Abstract
Vancomycin-resistant Enterococcus faecalis (VREfs) is an important nosocomial pathogen1,2. We undertook whole genome sequencing of E. faecalis associated with bloodstream infection in the UK and Ireland over more than a decade to determine the population structure and genetic associations with hospital adaptation. Three lineages predominated in the population, two of which (L1 and L2) were nationally distributed, and one (L3) geographically restricted. Genome comparison with a global collection identified that L1 and L3 were also present in the USA, but were genetically distinct. Over 90% of VREfs belonged to L1–L3, with resistance acquired and lost multiple times in L1 and L2, but only once followed by clonal expansion in L3. Putative virulence and antibiotic resistance genes were over-represented in L1, L2 and L3 isolates combined, versus the remainder. Each of the three main lineages contained a mixture of vancomycin-resistant and -susceptible E. faecalis (VSEfs), which has important implications for infection control and antibiotic stewardship.
Enterococci are the second and third most frequent cause of nosocomial infections in the USA and Europe, respectively1,2, with Enterococcus faecalis the most commonly isolated species2. Vancomycin is the first-line antimicrobial drug for enterococci with high-level resistance to ampicillin or for patients with penicillin allergy. Vancomycin resistance was first reported in 19883 and has subsequently increased in prevalence. This rise was predominantly due to E. faecium, but E. faecalis accounted for 11% of vancomycin-resistant Enterococcus (VRE) bacteraemias in the UK and Ireland (UK&I) between 2001 and 2013 (http://www.bsacsurv.org). Based on multi-locus sequence typing (MLST)4, it has been determined that vancomycin resistance in E. faecalis has arisen in multiple genetic backgrounds5,6 and is associated with epidemic lineages7. Microbial genome sequencing provides the opportunity to gain a detailed understanding of the molecular basis for hospital adaptation. The first whole-genome sequence of E. faecalis was published in 20038. Subsequent genome studies have compared 18 E. faecalis strains, demonstrating the contribution of mobile genetic elements to the diversity of the species9, and 25 clinical and 7 non-clinical isolates, which were found to be comparable in gene content10. The ability to sequence large bacterial collections means that the molecular epidemiology and gene content of epidemic and sporadic lineages can now be defined systematically.
We sequenced 168 E. faecalis isolates (58 vancomycin-resistant E. faecalis (VREfs) and 110 vancomycin-susceptible E. faecalis (VSEfs)) from national (British Society for Antimicrobial Chemotherapy (BSAC) n = 94), local (Cambridge University Hospitals NHS Foundation Trust (Addenbrooke’s Hospital and The Rosie Hospital) (CUH) n = 60) and reference (National Collection of Type Cultures (NCTC) n = 14) collections (Supplementary Table 1). BSAC isolates originated from 21 UK&I hospitals between 2001 and 2011 (see Supplementary Fig. 1 for the geographical and temporal distributions) and CUH isolates were collected between 2006 and 2012. All BSAC and CUH isolates were associated with bloodstream infection. NCTC isolates were from humans, livestock and food products, and were predominantly isolated before 1951 (10/14 isolates).
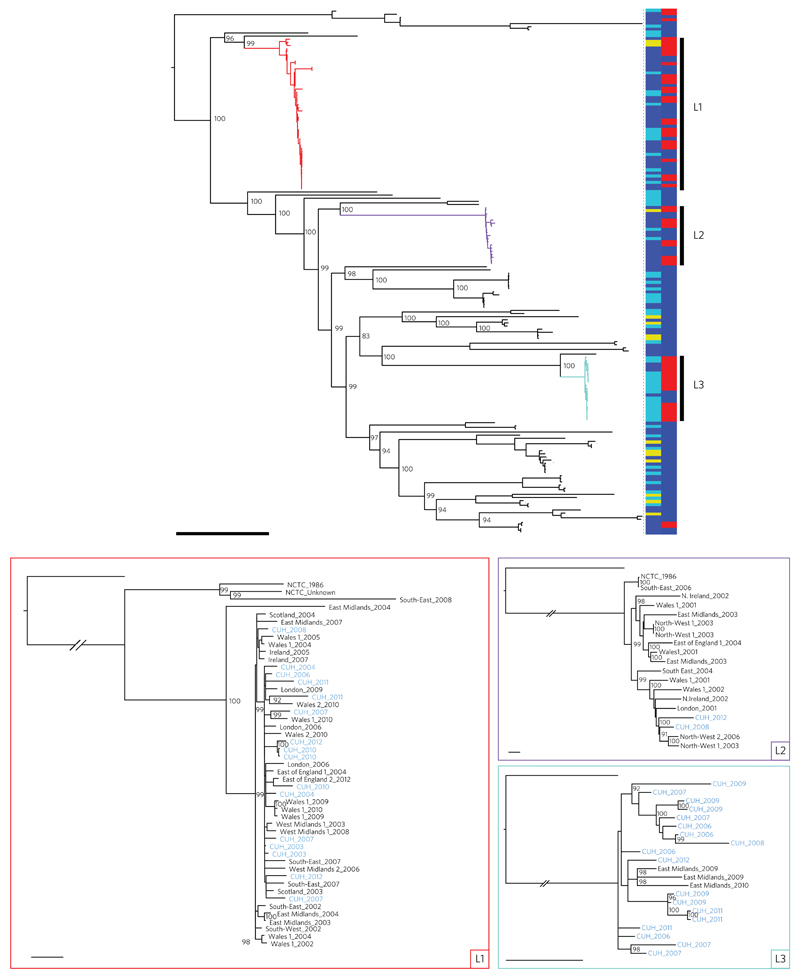
A comparison of these genomes against the core genome of E. faecalis V583 identified 124,194 single nucleotide polymorphisms (SNPs) over 2,886,189 nucleotides (Fig. 1). A striking feature of the phylogenetic tree based on these SNPs was that 53% of isolates clustered into three distinct lineages (termed L1, L2 and L3). L1 was represented in each year of the collection, while L2 was most frequently represented between 2001 and 2006, after which there were only two isolates identified that belonged to this lineage (Supplementary Fig. 1). This suggests clonal replacement, a phenomenon observed for other hospital-related pathogens such as methicillin-resistant Staphylococcus aureus11. Annotation of lineage-specific trees with geographical location demonstrated that L1 and L2 were nationally distributed (epidemic) clones, while L3 was only isolated in two locations (CUH and a hospital in the East Midlands referral network12) (Fig. 1). L3 was the dominant lineage at CUH (17/60 study isolates) and accounted for 3/13 isolates from a hospital in the East Midlands, the phylogenetic tree supporting a single introduction into this hospital followed by local diversification. We also explored the phylogenetic origins of VREfs by including all available E. faecalis isolates from the NCTC collection. Eleven of 14 NCTC isolates clustered with recent clinical isolates (Fig. 1), including seven isolated before 1951 and two VREfs from 1986 (the first year that VREfs was recognized) that belonged to L1 or L2, which in the case of L2 may represent a founder of the circulating vancomycin-resistant E. faecalis lineage.
Figure 1. Phylogeny of E. faecalis isolates drawn from across the UK and Ireland.
Top: midpoint rooted maximum likelihood tree of 168 E. faecalis isolates based on SNPs in the core genome. Coloured branches indicate the three dominant lineages (L1, red; L2, purple; L3, turquoise). Vertical bars show the source of each isolate on the left (dark blue, BSAC; light blue, CUH; yellow, NCTC) and the presence (red) or absence (blue) of vancomycin resistance determinants on the right. Bootstrap supports over 90% are labelled for the major nodes. Scale bar, 10,000 SNPs. Bottom: maximum likelihood trees of the three dominant lineages (L1, red; L2, purple; L3, turquoise) based on SNPs in the core genome after recombination was removed, and rooted on an outlier. The trees are labelled by referral network, with ‘1’ and ‘2’ indicating different hospitals within the referral network if more than one contributed to the BSAC study collection, and year of isolation with CUH isolates highlighted in blue. Bootstrap supports over 90% are labelled. Scale bars, 25 SNPs.
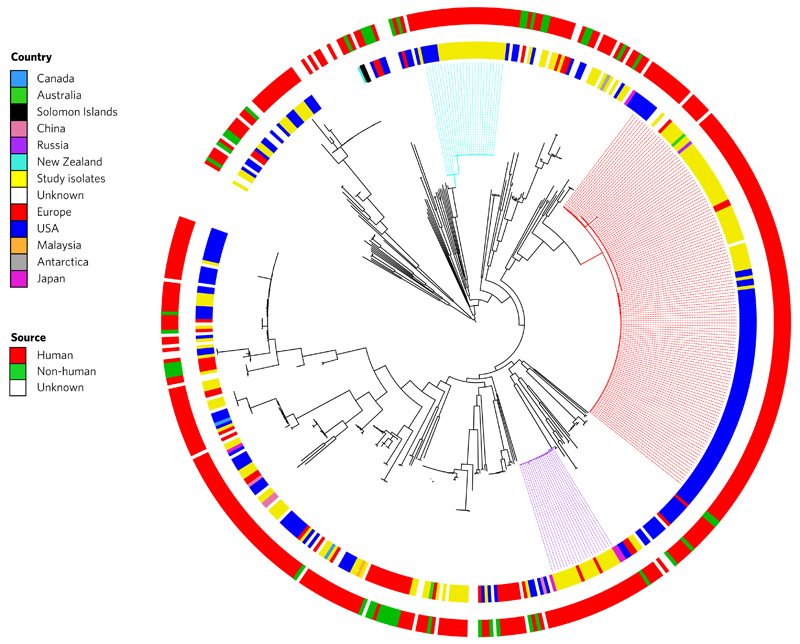
To place our collection into a global context we compared these to the E. faecalis genomes of isolates from around the world. This was achieved by retrieving all of the E. faecalis genomes (n = 353) held by the European Nucleotide Archive (ENA) as of 10 September 2015, and combining our data with 347 of these (excluding six based on data quality). The phylogenetic tree based on 1,293 genes conserved in 99% of these 515 isolates revealed that isolates contained within L1 and L3 that originated from the UK and USA were genetically distinct (Fig. 2). This indicates independent clonal expansion of dominant lineages with limited international dissemination. One explanation for this is that these lineages are hospital-associated, with limited carriage beyond hospitals. Studies investigating community carriage of VRE in the USA and UK have failed to identify VREfs13,14. By contrast, global isolates from the non-dominant STs were closely related to UK isolates.
Figure 2. Global population structure of E. faecalis.
Phylogeny of 168 study isolates combined with 347 isolates from geographically diverse locations downloaded from the European Nucleotide Archive (ENA). Maximum likelihood tree is based on SNPs in the 1,293 genes conserved in 99% of isolates. Coloured branches indicate the three dominant lineages (L1, red; L2, purple; L3, turquoise). The inner coloured ring indicates the country of isolation, and the outer coloured ring indicates the source of the isolate.
To compare our findings with those of published studies based on MLST, we assigned sequence types (STs) to all 168 study isolates (Supplementary Table 1). Isolates in L1 were assigned to ST6, ST384 and ST642 (clonal complex (CC)2), and L2 isolates were ST28 and ST640 (CC87). Both CCs have been described as high-risk lineages based on their association with hospital-derived isolates in Europe5–7. L3 isolates were ST103 (CC388), which has only been reported previously in relation to five clinical and two faecal isolates, all from the Americas10.
Comparison of the number of core genome SNPs for L1, L2 and L3 revealed a lower genetic diversity for L3 (range 3–60 SNPs, median 33 SNPs) than for L1 (range 2–375 SNPs, median 30 SNPs) and L2 (range 0–237 SNPs, median 139 SNPs). This led us to use Bayesian Evolutionary Analysis Sampling Trees (BEAST) to date these lineages15. The last common ancestor of L3 was estimated to be 1998 (95% highest posterior density (HPD) interval, 1980–2004) (Supplementary Fig. 2a), consistent with the earliest reported isolation of ST103 in the literature of 200210. The last common ancestor of L1 was predicted to be 1918 (95% HPD interval, 1868–1960), with a clonal expansion in 1997 (95% HPD interval, 1992–2000) (Supplementary Fig. 2b). The early estimate for the last common ancestor of L1 relied on just three outlying isolates, so a second algorithm was used to detect and remove recombination events, and the BEAST analysis was repeated to rule out the role of undetected recombination. This predicted a last common ancestor in 1852 (95% HPD interval, 1811–1956). It has been proposed previously that CC2 (L1) emerged recently, based on the lack of isolates identified prior to the 1980s16. Our analysis indicates that this lineage may have been in existence since the mid-1850s to early 1900s, with a clonal expansion in the 1990s. BEAST analysis of L2 failed, probably because of a limited number of isolates with high genetic diversity and wide temporal spread.
Establishing the rate of mutation in the core genome provides a molecular clock that contextualizes analyses of bacterial genomes during putative outbreak investigations17–19, but has not been defined previously for E. faecalis. The rate of evolution was estimated to be 8.18 × 10−7 SNPs/site/year (approximately 2.5 SNPs/year) for L1 and 1.14 × 10−6 SNPs/site/year (approximately 3.4 SNPs/year) for L3. Based on these mutation rates and patient ward locations, we excluded direct patient-to-patient transmission of the CUH study isolates.
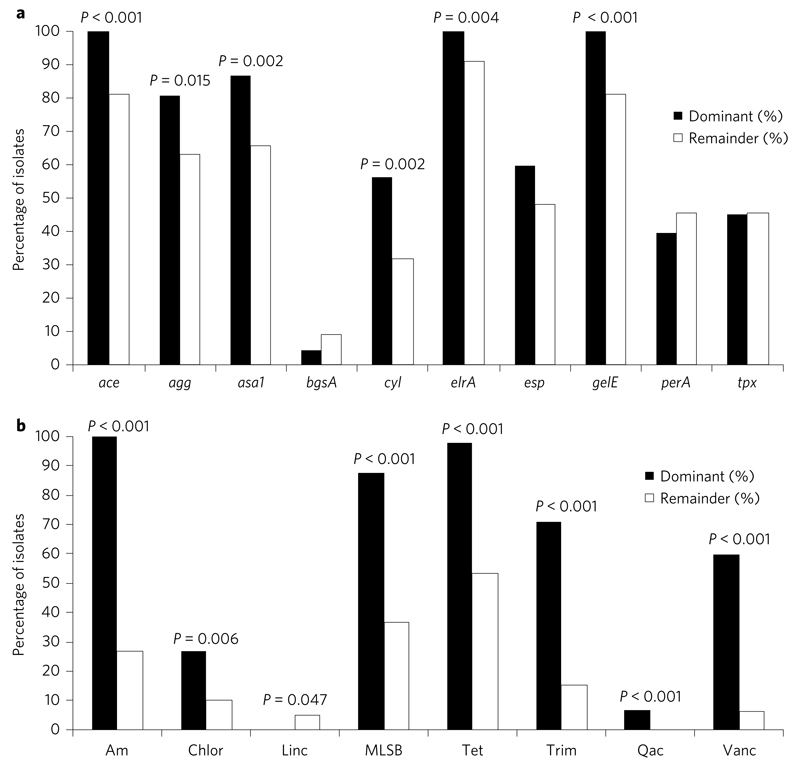
We explored the genetic basis for the success of the dominant E. faecalis lineages using a candidate gene approach by comparing the prevalence of putative virulence and antibiotic resistance genes in L1, L2 and L3 isolates combined, versus the remainder. ace, gelE, asa1, agg, cyl, elrA and genes conferring resistance to tetracyclines, aminoglycosides, trimethoprim, chloramphenicol, macrolides/lincosamides/streptogramin B (MLSB), quaternary ammonium compounds (qacs) and vancomycin were over-represented in L1–L3 compared to the rest (Fig. 3). There was a striking difference in the prevalence of genes encoding aminoglycoside and vancomycin resistance, two commonly used antibiotics for enterococcal infection, in dominant versus non-dominant lineages. Our findings extend previous reports that epidemic lineages are enriched for multi-drug resistance and specific virulence determinants6,7. We then compared the prevalence of the candidate virulence genes in VREfs versus VSEfs contained in L1, L2 and L3 (Supplementary Fig. 3). This showed no significant difference, indicating that over-representation of virulence genes is lineage- rather than VRE-specific.
Figure 3. Prevalence of virulence and antibiotic resistance genes in the dominant lineages (L1–L3, n = 89) and remainder (n = 79).
a,b, Graphs showing percentage of isolates for which putative virulence genes (a) or antibiotic resistance genes (grouped by antibiotic class) (b) were detected. Genes that were ubiquitous in the collection are not shown. P values are shown when a significant difference was observed using Fisher’s exact test. Virulence genes: ace = collagen adhesion protein; agg = aggregation substance; asa1 = aggregation substance; bgsA = biofilm-associated glycolipid synthesis A; cyl = cytolysin; elrA = enterococcal leucine-rich protein A; esp = enterococcal surface protein; gelE = gelatinase; perA = pathogenicity island-encoded regulator; tpx = thiol peroxidase. Antibiotic resistance genes: Am = aminoglycosides (comprising one or more of aac6′-2″, aph3″-III, aacA, ant-6-Ia, str); Chlor = chloramphenicol (cat); Linc = lincosamides (lnuB); MLSB = macrolide, lincosamide, streptogramin B (comprising ermB or ermT); Tet = tetracycline (comprising one or more of tetL, tetM, tetO, tetS); Trim = trimethoprim (comprising dfrC, dfrD, dfrF or dfrG); Qac = quaternary ammonium compounds and other antiseptics (qacZ); Vanc = vancomycin.
We then analysed the pangenome20 of the 168 isolates to obtain a more detailed understanding of their entire genomic repertoire. This indicated that E. faecalis has an open genome with a gamma value of 0.21 (Supplementary Fig. 4), corroborating results derived previously from the analysis of five genomes21. The pangenome contained 8,202 genes, of which 1,967 were conserved across the collection. Of the 6,235 genes in the accessory genome, 1,687 were present just once. The most common accessory genes encoded hypothetical proteins (n = 2,558), insertion sequence (IS) elements or transposons (n = 177), phage or plasmid-associated proteins (n = 462 and n = 113 respectively), transcriptional regulators (n = 225), ABC transporters or cassettes (n = 124), and phosphotransferase systems (n = 118). A total of 819 genes were only found in the three dominant lineages, of which 109 were present in more than 10 isolates (Supplementary Table 2), including a WxL domain surface protein unique to L1. Comparison of the amino acid sequence of the WxL protein from L1 to the proteome of V583 revealed a 100% match to EF_3248, one of 27 WxL proteins identified by Brinster and co-authors22. No genes or homoplasic non-synonymous SNPs were ubiquitous in the dominant lineages and absent from all sporadic lineages, suggesting that there is no single factor that contributed to the emergence of these dominant clones, although antibiotic resistance and virulence determinants are likely to represent multifactorial contributory factors. Analysis of non-synonymous SNPs unique to L3 revealed 122 SNPs in 95 genes (Supplementary Table 3), but no single genetic event was identified that might explain the geographically constrained success of this lineage.
Recombination is thought to be a major mechanism by which the E. faecalis genome evolves, which led us to estimate sites of recombination in the core genome23 for L1, L2 and L3 (Supplementary Fig. 5). Recombination accounted for 12.3% of the core genome in L1 (6.5% related to a large recombination event in two isolates) and 3.9% in L2, with a single predicted 4 bp recombination event in one L3 isolate. This contrasts with reports that recombination across the species is high4, which led us to use an alternative algorithm (BratNextGen24) to detect recombination. This revealed similarly low levels of recombination in L2 and L3 (6.2% and 0.3%, respectively) but higher rates in L1 (37%), although most of this (93%) was contained within two large recombination events (Supplementary Fig. 5). One possible explanation for the low levels of recombination is that this drove the initial diversification of the species but subsequently contributed little to short-term evolution.
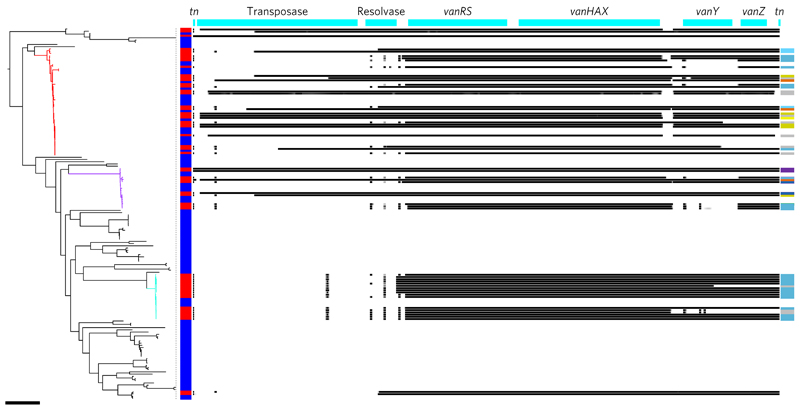
Finally, we analysed the genetic basis of vancomycin resistance in the collection. Nearly all VREfs (57/58) carried vanA, with a single NCTC isolate carrying vanB. Annotation of the tree with resistance to vancomycin showed that all three dominant lineages contained a mixture of VREfs and VSEfs, with 89% of BSAC and 95% of CUH VREfs belonging to L1–L3. Based on mapping to a reference vanA transposon (Tn1546), there was no SNP-based variation between transposons, with the exception of one that had a C→T substitution at position 5745. However, there was substantial variation in gene content. The transposase, resolvase, vanY and vanZ genes were not detected in some isolates, but despite this the minimum inhibitory concentration (available for the 35 vanA positive BSAC isolates) was consistently very high (≥256 mg l−1). There was considerable variation in genetic content within and between the L1 and L2 transposon, while L3 had limited variation, with two variants relating to VREfs isolated in 2006–2009 and 2009–2012, respectively, and three partial deletions (Fig. 4). Analysis of the insertion sites for Tn1546 revealed multiple insertion sites for L1 and L2, but only one site was identified for L3 in the 11/14 genomes for which this analysis proved possible (Fig. 4 and Supplementary Table 4). Analysis using BLAST revealed that these insertion sites were best matched to plasmids, a finding corroborated using plasmid extraction and vanA hybridization for insertion site types 1A, 1B, 2B and 3 (data not shown). These data indicate multiple acquisition and loss of the vanA transposon in L1 and L2, suggesting a significant fitness cost. By contrast, the single acquisition followed by clonal expansion in L3 suggests that the transposon has negligible cost or confers a benefit in this lineage. Foucault et al.25 demonstrated that its integration site in the chromosome predominantly determined the fitness cost of the vanB transposon. One possible reason for the retention of vanA in L3 is that the transposon has inserted into the plasmid at a location that lacks a fitness cost to the bacterium. However, vanA is inserted at the same site in ten isolates from L1 and L2, and there is limited evidence for retention of vanA in these isolates.
Figure 4. Mapping variation in the vancomycin resistance transposon.
Midpoint rooted maximum likelihood tree of all 168 E. faecalis with the three dominant lineages highlighted (L1, red; L2, purple; L3, turquoise) and the presence (red) or absence (blue) of a van transposon indicated in the vertical bar. Right: coverage plot (number of sequence reads that map to that location) of the vanA transposon in each isolate, with black indicating presence (30× coverage or above), graduating to white indicating absence (less than 10× coverage). The genes are labelled in the top bar (tn = inverted repeat). Colours in the vertical bar on the right indicate the different insertion sites in the three dominant lineages, with a description of each colour provided in Supplementary Table 4. Scale bar, 10,000 SNPs.
In conclusion, whole genome sequencing of E. faecalis has highlighted the dominance of epidemic lineages in the UK&I, but has also shown that a lineage with features of an epidemic lineage was confined to two hospitals. Additionally, we identified that the UK and USA have genetically distinct populations belonging to two of these lineages, suggesting a lack of international transmission. The mutation rate defined here will have utility in clinical practice as sequencing technology is introduced into the investigation of putative outbreaks. Genome-level data provided comprehensive insights into the gene content of dominant versus sporadic lineages and allowed us to describe the evolution of vancomycin resistance in this collection, which included multiple loss and acquisition events. The observation that the major VREfs lineages were also the common lineages for VSEfs has important implications for infection control and antibiotic stewardship, because the control of VREfs is likely to depend on defining and addressing drivers for VSEfs and its transmission.
Methods
Ethical approval
This study was approved by the National Research Ethics Service (ref. 12/EE/0439) and the Cambridge University Hospitals NHS Foundation Trust (CUH) Research and Development (R&D) Department.
Isolate collection
The 168 E. faecalis isolates used in this study were selected from three collections: NCTC (n = 14, deposited between 1927 and 2007), BSAC (n = 94, isolated between 2001 and 2011) and CUH (n = 60, isolated between November 2006 and December 2012). The collection was enriched for vancomycin-resistant isolates by selecting all of the available VREfs from NCTC (n = 3) and BSAC (n = 35) and the first stored isolate from all cases of VREfs bacteraemia at CUH (n = 20). To relate this to the underlying VSEfs population, 110 VSEfs were selected as follows: (1) all available VSEfs from the NCTC (n = 11), (2) 59 VSEfs from BSAC (35 matched to the BSAC VREfs cases by hospital and year of isolation, where available, and an additional 24 VSEfs to gain a greater representation of the VSEfs population); (3) 40 VSEfs from CUH (the first stored bacteraemia-associated isolate matched to CUH VREfs cases by isolation date (n = 17), or that occurred 30 days or more after admission (n = 19), and four additional VSEfs that were available to increase the representation of the local population). BSAC hospitals were assigned to referral networks described previously12, which are clusters of hospitals more likely to exchange patients within the cluster than outside of that cluster.
Microbiology and sequencing
Bacterial isolates were cultured on Columbia Blood Agar (Oxoid) and incubated at 37 °C for 48 h in air. Vancomycin susceptibility was determined using the agar dilution method26 (BSAC isolates) or the Vitek2 instrument (Biomerieux) with the AST-P607 card (CUH and NCTC VREfs isolates). DNA was extracted using QIAxtractor (QIAgen), according to the manufacturer’s instructions. Library preparation was conducted according to the Illumina protocol and sequencing was performed on an Illumina HiSeq2000 with 100-cycle paired-end runs. Sequence data for all isolates have been submitted to the ENA (www.ebi.ac.uk/ena) with the accession numbers shown in Supplementary Table 1.
Phylogenetic analyses
Sequence reads were mapped using SMALT (http://www.sanger.ac.uk/resources/software/smalt/) to the E. faecalis reference genome V583 (ENA accession number AE016830) for collection-wide analysis. This reference was selected because it is one of only two finished E. faecalis genomes from clinical isolates and has been used in multiple studies (the second complete genome having only been published in 2014). For analysis of lineages L1, L2 and L3, the oldest isolate from each lineage was selected as a reference for mapping and an assembly created using Velvet. Mobile genetic elements were identified using gene annotation, PHAST27 (phast.wishartlab.com), WebACT28 (http://www.webact.org) and BLAST29 (blast.ncbi.nlm.nih.gov) and were excluded in addition to contigs less than 500 bp in length to create a ‘core’ genome. The core genome sizes were 2,886,189 bp, 2,698,500 bp, 2,372,434 bp and 2,707,007 bp for V583, L1, L2 and L3, respectively. SNPs in the core genome were determined using an in-house script and were used to estimate maximum likelihood trees using RAxML30 with 100 bootstraps. Recombination was removed from the lineage-specific analyses using Gubbins23. To place the isolates into a global context, all of the available E. faecalis sequences listed in GenBank were downloaded from the ENA (n = 353). Six isolates were excluded due to poor assemblies/annotation. The assemblies of the remaining 347 isolates were combined with the assemblies of the study isolates (created using Velvet), annotated with Prokka, and a pangenome estimated using Roary20. A 90% identity cutoff was used, and core genes were defined as those in 99% of isolates. A maximum likelihood tree of the 25,294 SNPs in the 1,416 core genes was created using RAxML and 100 bootstraps. iTOL31 and FigTree were used to visualize the trees. Assemblies were compared to the MLST database (pubmlst.org/efaecalis/) sited at the University of Oxford32 using an in-house script.
Population history and mutation rate
Genetic diversity was calculated based on pairwise SNP differences. BEAST15 was used to date the phylogeny and estimate a mutation rate for L1 and L3 using the core genome after removal of regions of recombination using Gubbins. BratNextGen24 was used to verify the results of Gubbins for L1 using the following parameters: 10 iterations, 100 permutation runs and a significance threshold of 0.05. One NCTC isolate was excluded from the analysis for L1 because the isolation date was unknown. A Hasegawa, Kishino and Yano (HKY) model and gamma distribution were used, and the best molecular clock and tree selected based on Bayes factors calculated from path sampling and stepping stone sampling33,34. For L3, an exponential clock and constant tree were used, for L1 a lognormal clock and Bayesian skyline tree were used, and for the repeat analysis of L1 (with recombination events identified and removed based on BratNextGen) a lognormal clock and constant tree were used.
Detection of candidate genes
Virulence genes were chosen based on evidence from experimental mammalian models35, and their presence was determined by in silico PCR using previously published primers: ace36, esp37, gelE38, asa139, agg36 and cyl40, elrA (OEF2 and OEF8)41, gls2442, tpx (ef1933for and tpxrev)43, bgsA (bgsA for and bgsArev)44, srtA (EF3056F and EF3056R)45, sigV (SVRT1-2)46, epaA (AB270_epa_F and AB271_epa_R)47, epaB (AB272_epaB_F and AB273_epaB_R)47, epaE (AB276_epaE_F and AB277_epaE_R)47, epaN (AB288_epaN_F and AN289_epaN_R)47 and perA (perA-FF and perA-RR)48. The presence of msrA and msrB was determined by coverage of EF1681 and EF3164, respectively, when mapped to the V583 reference genome. The presence of vanA and vanB was established by in silico PCR using published primers35,49. Genes encoding resistance to additional antimicrobial drugs were detected by comparing the whole genome of each isolate with the ResFinder database (compiled in 2012)50, which has been manually curated since publication. Sequences were compared using an in-house script, and genes with 100% match to length and >90% identity match were classified as present. In silico PCR using previously published primers was used for genes not in the ResFinder database: dfrF51 and qacZ52. Statistical significance was determined using Fisher’s exact test.
Pangenome and recombination
The pangenome was estimated using Roary20. Core genes were defined as those present in all 168 isolates with a 90% ID cutoff. The proteome of E. faecalis strain V583 was downloaded from the ENA and interrogated with the WxL protein described in this study using the protein version of BLAST. Recombination was identified within L1–L3 using Gubbins and verified using BratNextGen as described above.
Characterizing the Tn1546 transposon
Sequence reads for each isolate were mapped to Tn1546 (accession number M97297) from E. faecium strain BM4147 using SMALT. The depth of coverage was between ~30× and ~500× for all isolates, with 53/57 (93%) at a depth of greater than ~50×. To identify the insertion sites, the sequences adjacent to the start and end of the Tn1546 transposons were extracted from the assemblies up to a maximum of 10,000 bp or the end of the contig. The sequences adjacent to the start of Tn1546 were too short to analyse, but sequences adjacent to the end of Tn1546 (termed ‘insertion site sequences’) were compared between isolates. Insertion site sequences that were identical for more than 200 bp were grouped (groups 1–3 in Supplementary Table 4), and subgroups were then defined if there were any changes in the downstream sequence, with no evidence that an insertion or deletion explained this change (changes are described in Supplementary Table 4). Where the available insertion site sequence was too short to determine to which subgroup it belonged, this was categorized into subgroup A (the most prevalent subgroup) for simplification of Fig. 4. Each subgroup was identified as plasmid- or chromosome-based using BLAST, with transposons considered plasmid-borne if the highest match was to a plasmid and there was no match above 25% coverage to an E. faecalis chromosome. Insertion site sequences less than 250 bp were not considered long enough for accurate identification. To verify whether the vanA transposons were located on plasmids, plasmid extraction followed by vanA hybridization was performed. Representative isolates were selected for each of the transposon insertion sites defined using the sequence data. Plasmids were extracted using the Kado and Liu53 method, except that 100 mg ml−1 lysozyme was added with the E buffer followed by incubation for 1 h. Extracts were run on a 0.7% agarose gel and blotted using capillary transfer onto Hybond N+ (Amersham). Hybridization was performed using the DIG-high prime labelling and detection starter kit I (Roche Applied Science), and luminescence was detected using CSPD (Roche Applied Science). E. faecium BM4147 and NCTC 8132 were used as positive and negative controls, respectively, and two isolates with known plasmid sizes were used as size markers (Yersinia enterolitica YE212/92 (BT 2, O:9) and YE53/03 (BT 1A, O:5)). Probes were created using the following primers: vanA-1, 5´-GGGAAAACGACAATTGC-3´; vanA-2, 5´GTACAATGCGGCCGTTA-3´49.
Accession numbers
The sequence data for the study isolates have been deposited in the ENA under study accession numbers PRJEB4344, PRJEB4345 and PRJEB4346, with the accession numbers for individual isolates listed in Supplementary Table 1. Additional sequences used in this study were the E. faecalis reference genome V583 (ENA accession number AE016830) and Tn1546 from E. faecium strain BM4147 (ENA accession number M97297).
Additional information
Supplementary information is available online. Reprints and permissions information is available online at www.nature.com/reprints. Correspondence and requests for materials should be addressed to K.E.R.
Acknowledgements
The authors thank the Wellcome Trust Sanger Institute library construction, sequence and core informatics teams, the staff at BSAC and the Cambridge Public Health England Microbiology and Public Health Laboratory, and H. Brodrick, A. Cain, D. Pickard, K. Judge and E. Blane for their technical support. The authors acknowledge BSAC for allowing the use of isolates from the BSAC Resistance Surveillance Project. This publication presents independent research supported by the Health Innovation Challenge Fund (HICF-T5-342 and WT098600), a parallel funding partnership between the UK Department of Health and the Wellcome Trust. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health or the Wellcome Trust. This project was also funded by a grant awarded to the Wellcome Trust Sanger Institute (098051). M.E.T. is a Clinical Scientist Fellow supported by the Academy of Medical Sciences, the Health Foundation and the NIHR Cambridge Biomedical Research Centre.
Footnotes
Author contributions
S.J.P. designed the study. K.E.R. performed bacterial identification, susceptibility testing and DNA extraction, and analysed the data. S.R. assisted with bioinformatic analysis. T.G., R.R., J.E.R., N.M.B. and J.P. contributed materials and data. M.E.T. completed ethical approvals. J.P. and S.J.P. were responsible for supervision and management of the study.
Competing interests
The authors declare no competing financial interests.
References
- 1.Sievert DM, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 2.Suetens C, Hopkins S, Kolman J, Diaz Högberg L. Point prevalence survey of healthcare-associated infections and antimicrobial use in the European acute care hospitals. ECDC. 2013 [Google Scholar]
- 3.Uttley AHC, Collins CH, Naidoo J, George RC. Vancomycin-resistant Enterococci. Lancet. 1988;2:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz-Garbajosa P, et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol. 2006;44:2220–2228. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freitas AR, Novais C, Ruiz-Garbajosa P, Coque TM, Peixe L. Clonal expansion within clonal complex 2 and spread of vancomycin-resistant plasmids among different genetic lineages of Enterococcus faecalis from Portugal. J Antimicrob Chemother. 2009;63:1104–1111. doi: 10.1093/jac/dkp103. [DOI] [PubMed] [Google Scholar]
- 6.Kuch A, et al. Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. J Antimicrob Chemother. 2012;67:551–558. doi: 10.1093/jac/dkr544. [DOI] [PubMed] [Google Scholar]
- 7.Kawalec M, et al. Clonal structure of Enterococcus faecalis isolated from Polish hospitals: characterization of epidemic clones. J Clin Microbiol. 2007;45:147–153. doi: 10.1128/JCM.01704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulsen IT, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 9.Palmer KL, et al. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio. 2012;3:1–11. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim EB, Marco ML. Nonclinical and clinical Enterococcus faecium strains, but not Enterococcus faecalis strains, have distinct structural and functional genomic features. Appl Environ Microbiol. 2014;80:154–165. doi: 10.1128/AEM.03108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu L-Y, et al. Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biol. 2015;16:81. doi: 10.1186/s13059-015-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donker T, Wallinga J, Slack R, Grundmann H. Hospital networks and the dispersal of hospital-acquired pathogens by patient transfer. PLoS ONE. 2012;7:e35002. doi: 10.1371/journal.pone.0035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coque TM, Tomayko JF, Ricke SC, Okhyusen PC, Murray BE. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob Agents Chemother. 1996;40:2605–2609. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordens JZ, Bates J, Griffiths DT. Faecal carriage and nosocomial spread of vancomycin-resistant Enterococcus faecium. J Antimicrob Chemother. 1994;34:515–528. doi: 10.1093/jac/34.4.515. [DOI] [PubMed] [Google Scholar]
- 15.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer KL, et al. Enterococcal genomics. In: Gilmore MS, et al., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary; 2014. http://www.ncbi.nlm.nih.gov/books/NBK190425/ [PubMed] [Google Scholar]
- 17.Köser CU, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2013;366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker TM, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Human Microbiome Jumpstart Reference Strains Consortium. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinster S, Furlan S, Serror P. C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J Bacteriol. 2007;189:1244–1253. doi: 10.1128/JB.00773-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2014;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marttinen P, et al. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012;40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foucault M, Depardieu F, Courvalin P, Grillot-Courvalin C. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci USA. 2010;107:16964–16969. doi: 10.1073/pnas.1006855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG. WebACT—an online companion for the Artemis Comparison Tool. Bioinformatics. 2005;21:3665–3666. doi: 10.1093/bioinformatics/bti601. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 31.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 32.Jolley KA, Maiden MCJ. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baele G, et al. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 2012;29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baele G, Li WLS, Drummond AJ, Suchard MA, Lemey P. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol. 2013;30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Depardieu F, Perichon B, Courvalin P. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J Clin Microbiol. 2004;42:5857–5860. doi: 10.1128/JCM.42.12.5857-5860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nallapareddy SR, Singh KV, Duh R-W, Weinstock GM, Murray BE. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of ace during human infections. Infect Immun. 2000;68:5210–5217. doi: 10.1128/iai.68.9.5210-5217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vankerckhoven V, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42:4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurkovic D, et al. Identification and characterization of enterococci from bryndza cheese. Lett Appl Microbiol. 2006;42:553–559. doi: 10.1111/j.1472-765X.2006.01918.x. [DOI] [PubMed] [Google Scholar]
- 41.Brinster S, et al. Enterococcal leucine-rich repeat-containing protein involved in virulence and host inflammatory response. Infect Immun. 2007;75:4463–4471. doi: 10.1128/IAI.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nallapareddy SR, Wenxiang H, Weinstock GM, Murray BE. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J Bacteriol. 2005;187:5709–5718. doi: 10.1128/JB.187.16.5709-5718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.La Carbona S, et al. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, alkyl hydroperoxide reductase and thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol Microbiol. 2007;66:1148–1163. doi: 10.1111/j.1365-2958.2007.05987.x. [DOI] [PubMed] [Google Scholar]
- 44.Theilacker C, et al. Glycolipids are involved in biofilm accumulation and prolonged bacteraemia in Enterococcus faecalis. Mol Microbiol. 2009;71:1055–1069. doi: 10.1111/j.1365-2958.2008.06587.x. [DOI] [PubMed] [Google Scholar]
- 45.Kemp KD, Singh KV, Nallapareddy SR, Murray BE. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect Immun. 2007;75:5399–5404. doi: 10.1128/IAI.00663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Jeune A, et al. The extracytoplasmic function sigma factor SigV plays a key role in the original model of lysozyme resistance and virulence of Enterococcus faecalis. PLoS ONE. 2010;5:e9658. doi: 10.1371/journal.pone.0009658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teng F, Singh KV, Bourgogne A, Zeng J, Murray BE. Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect Immun. 2009;77:3759–3767. doi: 10.1128/IAI.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coburn PS, Baghdayan AS, Dolan GT, Shankar N. An AraC-type transcriptional regulator encoded on the Enterococcus faecalis pathogenicity island contributes to pathogenesis and intracellular macrophage survival. Infect Immun. 2008;76:5668–5676. doi: 10.1128/IAI.00930-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:1434. doi: 10.1128/jcm.33.5.1434-1434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cattoir V, Huynh TM, Bourdon N, Auzou M, Leclercq R. Trimethoprim resistance genes in vancomycin-resistant Enterococcus faecium clinical isolates from France. Int J Antimicrob Agents. 2009;34:390–392. doi: 10.1016/j.ijantimicag.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 52.Braga TM, Marujo PE, Pomba C, Lopes MFS. Involvement, and dissemination, of the enterococcal small multidrug resistance transporter QacZ in resistance to quaternary ammonium compounds. J Antimicrob Chemother. 2011;66:283–286. doi: 10.1093/jac/dkq460. [DOI] [PubMed] [Google Scholar]
- 53.Kado CI, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.