic domain, intracellular trafficking and tissue distribution which suggests functional specialization (59). CD1a-autoreactive T cells were found to infiltrate human skin where they could recognize skin derived self-lipids, including fatty acids, presented by primary CD1a-expressing antigen presenting cells (35). We have recently shown that wasp and bee venom derived phospholipase can generate self-lipids for presentation by CD1a protein suggesting shared pathways of skin inflammation, albeit with differing clinical phenotypes dependent on depth of antigen delivery (44). Furthermore phospholipase activity is required for generation of CD1d-restricted NKT cell ligands in a model of hepatitis virus infection (62, 63), and thymic PLA contributes to the control of NKT cell selection (64). It is therefore possible that other CD1s may present shared antigens and contribute to inflammatory skin disease in the presence or absence of CD1a deficiency (65). However no studies have linked CD1a to biological processes relevant in human disease.
Secretory phospholipase A2 cleaves phospholipid to lysophospholipid and the sn-2 acyl chain, and while mechanisms have not been clear, it has long been implicated as having a role in atopic disease (66–73). Although PLAs are likely to have many roles, here we show that PLA-derived lipid products are presented by CD1a for recognition by T cells in the skin. Indeed, we also show that recombinant filaggrin can inhibit the PLA2 activity of HDM and can inhibit the generation of CD1a neolipid ligands for presentation to T cells. This is a hitherto unappreciated function of filaggrin, which may help link the presence of filaggrin insufficiency to cutaneous inflammation. The data also provide a potential resolution to the seemingly contrasting “inside-out” and “outside-in” hypotheses of atopic dermatitis, where instead of two independent possibilities, filaggrin can act in both a barrier function/hydration capacity and also as a direct inhibitor of cutaneous lipid-specific immune responses. However clinical disease may depend on a multi-hit process where filaggrin insufficiency combines with modulations in innate and adaptive immune responses. By inhibiting down-stream innate and T cell effector functions, for example through anti-IL-4Rα, acquired down-regulation of filaggrin may be reversed, leading to enhanced filaggrin inhibition of PLA2, and less barrier distress signals to CD1a-reactive T cells. 20-40% of individuals with moderate-severe atopic dermatitis have filaggrin null mutations, yet acquired filaggrin insufficiency and barrier impairment are common (3, 9). This supports the findings presented herein in which specific aspects related to the downstream immunological events are also important in contributing to the filaggrin insufficiency and compounded atopic cutaneous inflammation through a multi-hit model of disease pathogenesis (9, 74). The data further substantiate the pursuit of therapeutic strategies that modulate relevant immune responses.
Given that PLA2 generation of CD1a ligands has now been shown to be present in three allergens of relevance to humans (44), namely wasp venom, bee venom and HDM, the data suggest that this is part of a broader hypersensitivity system. Indeed phospholipases are known to be present in many other allergens including pollens and fungal allergens (75, 76) and therefore activation of this pathway may facilitate the allergenic process. By producing type 2 cytokines, CD1a-reactive lipid-specific T cells may lead to down-regulation of filaggrin and anti-microbial peptide expression and thus compound physical and antimicrobial barrier dysfunction. Furthermore they may license skin dendritic cells to amplify peptide-specific Th2 responses and subsequent IgE generation. For example, it is of interest that GM-CSF and IL-4 are routinely used to mature monocytes towards cells with CD1 and class I/II antigen presenting capacity. If CD1a-reactive responses do contribute to initiation of the allergic process, then we might predict that allergens delivered to anatomical sites that do not contain antigen presenting cells with high levels of CD1a protein would lead to differing systemic responses. This is indeed the case as wasp venom, bee venom and grass pollen subcutaneous immunotherapy are all known to be highly effective (77, 78). In contrast to the epidermis and dermis, subcutaneous tissue has few CD1a-expressing cells. The findings therefore have potential therapeutic implications. It is of interest that corticosteroids are known to inhibit PLA (79), and it may be that in the skin, this is an important mechanism for controlling CD1a-reactive T cell activity. The development of PLA inhibitors that target individual relevant allergen PLAs may enhance treatment efficacy whilst reducing side effects of broad host PLA inhibition seen with current corticosteroids.
While skin suction blisters offer access to human skin fluid and cells directly ex vivo without the need for further processing, they do add a potential limitation of the study. They are time consuming for donors, and so participant numbers become limiting. Furthermore skin suction blisters inevitably introduce physical trauma to the skin and so it is important to use control comparisons of unchallenged or non-lesional skin when examining challenged or lesional skin respectively. The current study is a cross-sectional analysis of affected individuals, and it will be important to validate the findings in other cross-sectional cohorts and to examine changes longitudinally. Lastly, although it is recognized that translational work has to pass through stages involving human subjects at some point during development, it can be difficult to prove causality in humans. Human skin antigenic challenge does offer temporal associations with clinical and immunological findings, lending support of causality, but CD1a transgenic models and human skin grafts in immunodeficient models may offer further evidence in the future.
In conclusion, we identify a pathway of human skin inflammation where HDM-derived PLA2 generates neolipid antigens for presentation to CD1a-reactive T cells. By also defining a function of filaggrin in inhibiting PLA2, we are able to potentially unify the conflicting “outside-in” and “inside-out” hypotheses of atopic dermatitis. The data would support therapeutic approaches to inhibit allergen-derived PLA2 activity, together with treatments that target the downstream immunological effector pathways.
Materials and Methods
Study design
The study was a laboratory analysis of human blood and skin T cell responses designed to test the hypothesis that CD1a-reactive lipid-specific T cells contribute to the human response to dust mites and atopic dermatitis. Atopic dermatitis was diagnosed according to the UK refinements of the Hanifin and Rajka diagnostic criteria, and adult participants were only excluded if on systemic immunosuppression or topical calcineurin inhibitors. Participants were recruited sequentially; blinding and randomization were not required as there was no intervention. Sample size was determined based on previous studies of CD1a-reactive T cell response frequencies in humans (80). All experiments were replicated as presented in the figure legends.
Isolation of human T-cells
Peripheral blood mononuclear cells (PBMC) were isolated from healthy adult donors and atopic dermatitis patients under local ethics approval (09/H0606/71). Atopic dermatitis was diagnosed using the UK refinements of the Hanifin and Rajka diagnostic criteria and disease severity was assessed using SCORAD. Donors aged 18-67 were recruited with disease severity SCORAD range 5-70. Patients were using topical corticosteroids, but were not on systemic immunosuppression nor topical calcineurin inhibitors. Adult (18-67 years) patients were recruited with moderate severity psoriasis who were not on systemic therapy. T-cells were purified from ficollized peripheral blood mononuclear cells using CD3 MACs beads (Miltenyi Biotec). CD1a reactivity was assessed by IFN-γ ELISPOT (Mabtech AB). ELISpot plates (Millipore Corp) were coated with anti-IFN-γ, anti-GM-CSF or anti-IL-13 antibody overnight (Mabtech AB). K562 cells were pulsed with HDM extract (7μg/ml unless otherwise stated) or 1μg/ml purified bee venom PLAs (Sigma) overnight, and were then washed and re-suspended in R5* (RPMI supplemented with 2mM L-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin plus 5% human serum). The plates were washed six times with RPMI and blocked for 1h with RPMI supplemented with 2 mM L-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin plus 10% human serum (R10*). 20-50,000 T-cells were added per well to which 10-25,000 K562 or primary cells were added. Wells were set-up in duplicate or triplicate. Phorbol myristate acetate 10 ng/ml and Ionomycin 500 ng/ml was included as a positive control, and T-cells alone in the absence of K562 was included as a negative control. After overnight incubation at 37°C and 5% CO2, plates were washed x6 in PBS-Tween 0.05% and incubated with 1 μg/ml of biotin-linked anti-IFN-γ, anti-GM-CSF or anti-IL-13 monoclonal antibody (Mabtech AB) for 2 h. After washing x 6 in PBS-Tween 0.05%, the plates were incubated for a further 1 hour with streptavidin-alkaline phosphatase (Mabtech AB). Spots were visualized using an alkaline phosphatase conjugate substrate kit (Biorad) and enumerated using an automated ELISpot reader (Autimmun Diagnostika gmbh ELISpot Reader Classic). In some experiments 10 μg/mL anti-CD1a blocking antibody (OKT-6), or 10 μg/mL IgG1 isotype control (P3), or 10 μM manoalide, an inhibitor of PLA2 that acts by covalently modifying lysine residues (Enzo Life Sciences), or 0.5 μg/mL Filaggrin monomers were added at to K562 or primary cells before addition of T cells. The frequency of the specific T-cell response was determined in polyclonal T-cell cultures derived from healthy controls (HC) and atopic dermatitis patients (AD) by calculating the mean number of spots/well in the K562-CD1a-HDM pulsed ELISpot wells and subtracting the mean number of spots/well in the K562-EV-HDM pulsed ELISpot wells. The frequency of the CD1a-autoreactive IFNγ T-cell response was determined by calculating the mean number of spots/well in the K562-CD1a unpulsed ELISpot wells and subtracting the mean number of spots/well in the K562-EV unpulsed ELISpot wells. For the Varicella zoster virus (VZV) T-cell responses, T-cells were washed and re-suspended in R5* and 1 vial (103.3 PFU/0.5ml) of Varilrix (live VZV vaccine, GlaxoSmithKline) was resuspended in 500μl R5*. 50μl of the reconstituted virus was incubated with 100,000 T-cells overnight. 50μl R5* was incubated with 100,000 T-cells overnight as a negative control. IFNγ and IL13 production were determined by ELISpot.
Viral-specific T cell lines
We generated HLA-A*0201-restricted GILGFVFTL (influenza A matrix), NLVPMVATV (CMV pp65) and GLCTLVAML (EBV BMLF1)-specific T cells by sorting from peripheral blood mononuclear cells ex vivo with relevant class I tetrameric complexes with maintenance as previously described (81). Cells were cultured in R10 at 2 × 106 cells per well in a 24-well Costar plate. IL-2 was added to a final concentration of 100 U/ml on day 3. The cultures were restimulated after 14 d using HLA-A*0201-positive BCLs pulsed with the appropriate peptides. For the functional assays, cells were pulsed with 200 ng/ml peptide and incubated with 10 μM manoalide, 0.5μg/ml filaggrin monomers or 0.5 μg/mL Sumo recombinant protein control before IFNγ ELISpot as above. ELISpots were performed using a ratio of 40,000 : APC cells (JY) : 5,000 T-cells.Isolation and generation of human APCs. We generated autologous mDC-DCS using CD14+ monocytes. CD14+ human cells were isolated using MACS cell separation (Miltenyi Biotec Inc) according to the supplier’s instructions. These cells were then differentiated in medium containing 50ng/ml GM-CSF and 1000iu/ml IL-4. After 4 days, CD1a expression was confirmed by Flow Cytometry, and the differentiated mDCS were used as antigen presenting cells for the ELISpot assay. The in vitro mDCs were incubated with 10 µg/ml anti-HLA-ABC and anti-HLA-DR blocking antibodies (W6/32 and L243 respectively) for one hour prior to co-culture with T-cells, in order to minimize HLA-restricted responses. We generated autologous LC-like cells in vitro using CD14+ monocytes. CD14+ human cells were isolated using MACS cell separation (Miltenyi Biotec Inc) according to the suppliers’ instructions. Briefly, such in vitro LC-like cells were prepared as previously described (51, 52) from CD14+ cells, which were cultured in 6-well plates in complete medium in the presence of IL-4 (250 ng/ml; PeproTech), GM-CSF (100 ng/ml) and TGF-β1 (10 ng/ml; PeproTech). At days 2 and 4, cultures were re-plated in the presence of the above cytokines to generate cells which were 24.9-26.5% CD1a+CD207+. The in vitro LC-like cells were incubated with 10 µg/ml anti-HLA-ABC and anti-HLA-DR blocking antibodies (W6/32 and L243 respectively) for one hour prior to co-culture with T-cells, in order to minimize HLA-restricted responses.
HDM lipid extraction
HDM lipids were extracted with chloroform, methanol and water using a modified Bligh-Dyer method (82). Briefly, HDM extract was resuspended in a solution of (4:2:1) methanol:chloroform:sample and vortexed before being heated for 30 minutes at 37-40°C. 2 volumes of chloroform and 3 volumes of water were added and the sample was vortexed again and centrifuged at 2-3000 RPM for 5 minutes resulting in separation of the aqueous and organic phases. This process was repeated twice on the aqueous phase to achieve maximal yield. The organic phase was then aspirated, dried by dessication, weighed and dissolved in 0.5% PBS-Tween. The aqueous phase was aspirated, centrifuged at 3000 RPM for 5 minutes and protein-enriched precipitates were resuspended in sterile PBS and concentration determined by Nanodrop.
Blister fluids
Health donor epidermis was injected with 0.7 μg of Dermatophagoides pteronissimus HDM extract (ALK) or saline. After 30 minutes, suction was applied to the skin at 200 mmHg for 1h, which induces a split between the epidermis and dermis (18). Blister fluid was isolated by needle aspiration at 24 hours and the cells were separated by centrifugation. The blister fluid phase was immediately stored at -20oC. Cytokines were measured by multiplex bead array. Blister derived T cells were FACS sorted and expanded using the rapid expansion method. Specifically, T-cells were plated out at 100-150 cells/well into T-cell media in a round-bottom 96-well plate and 50ng/ml of anti-CD3 (OKT3) antibody and irradiated PBMC and EBV transformed B-cells at 150-200,000 cells/well and 40,000 cells/well respectively were added. Blister T-cells were examined and split regularly and on expansion were maintained at a density of approximately 0.5-1 million cells/ml.
PLA2 biochemical activity experiments
PLA2 activity in HDM and blister fluids were detected using site specific substrate kit (Cayman Chemicals), according to manufacturers’ instructions. In a flat-bottom 96-well plate, 10 µl (0.7 μg ) HDM extract (ALK) or blister fluid plus 5 µl assay buffer and 10 µl DTNB, were incubated at RT with 200 µl substrate solution (diheptanoyl thio-PC). For inhibitor/filaggrin studies, HDM/blister fluid was incubated with the appropriate concentration of manoalide or filaggrin for 30 minutes at RT prior to plating out. In the presence of PLA2, cleavage of the substrate at the sn-2 position results in release of the thiol group, which reacts with DTNB to produce a colored precipitate. This is measured with a spectrophotometer over time (415nm) to give a measure of PLA2 activity (Bio-Rad iMark™ Microplate Reader).
Filaggrin synthesis
The expression plasmid was constructed using a sequence- and ligation-independent cloning (SLIC) method (83). Nucleotide sequence encoding 7th filaggrin repeat domain was cloned into pET28 vector using BamHI i XhoI restrictions sites. The construct contains N-terminal His6-tagged SUMO protein sequence which can be enzymatically cleaved by SUMO protease. The construct was transformed into E. coli BL21-CodonPlus-RIL and propagated overnight in LB liquid media containing kanamycin (50 µg/ml) and chloramphenicol (37.5 µg/ml) at 37°C. The bacterial cultures were diluted 1:50 in autoinduction media (Formedium AIM- Super Broth) supplemented with kanamycin and chloramphenicol and incubated for 48 h at 18°C with shaking. The cells were harvested by centrifugation (10 min, 5000 × g, 4°C). The bacteria pellet was mixed with lysis buffer (10 mM Tris pH 8, 150 mM NaCl, 10 mM imidazole) supplemented with protease inhibitors cocktail and lysed by sonication. The cell lysate was clarified by centrifugation (45 min, 70000 × g, 4°C). The following described purification procedure was performed on an ÄKTA™xpress chromatography system. The supernatant after centrifugation was loaded on Ni–NTA Agarose column (Qiagen). Unbound material was washed from the column with lysis buffer. Enriched proteins were subjected to on-column cleave by SUMO protease (20 μg protease for 5 ml resin) in elution buffer (10 mM Tris pH 8, 150 mM NaCl, 300 mM imidazole) for 8 h at 10°C. Realised protein was further purified by combinations of desalting and Ni-NTA Agarose columns equilibrated with the lysis buffer. Flow though fractions containing the purified protein were collected and automatically loaded into a pre-equilibrated column with buffer (10mM Tris pH 8, 150mM NaCl) Superdex 200 column (GE Healthcare). Fractions containing the purified filaggrin were collected and analyzed by SDS-PAGE. Identical purification from cells without the expression plasmid were processed in parallel as a control. In both cases the same fractions from gel filtration column were collected. All experiments were performed in parallel using control SUMO protein.
Filaggrin genotyping
The FLG mutations R501X and 2282del4 were genotyped using Taqman allelic discrimination assays (Life Technologies), as previously described(3). R501X was screened using forward primer 5’ CAC TGG AGG AAG ACA AGG ATC G 3’, reverse primer 5’ CCC TCT TGG GAC GCT GAA 3’ and the probes VIC-CAC GAG ACA GCT C and 6-FAM-CAT GAG ACA GCT CC. 2282del4 was screened using forward primer 5’ CCA CTG ACA GTG AGG GAC ATT CA 3’, reverse primer 5’ GGT GGC TCT GCT GAT GGT GA 3’ and the probes 6-FAM- CAC AGT CAG TGT CAG GCC ATG GAC A and VIC-AGA CAC ACA GTG TCA GGC CAT GGA CA alleles. Assays were performed in 384-well plates with each reaction comprising of 20ng DNA, 2.5 µl Universal PCR master mix and 0.125 µl 40X assay mix in a final reaction volume of 6 µl. Assays were run on an Applied Biosystems 7900HT Fast Real-Time PCR system under the following conditions: 1 cycle at 50°C for 2 minutes followed by 1 cycle at 95°C for 10 minutes then 40 cycles of 95°C 15 sec; 60°C 1 minute. Samples heterozygous for the 2282del4 mutation were also confirmed by Sanger sequencing using the published primers RPT1P7 (5' – AAT AGG TCT GGA CAC TCA GGT - 3') and RPT2P1 (5' – GGG AGG ACT CAG ACT GTT T - 3') (84). PCR conditions were 94°C for 5 min; 35 cycles of 94°C for 40 s, 57°C for 1 min, 72°C for 2 min; final extension step at 72°C for 7 min. PCR clean up and sequencing was performed by Source Bioscience plc.
Statistics
Cohort of healthy donors and atopic dermatitis patients investigated for CD1a-reactive HDM-specific responses were analyzed using the one-tailed unpaired and paired t test, Chi-squared and Pearson correlation. All other polyclonal T-cells responses were analyzed using the unpaired t test. The number of biological replicates for each data point is included in the figure legends. Statistical analyses were performed using Prism 6 (GraphPad software Inc.)
List of Supplementary Materials
Fig. S1. mDC and LC-like cell expression of CD1a and langerin.
Fig. S2. HDM-responsive CD1a-reactive T cells can produce IFNγ and/or IL-13 and are not enriched in patients with psoriasis.
Fig. S3. HDM-responsive CD1a-reactive T cells associate with FLG mutations.
Fig. S4. Viral-specific T cell responses in the presence of manoalide and filaggrin.
Table S1. Source Data
One sentence summary.
Lipid-specific CD1a-reactive T cells are enriched in atopic dermatitis skin, infiltrate after allergen challenge and show increased responses to allergen phospholipase derived neolipid antigens in the context of filaggrin insufficiency.
Acknowledgements
We thank Vanderson Rocha and NHSBT for cord blood samples.
Funding
This work was funded by the MRC and NIHR Biomedical Research Centre, the NIH (NIAMS R01 048632) and the Burroughs Wellcome Foundation Program in Translational Medicine. RJ is funded through a British Association of Dermatologists/British Skin Foundation/MRC Clinical Training Fellowship. GO also acknowledges the support of the National Institute for Health Research Clinical Research Network and support from Janssen Pharmaceuticals. J.B.S. acknowledges the support of a Marie Curie Career Integration Grant. M.S. is supported by Cancer Research UK (Programme Grant C399/A2291 to V.C). PF is supported by Science Foundation Ireland and National Children’s Research Centre.
Footnotes
Author contributions
RJ MS AL SS EB CA KLC CH DC MS DGO JBS HJ AD EP DJ performed experiments, and contributed to writing the paper, and PF AM WB BM VC GO designed experiments and wrote the paper.
Competing interests
The authors declare no competing interests.
Data and materials availability
Data and materials are available through the authors.
References
- 1.Asher MI, Montefort S, Bjorksten B, Lai CK, Strachan DP, Weiland SK, Williams H. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet. 2006;368:733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 2.Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J Allergy Clin Immunol. 2010;125:16–29 e11-11. doi: 10.1016/j.jaci.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, O'Regan GM, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 4.Rodríguez E, Baurecht H, Herberich E, Wagenpfeil S, Brown SJ, Cordell HJ, Irvine AD, Weidinger S. Meta-analysis of filaggrin polymorphisms in eczema and asthma: Robust risk factors in atopic disease. Journal of Allergy and Clinical Immunology. 2009;123:1361–1370.e1367. doi: 10.1016/j.jaci.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 5.Sandford AJ, Shirakawa T, Moffatt MF, Daniels SE, Ra C, Faux JA, Young RP, Nakamura Y, Lathrop GM, Cookson WO, et al. Localisation of atopy and beta subunit of high-affinity IgE receptor (Fc epsilon RI) on chromosome 11q. Lancet. 1993;341:332–334. doi: 10.1016/0140-6736(93)90136-5. [DOI] [PubMed] [Google Scholar]
- 6.Gutowska-Owsiak D, Ogg GS. Cytokine regulation of the epidermal barrier. Clin Exp Allergy. 2013;43:586–598. doi: 10.1111/cea.12023. [DOI] [PubMed] [Google Scholar]
- 7.Gutowska-Owsiak D, Schaupp AL, Salimi M, Selvakumar TA, McPherson T, Taylor S, Ogg GS. IL-17 downregulates filaggrin and affects keratinocyte expression of genes associated with cellular adhesion. Exp Dermatol. 2012;21:104–110. doi: 10.1111/j.1600-0625.2011.01412.x. [DOI] [PubMed] [Google Scholar]
- 8.Gutowska-Owsiak D, Schaupp AL, Salimi M, Taylor S, Ogg GS. Interleukin-22 downregulates filaggrin expression and affects expression of profilaggrin processing enzymes. Br J Dermatol. 2011;165:492–498. doi: 10.1111/j.1365-2133.2011.10400.x. [DOI] [PubMed] [Google Scholar]
- 9.Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, DeBenedetto A, Schneider L, Beck LA, Barnes KC, Leung DYM. Cytokine modulation of atopic dermatitis filaggrin skin expression. Journal of Allergy and Clinical Immunology. 2007;120:150–155. doi: 10.1016/j.jaci.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, Darst MA, Gao B, Boguniewicz M, Travers JB, Leung DY. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol. 2003;171:3262–3269. doi: 10.4049/jimmunol.171.6.3262. [DOI] [PubMed] [Google Scholar]
- 11.Beck LA, Thaci D, Hamilton JD, Graham NM, Bieber T, Rocklin R, Ming JE, Ren H, Kao R, Simpson E, Ardeleanu M, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med. 2014;371:130–139. doi: 10.1056/NEJMoa1314768. [DOI] [PubMed] [Google Scholar]
- 12.Leung DY. Role of IgE in atopic dermatitis. Curr Opin Immunol. 1993;5:956–962. doi: 10.1016/0952-7915(93)90112-6. [DOI] [PubMed] [Google Scholar]
- 13.Seneviratne SL, Jones L, King AS, Black A, Powell S, McMichael AJ, Ogg GS. Allergen-specific CD8(+) T cells and atopic disease. J Clin Invest. 2002;110:1283–1291. doi: 10.1172/JCI15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ardern-Jones M, Black A, Ogg G. Anti-LFA-1 inhibits Th2 function of human allergen-specific CD4+ T cells. British Journal of Dermatology. 2008;158:456–462. doi: 10.1111/j.1365-2133.2007.08393.x. [DOI] [PubMed] [Google Scholar]
- 15.Ardern-Jones M, Black A, Bateman E, Ogg G. Bacterial superantigen facilitates epithelial presentation of antigen to Th2 cells. Proc Natl Acad Sci U S A. 2007;104:5557–5562. doi: 10.1073/pnas.0700733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bateman E, Ardern-Jones M, Ogg G. Persistent central memory phenotype of circulating Fel d 1/DRB1*0101 tetramer-binding CD4+ T cells in adults with severe atopic dermatitis. Journal of Allergy and Clinical Immunology. 2006;118:1350–1356. doi: 10.1016/j.jaci.2006.07.040. [DOI] [PubMed] [Google Scholar]
- 17.Woodfolk JA. T-cell responses to allergens. Journal of Allergy and Clinical Immunology. 2007;119:280–294. doi: 10.1016/j.jaci.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska-Owsiak D, Wang X, Huang LC, Johnson D, Scanlon ST, McKenzie AN, Fallon PG, et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med. 2013;210:2939–2950. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue L, Salimi M, Panse I, Mjosberg JM, McKenzie AN, Spits H, Klenerman P, Ogg G. Prostaglandin D2 activates group 2 innate lymphoid cells via CRTH2. Journal Allergy Clinical Immunology. 2014;133:1184–1194. doi: 10.1016/j.jaci.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Indrasingh I, Chandi G, Jeyaseelan L, Vettivel S, Chandi SM. Quantitative analysis of CD1a (T6) positive Langerhans cells in human tonsil epithelium. Ann Anat. 1999;181:567–572. doi: 10.1016/S0940-9602(99)80066-1. [DOI] [PubMed] [Google Scholar]
- 21.van Haarst JM, Verhoeven GT, de Wit HJ, Hoogsteden HC, Debets R, Drexhage HA. CD1a+ and CD1a- accessory cells from human bronchoalveolar lavage differ in allostimulatory potential and cytokine production. Am J Respir Cell Mol Biol. 1996;15:752–759. doi: 10.1165/ajrcmb.15.6.8969270. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki A, Masuda A, Nagata H, Kameoka S, Kikawada Y, Yamakawa M, Kasajima T. Mature dendritic cells make clusters with T cells in the invasive margin of colorectal carcinoma. J Pathol. 2002;196:37–43. doi: 10.1002/path.1018. [DOI] [PubMed] [Google Scholar]
- 23.Prakash M, Kapembwa MS, Gotch F, Patterson S. Chemokine receptor expression on mucosal dendritic cells from the endocervix of healthy women. J Infect Dis. 2004;190:246–250. doi: 10.1086/422034. [DOI] [PubMed] [Google Scholar]
- 24.Masten BJ, Olson GK, Tarleton CA, Rund C, Schuyler M, Mehran R, Archibeque T, Lipscomb MF. Characterization of myeloid and plasmacytoid dendritic cells in human lung. J Immunol. 2006;177:7784–7793. doi: 10.4049/jimmunol.177.11.7784. [DOI] [PubMed] [Google Scholar]
- 25.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8- cytolytic T lymphocytes. Nature. 1989;341:447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 26.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O'Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, et al. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 27.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nat Immunol. 2010;11:1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saaf AM, Tengvall-Linder M, Chang HY, Adler AS, Wahlgren CF, Scheynius A, Nordenskjold M, Bradley M. Global expression profiling in atopic eczema reveals reciprocal expression of inflammatory and lipid genes. PLoS ONE. 2008;3:e4017. doi: 10.1371/journal.pone.0004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imokawa G. A possible mechanism underlying the ceramide deficiency in atopic dermatitis: expression of a deacylase enzyme that cleaves the N-acyl linkage of sphingomyelin and glucosylceramide. J Dermatol Sci. 2009;55:1–9. doi: 10.1016/j.jdermsci.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa J, Narita H, Kondo N, Hotta M, Takagi Y, Masukawa Y, Kitahara T, Takema Y, Koyano S, Yamazaki S, Hatamochi A. Changes in the ceramide profile of atopic dermatitis patients. J Invest Dermatol. 2010;130:2511–2514. doi: 10.1038/jid.2010.161. [DOI] [PubMed] [Google Scholar]
- 31.Di Nardo A, Wertz P, Giannetti A, Seidenari S. Ceramide and cholesterol composition of the skin of patients with atopic dermatitis. Acta Derm Venereol. 1998;78:27–30. doi: 10.1080/00015559850135788. [DOI] [PubMed] [Google Scholar]
- 32.Scharschmidt TC, Man MQ, Hatano Y, Crumrine D, Gunathilake R, Sundberg JP, Silva KA, Mauro TM, Hupe M, Cho S, Wu Y, et al. Filaggrin deficiency confers a paracellular barrier abnormality that reduces inflammatory thresholds to irritants and haptens. J Allergy Clin Immunol. 2009;124:496–506. 506 e491–506 e496. doi: 10.1016/j.jaci.2009.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gros E, Bussmann C, Bieber T, Forster I, Novak N. Expression of chemokines and chemokine receptors in lesional and nonlesional upper skin of patients with atopic dermatitis. J Allergy Clin Immunol. 2009;124:753–760 e751. doi: 10.1016/j.jaci.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 34.de Lalla C, Lepore M, Piccolo F, Mori L, Dellabona P, Casorati G. High frequency and adaptive-like dynamics of human CD1 self-reactive T cells. European Journal of Immunology. 2011;41:602–610. doi: 10.1002/eji.201041211. [DOI] [PubMed] [Google Scholar]
- 35.de Jong A, Cheng TY, Huang S, Gras S, Birkinshaw RW, Kasmar AG, Van Rhijn I, Pena-Cruz V, Ruan DT, Altman JD, Rossjohn J, Moody DB. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol. 2014;15:177–185. doi: 10.1038/ni.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manolova V, Kistowska M, Paoletti S, Baltariu GM, Bausinger H, Hanau D, Mori L, De Libero G. Functional CD1a is stabilized by exogenous lipids. Eur J Immunol. 2006;36:1083–1092. doi: 10.1002/eji.200535544. [DOI] [PubMed] [Google Scholar]
- 37.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 38.Zajonc DM, Crispin MD, Bowden TA, Young DC, Cheng TY, Hu J, Costello CE, Rudd PM, Dwek RA, Miller MJ, Brenner MB, et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22:209–219. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 40.Birkinshaw RW, Pellicci DG, Cheng TY, Keller AN, Sandoval-Romero M, Gras S, de Jong A, Uldrich AP, Moody DB, Godfrey DI, Rossjohn J. alphabeta T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol. 2015;16:258–266. doi: 10.1038/ni.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–752. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 42.McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, Tarlton A, et al. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204:1131–1144. doi: 10.1084/jem.20062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wun KS, Borg NA, Kjer-Nielsen L, Beddoe T, Koh R, Richardson SK, Thakur M, Howell AR, Scott-Browne JP, Gapin L, Godfrey DI, et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bourgeois E, Subramaniam S, Cheng T-Y, De Jong A, Layre E, Ly D, Salimi M, Legaspi A, Modlin RL, Salio M, Cerundolo V, et al. Bee venom processes human skin lipids for presentation by CD1a. Journal of Experimental Medicine. 2015;212:149–163. doi: 10.1084/jem.20141505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agea E, Russano A, Bistoni O, Mannucci R, Nicoletti I, Corazzi L, Postle AD, De Libero G, Porcelli SA, Spinozzi F. Human CD1-restricted T cell recognition of lipids from pollens. Journal of Experimental Medicine. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werfel T, Morita A, Grewe M, Renz H, Wahn U, Krutmann J, Kapp A. Allergen specificity of skin-infiltrating T cells is not restricted to a type-2 cytokine pattern in chronic skin lesions of atopic dermatitis. J Invest Dermatol. 1996;107:871–876. doi: 10.1111/1523-1747.ep12331164. [DOI] [PubMed] [Google Scholar]
- 47.Suarez-Farinas M, Tintle SJ, Shemer A, Chiricozzi A, Nograles K, Cardinale I, Duan S, Bowcock AM, Krueger JG, Guttman-Yassky E. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127:954–964 e951-954. doi: 10.1016/j.jaci.2010.12.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebane A, Zimmermann M, Aab A, Baurecht H, Koreck A, Karelson M, Abram K, Metsalu T, Pihlap M, Meyer N, Folster-Holst R, et al. Mechanisms of IFN-gamma-induced apoptosis of human skin keratinocytes in patients with atopic dermatitis. J Allergy Clin Immunol. 2012;129:1297–1306. doi: 10.1016/j.jaci.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 49.Gittler JK, Shemer A, Suarez-Farinas M, Fuentes-Duculan J, Gulewicz KJ, Wang CQ, Mitsui H, Cardinale I, de Guzman Strong C, Krueger JG, Guttman-Yassky E. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J Allergy Clin Immunol. 2012;130:1344–1354. doi: 10.1016/j.jaci.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haugaard L, Dahl R, Jacobsen L. A controlled dose-response study of immunotherapy with standardized, partially purified extract of house dust mite: clinical efficacy and side effects. J Allergy Clin Immunol. 1993;91:709–722. doi: 10.1016/0091-6749(93)90190-q. [DOI] [PubMed] [Google Scholar]
- 51.Caux C, Massacrier C, Vanbervliet B, Dubois B, Durand I, Cella M, Lanzavecchia A, Banchereau J. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to granulocyte-macrophage colony-stimulating factor plus tumor necrosis factor alpha: II. Functional analysis. Blood. 1997;90:1458–1470. [PubMed] [Google Scholar]
- 52.Geissmann F, Prost C, Monnet J-P, Dy M, Brousse N, Hermine O. Transforming Growth Factor beta 1, in the Presence of Granulocyte/Macrophage Colony-stimulating Factor and Interleukin 4, Induces Differentiation of Human Peripheral Blood Monocytes into Dendritic Langerhans Cells. J Exp Med. 1998;187:961–966. doi: 10.1084/jem.187.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4 - 8 - T lymphoyctes to a microbial antigen. Nature. 1992;360:593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- 54.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumour necrosis factor alpha. Journal of Experimental Medicine. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richards S, Scott IR, Harding CR, Liddell JE, Powell GM, Curtis CG. Evidence for filaggrin as a component of the cell envelope of the newborn rat. Biochem J. 1988;253:153–160. doi: 10.1042/bj2530153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.E. G. a. L. E. E Consortium. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–1456. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gell PG, Coombs RR. Aspects of Immunology. ed. 1. Blackwell; Oxford: 1963. [Google Scholar]
- 58.Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, Bharadwaj M, Miles JJ, Kjer-Nielsen L, Gras S, Williamson NA, Burrows SR, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554–558. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
- 59.Kasmar A, Van Rhijn I, Moody DB. The evolved functions of CD1 during infection. Curr Opin Immunol. 2009;21:397–403. doi: 10.1016/j.coi.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annu Rev Immunol. 2014;32:323–366. doi: 10.1146/annurev-immunol-032713-120243. [DOI] [PubMed] [Google Scholar]
- 61.Strid J, Sobolev O, Zafirova B, Polic B, Hayday A. The intraepithelial T cell response to NKG2D-ligands links lymphoid stress surveillance to atopy. Science. 2011;334:1293–1297. doi: 10.1126/science.1211250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, Trott DL, Ndonye RM, Veerapen N, Besra GS, Howell AR, et al. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biology. 2099;7:e1000228. doi: 10.1371/journal.pbio.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeissig S, Murata K, Sweet L, Publicover J, Hu Z, Kaser A, Bosse E, Iqbal J, Hussain MM, Balschun K, Rocken C, et al. Hepatitis B virus-induced lipid alterations contribute to natural killer T cell-dependent protective immunity. Nat Med. 2012;18:1060–1068. doi: 10.1038/nm.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paduraru C, Bezbradica JS, Kunte A, Kelly R, Shayman JA, Veerapen N, Cox LR, Besra GS, Cresswell P. Role for lysosomal phospholipase A2 in iNKT cell-mediated CD1d recognition. Proc Natl Acad Sci U S A. 2013;110:5097–5102. doi: 10.1073/pnas.1302923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seshadri C, Shenoy M, Wells RD, Hensley-McBain T, Andersen-Nissen E, McElrath MJ, Cheng TY, Moody DB, Hawn TR. Human CD1a deficiency is common and genetically regulated. J Immunol. 2013;191:1586–1593. doi: 10.4049/jimmunol.1300575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chilton FH, Averill FJ, Hubbard WC, Fonteh AN, Triggiani M, Liu MC. Antigen-induced generation of lyso-phospholipids in human airways. J Exp Med. 1996;183:2235–2245. doi: 10.1084/jem.183.5.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sane AC, Mendenhall T, Bass DA. Secretory phospholipase A2 activity is elevated in bronchoalveolar lavage fluid after ovalbumin sensitization of guinea pigs. J Leukoc Biol. 1996;60:704–709. doi: 10.1002/jlb.60.6.704. [DOI] [PubMed] [Google Scholar]
- 68.Bowton DL, Seeds MC, Fasano MB, Goldsmith B, Bass DA. Phospholipase A2 and arachidonate increase in bronchoalveolar lavage fluid after inhaled antigen challenge in asthmatics. Am J Respir Crit Care Med. 1997;155:421–425. doi: 10.1164/ajrccm.155.2.9032172. [DOI] [PubMed] [Google Scholar]
- 69.Chung YW, Oh HY, Kim JY, Kim JH, Kim IY. Allergen-induced proteolytic cleavage of annexin-1 and activation of cytosolic phospholipase A2 in the lungs of a mouse model of asthma. Proteomics. 2004;4:3328–3334. doi: 10.1002/pmic.200400895. [DOI] [PubMed] [Google Scholar]
- 70.Hallstrand TS, Chi EY, Singer AG, Gelb MH, Henderson WR., Jr Secreted phospholipase A2 group X overexpression in asthma and bronchial hyperresponsiveness. Am J Respir Crit Care Med. 2007;176:1072–1078. doi: 10.1164/rccm.200707-1088OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Granata F, Nardicchi V, Loffredo S, Frattini A, Ilaria Staiano R, Agostini C, Triggiani M. Secreted phospholipases A(2): A proinflammatory connection between macrophages and mast cells in the human lung. Immunobiology. 2009;214:811–821. doi: 10.1016/j.imbio.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 72.Triggiani M, Giannattasio G, Calabrese C, Loffredo S, Granata F, Fiorello A, Santini M, Gelb MH, Marone G. Lung mast cells are a source of secreted phospholipases A2. J Allergy Clin Immunol. 2009;124:558–565. doi: 10.1016/j.jaci.2009.04.035. 565 e551-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoder M, Zhuge Y, Yuan Y, Holian O, Kuo S, van Breemen R, Thomas LL, Lum H. Bioactive lysophosphatidylcholine 16:0 and 18:0 are elevated in lungs of asthmatic subjects. Allergy Asthma Immunol Res. 2014;6:61–65. doi: 10.4168/aair.2014.6.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gutowska-Owsiak D, Ogg G. Cytokine regulation of the epidermal barrier. Clinical & Experimental Allergy. 2012;21:104–110. doi: 10.1111/cea.12023. [DOI] [PubMed] [Google Scholar]
- 75.Iorio RA, Del Duca S, Calamelli E, Pula C, Lodolini M, Scamardella F, Pession A, Ricci G. Citrus allergy from pollen to clinical symptoms. PLoS ONE. 2013;8:e53680. doi: 10.1371/journal.pone.0053680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakahama T, Nakanishi Y, Viscomi AR, Takaya K, Kitamoto K, Ottonello S, Arioka M. Distinct enzymatic and cellular characteristics of two secretory phospholipases A2 in the filamentous fungus Aspergillus oryzae. Fungal Genet Biol. 2010;47:318–331. doi: 10.1016/j.fgb.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Durham SR, Walker SM, Varga EM, Jacobson MR, O'Brien F, Noble W, Till SJ, Hamid QA, Nouri-Aria KT. Long-term clinical efficacy of grass-pollen immunotherapy. N Engl J Med. 1999;341:468–475. doi: 10.1056/NEJM199908123410702. [DOI] [PubMed] [Google Scholar]
- 78.Diwakar L, Noorani S, Huissoon AP, Frew AJ, Krishna MT. Practice of venom immunotherapy in the United Kingdom: a national audit and review of literature. Clin Exp Allergy. 2008;38:1651–1658. doi: 10.1111/j.1365-2222.2008.03044.x. [DOI] [PubMed] [Google Scholar]
- 79.Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids--new mechanisms for old drugs. N Engl J Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 80.Subramaniam S, Aslam A, Misbah SA, Salio M, Cerundolo V, Moody DB, Ogg G. Elevated and cross-responsive CD1a-reactive T cells in bee and wasp venom allergic individuals. Eur J Immunol. 2015 doi: 10.1002/eji.201545869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salio M, Shepherd D, Dunbar PR, Palmowski M, Murphy K, Wu L, Cerundolo V. Mature dendritic cells prime functionally superior melan-A-specific CD8+ lymphocytes as compared with nonprofessional APC. J Immunol. 2001;167:1188–1197. doi: 10.4049/jimmunol.167.3.1188. [DOI] [PubMed] [Google Scholar]
- 82.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 83.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 84.Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, Liao H, Evans AT, Goudie DR, Lewis-Jones S, Arseculeratne G, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.