Abstract
Background
Shigellosis is a serious problem in Guizhou and Shigella sonnei is an increasingly prevalent etiologic agent of local shigellosis cases. No data, however, are available about the molecular characterization of the local isolates of S. sonnei. We have conducted this study to molecularly characterize the clinical isolates of S. sonnei in Guizhou Province.
Results
76 S. sonnei isolates, including four isolates from 1974–1982 and 72 isolates from 2008–2010, were used for analysis in this study. Pulsed-field gel electrophoresis (PFGE) based on XbaI digestion divided the 76 isolates into 38 PFGE patterns (PT) and 15 PTs were represented by more than one isolates with PT31 (N = 8) containing the most number of isolates, followed with PT2 (N = 6). Multiple-Locus Variable number tandem repeat (VNTR) Analysis (MLVA) based on seven VNTR loci discriminated them into 19 different MLVA types (MTs), and four MTs were represented by more than one isolate with MT4 (N = 39) containing the most number of isolates, followed with MT12 (N = 18). 15 Multilocus sequence typing (MLST) base on 15 loci differentiated the isolates into six sequence types (STs), among which four STs were novel. The most common STs are ST76 (N = 43) and ST116 (N = 25), accounting for 92.1%. Correlation between genetic relationships and geographical origins or isolation years was observed among the isolates studied. Majority of isolates were clustered in accordance with the origin of isolation years based on the genetic data, which were also from similar geographical origins.
Conclusions
Our results revealed the molecular characteristics including the specific genotypes such as four novel STs, clonal relationship, and genetic changes of local isolates from different years, which enhances our understanding of molecular characteristics of S. sonnei and contributes to the prevention and control of shigellosis in Guizhou Province.
Background
The shigellae are the causative agents of shigellosis, a common diarrheal disease in developing countries [1, 2]. The main symptom of this infection is bloody diarrhoea and the minimum infective dose is as low as 10–100 bacterial cells due to relative resistance to stomach acid [3]. The infection is most frequent in children, the elderly and the immunocompromised [4]. There are about 164.7 million cases of shigellosis annually worldwide, resulting in 1,100,000 deaths, most of which are children under five years of age [2, 5].
Shigella can be differentiated into four species or serogroups, S. dysenteriae, S. flexneri, S. boydii, and S. sonnei based on biochemical properties and group-specific O antigens in the outer membrane of the cell wall. There is a shift in Shigella dominance from S. flexneri to S. sonnei in developing and developed countries [3, 6–8]. In China, an previous study on serotype distribution of Shigella reported that S. flexneri, from 1989 to 2010, was always the most frequently isolated serogroup, followed by S. sonnei, but after 2000, there was a significant upward trend in the constituent ratio of group S. sonnei in south China [9]. Guizhou Province, with nearly 50 million people, is an impoverished province in the southwest of China. It has been reported that Shigella was isolated as early as 1950s in Guizhou Province [10], and 2,266 S. flexneri isolates were isolated in Guizhou Province from 1955 to 1987 [10]. However, S. sonnei is becoming the dominant serotype causing shigellosis in Guizhou in recent years. For instance, there were 48,222 cases of human shigellosis were reported from 2007 to 2010, and 146 isolates of Shigella, including 102 S. sonnei and 44 S. flexneri isolates, were isolated in Guizhou [11].
Molecular typing methods are powerful tools for epidemiological investigation of Shigella infections as well as other bacterial infections such as those caused by Shigatoxin-producing Escherichia coli O157: H7 [2]. PFGE is the gold standard method among the molecular typing methods with a standardized PulseNet protocol, which makes it possible to compare bacterial DNA fingerprints among laboratories, even internationally [2, 12, 13]. Nevertheless, more powerful and easier methods such as multilocus variable number tandem-repeat analysis (MLVA) and multilocous sequence typing (MLST) are newly devised. MLVA is based on the inherent variability of short sequences that are organized as tandem repeats at multiple VNTR loci, which has been used to study the genetic relatedness among S. sonnei [2, 3, 14, 15]. MLST is a housekeeping gene sequence-based genotyping technique first introduced by Maiden et al., which provides reproducibility, comparability and transferability between laboratories [16, 17]. The most important advantage of MLST is the unambiguity and electronic portability of nucleotide sequence data [18]. Besides, MLST sequence data can be held through a central database and queried through a web server [19, 20]. By MLST analysis, S. flexneri were categorized into two sequence type (ST) complexes based on seven house- keeping genes [21, 22], and then MLST based on 15 house-keeping genes were applied in Shigella genotyping and the protocol is shared at the EcMLST website [23, 24].
Although S. sonnei is becoming an important etiologic agent of shigellosis in Guizhou Province, there is limited information on the genetic background of local strains. Therefore, the objective of the study was to molecularly characterize the local S. sonnei strains with PFGE, MLVA and MLST. In addition, the prospect of using PFGE, MLVA and MLST for routine subtyping of local S. sonnei was compared and evaluated. This was intended to aid clinical laboratory diagnosis, active surveillance, outbreak investigation and source tracking for shigellosis the locality.
Materials and Methods
Bacteria isolates and serological confirmation
All the S. Sonnei isolates were serologically confirmed by slide agglutination with commercial Shigella monovalent antisera (Denka Seiken, Japan) according to the manufacturer’s instructions [25]. The confirmed isolates were kept in brain heart infusion broth with 50% glycerol at -80°C for further study.
PFGE
S. sonnei isolates were analyzed by PFGE with the digestion of XbaI according to the U.S. CDC PulseNet protocol [13, 26]. Briefly, XbaI-digested Salmonella serotype Braenderup H9812 was used as the molecular weight standard. Electrophoresis was performed with a CHEF-DR III System (Bio-Rad Laboratories, Hercules, CA) using 1% SeaKem Gold agarose. The interpretation of the PFGE patterns was performed with BioNumerics software (Applied Maths, St-Martens-Latern, Belgium) using the Dice similarity coefficient. The tree indicating relative genetic similarity was constructed on the basis of the unweighted pair group method of averages (UPGMA) and a position tolerance of 1%. A PFGE pattern with one or more DNA bands different from the others was taken to be a unique PFGE pattern. A dendrogram constructed using the PFGE patterns was generated by the UPGMA algorithm using the Dice-predicted similarity value of two patterns.
Preparation of DNA templates
DNA templates for PCR were prepared directly from bacterial colonies by the boiling method [25]. Briefly, a single colony from an overnight culture at 37°C on LB agar was suspended in 30 μl of distilled water and boiled at 100°C for 10 min. The sample was immediately cooled on ice for 5 min and centrifuged at 13,000 × g at 4°C for 10 min. The supernatant, containing DNA, was used as the template for PCR amplification.
MLVA
Seven VNTR loci, SS1, SS3, SS6, SS9, SS10, SS11, and SS13 and the primer sequences and conditions reported in previous studies were used in this study [3, 14]. Briefly, the forward primer for each primer set was labeled at its 5' end with an ABI compatible dye, HEX, FAM, TAMRA, or ROX by Tsingke (Beijing, China). Two multiplex polymerase chain reactions (mPCR) were carried out. The PCR products were analyzed by capillary electrophoresis on an ABI 3730XL sequencer with GeneScan 500 LIZ Size Standard (Applied Biosystems). Data were collected, and the lengths of the amplicons were determined with GeneScan data analysis software, v. 3.7 (Applied Biosystems). The copy number of the repeats of each VNTR locus was deduced from the length of the amplicons. This number was then converted into an allele designation, which in turn formed the allele string for the seven loci. The allele string was constructed in the following order: SS1-SS3-SS6-SS9-SS10-SS11-SS13. The data were incorporated into BioNumerics software and analyzed as described previously [27]. Each unique allelic string was designated a unique MLVA type (MT). A dendrogram was constructed by UPGMA clustering based on categorical coefficient. Minimum spanning tree algorithm was used to construct a minimum spanning tree (MST) to determine phylogenetic pattern.
MLST
15 loci including arcA, aroE, aspC, clpX, cyaA, dNaG, fadD, grpE, icdA, lysP, mdh, mtlD, mutS, rpoS and uidA were selected based on primers as previous study described [23, 24], which can be accessed on the sharing EcMLST website (http://www.shigatox.net/ecmlst/cgi-bin/index). Following the standard MLST protocol, the PCR products were detected by electrophoresis of 3μl of each reaction on a 1.5% agarose gel for 30 min at 100 V, and were sequenced by ABI 3730XL DNA sequencer. Each allele was assigned a different allele number and the allelic profile (string of fifteen integers) was used to define the sequence type (ST). The EcMLST website was established to provide a resource to investigators who can use this to assign the ST of further strains. New allele numbers and sequence type were submitted to the EcMLST curator for allocation. The data were incorporated into BioNumerics software and a dendrogram was constructed by UPGMA clustering based on categorical coefficient, and a minimum spanning tree (MST) analysis was used to infer relationships among the isolates and was done using BioNumerics (Applied Maths, Belgium) [23].
Results
Distribution of isolates and serotypes
A total of 76 strains of S. sonnei isolated in Guizhou Province were used for analysis in this study. Among the 76 isolates used in this study, four isolates were collected in 1974–1982, while the remaining 72 isolates were from 2008–2010. All the isolates originated from five prefectures (including Guiyang, Anshun, Qianxinan, Qiandongnan, Zunyi) of the nine prefectures in Guizhou Province, with the exception of prefecture Bijie, Qiannan Tongren, and Liupanshui. The results of slide agglutination test showed that only 2.63% (2/76) of the isolates belonged to S. sonnei phase I isolates, while 97.37% (74/76) of the isolates were serologically identified as S. sonnei phase II isolates.
PFGE based Genotypes
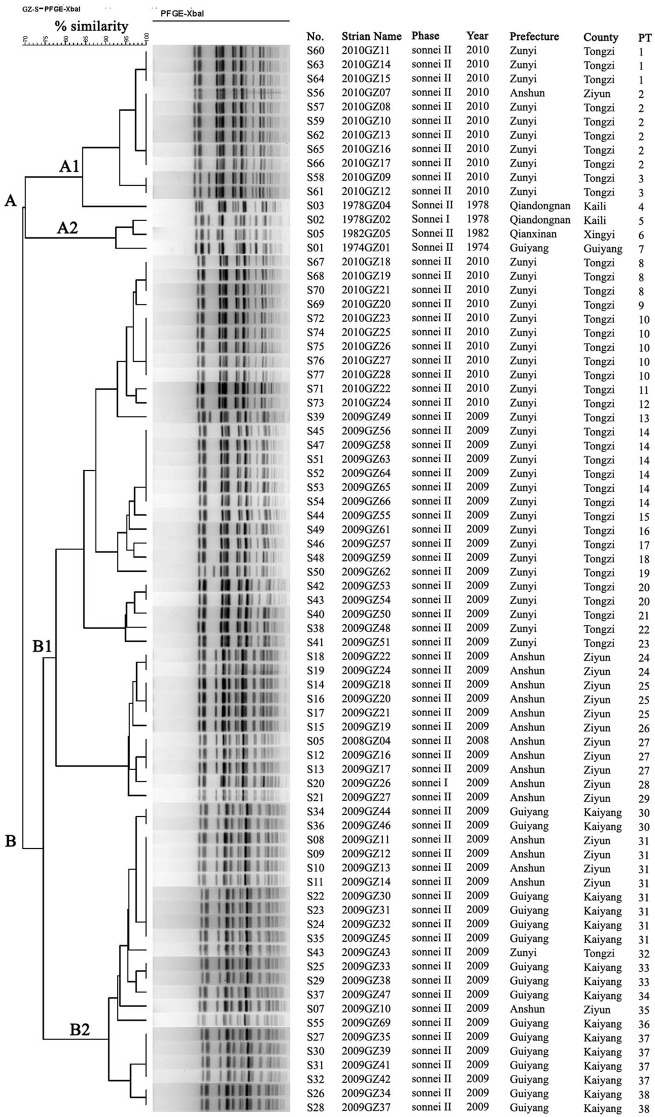
The genotypes and genetic relatedness of the 76 isolates were determined by PFGE. XbaI-digested S. sonnei DNA generated 38 reproducible unique PFGE patterns (PT). 15 patterns were represented by more than one isolates with PT31 (N = 8) containing the most number of isolates, followed with PT2 (N = 6). The dentrogram (Fig 1) of all the 76 isolates showed a 65% coefficient of similarity and can be classed into two gross clusters on the basis of their serotypes (cluster A and B). All the four isolates from 1974–1982 and part of the isolates from 2010 formed the cluster A, while all the isolates from 2009, part of isolates from 2012 and the one strain from 2008 were distributed in cluster B. Cluster A is further divided into cluster A1 and A2. Isolates from 2010 (cluster A1) and 1974–1982 (cluster A2) were further closely clustered in accordance with the isolation time, respectively. Cluster B was also grouped into B1 and B2 cluster. Isolates in cluster B1 included part isolates from 2009 and 2010, which were further clustered based on the origin of isolation year, whereas the one isolate (2008GZ04) from 2008 and part of isolates from 2009 were included in cluster B2. Besides, most of the isolates from the same year were closely clustered in accordance with the region origin in the dentrogram.
Fig 1. Relationships of 76 Shigella sonnei isolates from Guizhou Province based on Pulsed Field Gel Electrophoresis (PFGE).
The 76 S. sonnei isolates from Guizhou province were analyzed by PFGE using XbaI. The interpretation of the PFGE patterns was performed with BioNumerics software using the Dice similarity coefficient. The tree indicating relative genetic similarity was constructed on the basis of the unweighted pair group method of averages (UPGMA) and a position tolerance of 1%. A PFGE pattern with one or more DNA bands different from the others was taken to be a unique PFGE pattern. The dendrogram constructed using the PFGE patterns was generated by the UPGMA algorithm using the Dice-predicted similarity value of two patterns. The corresponding PFGE pattern, serotype and background information were shown alongside the dendrogram on the right.
MLVA based Genotypes
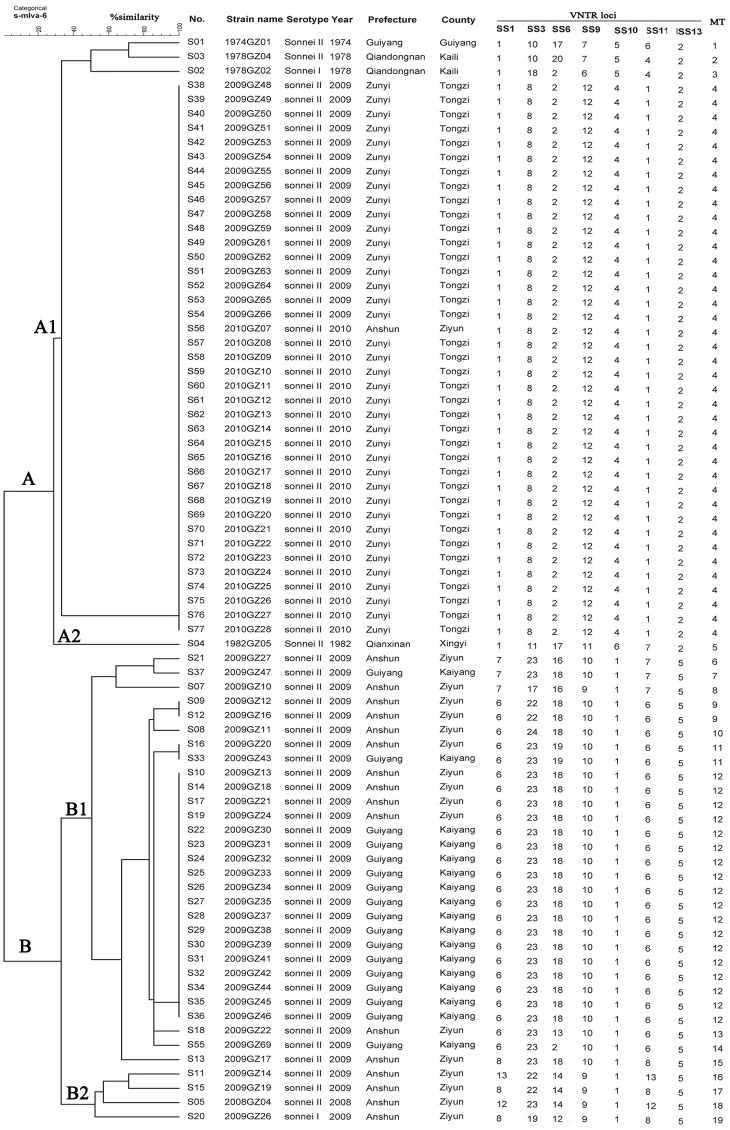
MLVA based on seven VNTR loci were performed to further characterize the S. sonnei isolates. All the 76 isolates showed a low coefficient of similarity (about 25.00%). The copy numbers of each VNTR locus are listed in Fig 2. The 76 S. sonnei isolates, based on the unique MLVA profiles, were discriminated into 19 different MLVA types (MTs). Four MTs were represented by more than one isolates with MT4 (N = 39) containing the most number of isolates, followed with MT12 (N = 18). M4 included 36 isolates come from 2009 and 2010, accounting for 47.37% (36/76) of the total number of isolates, while MT12 contains 18 isolates, making up 23.68% (18/76) of the total. Two main clusters, cluster A and B, were observed from the dendrogram generated (Fig 2). Cluster A was further divided into cluster A1 and A2. Three isolates (1974GZ01, 1978GZ02 and 1978GZ04) and part of isolates from 2009 and 2010 constitute the cluster A1 cluster, in which the isolates were further clustered based on the isolation years, while isolate 1982GZ05 separately formed the clade A2. All the isolates in cluster B were come from 2009, with exception of isolate 2008GZ04 from 2008. Isolates in cluster B were further gouped into cluster B1 and B2, which was further grouped in to subclusters in accordance with the region origin. The MST based on the MLVA data of 76 isolates indicated that the 19 MLVA profiles were divided in to two MLVA clusters (MC12 and MC7) and 11 singletons (Fig 3). MC12 contained isolates of six MTs (including MT9-14), while MC6 contains isolates belonged to MT6 and MT7.
Fig 2. Relationships of 76 Shigella sonnei isolates from Guizhou Province based on the Multiple-Locus Variable number tandem repeat (VNTR) Analysis (MLVA).
The 76 S. sonnei isolates from Guizhou Province were analyzed by MLVA based on seven VNTR loci. The copy number of the repeats of each VNTR locus was deduced from the length of the amplicons. This number was then converted into an allele designation, which in turn formed the allele string for the seven loci. The allele string was constructed in the following order: SS1-SS3-SS6-SS9-SS10-SS11-SS13. The data were incorporated into BioNumerics software and analyzed. Each unique allelic string was designated a unique MLVA type (MT). A dendrogram constructed using the PFGE patterns was generated by the UPGMA algorithm using the Dice-predicted similarity value of two patterns. The corresponding MLVA type with copy numbers for the seven VNTRs, serotype, and background information were shown alongside the dendrogram on the right.
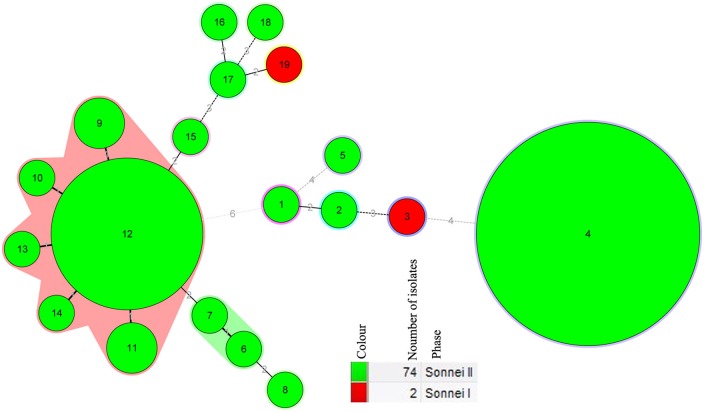
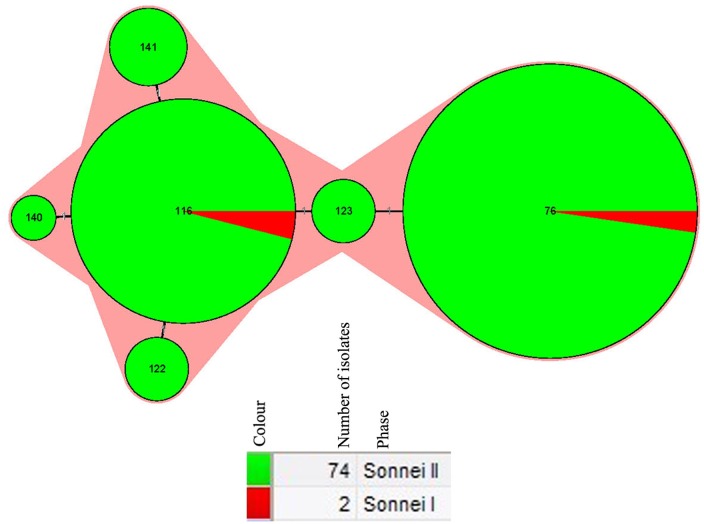
Fig 3. Minimum spanning tree of the isolates of 76 S. sonnei isolates from Guizhou Province based on the Multiple-Locus Variable number tandem repeat (VNTR) Analysis (MLVA).
The minimum spanning tree was constructed with the MLVA profiles of 76 isolates from Guizhou Province using BioNumerics software. Minimum spanning tree algorithm of BioNumerics software was used to construct a minimum spanning tree (MST) to determine phylogenetic pattern. Each circle corresponds to a MLVA type. The shadow zones in different color correspond to different MLVA clusters (MC), including MC12 and MC7. The size of the circle is proportional to the number of the isolates, and the color within the cycles represents the serotypes of the isolates. The corresponding color, number of isolates and serotypes were shown alongside the minimum spanning tree on the right.
MLST based genotypes
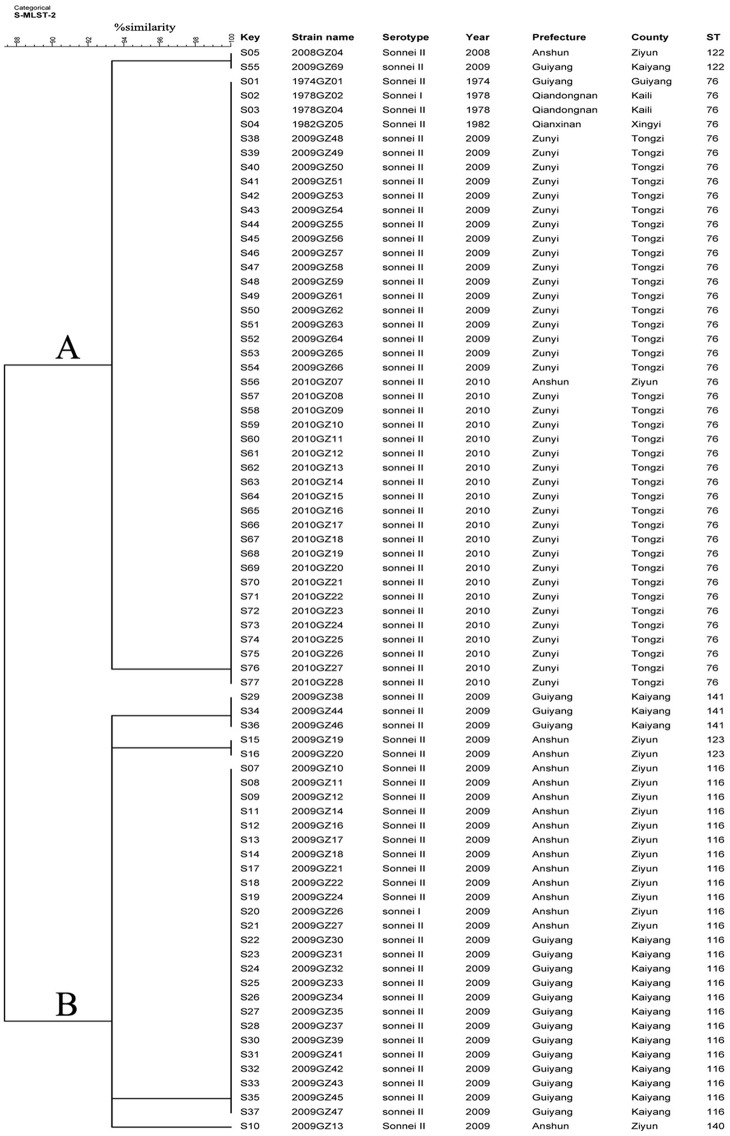
The 76 isolates were divided into six STs, including ST76, ST116, ST122, ST123, ST140 and ST141. Among the six STs, ST76 and ST116 were previously reported, while four STs including ST122, ST123, ST140 and ST141 were novel. The allele number for each loci and the designation of ST are listed in Table 1 and shared in the EcMLST website. The most common STs are ST76 (N = 43) and ST116 (N = 25), accounting for 92.1%. ST76 contained the most number of isolates, represented by 42 isolates of phase II from different years and one isolate (1978GZ02) of phase I from 1978, accounting for 55.26% of the total isolates. ST116 includes 23 isolates of phase II and one phase I isolate (2009GZ26). All the isolates of ST116 were come from 2009, making up for 31.58% (24/76) of the total isolates. Clustering tree (Fig 4) based on the MLST data shows that all the 76 isolates were divided into two gross clusters (Cluster A and B). All the four isolates from 1974–1982, parts of isolates from 2009 and the one isolated from 2008 were grouped in cluster A, while all the isolates in cluster B were come from 2009. Both of cluster A and B were further divided into subclusters. Besides, according to the MST (Fig 5) based on the eBURST algorithm with member STs differing by only one of the 15 loci, all the six STs belonged to one clonal complexes (CCs).
Table 1. Results obtained using MLST for 76 S. sonnei isolates from Guizhou Province.
| Strain No. | Isolates Name | Alle profiles | ST | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| arcA | aroE | aspC | clpX | cyaA | dNaG | fadD | grpE | icdA | lysP | mdh | mtlD | mutS | rpoS | uidA | |||
| S01 | 1974GZ01 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S02 | 1978GZ02 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S03 | 1978GZ04 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S04 | 1982GZ05 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S05 | 2008GZ04 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 122 |
| S07 | 2009GZ10 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S08 | 2009GZ11 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S09 | 2009GZ12 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S10 | 2009GZ13 | 9 | 13 | 10 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 140 |
| S11 | 2009GZ14 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S12 | 2009GZ16 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S13 | 2009GZ17 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S14 | 2009GZ18 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S15 | 2009GZ19 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 123 |
| S16 | 2009GZ20 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 123 |
| S17 | 2009GZ21 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S18 | 2009GZ22 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S19 | 2009GZ24 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S20 | 2009GZ26 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S21 | 2009GZ27 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S22 | 2009GZ30 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S23 | 2009GZ31 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S24 | 2009GZ32 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S25 | 2009GZ33 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S26 | 2009GZ34 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S27 | 2009GZ35 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S28 | 2009GZ37 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S29 | 2009GZ38 | 9 | 13 | 171 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 141 |
| S30 | 2009GZ39 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S31 | 2009GZ41 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S32 | 2009GZ42 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S33 | 2009GZ43 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S34 | 2009GZ44 | 9 | 13 | 171 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 141 |
| S35 | 2009GZ45 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S36 | 2009GZ46 | 9 | 13 | 171 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 141 |
| S37 | 2009GZ47 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 68 | 1 | 116 |
| S38 | 2009GZ48 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S39 | 2009GZ49 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S40 | 2009GZ50 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S41 | 2009GZ51 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S42 | 2009GZ53 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S43 | 2009GZ54 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S44 | 2009GZ55 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S45 | 2009GZ56 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S46 | 2009GZ57 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S47 | 2009GZ58 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S48 | 2009GZ59 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S49 | 2009GZ61 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S50 | 2009GZ62 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S51 | 2009GZ63 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S52 | 2009GZ64 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S53 | 2009GZ65 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S54 | 2009GZ66 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S55 | 2009GZ69 | 9 | 13 | 18 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 122 |
| S56 | 2010GZ07 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S57 | 2010GZ08 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S58 | 2010GZ09 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S59 | 2010GZ10 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S60 | 2010GZ11 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S61 | 2010GZ12 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S62 | 2010GZ13 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S63 | 2010GZ14 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S64 | 2010GZ15 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S65 | 2010GZ16 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S66 | 2010GZ17 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S67 | 2010GZ18 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S68 | 2010GZ19 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S69 | 2010GZ20 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S70 | 2010GZ21 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S71 | 2010GZ22 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S72 | 2010GZ23 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S73 | 2010GZ24 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S74 | 2010GZ25 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S75 | 2010GZ26 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S76 | 2010GZ27 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
| S77 | 2010GZ28 | 9 | 13 | 4 | 19 | 13 | 3 | 18 | 3 | 21 | 14 | 23 | 13 | 17 | 17 | 1 | 76 |
Note: 15 loci including arcA, aroE, aspC, clpX, cyaA, dNaG, fadD, grpE, icdA, lysP, mdh, mtlD, mutS, rpoS and uidA and the primers can be accessed on the sharing EcMLST website (http://www.shigatox.net/ecmlst/cgi-bin/index). Each allele was assigned a different allele number and the allelic profile (string of fifteen integers) was used to define the sequence type (ST) based on the EcMLST website, which was established to provide a resource to investigators who can use this to assign the ST of further strains. New allele numbers and sequence type were submitted to the EcMLST curator for allocation. ST = Sequence Type.
Fig 4. Relationships of 76 Shigella sonnei isolates from Guizhou Province based on Multilocus sequence typing (MLST).
The 76 S. sonnei isolates from Guizhou province were analyzed by MLST based on 15 loci including arcA, aroE, aspC, clpX, cyaA, dNaG, fadD, grpE, icdA, lysP, mdh, mtlD, mutS, rpoS and uidA. The primers sequence for the 15 alles can be accessed on the sharing EcMLST website (http://www.shigatox.net/ecmlst/cgi-bin/index). The clustering tree was constructed based on the allele number of 15 loci using BioNumerics Software. The corresponding ST, serotype, and background information were shown alongside the dendrogram on the right.
Fig 5. Minimum spanning tree of the isolates of 76 S. sonnei isolates from Guizhou Province based on the Multilocus sequence typing (MLST).
The minimum spanning tree was constructed with the six STs of the 76 isolates from Guizhou Province using BioNumerics Software. Minimum spanning tree algorithm of BioNumerics software was used to construct a minimum spanning tree (MST) to determine phylogenetic pattern. Each circle corresponds to a sequence type. The shadow zone indicates the six STs belonging to one clonal complex. The size of the circle is proportional to the number of the isolates, and the color within the cycles represents the serotypes of the isolates. The corresponding colour, serotype, number of isolates and back ground information were shown below the minimum spanning tree.
Discussion
S. flexneri was the predominant species in Guizhou Province in 1970s and 1980s, followed with S. sonnei [10], but S. sonnei is becoming the most important causative agent for shigellosis in recent decades in Guizhou Province [11]. No data, however, are available about the molecular characterization of the local isolates of S. sonnei. In order to systematically understand the molecular characteristics, four local isolates of S. sonnei from patients of shigellosis in 1974–1982 and 72 isolates from patients of shigellosis in 2008–2010 were used for analysis in this study. The serotypes of the isolates used in this study including two isolates of phase I and 74 isolates of phase II. The locality origin of these isolates covers five prefectures of the nine Profectures in Guizhou Province.
PFGE is a broadly applicable typing method with a high degree of intra- and inter laboratory reproducibility when standardized protocols are followed [13]. It was proved to be a powerful tool in the laboratory for discriminating Shigella isolates during an outbreak [28]. In this study, PFGE discriminate the 76 isolates of S. sonnei into 38 PTs, which indicates it is a powerful tool in discriminate S. sonnei isolates. PFGE based clustering tree branched out extensively, which suggests that the 76 isolates of S. sonnei in this study were heterogeneous.
The dentrogram of all the 76 isolates showed a 65% coefficient of similarity, which shared similar discriminatory power with S. sonnei isolates from other provinces [29, 30] and other countries [3, 31]. Isolates from close years were almost clustered closely and further divided into sub-clusters based on the region of isolation (Fig 1), indicating the genetic diversity and the genetic changes over the timescales.
MLVA is a prominent typing tool which has been developed for S. sonnei [2, 3, 15, 30]. In the present study, the 76 S. sonnei isolates were discriminated into 19 different MTs and showed a low (about 25%) coefficient of similarity, indicating the high discriminatory power of MLVA in subtyping S. sonnei isolates. Compared with the seven loci-based MLVA typing data of isolates from other conutries such as Malaysia[3], Japan [2], isolates of this study shared a lower overall similarity, which indicated the greater diversity and unique characteristics of S. sonnei isolates of Guizhou Province. Phylogenetic tree based on MLVA indicated that majority of isolates were clustered in accordance with the origin of isolation years, which were further closely clustered based on the geographical origin (Fig 2). The MST based on the MLVA data revealed two MCs and 11singletons among the 76 isolates, which further displayed the clonal relationships in the local isolates of Guizhou Province (Fig 3).
MLST based on 15 house-keeping genes were applied in Shigella genotyping and the protocol is shared at the EcMLST website [23, 24]. In this study, by using 15 house-keeping gens, 76 local isolates of S. sonnei from Guizhou Province, were divided into six STs, of which 4 STs including ST122, ST123, ST140 and ST141 were novel. Clustering tree based on the MLST data shows that all the 76 isolates were divided into two gross clusters, with a high coefficient of similarity (>90%), indicating the low discriminative ability in subtyping S. nnei isolates. Besides, MST based on the eBURST algorithm revealed that all the 76 isolates belong to only one CC with no singletons, indicating the relatively close genetic relationship among these isolates.
Additionally, according to the genotyping results of local isolates in this study, both of PFGE and MLVA displayed excellent discriminative ability in subtyping the S. sonnei isolates from Guizhou Province. Both techniques further differentiated the S. sonnei Phase I and II strains. However, MLVA was better at grouping the strains on the basis of isolation years, and the overall percentage of similarity among strains was low when subtyping using MLVA, which suggested MLVAwas of more excellent discriminative ability than PFGE in subtyping the S. sonnei isolates. Furthermore, MLVA subtyping was more rapid and less laborious compared to PFGE. Interpretation of result was less subjective and results were more readily comparable between laboratories. Compared with PFGE and MLVA, MLST divided the 76 isolates in to six STs, and all the STs belonged to one CC, which indicates the poor discriminative ability in distinguishing S. sonnei. All these suggest that MLVA may be a suitable complement to PFGE or even an alternative for routine subtyping of S. sonnei, and MLVA which is consistent with the early study by Koh, et al. in Malaysia [3], and use of a combination of molecular typing techniques is effective for epidemiological investigation.
Conclusions
In the present study, we apply PFGE, MLVA and MLST methods to genetically characterize the 76 local isolates of S. sonne isolated in Guizhou Province in different years. PFGE based on XbaI digestion divided the 76 isolates into 38 PTs and MLVA based on seven VNTR loci discriminated them into 19 different MTs, while MLST based on 15 loci differentiated the isolates into six STs, with four STs being novel. Phylogenetic tree based on the three genotyping methods showed that majority of isolates were clustered in accordance with the years of isolation, which were further closely clustered based on the geographical origin. PFGE and MLVA displayed more excellent resolution than MLST in discriminating the isolates and reveal the changing of genetic characteristics over different rears. Our study enhances our understanding of genetic characteristics of S. sonnei in Guizhou Province, and provides a scientific basis for control and prevention of Shigellosis in the locality.
Acknowledgments
This work was supported by the grant of special founds for outstanding youth talents of science and technology of Guizhou Province (Grant No. Qian Ke He Ren Word (2015) 09), Guizhou Province Government Founds for Talent Base Construction for Infectious Disease Control and Prevention (No. Qian Ren Ling Fa [2013] 15), High-Level Creative Talents Cultivation Funds in Guizhou Province (Qian Ke He (2016)4021) and national science and technology major project (No.2012ZX10004212-005) from the Ministry of Science and Technology of China, National Basic Research Priorities Program (No. 2011CB504901) and National Key Program for Infectious Diseases of China (No. 2013ZX10004221, 2013ZX10004216-001-002 and 2ZX10004215). We acknowledge the contributions of bacterial collection work of Kecheng Tian and the use of the EcMLST database which is operated by the Microbial Evolution Laboratory at Michigan State University.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the grant of special founds for outstanding youth talents of science and technology of Guizhou Province (Grant No. Qian Ke He Ren Word (2015) 09), Guizhou Province Government Founds for Talent Base Construction for Infectious Disease Control and Prevention (No. Qian Ren Ling Fa [2013] 15), High-Level Creative Talents Cultivation Funds in Guizhou Province (Qian Ke He (2016)4021) and national science and technology major project (No.2012ZX10004212-005) from the Ministry of Science and Technology of China, National Basic Research Priorities Program (No. 2011CB504901) and National Key Program for Infectious Diseases of China (No. 2013ZX10004221, 2013ZX10004216-001-002 and 2ZX10004215). The authors acknowledge the use of the EcMLST database, which is operated by the Microbial Evolution Laboratory at Michigan State University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Niyogi SK. Shigellosis. Journal of microbiology. 2005;43(2):133–43. Epub 2005/05/10. . [PubMed] [Google Scholar]
- 2.Izumiya H, Tada Y, Ito K, Morita-Ishihara T, Ohnishi M, Terajima J, et al. Characterization of Shigella sonnei isolates from travel-associated cases in Japan. J Med Microbiol. 2009;58(Pt 11):1486–91. Epub 2009/07/11. 10.1099/jmm.0.011809-0 . [DOI] [PubMed] [Google Scholar]
- 3.Koh XP, Chiou CS, Ajam N, Watanabe H, Ahmad N, Thong KL. Characterization of Shigella sonnei in Malaysia, an increasingly prevalent etiologic agent of local shigellosis cases. BMC Infect Dis. 2012;12:122 Epub 2012/05/23. 10.1186/1471-2334-12-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Future needs and directions for Shigella vaccines. Wkly Epidemiol Rec. 2006;81(6):51–8. Epub 2006/05/05. . [PubMed] [Google Scholar]
- 5.Knirel YA, Sun Q, Senchenkova SN, Perepelov AV, Shashkov AS, Xu J. O-antigen modifications providing antigenic diversity of Shigella flexneri and underlying genetic mechanisms. Biochemistry Biokhimiia. 2015;80(7):901–14. Epub 2015/11/07. 10.1134/S0006297915070093 . [DOI] [PubMed] [Google Scholar]
- 6.Chompook P, Samosornsuk S, von Seidlein L, Jitsanguansuk S, Sirima N, Sudjai S, et al. Estimating the burden of shigellosis in Thailand: 36-month population-based surveillance study. Bull World Health Organ. 2005;83(10):739–46. Epub 2005/11/12. /S0042-96862005001000010. [PMC free article] [PubMed] [Google Scholar]
- 7.Vinh H, Nhu NT, Nga TV, Duy PT, Campbell JI, Hoang NV, et al. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis. 2009;9:204 Epub 2009/12/17. 10.1186/1471-2334-9-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Organization WH. Initiative for Vaccine Research (IVR): Diarrhoeal Diseases. Shigellosis. (Updated February 2009).http://www.who.int/vaccine_research/diseases/diarrhoeal/en/index6.html.
- 9.Cao Y, Wei D, Qi S. Retrospective Analysis of Antibiotic Resistance and Serotype Distribution of Shigella spp. in China from 1989 to 2010. Tianjin Med J. 2013;41(1):8–82. [Google Scholar]
- 10.Wang D, Zhang Q, Li S. Serotype distribution of Shigella and the dynamical change of antibiotic esistance in Guizhou Province. Guizhou Medical Journal 1991;15(2): 114–116. [Google Scholar]
- 11.Wei X, Tian K, You L, Ma Q, Liu Y, Tang G. Epidemic Characteristics and Etiological Analysis of Bacillary Dysentery in Guizhou Province During the Period of 2007–2010. Practical Preventive Medicine. 2012;19(8):1185. [Google Scholar]
- 12.Gerner-Smidt P, Hise K, Kincaid J, Hunter S, Rolando S, Hyytia-Trees E, et al. PulseNet USA: a five-year update. Foodborne Pathog Dis. 2006;3(1):9–19. Epub 2006/04/11. 10.1089/fpd.2006.3.9 . [DOI] [PubMed] [Google Scholar]
- 13.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59–67. Epub 2006/04/11. 10.1089/fpd.2006.3.59 . [DOI] [PubMed] [Google Scholar]
- 14.Chiou CS, Watanabe H, Wang YW, Wang WL, Terajima J, Thong KL, et al. Utility of multilocus variable-number tandem-repeat analysis as a molecular tool for phylogenetic analysis of Shigella sonnei. J Clin Microbiol. 2009;47(4):1149–54. Epub 2009/02/20. 10.1128/JCM.01607-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang SY, Watanabe H, Terajima J, Li CC, Liao JC, Tung SK, et al. Multilocus variable-number tandem-repeat analysis for molecular typing of Shigella sonnei.J Clin Microbiol. 2007;45(11):3574–80. Epub 2007/09/21. 10.1128/JCM.00675-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(6):3140–5. Epub 1998/04/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urwin R, Maiden MC. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 2003;11(10):479–87. Epub 2003/10/15. . [DOI] [PubMed] [Google Scholar]
- 18.Nemoy LL, Kotetishvili M, Tigno J, Keefer-Norris A, Harris AD, Perencevich EN, et al. Multilocus sequence typing versus pulsed-field gel electrophoresis for characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates.J Clin Microbiol. 2005;43(4):1776–81. Epub 2005/04/09. 10.1128/JCM.43.4.1776-1781.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan MS, Maiden MC, Spratt BG. Database-driven multi locus sequence typing (MLST) of bacterial pathogens. Bioinformatics. 2001;17(11):1077–83. Epub 2001/11/29. . [DOI] [PubMed] [Google Scholar]
- 20.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186(5):1518–30. Epub 2004/02/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi SY, Jeon YS, Lee JH, Choi B, Moon SH, von Seidlein L, et al. Multilocus sequence typing analysis of Shigella flexneri isolates collected in Asian countries. J Med Microbiol. 2007;56(Pt 11):1460–6. Epub 2007/10/30. 10.1099/jmm.0.47322-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136–51. Epub 2006/05/13. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye C, Lan R, Xia S, Zhang J, Sun Q, Zhang S, et al. Emergence of a new multidrug-resistant serotype X variant in an epidemic clone of Shigella flexneri.J Clin Microbiol. 2010;48(2):419–26. Epub 2009/12/04. 10.1128/JCM.00614-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu S, Wang Z, Chen C, Liu N, Jia L, Liu W, et al. Emergence of a novel Shigella flexneri serotype 4s strain that evolved from a serotype X variant in China.J Clin Microbiol. 2011;49(3):1148–50. Epub 2010/12/24. 10.1128/JCM.01946-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Q, Lan R, Wang Y, Zhao A, Zhang S, Wang J, et al. Development of a multiplex PCR assay targeting O-antigen modification genes for molecular serotyping of Shigella flexneri.J Clin Microbiol. 2011;49(11):3766–70. Epub 2011/09/02. 10.1128/JCM.01259-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7 Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59–67. 10.1089/fpd.2006.3.59 ISI:000238252200009. [DOI] [PubMed] [Google Scholar]
- 27.Hyytia-Trees E, Smole SC, Fields PA, Swaminathan B, Ribot EM. Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157). Foodborne Pathog Dis. 2006;3(1):118–31. Epub 2006/04/11. 10.1089/fpd.2006.3.118 . [DOI] [PubMed] [Google Scholar]
- 28.Ahmed AM, Shimamoto T. Isolation and molecular characterization of Salmonella enterica, Escherichia coli O157:H7 and Shigella spp. from meat and dairy products in Egypt. Int J Food Microbiol. 2014;168–169:57–62. Epub 2013/11/19. 10.1016/j.ijfoodmicro.2013.10.014 . [DOI] [PubMed] [Google Scholar]
- 29.Xia S, Xu B, Huang L, Zhao JY, Ran L, Zhang J, et al. Prevalence and characterization of human Shigella infections in Henan Province, China, in 2006.J Clin Microbiol. 2011;49(1):232–42. Epub 2010/11/12. 10.1128/JCM.01508-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng T, Shi X, Yong W, Wang J, Xie G, Ding J. Molecular typing of Shigella sonnei isolates circulating in Nanjing, China, 2007–2011. Journal of infection in developing countries. 2014;8(12):1525–32. Epub 2014/12/17. 10.3855/jidc.4933 . [DOI] [PubMed] [Google Scholar]
- 31.Jin YH, Oh YH, Jung JH, Kim SJ, Kim JA, Han KY, et al. Antimicrobial resistance patterns and characterization of integrons of Shigella sonnei isolates in Seoul, 1999–2008. Journal of microbiology. 2010;48(2):236–42. Epub 2010/05/04. 10.1007/s12275-010-9220-z . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.