Summary
Single stem cells, including those in human epidermis, have a remarkable ability to reconstitute tissues in vitro, but the cellular mechanisms that enable this are ill defined. We used live imaging to track the outcome of thousands of divisions in clonal cultures of primary human epidermal keratinocytes. Two modes of proliferation were seen. In ‘balanced’ mode, similar proportions of proliferating and differentiating cells were generated, achieving the ‘population asymmetry’ that sustains epidermal homeostasis in vivo. In ‘expanding’ mode, an excess of cycling cells was produced, generating large expanding colonies. Cells in expanding mode switched their behaviour to balanced mode once local confluence was attained. However when a confluent area is wounded in a scratch assay, cells near the scratch switch back to expanding mode until the defect is closed. We conclude that the ability of a single epidermal stem cell to reconstitute an epithelium is explained by two interconvertible modes of proliferation regulated by confluence.
Recent advances in cell culture have highlighted the capacity of single adult stem cells to generate self-assembling organ like structures1–5. This property was first demonstrated in cultures of primary human epidermal keratinocytes. A subset of proliferating keratinocytes has the potential to generate large colonies that subsequently fuse to form multilayered sheets, recapitulating the organisation of the epidermis1, 2, 6, 7. In such cultures, proliferation is confined to the basal cell layer, with differentiating cells stratifying into overlying cell layers when local confluence is attained. This culture system has been widely used to study epidermal stem cells and their regulation8–15. In addition, epithelial sheets generated in vitro are used for autologous transplantation in burns patients. The cultured grafts persist as a histologically normal epidermis for many years16, 17.
There is marked heterogeneity in the proliferative potential of cultured keratinocytes, with colony sizes ranging from two to hundreds of cells one week after plating6, 7, 18. Sub-cloning suggests there are at least three kinds of clonogenic cell. ‘Holoclones’ have extensive cell renewal potential and generate large proliferating colonies, while small, differentiated colonies arise from ‘paraclones’ with minimal proliferative potential6, 7. A third class of ‘meroclone’ cell, produces colonies with intermediate properties.
These results may reflect a proliferative hierarchy of three or more cell types19. However, this seems at odds with the results of in vivo lineage tracing in homeostatic murine epidermis, where only one or two progenitor populations have been identified20–24. We reasoned that live imaging of a large sample of cells to resolve the dynamics of individual cells may reveal the basis of clonal diversity and how single cells can reconstitute epidermal sheets25.
Results
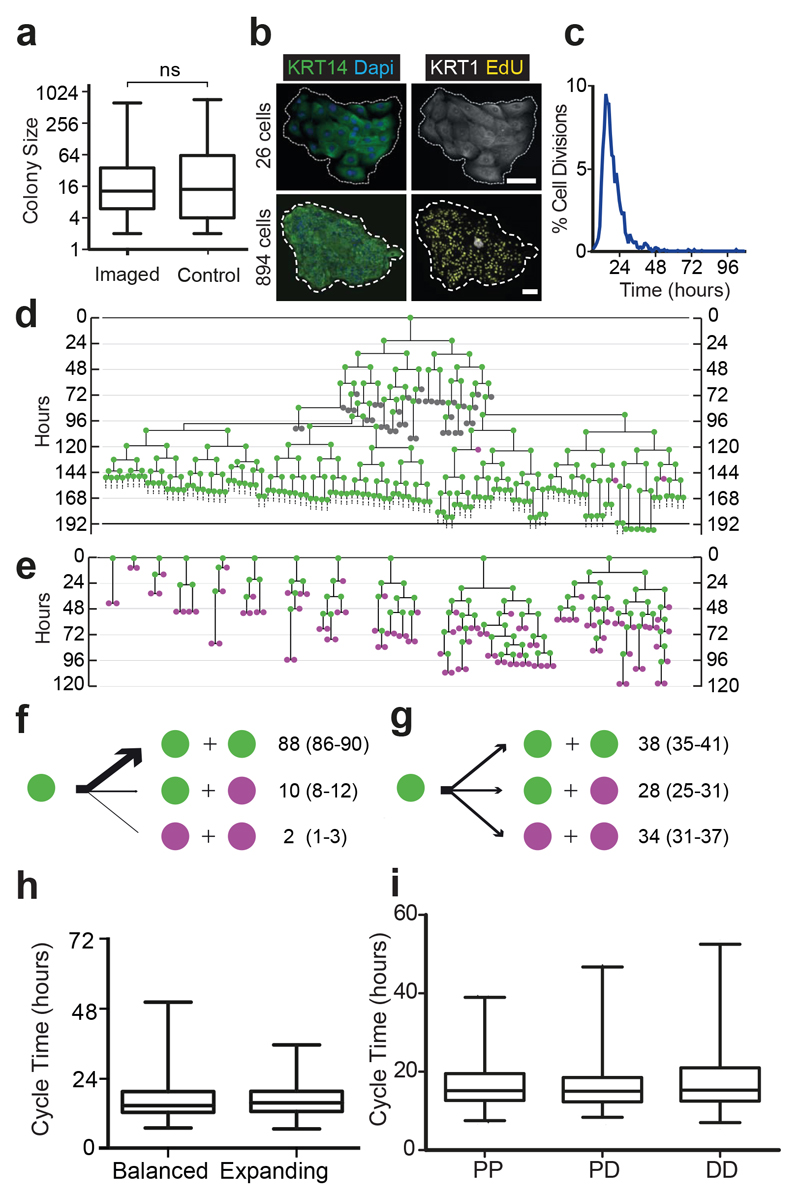
We used a high definition timelapse microscopy system to image clonal cultures of human neonatal foreskin epidermal keratinocytes (NFSK). Imaging did not alter the colony size distribution at 7 days (Fig. 1a)26. Staining revealed both microscopic differentiated colonies and large colonies containing numerous proliferating cells, indistinguishable from non-imaged controls (Fig. 1b). We next tracked 2208 complete cell cycles over 7 days of culture. Median cycle length, excluding the first division after plating, was 15.7 hours (range 4.7-100.2 hours, n=2127, Fig. 1c). We constructed lineage trees for 81 colonies, with a final size between 2-722 cells (Fig. 1d,e; Fig. 2; Supplementary Table 1). In the largest colonies it was only feasible to track cells over four sequential rounds of cell division so multiple sets of subclones spanning four cellular generations were tracked within each colony.
Figure 1. Live imaging of cultured keratinocytes.
a: Size distribution of live imaged (n=81) and non imaged control (n=1487) colonies after 7 days culture, in 3 independent experiments. Box boundaries indicate the 25th and 75th percentiles. Line across box is the median. Whiskers indicate 1st and 99th percentiles. There is no statistically significant difference between the distributions (p=0.15 Kolmogorov-Smirnov test). b: Typical colonies cultured for 6 days, treated with EdU and fixed 24 hours later,. White, differentiation marker KRT1; yellow, EdU; green, keratinocyte marker KRT14; blue, DAPI. Images representative of 3 independent experiments. Scale bar 100μm. c: Cycle times of 2127 live imaged cells from 3 independent experiments, median 15.7 hours, 99% of all divisions occur within 48 hours. d,e: Representative examples of two types of lineage trees, expanding, d, and balanced e, from 3 independent experiments. Dividing cells are green, non-dividing cells magenta and cells observed for <48 hours grey. See Figure 2a,b and Supplementary Table 1 for complete data set and Supplementary Videos 1 and 2 for example videos. f,g: Division outcomes in expanding (f, 928 divisions) and balanced colonies (g, 930 divisions), expressed as percentages with 95% confidence intervals. h Cell cycle time distributions in balanced and expanding colonies Box boundaries indicate the 25th and 75th percentiles. Line across box is the median. Whiskers indicate 1st and 99th percentiles. i The length of the preceding (maternal) cell cycle for daughter cells with each division outcome. Box boundaries indicate the 25th and 75th percentiles, line across box is the median. Whiskers indicate 1st and 99th percentiles. There is no significant difference between cycle time distributions for any division outcome (P=0.18 Kruskal-Wallis Test, n=1109 divisions for PP, 338 PD and 330 DD).
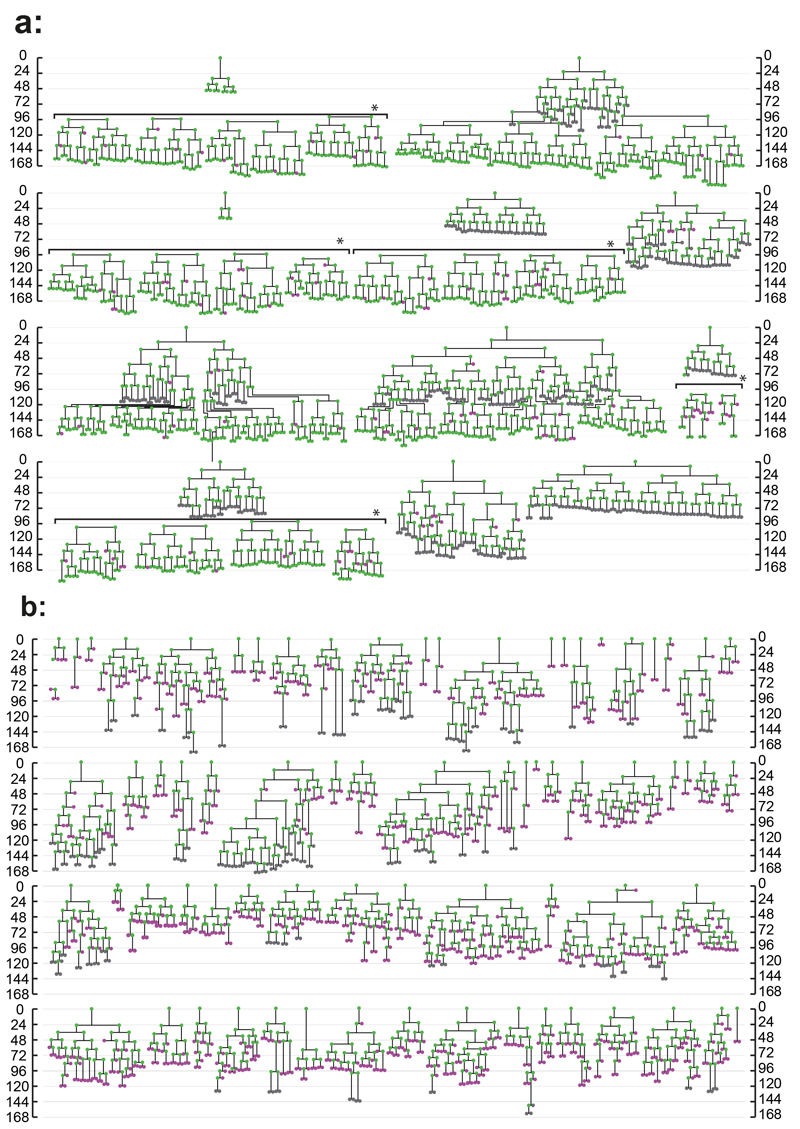
Figure 2. Lineage trees of Neonatal Foreskin Keratinocytes cultured at clonal density.
Scale indicates time since plating in hours. Magenta indicates cells that did not divide within 48 hours, green cells which were observed to divide and grey cells those which could not be tracked for at least 48 hours. Horizontal brackets in a, marked by *, indicate representative cells tracked within a single colony. a: expanding trees, b: balanced trees, see text for details. A total of 81 trees from 3 independent experiments is shown.
Keratinocyte division generates cells that go on to either exit the cell cycle and differentiate or divide6. Over 99% of divisions occurred within 48 hours (Fig. 1c). We therefore classified cells that did not divide within 48 hours as differentiating (D) and those that divided as proliferative (P, Fig. 1d,e). Cells that could not be tracked for 48 hours were classified as unknown (U) and excluded from further analysis (n=288) (Fig. 1d,e; Fig. 2; Supplementary Table 1). The validity of these assignments was supported by staining for proliferation and differentiation markers at the end of the experiment (Fig. 1b).
Three outcomes of cell division were observed, symmetric divisions producing two proliferating or two differentiating cells (PP or DD) and asymmetric PD divisions. After classifying division outcomes, two groups of lineage trees were apparent (Fig. 1d,e; Fig. 2a,b). In 11 colonies the first three rounds of division were exclusively PP, and subsequently PP divisions predominated (88% of all divisions, n=819) with occasional PD (10%, n=91) and rare DD (2%, n=18) divisions (Fig. 1d,f; Fig. 2a; Supplementary Video 1). There was no discernible pattern to the occurrence of PD or DD divisions within each colony. These colonies typically contained hundreds of cells by 7 days (median 422, range 108-722 cells). On average, 13 P cells were produced for each D cell, leading us to term this group of colonies ‘expanding’.
In the remaining 70 colonies, PP, PD and DD divisions were apparent at each round of cell division, resulting in a wide diversity of lineage trees (Fig. 1e,g; Fig. 2b; Supplementary Video 2). The final size of these colonies was far smaller than the expanding group (median 12, range 2-60 cells).
What does the varied nature of the lineage trees in the second group of colonies reveal about the proliferative properties of the cells that generate them? It might be that each variety of tree derives from a distinct cell type specified to undergo a particular series of PP, PD and DD divisions, but this seems unfeasibly complex. A far simpler explanation is that the diversity of trees arises from a single population in which the outcome of an individual cell division is unpredictable, but the distribution of PP, PD or DD outcomes are maintained. The latter interpretation is consistent with genetic lineage tracing in homeostatic mouse epidermis, where modelling argues the tissue is maintained by a single progenitor population which undergoes PP, PD and DD divisions, with the likelihood of PP and DD divisions being equal15–18,21,22. Strikingly, across the 70 trees, the frequency of PP divisions (38%, n= 353) was not significantly different from DD divisions (34%, n=318; Fig. 1g). We therefore termed these trees ‘balanced’ as, on average, approximately equal proportions of P and D cells are produced per cell division.
The observation of such ‘population asymmetry’ in dissociated cells in culture is remarkable and suggests the balanced likelihood of PP and DD divisions is highly robust20, 27.
We next examined whether there was a relationship between cell cycle time and division outcome. There was only a modest difference in median cycle time between balanced (14.8 hours, n=860 divisions) and expanding (15.8 hours, n=917 divisions) colonies (p=0.06, Mann Whitney test, Fig. 1h). We found no correlation between the length of the preceding cell cycle and division outcome (median 15.2 hours for 1109 PP divisions, 15.0 hours for 338 PD divisions and 15.3 hours for 330 DD divisions, p=0.18, Kruskal-Wallis test, Fig. 1i). This contrasts with retinal cells that have the same three types of division, but the preceding cell cycle time is found to be longer for DD than for PP and PD outcomes28.
We then imaged cells cultured directly from normal homeostatic adult abdominal epidermis (Supplementary Table 2). 1156 complete cell cycles were observed in 56 colonies (Supplementary Figs 1 & 2a-c; Supplementary Table 1). The median cycle length was 15.0 hours (range 5.5-62.3 hours), and 99.6% of divisions occurred within 48 hours (Supplementary Fig. 2a). The same three division outcomes were observed within 6 expanding and 50 balanced type trees (Supplementary Figs 1 & (2b,c). The median sizes of balanced and expanding colonies at 168 hours were 4 (range 2-93) and 299 (range 175-876) respectively. We conclude that adult epidermal cells exhibit the same behaviours as NFSK and that these modes of proliferation do not arise from prolonged cell culture.
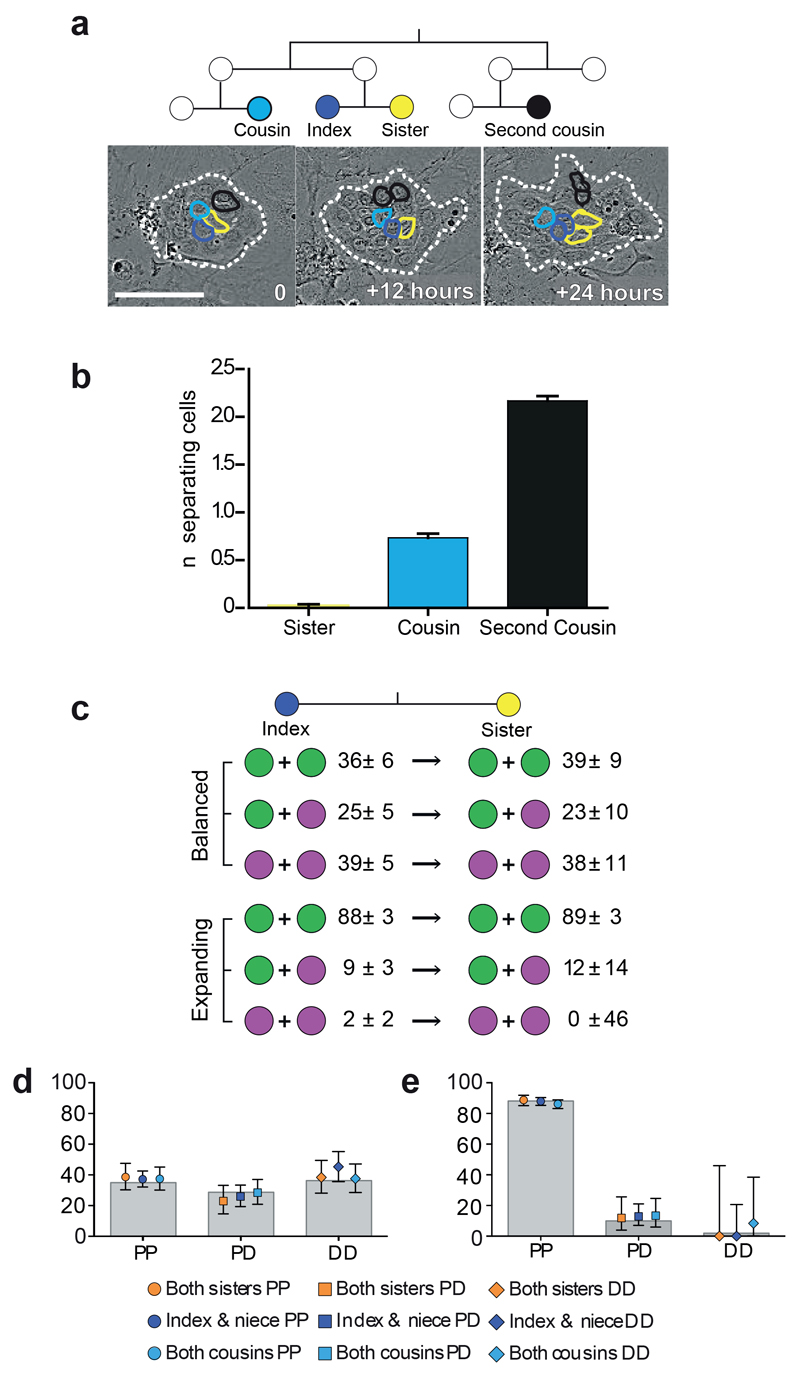
Are division outcomes dependent on the behaviour of neighbouring cells and/or the microenvironment? To investigate this we examined pairs of sister NFSKs in which both cells went on to divide. Sister cells remained adjacent to each other throughout their life span in 98% of cases (n=349), minimising micro-environmental differences between the cells (Fig. 3a,b). Comparing an index cell with its sister revealed no difference in division outcomes compared with the entire dividing cell population in either expanding or balanced colonies (Fig. 3c). We also analysed division outcomes in spatially separated index-‘niece’ and index-‘cousin’ pairs and again found no significant differences from the population average (Fig. 3b,d,e).
Figure 3. Division outcomes of sister cells.
a: Time lapse images of a typical colony from 81 independent videos in 3 independent experiments (dashed white line), with related cells outlined as dark blue=index cell; yellow=sister; cousin=cyan; black=second cousin. Scale bar 100μm. b: Mean number of cells separating sisters (n=345 sister pairs), cousins (456 pairs) and second cousins (390 pairs), error bars indicate 95% confidence intervals, data from 3 independent experiments. c: Division outcomes (%, 95% confidence interval) in sister pairs in balanced (n=305 divisions) and expanding (n=415 divisions) colonies. d,e: Division outcomes in sister, cousin (balanced 413, expanding 689) and niece (balanced 617, expanding 782) cell pairs, data from 3 independent experiments. Legend indicates relationship and outcome, overlaid onto population averages (grey bars) for balanced, d, and expanding, e, lineages. Error bars indicate 95% confidence intervals.
These results argue against extracellular factors as determinants of particular division outcomes, as if this was the case, the outcomes in adjacent cells would be expected to be similar. Furthermore, sister pair analysis excludes lateral inhibition, in which intracellular signalling drives adjacent cells into different fates, as a mechanism for achieving the similar likelihoods of DD and PP divisions observed in the balanced group29, 30. Thus, within colonies up to 7 days after plating, division outcomes appear cell autonomous. This observation supports a critical, but previously untested assumption made in the analysis of in vivo lineage tracing data20–23, 31, 32.
Collectively the results of the live imaging experiments suggest that cells within each colony are in one of two proliferative modes, balanced or expanding. The proliferative mode of the founder cell is inherited by its progeny, so the likelihood of a given outcome is the same for all divisions in the colony. This ‘two mode’ model, based directly on live imaging data makes testable quantitative predictions which we set out to validate.
First, we examined whether the wide distribution of colony sizes seen in 7 day NFSK cultures could be recapitulated by such a simple model. We performed a computer simulation of 7 days of growth of 30,000 virtual colonies based on the cell cycle time and probabilities of each division outcome observed by live imaging. Virtual founder cells were assigned to balanced or expanding modes in the observed proportions and each division was treated as an independent event. There was no significant difference between the size distribution of 1631 real 7 day colonies and that of 30,000 simulated colonies, indicating the model is consistent with experiment (Supplementary Fig. 2d).
To further challenge the model we performed cell tracking on a larger scale. NFSK were labelled with the fluorescent membrane dye, PKH26, plated at clonal density, and the number of cells per colony determined every 24 hours (n=333 colonies in total from 3 independent experiments; (Supplementary Fig. 3; Supplementary Table 3)33, 34. To classify colonies into balanced or expanding we drew on the live imaging data. Most balanced colonies contained only differentiated cells by 144 hours, but in those that retained proliferating cells, the proportionate increase in cell number between 144 and 168 hours was less than 50% whilst in expanding mode colonies it was significantly higher (p<0.0003 by Mann Whitney U test, Supplementary Fig. 4a,b). We therefore used the ratio of colony size at 168:144 hours to classify colonies as balanced or expanding (Methods online; Supplementary Fig. 4b-d).
This larger data set was then used to further challenge the predictions of the two mode model. Firstly, for a population of balanced mode colonies, the mean number of cells/colony is predicted to increase linearly, as on average, each division adds one differentiated and one proliferating cell to the colony. This linear growth signature discriminates balanced mode dynamics from other behaviours, for example transit amplifying cells that differentiate after a fixed number of divisions20. Across 304 balanced colonies, mean cells/colony did indeed increase linearly with time (r2=0.98), whilst the mean size of expanding mode colonies (n=29) rose exponentially as expected (Supplementary Fig. 4e,f).
A second prediction of the model, based on the computer simulation, was that colonies between 50-150 cells at 168 hours could arise from either of the two modes of division. Some colonies in expanding mode could, by chance, be have a longer than average cycle time for a few early rounds of division, while rare colonies in balanced mode may retain some proliferative cells (Supplementary Fig 2e). Examination of dye labelled colonies in this size range indicated that, as predicted, some were balanced whilst others were expanding (Supplementary Fig. 4c,d). This observation may explain why there appear to be three types of clonogenic cell when clonal 7 day cultures are sub-cloned7. The large colony forming ‘holoclone’ and small colony forming ‘paraclone’ cells are in expanding and balanced modes respectively. However, rather than reflecting a third cell type, the intermediate size ‘meroclone’ colonies may reflect their being a mixture of the largest balanced and the smallest expanding mode colonies7.
Taken together, the above results lead us to conclude the proliferative heterogeneity observed in clonal density 7 day human keratinocyte cultures is explicable by just two modes of cell proliferation tuned to either exponential proliferation or the balanced production of proliferating and differentiating cells.
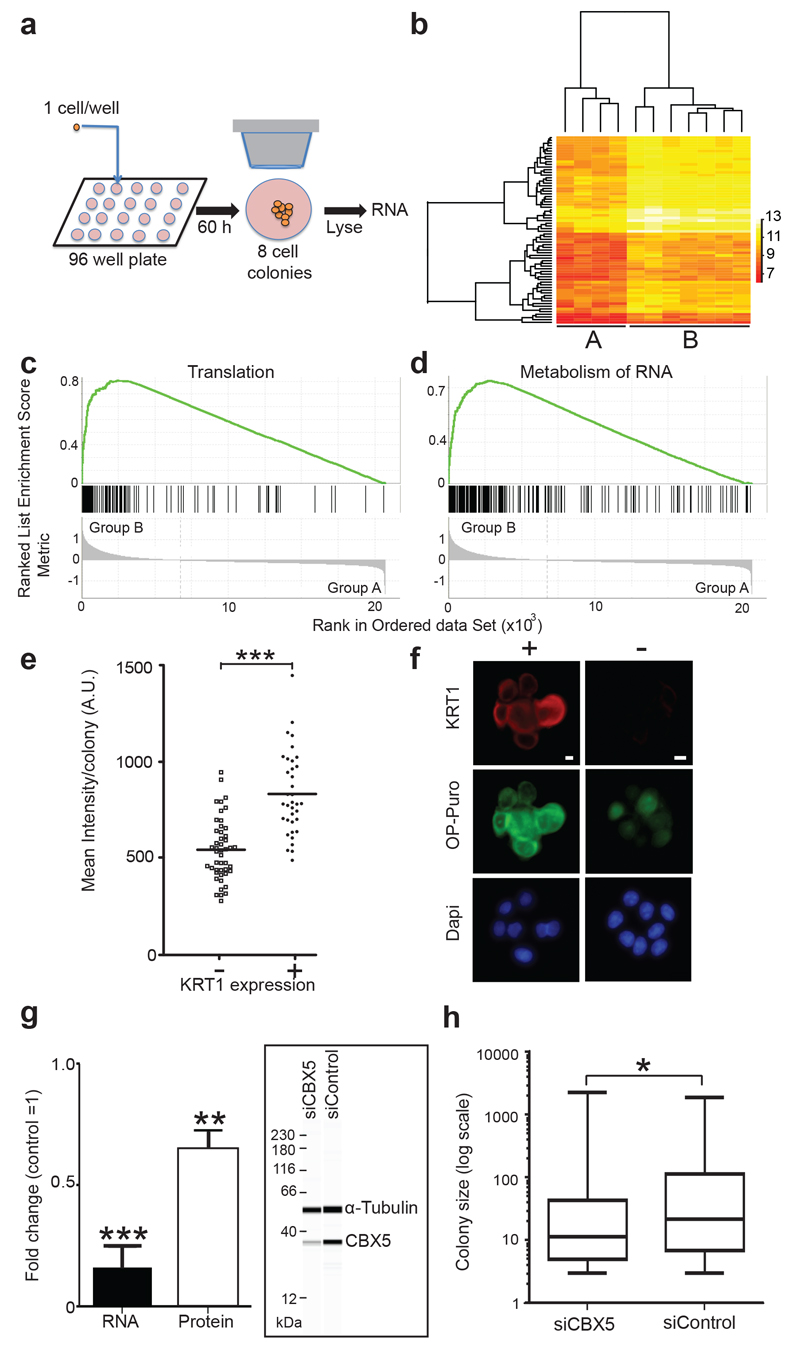
Each mode of proliferation would be expected to produce colonies with a distinct transcriptional profile. We therefore investigated global transcription in colonies containing 8 cells, 60 hours after plating (Fig. 4a,b). In expanding lineages, all 8 cells were in cycle, while a computer simulation indicated that balanced 8-cell colonies would be likely to contain 3-4 differentiating cells at this time point. Enzymatic digestion to isolate single cells grossly perturbed transcription, so we opted to analyse 8 cell colonies, selected on cell number alone. Colonies were lysed in situ and transcriptional analysis of the entire colony was performed (Fig. 4a). Hierarchical clustering analysis using all transcripts expressed above background levels revealed two groups of colonies, A and B (Fig. 4b). Differentially expressed transcripts are shown in Supplementary Tables 4 and 5. Group B had significantly higher levels of the S100 calcium binding proteins and their Annexin partners that are induced on differentiation, arguing they represented balanced colonies (Supplementary Table 6)13, 35, 36. Gene set enrichment analysis revealed Group B colonies also had higher levels of mRNAs associated with translation, RNA metabolism and biogenesis (Fig. 4c,d; Supplementary Table 7). To validate these results we measured global translation using a fluorescence based O-propargyl-puromycin assay in colonies cultured until the modal clone size was 8 cells37, 38. Balanced type colonies with one or more cells expressing the differentiation marker KRT1 had a significantly higher level of translation than those containing no positive cells (P<0.002 by t test in each of 3 experiments; Fig. 4e,f; Supplementary Table 8). This observation parallels reports of differential translation in stem and progenitor cells in other lineages37, 39–41. We also observed differences in expression of genes implicated in the regulation of differentiation. For example, mRNA encoding YY1, a transcription factor that supresses differentiation in a keratinocyte cell line is elevated in Group A colonies, while the Polycomb group protein SUZ12, expressed at higher levels in group B, is down regulated in hyperproliferative epidermis42–45. Motivated by a study showing the chromobox protein CBX4 regulates NFSK proliferation in culture we analysed the effect of knockdown of the related CBX5 protein that is expressed at higher levels in group B colonies9, 46, 47. CBX5 depletion significantly decreased colony size after 7 days of culture (Fig. 4g,h, Supplementary Table 9), paralleling the effect of CBX4 knockdown.
Figure 4. Transcriptional analysis of colonies.
a: Protocol: Single NFSK were flow sorted into individual wells of a cell culture plate, cultured for 60 hours, feeders removed, and entire 8 cell colonies lysed in situ. Following amplification cDNA was analysed by array analysis. b: Hierarchical clustering reveals two groups of colonies, A and B. Heat map shows transcripts with 2 fold or higher differential expression in 11 colonies (see also Supplementary Tables 4-6). c, d: Gene set enrichment analysis plots showing transcripts differentially expressed between groups A and B. Reactome ‘translation’ (c) and ‘metabolism of proteins’ (d) gene sets shown (nominal p values both <0.001, normalised enrichment scores 2.8 and 2.7 respectively, see also Supplementary table 7). e: Mean O-propargyl-puromycin (OP-Puro) fluorescence/colony after 60 hours of culture in colonies containing no (-) or one or more (+) cells expressing the differentiation marker KRT1. Typical example of 3 experiments shown, *** P<10-9 by t test (n=47 – and 36 + colonies, see also Supplementary Table 8). f: Appearance of typical colonies from one of 3 independent experiments stained for OP-Puro (green), KRT1 (red) and Dapi (blue), scale bars 10 μm. g: RNA and protein knockdown of CBX5 following siRNA transfection. Inset shows a typical quantitative capillary isoelectric focussing immunoassay68. Mean of four independent experiments shown, *** p=0.0004 by t-test, ** p=0.0028 by t-test, n=4 independent experiments for both protein and RNA. h: Colony size distribution 7 days post CBX5 knockdown. Typical example of three independent experiments is shown, *p=0.02 by Kolmogorov-Smirnov test, n=139 control and 164 CBX5 colonies, details of other experiments are shown in Supplementary Table 9.
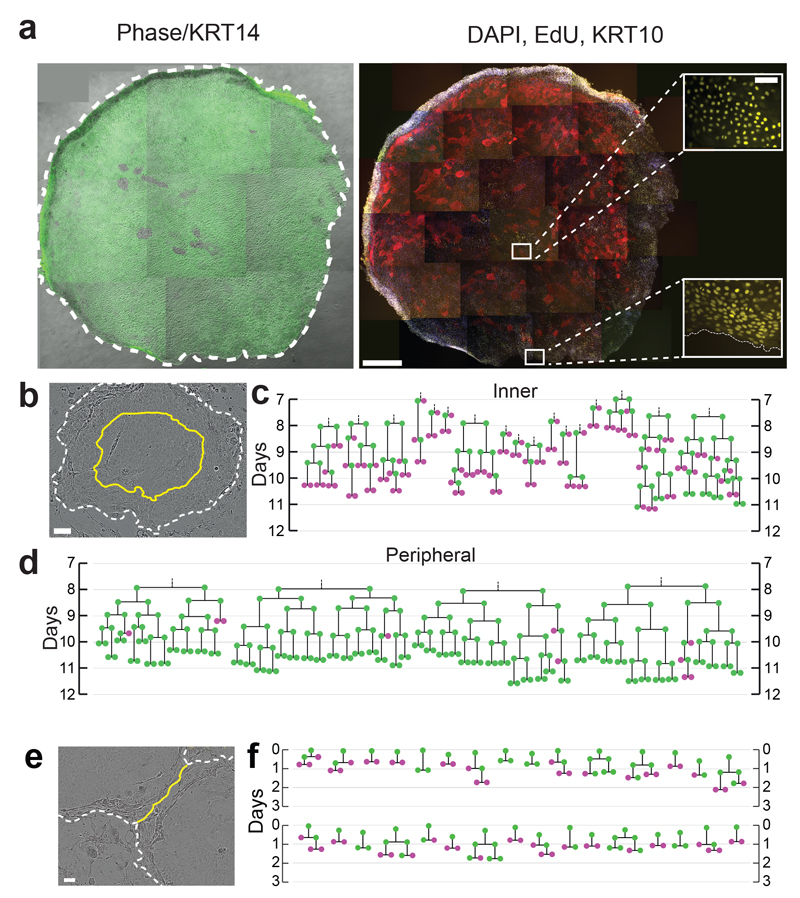
We went on to investigate cell behaviour at later time points. Beyond 7 days, differentiated cells in balanced colonies begin to detach from the culture dish, but large colonies continue to expand, becoming multilayered as differentiating cells stratify (Fig. 5a)48. By 12 days, EdU staining reveals a higher density of proliferating cells at the margin of large colonies than at the centre (Fig. 5a)48. This suggested a change in cell dynamics, so we tracked divisions within expanding NFSK colonies between 7-12 days.
Figure 5. Expanding colonies switch towards balance.
a: Typical large colony (12.5mm2 area) after 12 days of culture, treated with EdU for 24 hours prior to fixation. Left panel: Phase image overlaid with KRT14 staining green, right panel differentiation marker KRT10 (red), EdU (yellow), Dapi (Blue). Insets show EdU staining in centre and margin of colony. Image is representative of 3 independent experiments. Scale bars: main panels 0.5mm, insets 0.1mm. b: Still from typical video from three independent experiments showing colony after 9 days of culture. Inner third (by area) is bounded by yellow line, colony edge indicated by white dashed line, scale bar 100μm. c, d, lineage trees from inner third, c, and outer two thirds, d of colonies, derived from live imaging from 168-288 hours, dividing cells are green, non-dividing cells magenta. e: Still from typical video from 3 independent experiments showing margins of two fusing colonies (dashed lines). Yellow line indicates contact between keratinocytes in adjacent colonies, displacing overlying feeder cells, scale bar 100μm. f: lineage trees showing behaviour of cells within 7 cells of the colony edge at the time of fusion, derived from three independent videos of fusing colonies. Time is indicated from the first contact between colonies (5-9 days post plating), dividing cells are green, non-dividing cells magenta.
We found behaviour continued unchanged in the outer two thirds (by area) of the colony, with 92% PP, 6% PD and 2% DD divisions (n=104, Fig. 5b,d). However, in the stratified inner third of the colonies, behaviour switched from expanding towards balanced mode with 29% PP, 26% PD and 45% DD divisions (n=77, Fig. 5b,c). We also observed a switch from expanding to balanced mode at the margins of adjacent colonies as they fused (28% PP, 32% PD and 40% DD divisions, n=50, Fig. 5e,f).
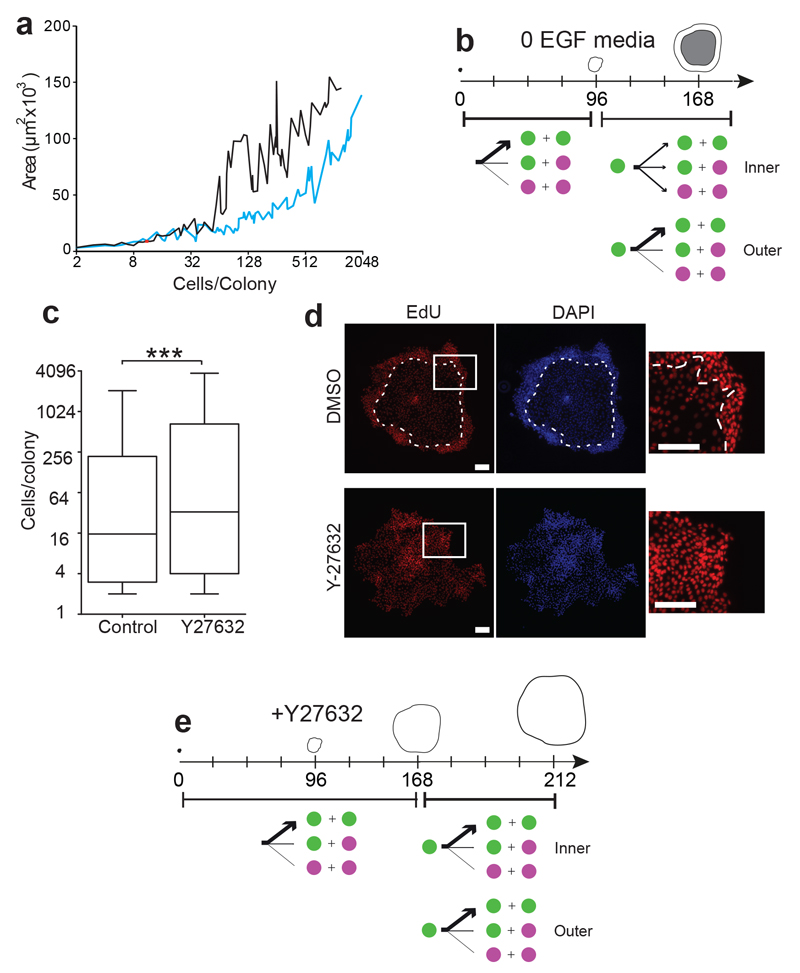
We speculated that local cell confluence may trigger the transition from expanding to balanced behaviour. To investigate this, we cultured NFSK in media without added EGF (EGF0)48, 49. This resulted in transient changes in EGFR activation, followed by adaptation, so that by 72 hours only minimal differences in the phosphorylation of EGFR and downstream signalling proteins were apparent (Supplementary Fig. 5a). However, consistent with previous reports, there was a marked increase in cell density in colonies of over 50 cells in size after 7 days of culture (Fig. 6a)48. Live imaging revealed the proportions of balanced and expanding lineage trees, cell cycle time distribution, and behaviour of cells in balanced colonies were similar in EGF0 and standard media with 10ng/mL EGF (Supplementary Fig. 5b-d; Supplementary Tables 1 & 10). However, in expanding colonies cultured in EGF0, the switch from expanding to balanced mode occurred several days earlier and over a larger area (the inner two thirds of the colony) (Fig. 6b; Supplementary Fig. 5d; Supplementary Tables 1 & 10). These findings are consistent with increased cell density promoting the transition from expanding towards balanced behaviour in large colonies.
Figure 6. Cell density and ROCK2 kinase activity regulate the switch from expanding to balanced mode of proliferation.
a: Colony size in cells and area after 7 days culture in media with (EGF10, black) or without (EGF0, blue) supplemental EGF, data from 3 independent experiments. b: Summary of cell dynamics in EGF0 media. From 0 to 96 hours, all cells grow in expanding mode, but between 96 and 168 hours, cells switch towards balanced mode in inner two thirds of colonies, remaining in expanding mode at the colony rim. See also and Supplementary Figure 5 c, d and Supplementary Table 1. c: Size distribution of colonies cultured for 7 days in 10 μM ROCK2 inhibitor Y27632 compared with controls. Typical example of three independent experiments shown, ***p=0.0007 by Mann-Whitney Test, n=302 control and 293 Y27632 treated colonies. Box boundaries indicate the 25th and 75th percentiles. Line across box is the median. Whiskers indicate 1st and 99th percentiles. d: Fluorescence micrographs of typical large colonies from three independent experiments cultured for 7 days in standard (EGF10) media with 10μM Y27632 (1307 cells) or control (1207 cells), treated with EdU (red) for the final 24 hours of culture. DAPI is blue, white dashed line indicates boundary of stratified cell layer in DMSO colony (no stratification was seen in Y27632 treated colonies). Insets show distribution of EdU positive cells. Scale bars 100μm. e: summary of effect of Y27632 on expanding mode colonies, no switch to balanced mode behaviour is seen in the inner third of colonies cultured for up to 212 hours post plating. See also Supplementary Figure 6 a, and Supplementary Table 1.
These findings led us to hypothesise that cytoskeletal signalling may play a role in the confluence regulated switch from expanding to balanced mode11, 50. We tested the effect of treating NFSK with Y27632 an inhibitor of the key regulator of the actin cytoskeleton, ROCK2 kinase, which is used in establishing primary cultures of adult stem cells from multiple lineages4, 5, 51, 52. ROCK2 inhibition in NFSK results in decreased terminal differentiation and increased proliferation, prolonging the lifetime of the cells in culture53, 54. As expected, we found that culture of NFSK in 10μM Y27632 for 7 days, increased colony size and altered the distribution of proliferating cells within expanding colonies (Fig. 6c,d). Live imaging for 10 days post plating revealed the switch from expanding mode in large colonies was blocked in the presence of Y27632, while balanced behaviour was unaffected (Fig. 6e, Supplementary Fig. 6; Supplementary Tables 1 & 11). These observations argue that the switch from expanding to balanced mode requires ROCK2 mediated cytoskeletal signalling.
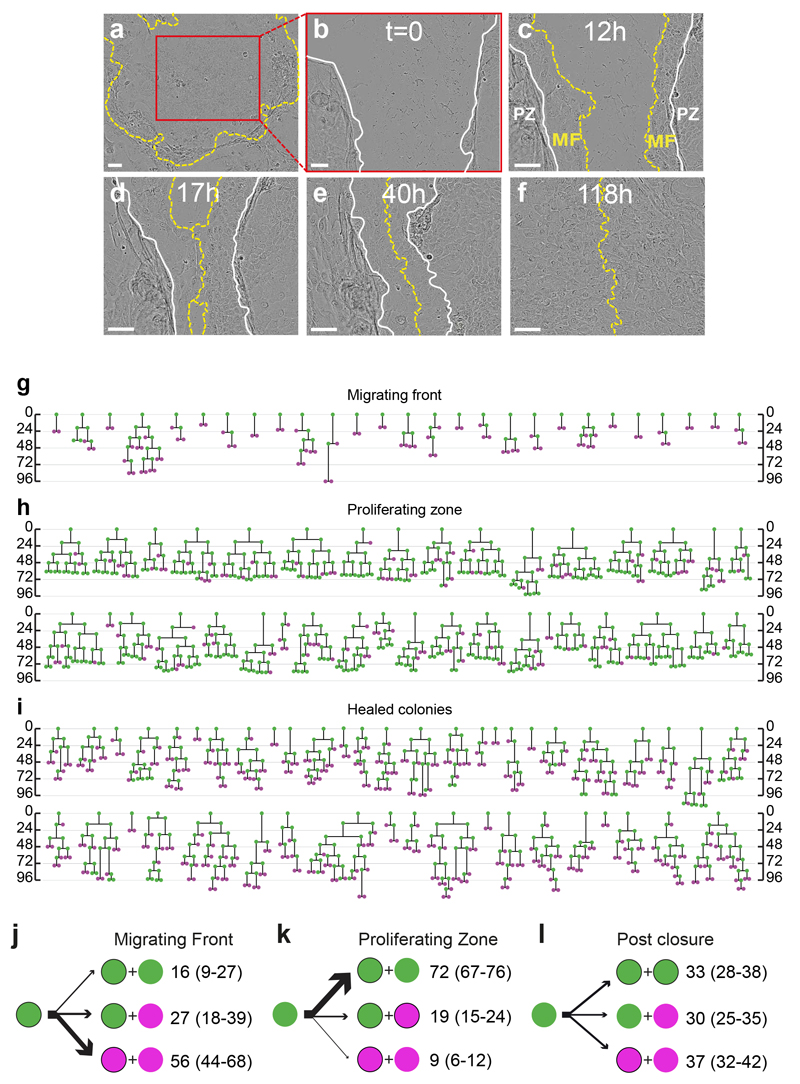
Finally we explored whether the confluence driven transition from expanding to balanced mode was reversible. The central regions of large colonies seeded 9 days previously, in which cells are in balanced mode, were scratched with a pipette tip and the cells close to the resulting ‘wound’ tracked by live imaging (Fig. 7a-f). We observed two discrete regions around the scratch, recapitulating keratinocyte behaviour at a wound margin in vivo31. Keratinocytes within 4 cell positions of the edge of the scratch formed a ‘migrating front’ (MF) from which collective migration of cells closed the defect within 72 hours. Cell divisions in the MF were rare (62 divisions in total in 27 colonies) and produced an excess of non-dividing cells (Fig. 7g, j). In contrast, in there were numerous divisions in the proliferative zone behind the MF (346 divisions in total in 36 colonies) where cells switched from balanced towards expanding mode, reverting to balanced mode when the defect had closed (Fig. 7h, i, k, l). These results indicate that keratinocytes can switch reversibly between the two modes of proliferation.
Figure 7. Effects of loss of confluence on mode of proliferation in large colonies.
a-f: Typical stills from large colonies (n=45 from 3 independent experiments), 9 days post seeding, scratched with a pipette tip. a: Immediately prior and b after scratch at T0. c: Typical appearance at 12 hours post scratch. Yellow dashed line indicates edge of scratch, MF migrating front, PZ proliferative zone, white solid line indicates boundary between MF and PZ. d: MF meet at 17 hours post scratch. e,f: Later images at 40 and 118 hours post scratch showing completion of repair. Scale bar 100⌠m. g-i: Lineage trees of proliferating cells in the centres of large colonies 9 days post plating subjected to scratching, g is from migrating front (62 divisions from 27 colonies in 3 independent experiments) and h proliferative zone pre scratch closure (346 divisions from 36 colonies in 3 independent experiments). 0 is time in hours since time of scratch in g and h. i shows trees of cell divisions post scratch closure (352 divisions from 45 colonies in 3 independent experiments, 0 hours is time of closure). j-l: Summary of cell behaviour, green indicates proliferating cells, magenta differentiating, non dividing cells, numbers are percentages of cells with a given division outcome with 95% confidence intervals in brackets. See also Supplementary Table 1 for full data set.
Discussion
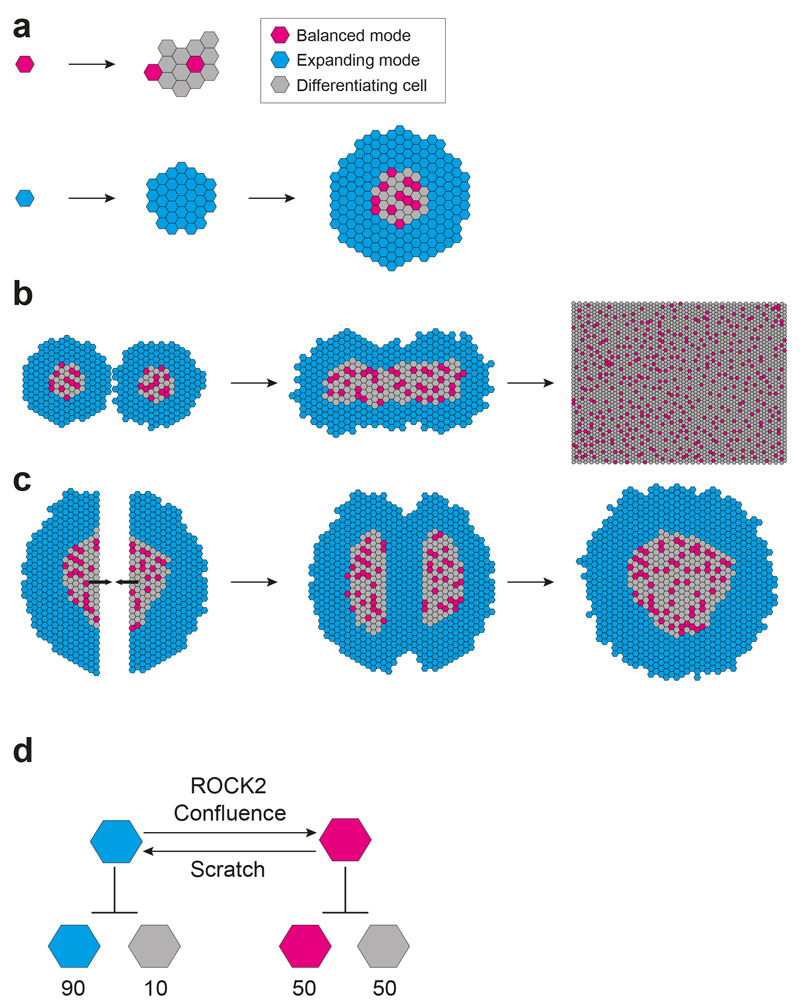
We conclude that clonal culture of human keratinocytes reveals two modes of proliferation, balanced and expanding, maintained across cellular generations (Fig. 8a). Once local confluence is attained, expanding mode cells switch to balanced mode, providing a simple and robust mechanism for generating an epithelium from a single cell (Fig. 8b). If confluence is removed by a scratch injury, balanced mode cells local to the defect transiently and reversibly switch to expanding behaviour until the defect is repaired (Fig. 8c). These results argue that rather than there being a hierarchy of stem and progenitor cells in human epidermal cultures, proliferating cells can switch between homeostatic and regenerative behaviour, a finding in keeping with recent lineage tracing studies in mice23, 31.
Figure 8. Two interconvertible modes of proliferation underpin epithelial reconstitution in vitro.
a: Keratinocyte proliferation in clonal culture. Small ‘paraclones’ and ‘meroclones’ with limited subclonal potential are derived from cells in balanced mode division (magenta cells) (2) large ‘holoclones’ and remaining ‘meroclones’ are derived from expanding mode divisions (cyan cells). Divisions at the centre of the holoclones switch from expanding to balanced divisions with local confluence. Grey cells represent differentiating keratinocytes, and suprabasal cells are omitted for clarity. b: Fusion of adjacent large colonies triggers a switch from expanding to balanced mode. As additional colonies fuse, a large sheet of keratinocytes results with the overall epithelium in balance. c: If the centre of a large colony is wounded, the central keratinocytes in balanced mode switch from balanced to expanding mode to reepithelialise the defect, and revert to balanced divisions when the gap is closed. d: Two interconvertible modes of proliferation. The average division outcomes of balanced and expanding mode cells are shown with the likelihood of each type of cell indicated as a percentage. Confluence promotes a switch from expanding to balanced mode, a process that requires ROCK2 activity. Loss of local confluence in a scratch assay results in a switch from balanced to expanding mode.
The observation that ROCK2 inhibition prevents the switch from expanding to balanced mode in the confluent centres of large colonies joins with a significant body of evidence linking the fate of keratinocytes to the actin signalling55–59. For example, inhibition of the β1 integrins that connect the extracellular matrix to the actin cytoskeleton substantially increases the number of small, differentiated balanced mode type colonies in clonal culture60. Restricting the contact area of keratinocytes with basement membrane also promotes their differentiation via SRF signalling as does inhibition of the actin regulator RAC1 or activation of ROCK211, 61, 62.
This study does not reveal whether cells commit to proliferation or differentiation before, during or after cell division or how the mode of proliferation is determined when cells are plated at clonal density. However, the transcriptional analysis of 8 cell colonies gives potential clues to the molecular basis of the modes. Increased translation is a strong signature of balanced mode dynamics, and Rapamycin treatment that suppresses translation, promotes the formation of large, expanding type colonies61, 63.
The two inter convertible modes of keratinocyte proliferation may be viewed within a wider perspective of behavioural plasticity of adult stem and progenitor cells64. Across evolution, tissue maintenance and repair by a hierarchy of cells with invariant fate is rare in comparison to population asymmetry, stochastic fate and behavioural plasticity in response to injury23, 31, 65–67. Switching between modes of proliferation provides a robust cellular mechanism for organ repair and tissue reconstitution by single cells in vitro31.
Methods Online
Ethics
Work with human material was carried out in compliance with the UK Human Tissue Act (2004) under a protocol approved by the UK National Research Ethics Service (08/H0306/128). All subjects gave written informed consent for the use of their surplus tissue in research.
Cell Authentication
Neonatal foreskin keratinocytes (NFSK) were obtained directly from ATCC (PCS-200-010). J2-3T3 fibroblasts were a gift from James Rheinwald (Department of Dermatology, Brigham and Women's Hospital, Boston USA). All cells were tested for mycoplasma on initial culture and at least 3 monthly thereafter using a Mycoprobe Mycoplasma Detection Kit (CUL001B, R&D systems) according to the manufacturer’s instructions. No mycoplasma positive cells were used in this work.
Feeder Layer
Keratinocytes were cultured with feeder cells. 3T3 J2 cells were maintained in Dulbecco’s Modified Eagle Medium (D-MEM) (Invitrogen 12320-032) supplemented with 10% donor bovine serum (Invitrogen 16030-074) at 37°C in an atmosphere of 5% CO2. Cultures were passaged twice per week by 5 minute treatment with 1X 0.25% trypsin - 3.4mM Ethylene-diamine-tetra acetic acid (EDTA, diluted from Sigma-Aldrich® T4174) to detach cells. Trypsin-EDTA solution was neutralised with culture medium and cells were seeded to new flasks at a dilution of 1 in 6. 3T3 cultures were maintained up to passage 16.
Feeder layers were prepared by incubating 3T3 cells for 3 hours with 4μg/ml mitomycin C (Sigma-Aldrich® M4287), followed by washing with phosphate buffered solution (PBS: 10mM Na2HPO4, 1.8mM KH2PO4, 137mM NaCl, 2.7mM KCl at pH 7.4). Cells were then detached by trypsinisation as above and seeded at densities of 2 × 106 cells into 75cm2 flasks (~25,000 cells/cm2) prior to the addition of keratinocytes.
Keratinocyte Culture
Keratinocytes were maintained in complete FAD medium (50:50 high glucose D-MEM (Invitrogen 11971-025): D-MEM F12 (Invitrogen 31330-038), supplemented with 5μg/ml insulin(Sigma-Aldrich® I5500), 1.8×10-4M adenine (Sigma-Aldrich® A3159), 0.5μg/ml hydrocortisone (Calbiochem 386698), 1×10-10M cholera toxin (Sigma-Aldrich® C8052), 10ng/ml Epidermal Growth Factor (EGF, PeproTech EC Ltd 100-15) and 5% foetal calf serum (FCS, PAA Laboratories A15-041)) with a 3T3 J2 feeder layer.
Cultures were maintained at 37°C in 5% CO2. Cells were passaged at around 80% confluence and split 1 in 20 following detachment by incubation for 15 minutes in 1X trypsin-EDTA (~8000 cells/cm2) at 37°C. Neonatal Foreskin Keratinocytes were used between passages 3 and 7.
Isolation of adult keratinocytes
Adult keratinocytes were obtained directly from discarded skin from abdominoplasties. Anonymised details of donors are shown in Supplementary Table 2. Excess human skin from abdominoplasty surgery was harvested as 350-400 μm thickness skingrafts with a handheld modified Watson-Braithwaite knife. Skin was washed in PBS three times, cut into pieces approximately 5×5mm, and treated with 5mM EDTA in PBS for 90-120 minutes, after which the epidermis was peeled from the dermis with forceps and minced with a scalpel69. The epidermal mince was treated with 1X trypsin–EDTA for periods up to 30 minutes on a magnetic stirrer at 37°C. Treated cells were passed through a 70μm strainer, before counting on a haemocytometer.
Colony forming assays
Mitomycin treated J2 feeder layers were prepared as above at 25,000 cells/cm2. Neonatal or adult keratinocytes were seeded at 25 cells/cm2 and maintained as above for clonal density cultures. For some experiments, the thymidine analogue 5-ethynyl-2’deoxyuridine (EdU) was added at 10μM concentration for 24 hours prior to fixation to label cells with active DNA synthesis (Invitrogen A10044). At time points indicated in the text, feeder cells were removed by gentle pipetting, and plates were fixed with 4% Paraformaldehyde (PFA, Fisher Scientific W0443L) in PBS for 5 minutes at room temperature. The plates were then washed thrice with PBS, and stored in PBS at 4°C until further analysis. Results presented are from a minimum of three dishes at each time point in each experiment.
For immunostaining macroscopic colonies at 12 days post seeding, individual colonies were detached from culture plates by 5 minute incubation in 5mM EDTA at 37°C. Colonies were fixed in 4% PFA for 30 minutes and stored in PBS at 4°C until processing for immunostaining described below.
Culture for live imaging
For live imaging NFSK passage 3-6 or adult keratinocytes isolated directly from skin were plated at 400 cells/cm2 onto J2-3T3 feeder cells seeded at 15,000 cells/cm2 in ImageLock 24 well plates (Fisher Scientific NC0443043). The higher seeding density of keratinocytes was used to ensure cells would attach within the fixed central area of each well of the culture plate (3.7% of the total well area) that was imaged. The lower density of feeder cells allowed better visualisation of small keratinocyte colonies that would otherwise be obscured by overlying 3T3 cells. The ImagelLock plates have fiducial markers at the bottom of the plate which allowed accurate referencing of imaging fields for timelapse capture by the Incucyte system, allowing media to be changed twice a week without disrupting imaging. To compensate for lower feeder cell density, keratinocyte media conditioned by 2 days of culture with 3T3 J2 cells and NFSKs was used. After collection, conditioned media was filtered through a 0.22μm membrane filter (Millipore GPWP02500) and diluted 1:1 in fresh keratinocyte media prior to use.
High definition phase contrast images were taken every 10 minutes for 168-278 hours after plating using an Incucyte Kinetic Imaging System (Essen Bioscience). The average exposure time per frame was 1.1 milliseconds, resulting in cumulative light exposure of 1.1 seconds over 168 hours. Images captured a fixed field of view of the central area of each well. All single cells that divided to produce colonies and remained in the field of view throughout imaging were included in the analysis.
To test the effect of ROCK2 inhibition, Keratinocytes were cultured as above in standard keratinocyte media containing 10μM Y-27632 (Sigma)53, 54. To alter colony morphology, some cultures used Keratinocyte media with 0ng/mL, 5ngl/mL, or 20 ng/mL of supplemental EGF as indicated in the text.
Lineage analysis
Lineage trees were generated by manual analysis of Incucyte videos. Colonies were identified in the final frame of videos were first viewed backwards to ensure the colony remained in field, and then forwards from the first division of the founder cell, noting each cell division on an Excel spreadsheet. For large colonies it proved infeasible to track all divisions. The divisions of the founder cell were tracked for up to 5 generations and representative progeny followed thereafter. Cell cycle time was recorded for the second and subsequent divisions. Cells observed to divide were scored proliferative. It was found that over 99% of divisions occurred within 48 hours, allowing cells which had not divided in this period to be scored as post mitotic differentiating cells. 10% of cells that could not be tracked for 48 hours were scored as unclassified and excluded from the analysis. In large colonies, the location of cells within the colony was also scored.
Immnuostaining and microscopy
For immunostaining, fixed cell plates were incubated in Fish Skin Gelatin (FSG) buffer, comprising 0.5% Triton X100 (BDH 28817.295), 0.25% FSG (Sigma-Aldrich G7765), 1 Bovine serum albumen (BSA, Calbiochem 126575), and 10% Donkey or Goat serum (Sigma-Aldrich® D9663 or G9023 respectively) depending on secondary antibody species) in PBS. Primary antibodies were diluted in FSG and incubated overnight at 4°C. Following three 5 minute washes in PBS secondary antibodies (Invitrogen Molecular Probes) were diluted in FSG, incubated for 3 hours at room temperature, plates washed three times with PBS and mounted in Vectashield® Hardset™ Mounting Medium with 4’,6-diamidino-2-phenylindole (DAPI, Vector labs H-1500).
Colonies were imaged on a Zeiss Axio-Observer D1 microscope with Zeiss Axiovision software. Colony area was quantified using Axiovision and cell counting for large colonies was automated using the ImageJ (1.40g, NIH) cell counter module.
Detection of EdU
Incorporated EdU was detected using the Click-iT® EdU Alexa Fluor® Imaging Kits (Invitrogen™ Molecular Probes® 10337-10340) according to the manufacturer’s instructions. Plates were permeabilised in 0.5% Triton PBS for 1 hour, washed three times in 3% BSA and then incubated in the Click-iT® reaction cocktail (containing buffer, CuSO4 and Alexa Fluor azide) for 1 hour at room temperature. Samples were then washed three times for 5 minutes each in 3% BSA prior to mounting with DAPI containing media.
When used in conjunction with standard immunofluorescence, the sequence of labelling was: block, primary antibody incubation in FSG buffer, three PBS washes, post fix with 4% PFA for 30 minutes at room temperature, three PBS washes, EdU detection protocol, secondary antibody incubation in FSG without Triton, PBS washes followed by mounting.
Scratch Assays
For scratch assays NFSK were cultured for 9 days at clonal density. Large colonies were then scratched with a plastic pipette tip creating a defect approximately 750⌠m wide across the centre of the colony. Live imaging was then performed in the Incucyte system as described above.
PKH26 Dye labelling of keratinocytes
Keratinocytes were labelled with PKH26, a lipid binding yellow-orange fluorescent dye, with a PKH26 Cell Linker Kit (Sigma-Aldrich PKH26GL), following the manufacturer’s instructions. A suspension containing 2×106 keratinocytes was washed in PBS to remove serum and resuspended in 1mL of Diluent C (labelling vehicle, an iso-osmotic aqueous solution). This cell suspension was added to a PKH26 dye solution to achieve a final dye concentration of 2μM. The mixed dye/cell suspension was incubated for 5 minutes at room temperature with periodic agitation. Adding serum and further incubation for a minute stopped the labelling reaction. The cell suspension was then centrifuged and washed three times in media.
To track cell growth, cells were then plated at 25 cells/cm2 on to a feeder layer and cultured as described above. Colonies were identified by fluorescence imaging and on a Zeiss Axiovert microscope and the x and y coordinates of each colony recorded with a Vernier graduated stage. Colonies were re examined every 24 hours and the number of cells counted, until 168 hours when dishes were fixed and stained as described above.
To resolve multicellular colonies into balanced or expanding mode, the ratio of the number of cells at 168 hours to that at 144 hours was calculated. Based on the properties of balanced and expanding mode cells we expected that this ratio would be <1.5 in balanced mode colonies due to the accumulation of differentiated cells and the likelihood of exclusively PP divisions decreasing as the number of progenitors in the colony increases (Fig. 2b, Supplementary Fig. 4a,b). In contrast the size of expanding mode colonies was expected to rise by more than 50% between 144 and 168 hours as differentiated cells are rare and PP divisions predominate (Fig. 2a, Supplementary Fig. 4b). 168:144 hour ratio in cell number effectively discriminated between balanced and expanded mode colonies identified by live imaging (Supplementary Fig. 4b). We also observed that colonies with <50 cells, predicted to be almost exclusively balanced by the computer simulation, the 168:144 ratio was <1.5, whilst for colonies of more than 150 cells, comprising mostly expanding mode cells in the simulation, the ratio was >1.5 (Supplementary Figure 4c). In the size range 50-150 cells contained some colonies with a ratio <1.5 and others with a ratio >1.5, consistent with colonies in this size range being either balanced or expanding.
Immunoblotting
NFSK were seeded at 103 cells/cm2 in media containing 0, 10 or 20 ng/ml supplemental EGF. At the times indicated, cells were scraped into lysis buffer comprising 20mM Hepes-NaOH pH 7.9, Glycerol 10%, NaCl 0.4M, NP-40 0.5%, EDTA 0.2mM, Dithiothreitol (DTT) 1mM, and 1x Halt Protease and Phosphatase inhibitor (Thermo Fisher 78440) and centrifuged at 13000rpm at 4°C for 10 minutes. Protein concentrations were measured using standard Bradford protein assays (BioRAD QuickSTART™ Bradford Dye Reagents 500-0202). Lysates were mixed 1:1 with loading buffer (2X concentration of Tris-HCl pH6.8, 4% SDS, 20% Glycerol, Bromophenol blue and 0.2% β-mercaptethanol). Samples were heated at 95°C for 5 minutes. 6 μg of each sample was loaded onto a 7.5% or 10% of SDS-polyacrylamide gel. Proteins were separated by electrophoresis and transferred onto Immobilon-P membrane (pore size 0.45 μm, Millipore IPVH00010). Membranes were first incubated in blocking buffer (5% dried skimmed milk, PBS, 0.1% Tween-20) at room temperature for 1 hour and then with primary antibodies diluted in blocking buffer for 1 hour at room temperature or overnight at 4°C on a rocking platform. After rinsing in PBS, 0.1% tween-20, membranes were placed on SNAP-id protein detection system (Millipore SNAP2MB1). Washing and incubation with HRP-conjugated secondary antibody were performed on the system according to the manufacturer’s instructions. Proteins were detected using Immobilon Western Chemiluminescent HRP substrate (Millipore WBKLS0500) or SuperSignal West Femto Chemiluminescent Substrate (ThermoFisher Scientific 34095) for high sensitivity.
Antibodies
Details of antibodies used are given in Supplementary Table 12.
Transcriptional analysis of 8 cell colonies
A single cell suspension of NFSK cells were sorted by flow cytometry into half area 96-well plates (Costar 3696), gating on forward and side scatter to isolate keratinocytes and excluding dead or dying cells by gating out the 7AAD positive population and then cultured in standard media with feeder cells6. After 2.5 days, feeders were removed by pipetting and 8-cell colonies were identified by phase microscopy. Colonies were lysed in TRIzol (Invitrogen) on the plate and RNA was extracted according to manufacturers instructions. RNA was precipitated with linear acrylamide (Ambion) and the RNA pellet was dissolved in Direct Lysis buffer from WT-Ovation One-direct Kit) (NuGEN), after which cDNA was amplified following the manufacturer’s protocol.
Amplified cDNA was hybridised to Affymetrix Human Gene 1.0 ST arrays. Data was analysed as follows. The Bioconductor R xps package was used to create an expression matrix from Affymetrix data by Robust Multi-array Average normalisation. The samples were then filtered based on detection above background. Hierarchical clustering was performed for values with a minimum log fold change of 0.5 and the Linear Models for Microarray Data Bioconductor package used to determine differentially expressed genes.
Functional analyses were carried out using DAVID (http://david.abcc.ncifcrf.gov) and GSEA software (Broad Institute of MIT and Harvard University, http://www.broad.mit.edu/gsea)70. For GSEA analysis the reactome gene set database was selected. Default parameters were maintained except that the permutation type was by gene set with 1000 permutations. We defined gene sets as being enriched if they had a FDR< 25% and a nominal p value<0.05.
Measurement of translation by O-propargyl-puromycin assay)
48 hours after seeding at clonal density, NFSK were cultured with 50μM O-propargyl-puromycin (Jena Bioscience, NU-931-05) in keratinocyte media for 1 hour. Plates were then fixed with 4% paraformaldehyde for 10 minutes on ice. Colonies were stained following the same protocol described above for immnostaining with EdU using a 1:500 dilution of a Keratin 1 antibody (AF-87, Covance) and an Alexa Fluor Click-IT kit (Invitrogen, C10337) to detect O-propargyl-puromycin. Colonies containing 6 to 10 cells were imaged using a Zeiss Axio-Observer D1 microscope with Zeiss Axiovision software and mean colony fluorescence quantified using NIH ImageJ software.
CBX5 knockdown
0.1x104 NFSK cells were seeded in a 12 well plate along with a J2 feeder layer. 48 hours post seeding, feeder cells were removed using gentle pipetting and 1ml fresh CFAD media was added. For RNA and protein knockdown experiments 80pmol/well SilencerSelect siRNA directed to CBX5 (S23885, Life technologies) or Negative control siRNA (Life technologies 4390843) was added to each well with 3μl Lipofectamine 3000 transfection reagent in 1ml CFAD as described by the manufacturers (Life Technologies). Cells were left to incubate for 24 hours before media was changed and a 3T3 feeder cell layer re-seeded. 48 hours post transfection cells were lysed in either RLT buffer (Qiagen) for RNA extraction or RIPA (25mM Tris-Hcl pH7.6, 150mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, ThermoFisher Scientific 89900) buffer for protein assays.
RNA was extracted using the RNEasy kit (Qiagen, 74104) and reverse transcribed using the Quantitec kit (Qiagen 205313). qPCR was carried out using a StepOne plus PCR machine (ThermoFisher Scientific) with Taqman probes (Hs01127577_m1 CBX5 and Hs99999905_m1 GAPDH) and reagents (Life Technologies).
For quantitative capillary isoelectric immunoassay, 4μl of protein was run on the WES system (Protein Simple) and hybridised using anti-CBX5 and anti-alpha tubulin as primary antibodies68. Run conditions were as recommended by the manufacturer. Peak areas were determined using Compass software (Protein Simple) and normalised to alpha-tubulin. RNA and protein knockdown experiments were performed in biological quadruplicate.
For assaying colony size post CBX5 knockdown, cells were transfected with CBX5 or control siRNA at 40pmol/well and cultured for 7 days before being fixed with 4% PFA and stained and imaged as described above. Colony size assays were performed in biological triplicate.
Reproducibility of experiments
Figure 1b shows typical images from each of 3 independent experiments. Figure 1d,e shows representative examples from a total of 81 lineage trees from 3 independent experiments, full data set is shown in Figure 2. Figure 3a shows typical images from one of 81 colonies videoed. Figure 4f shows typical colonies from one of 3 independent experiments. Figure 5a is representative of colonies in 3 independent experiments. Figures 5b and 5e show representative colonies from 3 independent experiments. Figure 6d shows representative colonies from 3 independent experiments. Figure 7a-f shows a representative colony from 3 independent experiments. Supplementary Figures 3b,c show representative colonies from 3 independent experiments. Supplementary Figure 5a shows a representative blot from 3 independent experiments. Supplementary videos 1 and 2 are representative of 81 videos from 3 independent experiments.
Statistics
Pairwise comparison of normally distributed data with similar variance was by unpaired two-tailed t test. Colony size distributions were analysed by Kolmogorov-Smirnov or Mann-Whitney unpaired tests. Division outcome data are presented with the 95% confidence interval of the proportion. GraphPad Prism 6 software was used for statistical testing and generating plots. Monte Carlo simulations based on live imaging data were run in Excel using a sequential random number generator.
Accession Codes
Microarray data were deposited in ArrayExpress with accession number E-MTAB-3128.
Supplementary Material
Acknowledgments
The initial association of holoclone and paraclone type behaviour in clonal cultures of NFSK with stem and balanced progenitor dynamics was due to BDS working in collaboration with PHJ, VN-N, David Doupé and Allon Klein, based on the quantitative analysis of published and unpublished colony size distributions6. We thank Gözde Akdeniz & David Doupé for experimental work that led up to the project that was analysed by Allon Klein and Genneth Zhang, Patrick Lombard at the Wellcome Trust-Medical Research Council Cambridge Stem Cell Institute for Bioinformatics analysis and Esther Choolun for technical assistance. We acknowledge the support of the Wellcome Trust, Cambridge Cancer Centre, Medical Research Council, the NC3Rs (National Centre for the Replacement, Refinement and Reduction of Animals in Research) and Cancer Research UK (Programme grant C609/A17257).
Footnotes
Author Contributions
AR, KM, JF, BNN and PHJ designed and performed experiments and analysed and interpreted data. BDS proposed using growth rate from 144 to 168 hours to classify division mode in dye labelled cells. AR, KM, JF and PHJ wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 2.Watt FM, Green H. Stratification and terminal differentiation of cultured epidermal cells. Nature. 1982;295:434–436. doi: 10.1038/295434a0. [DOI] [PubMed] [Google Scholar]
- 3.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 4.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. Embo J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 7.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janich P, et al. Human Epidermal Stem Cell Function Is Regulated by Circadian Oscillations. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Luis NM, et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell. 2011;9:233–246. doi: 10.1016/j.stem.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 10.Mulder KW, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14:753–763. doi: 10.1038/ncb2520. [DOI] [PubMed] [Google Scholar]
- 11.Connelly JT, et al. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol. 2010;12:711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 12.Tan DW, et al. Single-cell gene expression profiling reveals functional heterogeneity of undifferentiated human epidermal cells. Development. 2013;140:1433–1444. doi: 10.1242/dev.087551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li A, Simmons PJ, Kaur P. Identification and isolation of candidate human keratinocyte stem cells based on cell surface phenotype. Proc Natl Acad Sci U S A. 1998;95:3902–3907. doi: 10.1073/pnas.95.7.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowell S, Jones P, Le Roux I, Dunne J, Watt FM. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol. 2000;10:491–500. doi: 10.1016/s0960-9822(00)00451-6. [DOI] [PubMed] [Google Scholar]
- 15.Rochat A, Kobayashi K, Barrandon Y. Location of stem cells of human hair follicles by clonal analysis. Cell. 1994;76:1063–1073. doi: 10.1016/0092-8674(94)90383-2. [DOI] [PubMed] [Google Scholar]
- 16.Gallico GG, 3rd, O'Connor NE, Compton CC, Kehinde O, Green H. Permanent coverage of large burn wounds with autologous cultured human epithelium. N Engl J Med. 1984;311:448–451. doi: 10.1056/NEJM198408163110706. [DOI] [PubMed] [Google Scholar]
- 17.Compton CC, et al. Skin regenerated from cultured epithelial autografts on full-thickness burn wounds from 6 days to 5 years after grafting. A light, electron microscopic and immunohistochemical study. Lab Invest. 1989;60:600–612. [PubMed] [Google Scholar]
- 18.Jones PH, Harper S, Watt FM. Stem cell patterning and fate in human epidermis. Cell. 1995;80:83–93. doi: 10.1016/0092-8674(95)90453-0. [DOI] [PubMed] [Google Scholar]
- 19.Fortunel NO, et al. Exploration of the functional hierarchy of the basal layer of human epidermis at the single-cell level using parallel clonal microcultures of keratinocytes. Exp Dermatol. 2010;19:387–392. doi: 10.1111/j.1600-0625.2009.01046.x. [DOI] [PubMed] [Google Scholar]
- 20.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 21.Doupe DP, Klein AM, Simons BD, Jones PH. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev Cell. 2010;18:317–323. doi: 10.1016/j.devcel.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Mascre G, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 23.Lim X, et al. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science. 2013;342:1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alcolea MP, et al. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat Cell Biol. 2014;16:615–622. doi: 10.1038/ncb2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coutu DL, Schroeder T. Probing cellular processes by long-term live imaging--historic problems and current solutions. J Cell Sci. 2013;126:3805–3815. doi: 10.1242/jcs.118349. [DOI] [PubMed] [Google Scholar]
- 26.Thon JN, Devine MT, Jurak Begonja A, Tibbitts J, Italiano JE., Jr High-content live-cell imaging assay used to establish mechanism of trastuzumab emtansine (T-DM1)--mediated inhibition of platelet production. Blood. 2012;120:1975–1984. doi: 10.1182/blood-2012-04-420968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watt FM. Epidermal stem cells as targets for gene transfer. Hum Gene Ther. 2000;11:2261–2266. doi: 10.1089/104303400750035799. [DOI] [PubMed] [Google Scholar]
- 28.He J, et al. How variable clones build an invariant retina. Neuron. 2012;75:786–798. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson P. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development. 1990;109:509–519. doi: 10.1242/dev.109.3.509. [DOI] [PubMed] [Google Scholar]
- 30.Rangarajan A, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doupe DP, et al. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 2012;337:1091–1093. doi: 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alcolea MP, Jones PH. Tracking cells in their native habitat: lineage tracing in epithelial neoplasia. Nat Rev Cancer. 2013;13:161–171. doi: 10.1038/nrc3460. [DOI] [PubMed] [Google Scholar]
- 33.Cicalese A, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 34.van den Bogaerdt AJ, et al. Upside-down transfer of porcine keratinocytes from a porous, synthetic dressing to experimental full-thickness wounds. Wound Repair Regen. 2004;12:225–234. doi: 10.1111/j.1067-1927.2004.012115.x. [DOI] [PubMed] [Google Scholar]
- 35.O'Shaughnessy RF, Seery JP, Celis JE, Frischauf A, Watt FM. PA-FABP, a novel marker of human epidermal transit amplifying cells revealed by 2D protein gel electrophoresis and cDNA array hybridisation. FEBS Lett. 2000;486:149–154. doi: 10.1016/s0014-5793(00)02252-3. [DOI] [PubMed] [Google Scholar]
- 36.Eckert RL, et al. S100 Proteins in the Epidermis. J Investig Dermatol. 2003;123:23–33. doi: 10.1111/j.0022-202X.2004.22719.x. [DOI] [PubMed] [Google Scholar]
- 37.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shyh-Chang N, et al. Lin28 Enhances Tissue Repair by Reprogramming Cellular Metabolism. Cell. 2013;155:778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q, Shalaby NA, Buszczak M. Changes in rRNA Transcription Influence Proliferation and Cell Fate Within a Stem Cell Lineage. Science. 2014;343:298–301. doi: 10.1126/science.1246384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buszczak M, Signer RAJ, Morrison SJ. Cellular differences in protein synthesis regulate tissue homeostasis. Cell. 2014;159:242–251. doi: 10.1016/j.cell.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taguchi S, et al. Overexpression of the transcription factor Yin-Yang-1 suppresses differentiation of HaCaT cells in three-dimensional cell culture. J Invest Dermatol. 2011;131:37–45. doi: 10.1038/jid.2010.229. [DOI] [PubMed] [Google Scholar]
- 43.Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perekatt AO, et al. YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc Natl Acad Sci U S A. 2014;111:7695–7700. doi: 10.1073/pnas.1400128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, et al. Yin-yang 1 negatively regulates the differentiation-specific transcription of mouse loricrin gene in undifferentiated keratinocytes. J Invest Dermatol. 2004;123:1120–1126. doi: 10.1111/j.0022-202X.2004.23492.x. [DOI] [PubMed] [Google Scholar]
- 46.Somervaille TC, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wongtawan T, Taylor JE, Lawson KA, Wilmut I, Pennings S. Histone H4K20me3 and HP1alpha are late heterochromatin markers in development, but present in undifferentiated embryonic stem cells. J Cell Sci. 2011;124:1878–1890. doi: 10.1242/jcs.080721. [DOI] [PubMed] [Google Scholar]
- 48.Barrandon Y, Green H. Cell migration is essential for sustained growth of keratinocyte colonies: the roles of transforming growth factor-alpha and epidermal growth factor. Cell. 1987;50:1131–1137. doi: 10.1016/0092-8674(87)90179-6. [DOI] [PubMed] [Google Scholar]
- 49.Rheinwald JG, Green H. Epidermal growth factor and the multiplication of cultured human epidermal keratinocytes. Nature. 1977;265:421–424. doi: 10.1038/265421a0. [DOI] [PubMed] [Google Scholar]
- 50.Bellas E, Chen CS. Forms, forces, and stem cell fate. Curr Opin Cell Biol. 2014;31:92–97. doi: 10.1016/j.ceb.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barker N, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 52.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 53.Chapman S, McDermott DH, Shen K, Jang MK, McBride AA. The effect of Rho kinase inhibition on long-term keratinocyte proliferation is rapid and conditional. Stem Cell Res Ther. 2014;5 doi: 10.1186/scrt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman S, Liu X, Meyers C, Schlegel R, McBride AA. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J Clin Invest. 2010;120:2619–2626. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luxenburg C, et al. Wdr1-mediated cell shape dynamics and cortical tension are essential for epidermal planar cell polarity. Nat Cell Biol. 2015;17:592–604. doi: 10.1038/ncb3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee B, et al. Transcriptional mechanisms link epithelial plasticity to adhesion and differentiation of epidermal progenitor cells. Dev Cell. 2014;29:47–58. doi: 10.1016/j.devcel.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amelio I, et al. miR-24 triggers epidermal differentiation by controlling actin adhesion and cell migration. J Cell Biol. 2012;199:347–363. doi: 10.1083/jcb.201203134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nanba D, et al. Cell motion predicts human epidermal stemness. J Cell Biol. 2015;209:305–315. doi: 10.1083/jcb.201409024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lecuit T, Yap AS. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol. 2015;17:533–539. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- 60.Zhu AJ, Haase I, Watt FM. Signaling via beta1 integrins and mitogen-activated protein kinase determines human epidermal stem cell fate in vitro. Proc Natl Acad Sci U S A. 1999;96:6728–6733. doi: 10.1073/pnas.96.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nanba D, et al. Actin filament dynamics impacts keratinocyte stem cell maintenance. EMBO Mol Med. 2013;5:640–653. doi: 10.1002/emmm.201201839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McMullan R, et al. Keratinocyte differentiation is regulated by the Rho and ROCK signaling pathway. Curr Biol. 2003;13:2185–2189. doi: 10.1016/j.cub.2003.11.050. [DOI] [PubMed] [Google Scholar]
- 63.Iglesias-Bartolome R, et al. mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell. 2012;11:401–414. doi: 10.1016/j.stem.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wabik A, Jones PH. Switching roles: the functional plasticity of adult tissue stem cells. EMBO J. 2015;34:1164–1179. doi: 10.15252/embj.201490386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Centanin L, et al. Exclusive multipotency and preferential asymmetric divisions in post-embryonic neural stem cells of the fish retina. Development. 2014;141:3472–3482. doi: 10.1242/dev.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013;502:513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buczacki SJ, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 68.Aspinall-O'Dea M, et al. Antibody-based detection of protein phosphorylation status to track the efficacy of novel therapies using nanogram protein quantities from stem cells and cell lines. Nat Protocols. 2015;10:149–168. doi: 10.1038/nprot.2015.007. [DOI] [PubMed] [Google Scholar]
- 69.Jensen KB, Driskell RR, Watt FM. Assaying proliferation and differentiation capacity of stem cells using disaggregated adult mouse epidermis. Nat Protoc. 2010;5:898–911. doi: 10.1038/nprot.2010.39. [DOI] [PubMed] [Google Scholar]
- 70.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.