Abstract
Nucleic acids have emerged as an effective material for assembling complex nanoscale structures. To tailor the structures to function optimally for particular applications, a broad structural design space is desired. Despite the many discrete and extended structures demonstrated in the past few decades, the design space remains to be fully explored. In particular, the complex finite-sized structures produced to date have been typically based on a small number of structural motifs. Here, we perform a comprehensive study of the design space for complex DNA structures, using more than 30 distinct motifs derived from single-stranded tiles. These motifs self-assemble to form structures with diverse strand weaving patterns and specific geometric properties, such as curvature and twist. We performed a systematic study to control and characterize the curvature of the structures, and constructed a flat structure with a corrugated strand pattern. The work here reveals the broadness of the design space for complex DNA nanostructures.
Introduction
Self-assembly of nucleic acids (DNA and RNA) provides a powerful approach for constructing sophisticated synthetic molecular structures and devices. By encoding sequence complementarity into component DNA strands, prescribed structures can be assembled under the appropriate formation conditions.1 After three decades of development, the complexity of synthetic DNA structures has grown from simple branched junctions1 formed from just a few strands to complex 2D and 3D objects composed of hundreds or even thousands of distinct strands.2-22 Moreover, researchers have demonstrated the construction of dynamic systems, including switches,23 walkers,24-26 circuits,25,27,29 and triggered assembly systems.25
Two methods that are particularly effective for assembling discrete mega-Dalton structures with arbitrarily prescribed shape are DNA origami10,14-18,22, and single-stranded tiles (SST)20 and bricks.21 In DNA origami, hundreds of short, synthetic DNA strands fold a long scaffold (typically the M13 viral genome) into a desired structure.10,16-18,22 More recently, researchers have demonstrated finite complex 2D and 3D shapes self-assembled from hundreds to thousands of distinct single-stranded tiles and bricks.20,21
Unlike DNA origami, no scaffold strand is required for SST structures which are composed entirely of short synthetic DNA strands. The elimination of the scaffold strand makes SST an effective tool for systematic and rapid study of the geometry, sequence, and structural design space for complex DNA structures. First, as the SST-based structure has a modular architecture – each strand can be included, removed, or replaced independently – SST enables rapid prototyping of the shape space (more than 100 distinct, sophisticated 2D20 and 3D21 shapes were experimentally demonstrated). Second, as the strand sequences are no longer restricted to be derived from the biogenic scaffold sequence, SST enables rapid probing of the sequence space (structures were formed from both specifically designed and randomly generated sequences20,21). Lastly and importantly, by removing the restriction of scaffold routing through the shape and using diverse structural motifs derived from the canonical SST motif,20 we will demonstrate here a systematic study of the structural design space for forming complex DNA nanostructures.
To tailor the structures to function optimally for particular applications, a broad design space for DNA nanostructures is desired. Despite the many discrete and extended structures demonstrated in the past few decades, the structural design space has yet to be fully explored. In particular, complex, finite-sized structures formed by origami and SST are primarily based on the double- or single-stranded versions of the basic DAE (anti-parallel double-crossover molecules with an even number of half-helical turns) motif3 and their derivatives. Beyond origami and SST, however, a richer motif space has been explored for forming relatively simple discrete and extended structures. This has been achieved using various structural motifs, such as diverse double crossover motifs,3 PX based structures,7,37 structures with flexible linker joints,11,14,15,29,38 wireframe structures,1,2,7,8,11,38-41 and metal-DNA junctions.42
The next challenge is to develop a versatile method for creating complex, discrete-sized DNA nanostructures with diverse structural motifs, strand weaving patterns, and desired structural properties. Building on the richness of the previous structural motifs for building DNA nanostructures, and taking advantage of the designability and flexibility of SST-based assembly, we performed a comprehensive study of the design space for constructing complex, finite-sized DNA structures. We tested more than 30 distinct motifs derived from single-stranded tiles.20 Most motifs self-assembled to form structures with diverse strand weaving patterns and specific geometric properties, such as curvature and twist. Moreover, we performed a systematic study to control and characterize the curvature of the structures, and constructed a flat structure with a corrugated strand pattern. Our success in assembling a myriad of different motifs demonstrates the robustness and versatility of the single-stranded tile approach. More generally, it reveals the broadness of the design space for constructing complex DNA nanostructures.
Results and Discussions
Motif design parameters
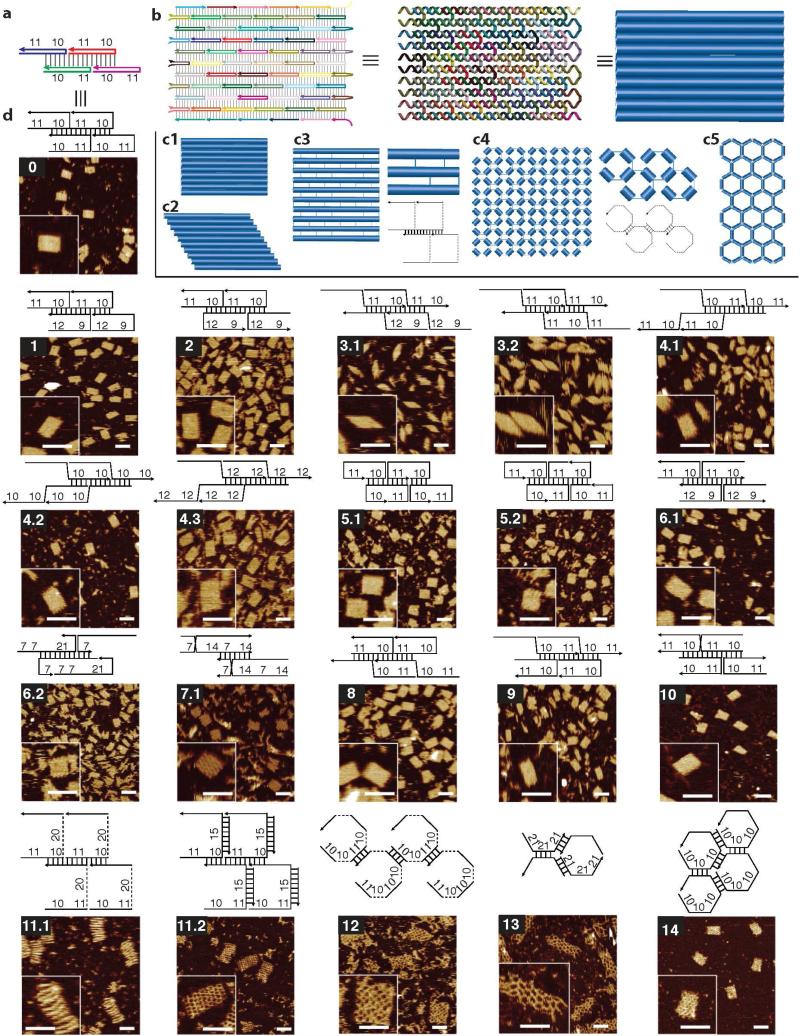
We began our study for the structural design space with the canonical U-shaped SST motif (motif 0, Fig. 1a,b) reported in our previous work.12,20 This 42-base SST motif has four consecutive domains (each measuring 10 or 11 nt).12,20 By pairing up complementary domains, individual tiles self-assemble into DNA lattices composed of parallelly-arranged double-helices connected by periodic single-stranded crossover linkages. Note that linkage is merely a phosphate in the DNA backbone. In the diagrams in Fig. 1, the linkage is artificially stretched to more clearly show the strand-weaving pattern. Starting with this simple U-shaped motif, we systematically explored important parameters for motif design. Below, we first give an overview of some important parameters and their putative effects on the geometrical, mechanical, and thermodynamic properties of the structures. In the next section, we describe the motif design and its implementation, in detail.
Figure 1.
Motif designs and AFM imaging. A representative subset of the motifs studied is shown here. See SI Sect. SI1.3 for the full repertoire. (a)A 2×2 unit (containing four SSTs) of the canonical motif (motif 0). Colors distinguish sequences. (b) Strand diagram (left), helix diagram (middle), and cylinder diagram (right) of a 10 helix × 11 turn (10H×11T) structure formed from motif 0. (c1-c5) Cylinder diagrams for structures formed from different motifs. A cylinder depicts a DNA duplex. A line depicts a single-strand DNA segment. Structures from motifs 0-2 and 4-10 have a rectangle shape containing parallelly-arranged double-helices (c1); structures from motifs 3.1 and 3.2 have a parallelogram shape containing parallelly-arranged double-helices (c2); structures from motif 11 have a rectangle shape containing parallelly-arranged double-helices connected by single-stranded linkers (c3); structures from motif 12 have a rectangle shape with a fish-net pattern (composed of short duplex segments connected by single-stranded linkers) (c4); structures from motif 13 and 14 have a rectangle shape with a honeycomb pattern composed of short DNA duplex segments (c5). (d) Strand diagrams and the corresponding AFM images for structures formed from different motifs. Top panel: a 2×2 repeating unit. Numbers indicate domain or linker lengths in nucleotide. Bottom panel: AFM image. Inset shows a magnified view. Scale bars: 50 nm. See SI Sect. SI1.5 for the measured dimensions and measured gel yields (5% to 44%).
Domain length
Motif 0 contains 10 and 11 nt domains. For most of the motifs explored in our study (including motif 0), we tested several different domain lengths. Changing domain length could affect (1) the geometrical properties of the assembled structure, such as curvature17,18 and twist;17,43,44 (2) thermodynamic properties, such as formation and melting temperature; (3) mechanical properties, such as structural rigidity and internal strain.17,45 Domain length is thus a basic design variable that we explored extensively in our study. Changing domain length also directly affects the crossover patterns described below.
Crossover type
Structures formed from motif 0 possess single-stranded, anti-parallel crossovers. In our study, we explored four combinations of single-12,46 vs. double-stranded crossovers, with parallel vs. anti-parallel orientations. Here we use “parallel” (or “anti-parallel”) to describe crossovers in which the orientation of the strand forming the crossover is the same (or opposite) on both sides of the crossover. The crossover type directly affects the weaving patterns of the strands and the geometrical properties of the structure, which may in turn affect their mechanical and thermodynamic properties. For example, geometrically, the antiparallel and parallel crossovers result in structures composed of helices that are connected in a parallel or anti-parallel fashion, referred to as parallelly-connected helices or anti-parallelly connected helices. Note that We also use the term parallelly-arranged double-helices to refer to both the double helices connected by either parallel or anti-parallel crossovers.. In the antiparallel crossover-based structure, all the crossovers display the major grooves on the same side of the structure47 and as a result, induce an accumulated curvature for the structure. In contrast, the parallel crossover based structure avoids such asymmetry and curvature. In the antiparallel crossover based structure, the major groove of one helix faces the minor groove of an adjacent helix;3 in contrast, the parallel crossover based structure has major-major and minor-minor groove pairings. Hence, the former should have less strain and more thermal stability.3 Compared with double-stranded crossover, single-stranded crossover based structures may be structurally less rigid, mechanically less strained, and in turn, thermodynamically more stable.
Crossover pattern
Motif 0 has a 21 nt crossover spacing for crossovers between the two adjacent helices; along one particular helix, a crossover appears every 10 or 11 nt. We systematically changed crossover patterns. Crossover patterns could also lead to a combination of geometric, thermodynamic, and mechanical effects. Geometrically, the curvature of the overall structure (as determined by the dihedral angle formed between three adjacent helices) can be tuned by translating all crossover points along the direction parallel to the helical axis while preserving the spacing;17,18 twist, on the other hand, depends on the crossover spacing.17,43,44 Mechanically, strain could accumulate in twisted and/or curved structures. 17,43,44 Such mechanical stress in turn could affect the thermodynamic properties of the structure.
Symmetry
By adjusting domain length, crossover type, and crossover patterns, we can design structures with more symmetric strand weaving patterns, such as corrugated patterns.29 Such symmetry can help to eliminate curvature in the structure.
Linkers
Structures formed from motif 0 are composed of compact parallelly-arranged double-helices connected by periodic single-stranded crossovers (composed of a single phosphate in the DNA backbone). In our study, we included single-stranded linkers of varying lengths between helices and between binding domains. This results in structures that are geometrically and structurally more flexible. Inclusion of linkers should also help to relieve the electrostatic repulsion between adjacent helices, hence stabilizing the structure.
Structures formed from diverse motifs
Overview
By systematically exploring the above as well as some other design parameters, we tested a total of 36 different motifs, 32 of which formed desired structures (an 89% success rate). A representative subset of all successful motif designs is shown in Fig. 1 (see SI Sect. SI1.3 for the full repertoire). Different morphologies were presented, including compact rectangle and parallelogram of packed DNA duplexes (Fig. 1c1 and Fig. 1c2), rectangles of spaced DNA duplexes (Fig. 1c3), and rectangles of square and hexagonal lattices (Fig. 1c4 and Fig. 1c5).
Structure assembly and characterization
The DNA structures were assembled with unpurified DNA strands with randomly designed sequences, and were mixed at roughly equal ratio without careful adjustment of stoichiometry. After single-step (one-pot) annealing from 90°C to 25°C over 17 h in 0.5× TE buffer supplemented with 15 mM Mg (see SI Sect. SI1.2 for the effects of buffer ion strength and annealing time on the assembly yields), the solution was subjected to 2% native agarose gel electrophoresis and produced one dominant product band, the ratio between the fluorescence intensity of the product band and that of the entire lane was used to approximate the yield of structure formation, which was measured to range from 5% to 44% (SI Fig. SI1).
Annealed samples were subjected to atomic force microscopy (AFM) imaging and showed the expected morphology and dimensions. For samples with relatively low yields (motifs 4.6, 7.2, and 10), structures were gel-purified from the product bands on a 2% agarose gel prior to AFM imaging. See SI Sect. SI1.3 for the larger AFM images. See SI Sect. SI1.5 for measurements of the sizes of the structures.
Changing domain lengths (motif 1)
First, we adjusted the domain lengths of two adjacent domains within a double-helix from 10-11 nt to 9-12 nt (motif 1). This changes the distance between adjacent crossovers along the same helix, resulting in the relative change of the dihedral angle formed by the three adjacent helices by 34.3°. This motif was later used to build a larger structure for curvature study.
Bi-directional motifs (motif 2)
Next, we flipped the orientation of the U-shaped motifs in every other row, resulting in a structure composed of U-shaped SSTs with alternating orientations along adjacent rows. Such a modification makes the overall structure more symmetric along the helical direction.
Z-shaped motifs (motifs 3.1, 3.2)
Structures formed from U-shaped motifs have anti-parallel crossovers. We then designed structures with two Z-shaped motifs and obtained structures containing parallel crossovers. Such parallel crossover weaving patterns are expected to cancel out the curvature resulting from the asymmetric arrangement of major and minor groves at the crossovers (see Curvature section for details) and was used for curvature study.
Corrugated structures (motif 4.1)
We further changed the orientation of every other row in the Z-shaped motif to form a symmetric structure with a corrugated29 strand pattern. This corrugation should in principle eliminate any curvature and result in a perfectly flat structure. This structure was used in the curvature study below.
Twisted structures (motifs 4.2 to 4.6)
Based on the corrugated structure from motif 4.1, we changed the crossover spacing from the canonical two full helical turns (i.e. 21 nt) to 18, 20, 22, 24, and 26 nt using motifs 4.2 to 4.6, respectively. This introduced a significant internal twist to the structure.17,43,44 The structures with 20 nt crossover spacing (motif 4.2) were used for the twist study and significant twist was observed (see below). The yield of the structure with the shortest spacing (motif 4.6) was relatively low (26%) compared to that of the rest of the group (33%-42%).
Double-stranded crossovers (motifs 5.x, 6.x, and 7.x)
Structures formed from all the above motifs contain single-stranded, half crossovers between adjacent helices. We next tested structures with double-stranded, full crossovers. We first tried to shift the nicking points in the original U-shaped motif to be positioned away from the crossovers (motifs 5.1 to 5.3). Then we continued to design H-shaped motifs by increasing crossover spacing from 2 helical turns to 4 helical turns (motifs 6.1 to 6.3). We also designed X-shaped motifs that were derived Z-shaped motifs (motifs 7.1 and 7.2). Structures with full crossovers have nicks on the duplex (rather than at the crossovers), which could permit the use of enzymatic ligation to covalently join the two ends of an SST molecule to form a ring.
Hybrid motifs (motifs 8 to 10)
We combined different motifs to form hybrid structures with both U-shaped and Z-shaped rows (motifs 8 and 9) or with H-shaped and X-shaped rows (motif 10).
Linkers at crossovers (motifs 11.1 to 11.4)
At the crossovers, we placed single-stranded DNA linkers with varying lengths (from 2 nt to 20 nt, motifs 11.1, 11.3, and 11.4). We also tested to form structures with 15 bp double-stranded linkers (motif 11.2). In addition to the 10H ×11T structure, we also used motif 11.1 to form a 24H ×29T structure (SI Fig. SI29), demonstrating the scalability of structures formed from linker motifs.
Linkers between neighboring domains (motif 12)
We formed structures from a motif with linkers between every pair of neighboring domains (motif 12). In the resulting structures, individual 10 or 11 bp double-stranded DNA duplexes were connected by flexible 10 nt, single-stranded poly-T linkers. The structure was thus likely loosely connected without well-defined morphology. AFM imaging was performed only after the addition of 10 nt poly-A strands, which were used to complement the poly-T linkers, and helped to result in a structure with better defined shape. After poly-A complementation, grid-like structures were observed.
Wireframe lattices (motifs 13, 14)
We constructed hexagonal lattices with different mesh sizes: one formed from a motif with three 21-nt domains (motif 13) and another from a motif with six 10-nt domains (motif 14). The hexagon pattern is similar to previous work on hexagon lattice based tubes.41 In contrast to the typical structures composed of parallelly-arranged double-helices linked by periodic (half-)crossovers, these wire-frame structures40,41 are composed of short duplex fragments connected with junction points. Interestingly, due to the electrostatic repulsions between adjacent DNA helices, a structure composed of parallelly-arranged double-helices typically possesses a gap (e.g. 0.5 nm to 1.5 nm in DNA origami10,14,16 and SST structures12,20,21) between the adjacent crossovers, and can be viewed as a wireframe structure with a rectangular mesh pattern. In SI Sect. SI1.4, we discuss the transformation between parallel pattern and rectangular wire-frame pattern in more detail.
Curvature: Modeling and design
The strand-weaving pattern of the strands can affect the curvature of the structure. Fig. 2 summarizes our strategies to control structure curvature. SI Sect. SI2.1 gives modeling and analysis details.
Figure 2.
Curvature models. (a) Curvature model (left) and corresponding strand diagram (right) of motif 0. Inset depicts the major groove side vs. minor groove side. (b) Curvature model of motif 3.1 without (left) and with (right) offset angle α.(c) Curvature model of motif 3.2 with offset angle α.(d) Curvature model of motif 4.1. In this corrugated design, the offset angle α cancels out independent of the actual value α. (e and f) show predicted cross-sectional views of 24-helix structures formed from motifs 0 and 4.1, respectively. See SI Sect. SI2.1 and Fig. SI39 for modeling and calculation details.
Domain length adjustment
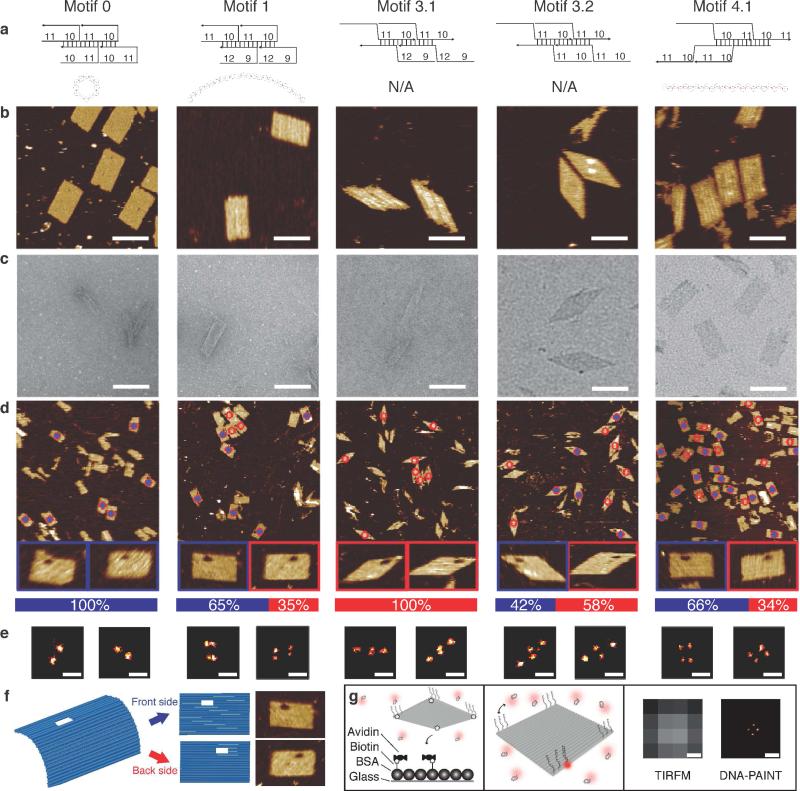
Working towards our goal of controlling curvature to design a flat structure, we first attempted to reduce curvature by adjusting the domain length of the canonical U-shaped SST motif. Assuming that the crossover points of adjacent double helices are collinear with the centers of the adjacent helices, our model predicts an average curvature of 30° per double helix for the motif 0, and 720° for the 24H×29T rectangle (see Figs. 2a and 2e for cross-sectional view of zoomed-in structure and the overall structure; see SI Sect. SI2.1.1 for detailed calculation). This curvature was confirmed by our experimental data (Fig. 3, column motif 0).
Figure 3.
Curvature characterization. See SI Sect. SI2.1 for curvature calculation and modeling details. (a) Strand diagrams (top) and curvature models in cross section views (bottom). (b) AFM images. Scale bars: 100 nm. See SI Sect. SI2.3 for larger images. (c) TEM images. Scale bars: 100 nm. See SI Sect. SI2.4 for larger images. (d) AFM-based landing assay results. A structure that landed with its front side facing up is marked with a blue dot inside a red circle; the one with back side facing up is marked by a hollow red circle. Magnified views show the structures with orientational markers (front side structures in blue boxes and back side structures in red boxes). The ratio between two landing orientations is shown in the bar chart in the bottom panel (blue: front side; red: back side). See SI Sect. SI2.5 for larger images and more details. (e) Representative DNA-PAINT super-resolution images of different SST structures labeled at four corners (scale bars: 100 nm). Measured point distances for motifs 0, 1, 3.1, 3.2, and 4.1 were 73 nm, 27 nm and 73 nm (short and long axis), 31 nm and 154 nm (parallelogram height and length), 45 nm and 148 nm (parallelogram height and length), and 45 nm and 77 nm (short and long axis), respectively. (f) Schematics of AFM-based landing assay. (g) Schematics of DNA-PAINT super-resolution imaging. Left panel: an SST structure was labeled on the top surface with 9 nt single-stranded “docking” sites for DNA-PAINT imaging strands. It was also labeled on the bottom surface with biotinylated strands for surface attachment. Center panel: Imager strands bind transiently to the docking sites in the four corners, producing apparent blinking. Right panel: diffraction-limited TIRFM image (left) and super-resolution image (right). Scale bars, 100 nm.
By adopting a domain length of 9/12 nt (motif 1) instead of the canonical 10/11 nt in motif 0, we were able to reduce the calculated average curvature per double helix from 30° to −4°, and −96° for the 24H rectangle (see SI Sect. SI2.1.1 for detailed calculation). This reduced curvature was confirmed by experimental data (Fig. 3, column motif 1).
However, a 2D structure (such as that formed by motif 0 or motif 1) designed with anti-parallel crossovers will always possess a side that displays minor grooves at crossovers and another side that displays major grooves at crossovers.47 As such, the curvature will not be perfectly counterbalanced for antiparallel crossover-based structures designed from U-shaped motifs (Fig. 2a).
Parallel crossover design
Next, we hypothesize that the curvature induced by the asymmetric arrangement of major and minor grooves at the crossovers on the two sides of the structures would be negated in parallel crossover-based structures formed from Z-shaped motifs: here, on either side of the structure, the crossovers display major and minor grooves in an alternating fashion. Like the U-shaped motif, we assume the crossover points of adjacent double helices are collinear with the centers of the adjacent helices, and designed a Z-shaped motif (motif 3.1) based structure that is expected to be perfectly flat (see Figs. 2b, left panel; see SI Sect. SI2.1.2 for detailed calculation). However, our experimental data revealed significant curvature in the structure (Fig. 3, column motif 3.1).
The discrepancy between our model and our experiments led us to revise the previous assumption that the crossover points of adjacent double helices are collinear with the centers of the adjacent helices. Although our experimental results from U-shaped motifs (motifs 0 and 1) did not invalidate such an assumption, the results from Z-shaped motifs were clearly incompatible with this collinear assumption. To reconcile this discrepancy between the model and the experimental results, we introduced an offset angle α formed between a base at the crossover and the two center points of the adjacent helices (middle panels of Fig. 2b and SI Fig. SI40c). In a structure formed from motif 3.1, this offset angle at the crossover always appears on the same side of the structure, resulting in an accumulative curvature for this otherwise flat structure.
Incidentally, another structure based on a Z-shaped motif with a different domain length (motif 3.2) appeared approximately flat in our experiments (Fig. 3, column motif 3.2). Assuming this structure (motif 3.2) has 0° curvature, we back-calculated an approximate value for the offset angle α (8.5°, Fig. 2c). Using this α value, the 24H×29T structure formed from motif 3.1 was expected to have significant curvature (see Figs. 2b, middle panel; see SI Sect. SI2.1.2 for detailed calculation), consistent with experimental observation of a curved structure.
Corrugation design
We next devised a corrugated design (motif 4.1) where the offset angle α appears alternately on the two sides of the structure, thus canceling out the curvature effect (independent of its value, Figs. 2d and 2f; see SI Sect. SI2.1.3 for detailed calculation). The predicted flatness of the structure was experimentally verified (Fig. 3, column motif 4.1).
Curvature: Experimental characterization
We performed a case study below to test the curvature of 24H×29T rectangle structures of a number of motifs.
Characterization by AFM and TEM
To study the curvature of the structures, we constructed 24H×29T structures with each of the above 5 motifs. Sequence design and structure assembly were conducted in a similar fashion as their 10H×11T counterparts. Then the annealed samples were subjected to 2% native gel electrophoresis (see SI Fig. SI41 for gel data), and the product band was extracted, purified by centrifugation, and imaged with AFM and TEM. Under AFM, all the structures appeared as flat, single-layer structures on mica surface (Fig. 3b); no curvature information was revealed. However TEM images more effectively reflected the structure configurations in solution (Fig. 3c): the structure generated from motif 0 (720° curvature expected) appeared as a dense, rolled up rectangle; the structure from motif 1 (−96° expected) appeared as a wider rectangle; rolled up and flat parallelograms were observed for the structure made from motifs 3.1 and 3.2, respectively; and a flat rectangle was observed for structure from motif 4.1 (0° curvature expected).
AFM-based landing essay
The different curvature configurations of the structures were also confirmed by two innovative assays. The first was the AFM-based landing assay in which a small hole was introduced to the top-left corners of the structure as an orientational marker (Fig. 3f). If the marker was more likely to show up at the top left corner rather than its mirror image (top right corner), it indicated the structure landed on the mica with its front side facing up, suggesting the structure curved up instead of down (motif 0). On the contrary, if the mirror image (back side facing up) was more likely, the structures had the tendency to curve down (motif 3.1). If the marker showed no bias for either side, the structure would be flat. Our experimental results (Fig. 3d) showed a perfect bias for the rolled up structures in the predicted curving direction (motif 0, 100% front side facing up (N=36); motif 3.1, 100% back side facing up (N=57)). However, likely due to a limited sample size, the assay did not effectively differentiate between other less curved structures (motif 1, 65%, front side facing up (N=74); motif 3.2, 42% front side facing up (N=42); motif 4.1, 66% front side facing up (N=203)). Note that only well-formed structures (defined as those showing no defects >15 nm in diameter in the expected boundary, nor >10 nm in diameter in the interior of the structure) were counted in our statistics. See SI Sect. SI2.5 for details.
Super-resolution fluorescence microscopy assay
Super resolution fluorescence microscopy was the second approach employed to assess the curvature of SST structures (Fig. 3g). Here we used DNA-PAINT32,35,48 to obtain sub-diffraction images of SST structures. DNA-PAINT exploits the repetitive, transient binding of short fluorescently-labeled oligonucleotides (“imager” strands) to complementary strands (“docking” sites) on a structure or molecule of interest. Using Total Internal Fluorescence Microscopy (TIRFM), binding events of imager strands to docking sites were observed as single-molecule events and the fluorescence emission was fitted to a 2D Gaussian function, yielding sub-diffraction resolution images. SST structures were “labeled” with DNA-PAINT docking sites by extending the three outmost strands in each of the four corners by a 9 nt long sequence at their 3′-end (Fig. 3g, center panel). The structures also carried four biotinylated strands pointing towards the opposite side of the docking strands for surface attachment (Fig. 3g, left panel). Once bound to a functionalized glass slide, fluorescently labeled DNA-PAINT imager strands were introduced, which transiently bound to the docking strands. The right panel of Fig. 3g includes a typical diffraction-limited TIRF image of a DNA nanostructure alongside the DNA-PAINT image after reconstruction. The increase in resolution is clearly visible.
The results of the DNA-PAINT study on the five different SST motifs are in good agreement with the TEM analysis as well as the AFM-based landing assay (Fig. 3e). For motif 0, only two apparent points along a distance corresponding to the long side of the rectangular structure were visible, suggesting a “rolled-up” structure where the two corners were placed right next to each other, or only one of the two corners was available to bind the imager strand. In contrast, in the structure from motif 1 we observed four separate points, two of which were in close proximity, suggesting a slightly curved structure. Images for motifs 3.1 and 3.2 were consistent with an almost rolled-up structure (3.1) and a flat parallelogram structure (3.2), respectively. Finally, the image for motif 4.1 (with four visible corners) suggests a flat rectangular structure.
Twist
Beyond assessing the curvature in different SST designs, we also studied the twist of the SST structures by creating homopolymers of motifs 4.1 and 4.2. We designed the leftmost column of the SST tiles to pair with the rightmost column of the rectangle. As a consequence, multiple units of rectangles align in tandem to form a long polymer with individual rectangles as monomer units (SI Fig. SI57, top). Polymerization helps to identify the small degrees of twist that would be difficult if not impossible to visualize using only monomeric structures. AFM images revealed that the homopolymers formed from motif 4.1 yielded ribbons with virtually no global twist (SI Fig. SI58a, bottom). This was expected, as the design used 21 bases between adjacent crossover points, thus obeying 10.5 bp spacing per helical turn as in the natural form of B-DNA. In contrast, polymers formed from motif 4.2 showed a global left-handed twist (SI Fig. SI58b, bottom). This is consistent with the fact that motif 4.2 used 20 bases between adjacent crossover points and was expected to produce a locally overwound helix. Our results agree with earlier studies of curvature and twist for DNA origami structures17,43,44 and suggest that the twist of the SST structures can be modulated by changing crossover spacing distances.
Conclusion
In summary, we evaluated more than 30 SST motifs that form lattice structures with diverse strand weaving patterns. Most of the motifs self-assembled into the designed structures with reasonable yields. Using some of the motifs, a systematic study was conducted to reduce the curvature in the assembled structure. The difference in curvature was characterized by TEM, an AFM-based landing assay, and super-resolution microscopy. A corrugated weaving pattern was predicted and experimentally verified to produce a flat structure.
The simple and robust nature of SSTs has enabled us to survey the structural space for forming complex structures in a rapid and comprehensive fashion. An unprecedented collection of diverse weaving patterns was demonstrated. We were able to define simple-to-implement design rules that lead to structures with specific geometric properties such curvature and twist. More broadly, the diverse collection of motifs will allow us to modulate the geometrical, mechanical, and thermodynamic parameters. We demonstrated the tuning of curvature and twist with the new motifs. In a related work,49 we demonstrated that by using the motifs reported here, we can form complex DNA nanostructures isothermally at any prescribed temperature between 15°C and 69°C, and under biocompatible conditions. The work here thus provides a new set of tools for modulating the structural properties of complex DNA structures, and reveals the broad design space for forming such structures.
Methods
DNA sequence design
Most DNA sequences were designed with the Uniquimer software by populating the motifs with random sequences50 while maintaining the required complementarity relations. The following design rules were applied for sequence generation: 1) Nucleotides (i.e. A, C, G, T) are generated one by one randomly. 2) Complementary nucleotides to one generated are matched following the base-pairing rule: A to T, C to G and vice versa. 3) Specific segments (e.g four consecutive A, C, G, T) are not allowed. When such segments emerge during design, the most recent generated nucleotide will be mutated until disallowance of specific segments is satisfied. Due to the difficulty of designing structures with wireframe motifs, a separate script was developed following the same design principles. Manual design and/or optimization was used for the linker segment sequences (e.g. motifs 12.1 to 12.4 and motif 13) and the handle segment sequences (e.g. handle segment sequences for structures of motifs 0, 1, 3.1, 3.2 and 4.1 in super-resolution study.
Sample preparation
DNA strands were synthesized by Integrated DNA Technology, Inc (http://www.idtdna.com). To assemble the structures, DNA strands were mixed to roughly equal molar concentration of 200 nM for smaller structures (all 10H×11T structures, structures with linkers and wireframe structures), and 100 nM for larger structures (24H×29T and 24H×28T structures for the curvature study) in 0.5×TE buffer (5 mM Tris, pH 7.9, 1 mM EDTA) supplemented with 15 mM MgCl2. The mixtures was then annealed in a PCR thermo cycler by cooling from 90°C to 25°C over a period of 17 hours with a specific cooling program.20 The annealed samples were then subjected to 2% agarose gel electrophoresis (gel prepared in 0.5×TBE buffer supplemented with 10 mM MgCl2 and pre-stained with SYBR safe) in an ice water bath. If necessary, the target gel bands were excised out and put into a Freeze N’ Squeeze column (Bio-Rad). The gel pieces were crushed using a microtube pestle in the column and the column was then directly subjected to centrifuge at 438 g for 3 minutes. Purified samples were collected in the eluate, and concentrations were determined by Nanodrop absorption at 260 m prior to AFM or TEM imaging.
AFM imaging
AFM images were obtained using an SPM Multi-mode with Digital Instruments Nanoscope V controller (Vecco). A 5 μL droplet (2 to 10 nM) of annealed (or purified) sample and then a 40 μL drop of 0.5×TE / 10mM MgCl2 solution were applied to a freshly cleaved mica surface and left for approximately 2 min. Sometimes, additional dilution of the sample was performed to achieve the desired sample density. As for the cases of 10H×11T structures, 20 μL supplemental 10mM NiCl2 was added to increase the strength of DNA-mica binding.51 The images were taken under the liquid tapping mode, with C-type triangular tips (resonant frequency, f0 = 40-75kHz; spring constant, k = 0.24Nm−1) from the SNL-10 silicon nitride cantilever chip (Bruker Corparation).
TEM imaging
For TEM imaging, a 3.5 μL sample (1-5 nM) was adsorbed onto glow discharged carbon-coated TEM grids for 4 minutes and then stained for 1 minute or a few seconds using a 2% aqueous uranyl formate solution containing 25 mM NaOH. Imaging was performed using a JEOL JEM-1400 TEM operated at 80 kV.
Super-resolution imaging
Super-resolution studies were performed with DNA PAINT.43 The samples were diluted in DNA-PAINT imaging buffer (5 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM EDTA, 0.05% Tween-20) to 50 pM and immobilized to a coverslip in a flow chamber via biotin streptavidin binding interaction. To fix the samples, solutions containing 1mg/mL BSA-biotin, 2mg/mL streptavidin, and biotin-labelled DNA nanostructures were flushed into the flow chamber and incubated in the above order. The chamber was then washed with DNA-PAINT imaging buffer containing 20 nM ATTO6553-labeled imager strands and sealed with 5 minutes epoxy before imaging. DNA-PAINT super-resolution images were acquired on an inverted Nikon Ti-E microscope (Nikon Instruments, Melville, NY) with the Perfect Focus System, applying an objective-type TIRF configuration using a Nikon TIRF illuminator with an oil-immersion objective (100× Plan Apo, NA 1.49, Oil, Nikon). A 647 nm laser (Agilent MLC400B, 80 mW at the objective) was used for TIRF excitation. The laser beam was filtered with a clean up filter (642/20 Chroma Technologies) and coupled into the microscope objective using a multi-band beam-splitter (zt405/488-491/561/638rpc, Chroma Technologies). Fluorescence was spectrally filtered with an emission filter (700/75 Chroma Technologies) and imaged on an EMCCD camera (Andor iXon 3, Andor Technologies, North Ireland). 5,000 frames were recorded at a frame rate of 10 Hz. Super-resolution images were reconstructed using spot-finding and 2D Gaussian fitting algorithms programmed in LabVIEW (National Instruments Corporation). Images were drift corrected and the channels aligned using DNA-origami drift markers.
Yield quantification by gel electrophoresis
Yield was estimated by analysis using native agarose gel electrophoresis, pre-stained with SYBR Safe DNA stain. The ratio between the fluorescence intensity of the product band and that of the entire lane was taken as an estimate of the gross yield of structural formation.
Structure size measurement and statistics
AFM measurements were obtained using Nanoscope Analysis (version 1.20; Vecco). The ‘cross-section’ function was used to measure distances (lengths and widths of the structures of different sizes). Well-formed structures were chosen for the measurements. TEM images were analyzed using ImageJ (version 1.46r; NIH). The straight line function was used to measure widths of certain structures. Six or ten sample points were collected for each distance measurement and the statistics (e.g. the mean and the standard deviation) were based on those 6 or 10 data points.
Supplementary Material
ACKNOWLEDGMENT
We thank Wei Sun and Hoang Lu for technical assistance and David Zhang, Erik Winfree, William Sherman, and Robert Barish for discussion. DNA-PAINT data were collected at the Nikon Imaging Center at Harvard Medical School. This work was funded by an Office of Naval Research (ONR) Young Investigator Program award N000141110914, an ONR grant N000141010827, an National Science Foundation (NSF) Faculty Early Career Development Award 1054898 and an NSF grant 1162459, an NIH Directors New Innovator Award 1DP2OD007292, and a Wyss Institute Faculty Startup Fund to P.Y.. M.D. acknowledges an international student research fellowship from the Howard Hughes Medical Institute. C.M. acknowledges a graduate research fellowship from the Fannie and John Hertz Foundation. R. J. acknowledges a postdoctoral fellowship from the Alexander von Humboldt Foundation.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available Experimental details, additional figures, and sequence data for each structure. This information is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Seeman NC. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Seeman NC. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 3.Fu TJ, Seeman NC. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 4.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 5.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 6.Rothemund PWK, Papadakis N, Winfree E. PLoS Biology. 2004;2:2041–2053. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 8.Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 9.Park SH, Pistol C, Ahn SJ, Reif JH, Lebeck AR, Dwyer C, Labean TH. Angew. Chem. Int. Ed. 2006;45:735–739. doi: 10.1002/anie.200503797. [DOI] [PubMed] [Google Scholar]
- 10.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 11.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 12.Yin P, Hariadi R, Sahu S, Choi HMT, Park SH, LaBean TH, Reif JH. Science. 2008;321:824–826. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- 13.Zheng JP, Birktoft J, Chen Y, Wang T, Sha RJ, Constantinou P, Ginell S, Mao C, Seeman NC. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H. Nano. Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 15.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 16.Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietz H, Douglas SM, Shih WM. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 19.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 20.Wei B, Dai M, Yin P. Nature. 2012;485:623–626. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke Y, Ong LL, Shih WM, Yin P. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han D, Pal S, Yang Y, Jiang S, Nangreave J, Liu Y, Yan H. Science. 2013;339:1412–1415. doi: 10.1126/science.1232252. [DOI] [PubMed] [Google Scholar]
- 23.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 24.Sherman WB, Seeman NC. Nano Lett. 2004;4:1203–1207. [Google Scholar]
- 25.Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 26.Omabegho T, Sha R, Seeman NC. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seelig G, Soloveichik D, Zhang DY, Winfree E. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 28.Qian L, Winfree E. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 29.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 30.Kuzyk A, Schreiber R, Fan Z, Pardatscher G, Roller E, Högele A, Simmel FC, Govorov AO, Liedl T. Nature. 2012;483:311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- 31.Choi HMT, Chang JY, Trinh LA, Padilla JE, Fraser SE, Pierce NA. Nature Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C, Jungmann R, Leifer AM, Li C, Levner D, Church GM, Shih WM, Yin P. Nature Chemistry. 2012;4:832–839. doi: 10.1038/nchem.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Z, Sun W, Ke Y, Shih C, Paulus GLC, Wang QH, Mu B, Yin P, Strano MS. Nature Communications. 2013;4:1663. doi: 10.1038/ncomms2690. [DOI] [PubMed] [Google Scholar]
- 34.Berardi MJ, Shih WM, Harrison SC, Chou JJ. Nature. 2011;476:109–113. doi: 10.1038/nature10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derr ND, Goodman BS, Jungmann R, Leschziner AE, Shih WM, Reck-Peterson SL. Science. 2012;338:662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Yan H, Shen Z, Seeman NC. J. Am. Chem. Soc. 2002;124:12940–12941. doi: 10.1021/ja026973b. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Su M, He Y, Zhao X, Fang PA, Ribbe A, Jiang W, Mao C. PNAS. 2008;105:10665–10669. doi: 10.1073/pnas.0803841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao C, Sun W, Seeman NC. J. Am. Chem. Soc. 1999;121:5437–5443. [Google Scholar]
- 40.Malo J, Mitchell JC, Venien-Bryan C, Harris JR, Wille H, Sherratt DJ, Turberfield AJ. Angew. Chem. Intl. Ed. 2005;44:3057–3061. doi: 10.1002/anie.200463027. [DOI] [PubMed] [Google Scholar]
- 41.Wilner OI, Orbach R, Henning A, Teller C, Yehezkeli O, Mertig M, Harries D, Willner I. Nature Communications. 2011;2:540. doi: 10.1038/ncomms1535. [DOI] [PubMed] [Google Scholar]
- 42.Yang H, Sleiman H. Angew. Chem. Int. Ed. 2008;47:2443–2446. doi: 10.1002/anie.200703741. [DOI] [PubMed] [Google Scholar]
- 43.Ke Y, Douglas SM, Liu M, Sharma J, Cheng A, Leung A, Liu Y, Shih WM, Yan H. J. Am. Chem. Soc. 2009;131:15903–15908. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jungmann R, Scheible M, Kuzyk A, Pardatscher G, Castro CE, Simmel FC. Nanotechnology. 2011;22:275301. doi: 10.1088/0957-4484/22/27/275301. [DOI] [PubMed] [Google Scholar]
- 45.Kim D, Kilchherr F, Dietz H, Bathe M. Nucleic Acids Res. 2011;40:2862–2868. doi: 10.1093/nar/gkr1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H, Chen Y, He Y, Ribbe A, Mao C. Angew. Chem. Int. Ed. 2006;45:1942–1945. doi: 10.1002/anie.200504022. [DOI] [PubMed] [Google Scholar]
- 47.Rothemund PWK, Ekani-Nkodo A, Papadakis N, Kumar A, Fygenson DK, Winfree E. J. Am. Chem. Soc. 2004;126:16344–16353. doi: 10.1021/ja044319l. [DOI] [PubMed] [Google Scholar]
- 48.Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P, Simmel FC. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 49.Myhrvold C, Dai M, Silver P, Yin P. Nano Lett. 2013;13:4242–4248. doi: 10.1021/nl4019512. [DOI] [PubMed] [Google Scholar]
- 50.Wei B, Wang Z, Mi YJ. Comput. Theor. Nanosci. 2007;4:133–141. [Google Scholar]
- 51.Hansma HG, Laney DE. Biophys. J. 1996;70:1933–1939. doi: 10.1016/S0006-3495(96)79757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.