Abstract
Genome-wide measurements of transcriptional activity in bacteria indicate that the transcription of successive genes is strongly correlated beyond the scale of operons. Here, we analyze hundreds of bacterial genomes to identify supra-operonic segments of genes that are proximal in a large number of genomes. We show that these synteny segments correspond to genomic units of strong transcriptional co-expression. Structurally, the segments contain operons with specific relative orientations (co-directional or divergent) and nucleoid-associated proteins are found to bind at their boundaries. Functionally, operons inside a same segment are highly co-expressed even in the apparent absence of regulatory factors at their promoter regions. Remote operons along DNA can also be co-expressed if their corresponding segments share a transcriptional or sigma factor, without requiring these factors to bind directly to the promoters of the operons. As evidence that these results apply across the bacterial kingdom, we demonstrate them both in the Gram-negative bacterium Escherichia coli and in the Gram-positive bacterium Bacillus subtilis. The underlying process that we propose involves only RNA-polymerases and DNA: it implies that the transcription of an operon mechanically enhances the transcription of adjacent operons. In support of a primary role of this regulation by facilitated co-transcription, we show that the transcription en bloc of successive operons as a result of transcriptional read-through is strongly and specifically enhanced in synteny segments. Finally, our analysis indicates that facilitated co-transcription may be evolutionary primitive and may apply beyond bacteria.
Introduction
Characterizing variations of gene expression genome-wide, understanding their relation to genome organization and explaining the underlying mechanisms are fundamental challenges in biology [1]. To achieve these goals, the description of the transcriptional landscape of genomes has been refined, revealing a complex architecture not only in eukaryotes [2, 3] but also in bacteria [4, 5]. Concomitantly, genome-wide analyses of co-transcription [6–10] have led to a system-level perspective of transcriptional regulation [9, 11–13].
From a mechanistic viewpoint, many regulatory processes are known to affect gene transcription [1], with the core architecture of regulatory networks generally associated with the regulation of operons by sigma factors (SFs) and transcription factors (TFs). Yet, as we review below, these three elements alone (operons, SFs and TFs) fail to account for the most prominent features of bacterial gene co-expression, both in the Gram-negative Escherichia coli and in the Gram-positive Bacillus subtilis. Additional regulatory elements, including small metabolites [14, 15], small RNAs [16, 17], transcriptional attenuators [18], global physiological effects [19, 20] and topological properties of chromosomes [21–23] are thus expected to play a role. Yet, the implications and specificities of these mechanisms are too poorly understood to yield an alternative, reliable decomposition of genomes.
Interestingly, several previous studies suggest that transcription is primarily coordinated above the scale of operons in bacteria. For instance, in B. subtilis, high-resolution micro-array data has revealed large supra-operonic transcriptional units, which are controlled by SFs and essential for the adaptative properties of the bacterium [9]. In E. coli, micro-array data obtained under a large panel of conditions [24] has also highlighted the presence of large supra-operonic domains of coordinated gene expression dedicated to specific transcriptional responses [10]. The systematic identification of such genomic units of transcriptional coordination in every bacterium and the investigation of the underlying regulatory mechanisms remain, however, problematic for at least three reasons. First, transcription is condition-dependent so that transcriptional units may differ from one condition to the other [9, 10, 13, 25]. Second, transcription is stochastic and, even under the same condition, different units may be transcribed; in particular, transcriptional termination is rarely as sharp as transcriptional initiation. Finally, transcription is not necessarily functional and not all transcriptional units are equally relevant. Assessing functional significance is in fact challenging as this notion ultimately refers to a measure of “fitness”, which is hardly accessible given our limited knowledge of the environmental conditions under which this fitness should be evaluated.
An indirect approach to identify functionally relevant transcriptional units utilises evolutionary conservation across species as a proxy for fitness. This approach relies on the principle that features shared among a large number of distinct species must be under strong selective pressures and, therefore, are functionally significant [26]. Past studies have exploited this principle to parse out the commonalities and differences of gene regulation in different species, mostly among eukaryotes [27–29]. The limited number of species for which extensive gene expression data is available has, however, precluded a precise comparison of co-expression units [30]. Here, we circumvent this difficulty by studying the evolutionary conservation of the clustering of genes along chromosomes. In bacteria, just as in eukaryotes [30–36], “synteny”, the conservation of chromosomal proximity between genes [37–39], has indeed been shown to be tightly related to co-expression properties [40, 41] and to be useful to the inference of functional associations [42–44].
From a comprehensive analysis of synteny across a thousand annotated bacterial genomes, we thus identify “synteny segments” in every annotated genome. To this end, we define synteny segments of a particular genome as groups of consecutive genes that are co-localized both in the genome and in a significant number of other, phylogenetically distant genomes. By studying the organization of these clusters in the thousand genomes and by examining their structural and regulatory properties in two of the best characterized bacteria, E. coli and B. subtilis, we demonstrate that these synteny segments reflect supra-operonic genomic units that lie at the core of the coordination of transcription. To explain our results, we propose that “facilitated co-transcription”, the transcription of a gene (or operon) induced by the transcription of the gene (or operon) located immediately upstream, sharing or not the same orientation, is at the evolutionary origin of transcriptional regulation and still constitutes today its main basis. We find evidence for this scenario in RNA-seq data in E. coli and high-resolution micro-array data in B. subtilis. Finally, we show that our hypothesis both disposes of controversies over the evolutionary origins of gene clusters in bacterial chromosomes and allows to better apprehend the striking evolutionary properties of regulatory networks. We also discuss the relevance of this scenario beyond the bacterial kingdom.
Results
Previous analyses have revealed similarities in the patterns of gene co-expression between E. coli and B. subtilis [45] despite their substantial evolutionary divergence (E. coli is a Gram-negative proteobacteria, B. subtilis is a Gram-positive firmicute). In this context, we first quantify the extent to which known regulatory mechanisms can explain gene co-expression in these two organisms by analyzing publicly available micro-array datasets. For E. coli, we use a compendium of micro-array data collected by different laboratories and normalized uniformly using a quantile normalization procedure [8]; this dataset covers 4320 genes (NC_000913 genome in NCBI reference) in 466 different conditions. For B. subtilis, we use the 22-base high-resolution micro-array dataset produced by the BaSysBio consortium, which covers 4162 genes (NC_000964 genome) in 104 different conditions [9]; for consistency, we first normalized this dataset using the same quantile normalization procedure as the E. coli dataset, even though this has no incidence on the results. For each dataset, we quantify the level of co-expression between two genes by the Pearson correlation coefficient of their transcriptomic profile (Materials and methods) and display the results in the form of a heat-map (panels B and F of Fig 1). To assess the role of operons and of TF and SF binding sites, we rely on public databases. For E. coli, we use the RegulonDB database [46] (last update: 02/05/2015). For B. subtilis, we respectively identify operons, TFs and SFs binding sites using the biocyc database (biocyc.org), the DBTBS database [47] (last update: 02/05/2015) and the comprehensive database provided by the BaSysBio consortium [9]. For the latter, we consider that an operon is directly regulated by a SF if its promoter region (up to 500 bases) contain at least one binding site of this SF (Table S2 in SOM of [9]).
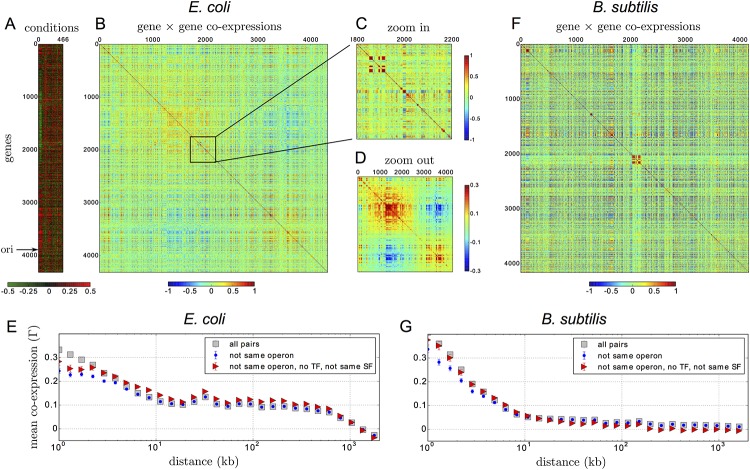
Fig 1. Spatial patterns of gene co-expression in E. coli and B. subtilis.
A. For E. coli, we use micro-array data reporting the expression levels of 4320 genes in 466 conditions. This data is represented as a matrix with genes along rows (following their order along the chromosome) and conditions along columns: high expression appears in red and low expression in green (the data is normalized so that the mean expression of a gene across conditions is zero). B. The co-expression between every pair ij of genes is computed from their profiles of expression in the micro-array data and represented as a square matrix Cij with the first gene i along the rows and the second j along the columns. The expression of two genes is positively correlated (in red) if they tend to be expressed in the same conditions and anti-correlated (in blue) if they tend to be expressed in different conditions. C. Zoom in the co-expression of 400 genes, showing in red small clusters of correlated genes on the scale of 10 kb (∼ 10 genes). D. “Zoom out” obtained by averaging the matrix on a scale of 10 kb (Gaussian filtering with a standard deviation of 10 genes), showing in red two large clusters of size ∼ 1 Mb, whose respective expressions are anti-correlated. E. These different features are recapitulated in the mean co-expression function Γ(d), defined as the average co-expression over pairs of genes at the same distance d along the chromosome. Γ(d) is computed for all pairs of genes (gray squares), for pairs of genes in distinct operons, irrespectively of their regulation by a common TF or SF (blue points), and for pairs of genes in distinct operons that are both not known to be regulated by any TF and not known to be regulated by a common SF (red triangles). F. Corresponding co-expression matrix for B. subtilis, with the presence of a highly correlated (red) cluster at the center due to the prophage SPβ genes. G. For distances below ∼ 20 kb, the mean co-expression function Γ(d) for B. subtilis is similar to that in E. coli, with in particular a poor impact of the direct action of TFs/SFs on the enhanced co-expression observed at short distances (Γ(d) is computed without including the prophage SPβ genes, which have a singular behavior).
Gene co-expression is enhanced beyond the scale of operons without the involvement of TFs or SFs
In E. coli, our analysis reveals a hierarchical genomic organization of transcriptional co-expression in agreement with previous works (Fig 1A–1E) [45, 48]. At the bottom of this hierarchical organization, for genomic scales up to 10 kilo-bases (kb, the scale of a single gene), small clusters of positively correlated genes can be distinguished (Fig 1C). At the top of the hierarchy, we observe a global pattern of anti-correlation between two large clusters that have a genomic extension of the order of 1 Mb (1/4 of the genome length; Fig 1D). These features are recapitulated in the shape of the co-expression function Γ(d), defined here as the mean co-expression Cij between pairs of genes separated by a given genomic distance d. As shown in Fig 1E (grey squares), Γ(d) presents a first decrease up to d ∼ 10 kb, which reflects the presence of the small correlated clusters. It is followed by a long plateau ending around d ∼ 1 Mb, which reflects the presence of the two globally anti-correlated clusters (S1 Fig). Remarkably, considering only pairs of genes in different operons in the computation of Γ(d) does reduce the degree of co-expression at very short scale but does not suppress its enhancement up to 10 kb (Fig 1E, blue dots).
As discussed in detail in [49], the global pattern of anti-correlation matches the genomic distribution of the main SF of E. coli, σ70, and correlates with the locations of the origin and terminus of replication (S1E Fig). Yet, retaining only the operons known to be transcribed with σ70, and with σ70 only, does not suppress the anti-correlations (S2A Fig). A similar conclusion is reached when considering Fis, a NAP whose activity is also associated with different phases of cell growth [23] (S1E Fig). More strikingly, considering only pairs of genes that are reported to be regulated by different SFs and not known to be regulated by any TF leaves intact the two patterns of short and long scale correlations (Fig 1E, red triangles). In fact, the majority of correlated pairs of genes does not appear to share a common TF or a common SF (S2B and S2C Fig).
As previously reported [45], at small scales B. subtilis has a similar pattern of co-expression (Fig 1F) with small clusters of positively correlated genes displaying enhanced co-expression at distances below d ∼ 10 kb (grey squares in Fig 1G). At larger scales, some major differences are apparent, including the presence of a large ∼ 200 kb cluster of strongly co-expressed genes corresponding to the expression of the subunits of a non-ribosomal peptide synthase, the PksX megacomplex. The two large anti-correlated domains found in E. coli are also absent, corroborating the specificity of this large-scale pattern to γ-proteobacteria [49]. More importantly, just as in E. coli, the excess of co-expression at distances below d ∼ 10 kb can neither be explained by the decomposition into operons (blue points in Fig 1G), nor by the direct action of TFs or SFs (red triangles).
Beyond operons: synteny segments as units of co-expression fitting in the hierarchy of chromosomal structures
Interestingly, synteny, the conservation of gene proximity across species, can be used as a proxy of co-expression as the two properties are well known to correlate [38]. In particular, for both E. coli and B. subtilis, the more co-expressed two proximal genes are, the more likely they are to remain proximal in other distant bacterial species, independently of whether the genes belong to a same operon (S3 Fig). Following this observation, we analyzed more than 1000 complete bacterial genomes publicly available in order to identify their synteny clusters.
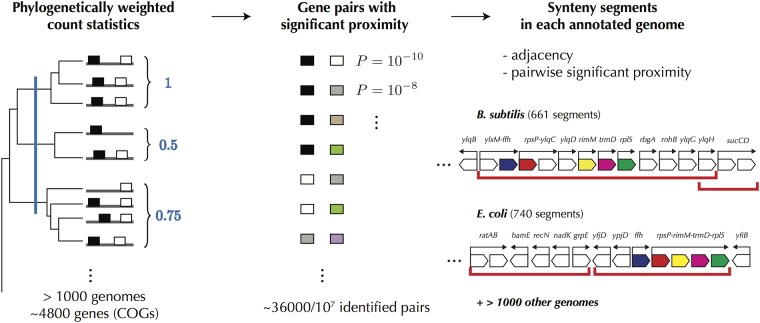
We thus define the “synteny segments” of every annotated bacterial genome as the sets of genes that are both consecutive along the chromosome and proximal in a significant number of other, phylogenetically distant bacterial chromosomes (Fig 2). To build the segments, we map the genes of all genomes to one of the 4764 orthology classes defined by the Cluster of Orthologous Genes (COG) annotation [50]. We then identify the pairs of COGs that tend to remain proximal by counting the number of genomes in which two COGs are below a certain distance, itself self-consistently defined (Materials and methods) and by comparing this number to what is expected from a null model where the positions of the genes are randomly drawn according to a uniform law. To this end, we explicitly consider the effect of multiple copies of COGs and mitigate the biases coming from the uneven phylogenetic distribution of available genomes (e.g. the presence of 62 different strains of E. coli) by down-weighting the contributions of genomes from over-represented clades. Specifically, we follow a procedure that proved its value in other contexts [51] and weight every genome in inverse proportion to the number of genomes within a certain phylogenetic distance δ (left column of Fig 2). Fixing the false discovery rate to 0.005, we finally obtain, among the ∼ 107 possible pairs of COGs, ∼ 36000 pairs with a significantly conserved proximity. From these pairs of genes in synteny, we define a synteny segment in a specific genome as a maximal genomic domain inside which every pair of genes is in synteny.
Fig 2. Definition of synteny segments from hundreds of genomes.
Left: Synteny refers to a conservation of gene proximity along the chromosome of different species. Our analysis is based on > 1000 complete annotated genomes of bacteria. Its principle is to compute for each pair of genes the fraction of genomes in which they are proximal: if this fraction is large, the pair is considered in synteny. In practice, the calculation needs to be corrected for phylogenetic relationships between genomes since finding two genes proximal in several closely related genomes is not as meaningful as finding them in distantly related genomes (the phylogenetic depth separating close genomes from distant genomes is schematically indicated by the vertical blue line). Middle: The result is a list of pairs of genes with a p-value indicating the significance of the conservation of their proximity. We say that the pair is in synteny if the p-value is small enough, with a cut-off accounting for multiple hypothesis testing (Materials and methods). Right: In each genome, we define as synteny segment a maximal cluster of consecutive genes where every pair of genes in the cluster is in synteny. A few examples of synteny segments, delineated by red lines, are shown in B. subtilis and E. coli with white boxes representing individual genes and arrows above them indicating operons. Boxes in color indicate known orthologous genes.
Taking a small false discovery rate and imposing every pair of genes in segment to be in synteny are very stringent statistical criteria to ensure that the segments that we define do reflect significant features. As a down side of taking very conservative statistical criteria, our approach is thus expected to miss not only many relevant genes in the clusters but also potentially relevant clusters entirely. This choice of minimizing type I errors (minimizing false positives) at the expense of type II errors (many false negatives) reflects our objective, which is to understand the nature of the fundamental regulatory units of bacterial genomes and not to systematically reannotate these genomes.
As a result, we find synteny segments of phylogenetically distant species that may contain similar genes but that differ in their composition (see Fig 2 for a few examples in E. coli and B. subtilis). We also observe that segments are distributed nearly uniformly along the chromosomes (S4 Fig) with a size distribution that follows, both in E. coli and B. subtilis, the size distribution of their polycistronic operons (S5 Fig). Altogether, this suggests that synteny segments represent different outcomes of a common stochastic evolutionary process [52].
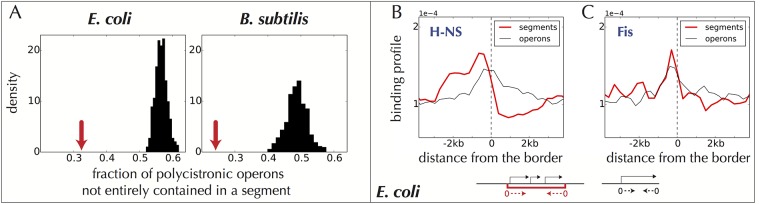
More specifically, we obtain 740 synteny segments in E. coli and 661 in B. subtilis respectively (S1 and S2 Files). These segments fit remarkably well within the known hierarchical architecture of bacterial chromosomes. At the lowest level, operons are rarely found to overlap only partially with a segment, meaning that segments contain operons (Fig 3A; see also Fig 2 for a few explicit examples). At a higher level, an analysis of the genome-wide binding profiles of various proteins onto the E. coli chromosome [53, 54] reveals a high preference for the nucleoid associated proteins (NAPs) Fis and H-NS to bind at the borders of synteny segments. Specifically, 359 out of 444 H-NS binding regions, and 866 out of 1246 Fis binding regions, are found within 3 kb of the border of a segment (p-values 7.10−5 and 5.10−6). In addition, we observe a strong enrichment of H-NS immediately outside synteny segments and a depletion inside them (red profile in Fig 3B); the resulting staircase-like binding profile notably differs from the binding profile around promoters of operons not located at a border (black profile). The same profile is obtained for the transcriptionally silenced extended protein occupancy domains (tsEPODs, of extension > 2 kb) identified in [53], in agreement with the fact that most of tsEPODs overlap with H-NS binding regions (S6 Fig). Fis also displays a tendency to bind immediately outside of the segments with a binding profile which, however, does not differ significantly from that of operons (Fig 3C). In contrast, the highly expressed extended protein occupancy domains (heEPODs, of extension > 2 kb, enriched in RNA polymerases) also identified in [53] are not enriched at the border of segments; instead, they tend to be located within the segments: 102 out of the 121 heEPODs overlap with the segments (p-value 4.10−9).
Fig 3. Relation of synteny segments to operons and to NAP binding sites.
A. Operons are contained within synteny segments: few operons are only partially included inside a segment (red arrow), as compared to situations in which the synteny segments are all translated by a finite number of genes. A translation by n genes is defined as follows: we first label all the genes as g0, g1, g2…, following their order along the chromosome; a translation by n genes of a segment gk, gk+1, gk+2 is then gk+n, gk+n+1, gk+n+2, where the additions k + n, k + n + 1, k + n + 2 are understood modulo the total number of genes. The black histograms report a statistics over all possible translations. The distances between the red arrows and the histograms are here indicative of high significances (low p-values). B. In E. coli, the NAP H-NS shows a staircase-like binding profile at the border of synteny segments (red line) that is markably different from the binding profile around the promoters of operons not located at a border (black line, whose shape is mainly due to the TSSs). C. In contrast, the binding profile of the NAP Fis at the border of segments is not significantly different from that at the borders of operons. In B and C, data is from [54] and the profiles are computed with respect to the boundaries of the segments/operons as indicated by the small drawings at the bottom.
TFs/SFs are not necessary for intra-segment co-expression but can couple the expression of distant segments
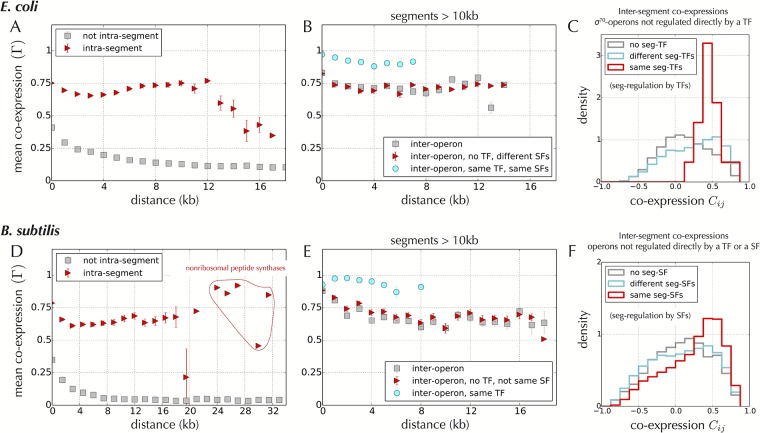
As respectively shown in Fig 4A and 4D for E. coli and B. subtilis, where the co-expression function Γ(d) of Fig 1E and 1G is compared for pairs of genes belonging to a same segment or for other pairs, co-expression occurs at high levels within synteny segments, and at low levels outside. Enhancement of co-expression within synteny segments hold equally for other phylogenetically distant bacterial species for which genome-wide transcriptional data is available in a large number of conditions (S7 Fig), corroborating the significance of synteny segments for co-expression properties in bacteria. Most notably, excluding pairs within a same operon (and segments < 10 kb, which contribute only at short distances; S8 Fig) reveals that the strong co-expression inside segments is not due to operons only, but occurs between different operons, independent of their genomic distance (Fig 4B and 4E and S7 Fig). In agreement with our analysis in Fig 1, we also find that this strong co-expression does not seem to be due to a co-regulation by TFs or SFs (red triangles in Fig 4B and 4E), although the presence of common TFs does enhance co-expression to levels close to the maximal possible value of 1 (cyan points).
Fig 4. Co-expression within synteny segments in E. coli and B. subtilis.
A. As in Fig 1E, the mean co-expression Γ(d) represents an average co-expression for pairs of genes at a given distance d along the chromosome of E. coli, but here it is computed for two distinct subsets of pairs, those belonging to a same segment (red triangles), and all others (gray squares). This shows that pairs of genes in a same synteny segment are significantly more co-expressed than pairs outside the segments. B. To discard the contribution from operons, we verify that the same results hold for pairs of genes in different operons and in segments larger than 10 kb (gray squares). The results also hold when further restricting to pairs of operons that do not share the same SF and are not regulated by any TF (red triangles), which indicates that co-regulation by TFs and SFs is not necessary to the co-expression of distinct operons inside a same segment. Considering pairs of genes regulated by the same TFs and SFs (cyan points), we observe, however, that these factors can raise the co-expression to its maximal value of 1. High levels of co-expression inside segments are observed irrespectively of the relative orientation of the operons (S8 Fig). The results also hold for small segments < 10 kb, although the average co-expression level is lower (S8 Fig). C. Distribution of the co-expression Cij between pairs of genes that are not directly regulated by a TF and that belong to different synteny segments—only the genes regulated by the SF σ70 are considered. Gray distribution: pairs in segments without seg-TFs. Cyan distribution: pairs in segments with different sets of TFs. Red distribution: pairs in segments with exactly the same seg-TFs. The peak of the red distribution at high values of co-expression provides evidence for seg-TF regulation in E. coli. D, E, F. Essentially the same results are obtained in B. subtilis with one major difference: seg-regulation occurs through SFs (panel F), not TFs, in agreement with the general observation that the effect of SFs dominates over that of TFs in B. subtilis [9].
Next, we find that operons that are not known to be directly regulated by a TF can be strongly co-expressed not only when they belong to the same segment but also when they belong to distant segments that share the same TF or SF signature. To demonstrate this novel layer of regulation, which we shall call “seg-regulation”, we define the “seg-TFs” (respectively, “seg-SFs”) of a segment as the set of TFs (respectively, SFs) that directly regulate at least one operon in the segment. In E. coli, Fig 4C shows that pairs of genes regulated by the SF σ70 but in different segments and with no TF of their own are significantly more co-expressed when they have exactly the same seg-TFs (red distribution) than when they have different seg-TFs (cyan distribution). Pairs of such genes are in fact also significantly more co-expressed when they both have a seg-TF, irrespectively of its identity, than when they both have no seg-TF, consistent with an indirect contribution from the regulation of TFs by other TFs [55]. No similar seg-TF regulation is observed in B. subtilis. Instead, the expression of distant segments in this bacterium appears to be coupled by SFs (Fig 4F). We find indeed that pairs of genes in different segments and with no SF of their own are significantly more co-expressed when they have exactly the same seg-SFs (red distribution) than when they have different seg-SFs (cyan distribution). This result corroborates the earlier report that most gene expression variation in B. subtilis is explained by changes in expression of the SFs [9].
The functional significance of seg-regulation is comforted by observing that the number of TF-regulated operons in both E. coli and B. subtilis segments is independent of the size of the segments, with on average one operon that is regulated (S9 Fig). This indeed suggests that no additional TF binding site is needed if a binding site is already present in the segment, which is therefore sufficient to regulate all operons of the segment—the same tendency is observed in B. subtilis for the SFs, with only a subpart of the segment that is directly regulated by a SF (S9 Fig).
Finally, let us mention a striking evolutionary link between short and long-range co-expression: pairs of genes that are distant in a genome, but in synteny in other genomes, are on average more co-expressed than those not in synteny. This phenomenon appears to be specific, in the sense that it does not apply to adjacent genes (S10 Fig). It suggests, as previously proposed in fungi [56], that operons that were previously proximal but later set apart evolve, or have evolved, similar cis-regulation.
Operons within synteny segments are specifically organized and subject to transcriptional read-through
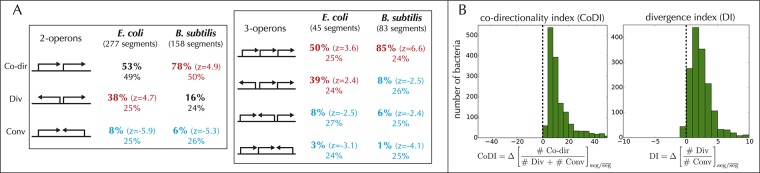
To identify the mechanisms behind the strong co-expression of operons in a same synteny segment, we compare the relative orientations of operons inside and outside the segments. Inside the synteny segments made of two operons of E. coli and B. subtilis, for which operon maps have been curated for many years (Materials and methods), we first observe that convergent orientations are strongly under-represented. More specifically, in B. subtilis operons are mostly co-directional (in ∼ 80% of the cases), while a significant fraction of them (38% instead of the expected 25%) are divergent in E. coli (Fig 5A). Similarly, inside synteny segments made of three operons, patterns of co-directionality are strongly over-represented, especially in B. subtilis. In E. coli, we also find an over-representation of patterns with divergent operons.
Fig 5. Relative orientations of operons inside synteny segments.
A. Statistics in E. coli and B. subtilis over all synteny segments made of 2 and 3 operons exactly, showing that some organizations are over-represented (in red, with z > 1.65, i.e., p-value < 0.05) or under-represented (in blue, z < −1.65). Percentages on the first and second line correspond, respectively, to statistics within the segments and overall, for the total of 2647 operons in E. coli and 3450 operons in B. subtilis. B. The over-representation of co-directional genes or divergent operons inside the segments applies to every bacterial genome. This is demonstrated by computing, in each genome, indexes that reflect the statistics of gene orientations; the histograms indicate the distribution of these indexes over all genomes. For co-directionality, we define an index (CoDI) as the difference, between genes inside and outside segments, of the ratio of co-directional over divergent or convergent orientations, and observe that this index is positive in all genomes (left histogram). For divergence, we compute the difference of the ratio of divergent over convergent orientations (divergence index DI), a ratio that must be equal to 1 over an entire circular chromosome, and observe that DI is positive in almost every genome (right histogram).
As expected from the evolutionary conservation of synteny segments, these features are not specific to E. coli and B. subtilis, but shared across all bacterial species. To demonstrate this universal behavior, we circumvent the difficulty of defining operons by comparing the relative number of co-directional, divergent and convergent adjacent genes along each genome (Fig 5B). We observe that the ratio of co-directional over divergent or convergent orientations is systematically larger for successive genes that lie inside the synteny segments. Similarly, the ratio of divergent over convergent gene orientations, which must be 1 over an entire circular chromosome, is larger inside than outside synteny segments for almost all bacterial genomes; the genomes that do not share this property consist, without exception, of > 90% co-directional gene pairs.
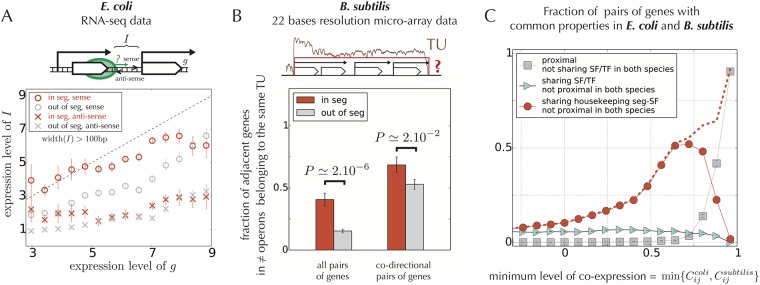
Is this singular organization of genes and operons inside synteny segments related to transcriptional co-expression? In E. coli, using strand-specific RNA expression profiles obtained by RNA-seq [24], we observe, indeed, that co-directional operons in a same segment are likely to be transcribed as a single large transcriptional unit. Specifically, for consecutive co-directional operons in synteny segments, we observe a correlation between the transcription of the first cistron in the downstream operon and the transcription of the upstream, non-coding inter-operonic sequence on the same strand, indicating that the gene is transcribed as a consequence of the RNA polymerase further proceeding after the transcription of the upstream operon (Fig 6A). As a control, the same analysis with the inter-operonic sequence on the opposite strand (anti-sense transcription) does not show this effect. To avoid false negatives in the identification of operons, we limited this analysis to pairs of genes separated by more than 100 bp, which is the maximal inter-gene distance considered in most operon predictions [24]. In the case of divergent operons, a previous study showed that adjacent bidirectionally transcribed genes tend to be functionally associated, with in E. coli many cases where one gene encodes a regulator that both controls the divergently transcribed operon and its own synthesis [43].
Fig 6. Evidence for the occurrence and evolutionary conservation of facilitated co-transcription.
A. Evidence from RNA-seq data that transcriptional read-through is wide-spread inside the synteny segments of E. coli. For pairs of consecutive co-directional operons, we measure the correlation between the transcription of the first gene g in the downstream operon and the sense or antisense transcription of the upstream inter-operonic sequence I (small drawing on top). The graph shows that the sense transcription of I (circles) correlates strongly with the transcription of g, much more than anti-sense transcription (crosses), and that this correlation is stronger for operons inside a same segment (in red), except at very high transcription levels (> 8), in which case we observe a high correlation in any case. The analysis is here restricted to inter-operonic regions longer than 100 bp to exclude possibly mis-annotated operons [24]. B. Evidence from high-resolution micro-array data that transcriptional read-through is wide-spread inside the synteny segments of B. subtilis. The fraction of adjacent genes that belong to a same transcriptional unit (TU) as identified experimentally in B. subtilis [9] is indeed significantly higher for genes in synteny segments (red bars) than for other genes (gray bars). This is consistent with a broad overlap between the TUs and synteny segments. The two bars on the left are based on all pairs of genes in different operons and those on the right on pairs of co-directional genes in different operons. C. Analysis of regulatory mechanisms involved in pairs of genes with high co-expression levels in both E. coli and B. subtilis (conserved co-expression). The fraction of pairs sharing the same regulatory properties in both bacteria is represented as a function of the minimum of the co-expression levels between B. subtilis and E. coli (x-axis). We analyze three different properties: proximity (distance d < 20 kb), shared direct regulation by TFs or SFs, and shared seg-regulation by seg-TFs or seg-SFs (without imposing the TFs in E. coli and B. subtilis to be orthologous). We observe that the conservation of high co-expression (> 0.75) mostly results from the conservation of gene proximity (gray squares) and not from the conservation of a direct co-regulation by TFs or SFs (cyan triangles). Below 0.75, the contribution of proximity vanishes. Instead, we observe a strong relationship between the level of conserved co-expression and the tendency for being seg-regulated by the housekeeping SF (red points). The dashed red curve indicates the fraction of pairs that are explained either by proximity or by this seg-regulation, which covers the majority of pairs for co-expression levels above ∼ 0.6.
In B. subtilis, a comparison of our synteny segments to the transcriptional units (TUs) identified by the BaSysBio consortium, which also often extend beyond known operons [9], leads to the same conclusion. Fig 6B indeed shows that successive operons are significantly more likely to belong to a same TU if they belong to the same segment. This result is not explained by an enrichment in co-directional operons in both sets since it is also observed when restricting to pairs of co-directional operons (as in E. coli, our analysis is limited to pairs of genes separated by more than 100 bp). Two types of TUs were in fact defined in [9]: “short TUs”, which are minimal TUs found in most conditions, and “long TUs”, which are maximal TUs found in at least one condition. Here, we find a more significant overlap with the long TUs (S11 Fig), in agreement with the fact that small TUs correspond more often to single operons and therefore are not accounted for in our analysis of successive operons.
Altogether, these results indicate that transcriptional read-through, the ability of RNA polymerases to override termination signals [18] and, hence, to transcribe multiple consecutive co-directional operons into the same mRNA, is ubiquitous in synteny segments and more limited outside segments. This phenomenon thus provides a simple mechanism for seg-regulation by TFs and SFs: if an operon without any binding site near its promoter is preceded by a co-directional operon with such a site, it can be regulated by the TF or SF. There is in fact a general association between TFs and the orientations of regulated operons in E. coli [57]: among operons of E. coli preceded by a co-directional operon, only 307 are regulated by a TF while 992 are not, a difference that is highly significant (P ≃ 10−7, binomial test). The situation is similar in B. subtilis for the SFs: among operons preceded by a co-directional operon, only 1090 out of 2234 are regulated by a SF (P ≃ 10−160), which is in agreement with the crucial role of SFs in the co-expression patterns of this bacterium [9] (see above). Given that operons in segments are preferentially orientated co-directionally or divergently (Fig 5), altogether this suggests that synteny segments represent supra-operonic co-expression units that are controlled only by a subset of “entry points” for RNA polymerases.
Conservation of co-expression is explained by proximity, not by the action of TFs or SFs
In contrast to relations of proximity, TFs are known to be poorly conserved [58], suggesting that pairs of genes that are co-regulated in a given species by a set of TFs may not be regulated by the same TFs, or by any TF, in a different species. The situation is somehow intermediate in the case of SFs with, on the one hand, the presence of highly conserved housekeeping SFs (σ70 in E. coli and SigA in B. subtilis) and, on the other hand, a rich diversity of stress-related SFs [59]. These considerations raise the question of the mechanisms behind the conservation of transcriptional co-expression in bacteria.
To address this problem, we consider pairs of genes that are highly co-expressed both in E. coli and in B. subtilis. We examine whether in both species these pairs of genes (i) are proximal, (ii) are directly regulated by a common TF or a SF, or (iii) share a common seg-TF or seg-SF (shared seg-regulation). To this end, we measure the fraction of pairs having one of these properties alone and report this fraction as a function of the minimum of their co-expression level in E. coli and in B. subtilis (x-axis in Fig 6C)—a large minimum co-expression indicates that co-expression is high in the two bacteria.
Our analysis reveals that strongly co-expressed pairs of genes in the two strains (Cij > 0.75) are mostly proximal in the two genomes, independent of whether the genes are co-regulated by a common TF or SF (gray squares in Fig 6C). In contrast, these pairs are not enriched in non-proximal genes regulated by a common SF or by a common TF, even without requiring the TFs to be orthologous between E. coli and B. subtilis (cyan triangles in Fig 6C). More strikingly, for lower, yet significantly conserved co-expression levels (e.g. Cij ∼ 0.5), we do not observe any contribution from these two mechanisms (proximity and direct co-regulation), but a strong relation to the tendency for being seg-regulated by housekeeping SFs (red points; see S12 Fig for further details). In particular, we observe that the majority of pairs of genes with conserved co-expression levels above ∼ 0.6 are either proximal or seg-regulated by these housekeeping SFs (red dashed curve).
Discussion
Following the principle that evolutionary conservation across distant species reflects functionally important biological processes, we identified from a comparison of > 1000 bacterial genomes supra-operonic genomic units of co-expression that we call “synteny segments”. These synteny segments are consistent with previously proposed concepts of uber-operons [38], superoperons [40], persistent genes [41], clusters of pathway-related operons [60] and cluster of statistically correlated genes [61]. Structurally, they contain the operons (Fig 3A) and, in E. coli, the nucleoid-associated protein H-NS binds at their border (Fig 3B). The operons within segments are most often oriented co-directionally or divergently (Fig 5). Functionally, distinct operons within a same segment are strongly co-expressed, irrespectively of the presence of common transcription factors (TFs) or sigma factors (SFs).
Facilitated co-transcription as a basic mode of regulation
Given the particular orientations of operons inside segments and the peripheral roles that TFs and SFs play in the coordination of their transcription, a parsimonious hypothesis, which we call “facilitated co-transcription”, is sufficient to summarize our findings: in absence of additional molecular factors or specific inter-gene sequence motifs, the transcription of a gene is facilitated by the transcription of the gene located immediately upstream. Under this hypothesis, the transcription of a gene facilitates the transcription of co-directional downstream genes and of divergent upstream genes.
Facilitated co-transcription may have different origins, depending on the relative orientation of the genes. For co-directional genes, a likely mechanism is transcriptional read-through, the overriding of termination signals by RNA polymerases [18], which is known to be a major source of transcripts in bacteria [62]. This conclusion is supported by our analyses of RNA-seq data in E. coli [24] and high-resolution micro-arrays in B. subtilis [9] (Fig 6A and 6B), where transcriptional read-through is found to be enhanced in synteny segments. Additional evidence for the frequent co-transcription of several successive operons is also found in a recent comprehensive analysis of transcriptional data in M. pneumoniae [25].
For divergent genes, the most likely physical mechanism is supercoiling [22, 23], which again does not require any factor beyond RNA polymerases and DNA: transcribing RNA polymerases generate torsional constraints that can affect the structural properties of nearby promoters [63, 64]. Bidirectional promoters are indeed known to be associated with pervasive antisense transcription [65, 66]. More generally, supercoiling is thought to affect the structuring of chromosomes over a broad range of length scales, from 10 kb to a few hundreds kb [67, 68] and is recognized as an important factor of genome-wide coordination of gene expression [21–23, 69]. In particular, the chromosome of E. coli has been shown to be organized into ∼ 10 kb-long independent domains of supercoiled DNA [67], the typical length scale of the co-expression clusters (Fig 1). The extended regions of DNA bound by H-NS have also been proposed to isolate these supercoiled domains from each other [53].
The co-transcription of several operons within a segment may thus not require any specific molecular machinery beyond RNA polymerases and DNA. Our hypothesis of facilitated co-transcription also implies the “seg-regulation” reported in Fig 4C and 4F: operons that are not co-regulated by TFs or SFs can nevertheless be co-expressed if their respective segments are coupled. More generally, the concept of seg-regulation provides a simple basis for understanding some of the long-range co-regulation that occurs between distant operons.
Facilitated co-transcription as an evolutionarily primitive mode of regulation
From an evolutionary standpoint, facilitated co-transcription may represent the most primitive form of gene regulation. In this scenario, gene clustering would have come first and TF- or SF-specific regulations would represent subsequent additions, tailored to the need of each specific lineage. In support for this view, we note that TFs and their network evolve quickly compared to other genetic networks [58, 70–72], while the clustering of genes may be highly conserved throughout evolution [38]. In E. coli, the rewiring of gene regulatory networks has been shown to have only a marginal impact both on the genome-wide transcription and on fitness of the bacterium [73], although the biophysics of transcriptional regulation is known to impose constraints on the organization of bacterial genomes [74, 75]. Along the same line, while transcription is known to be regulated at a global level by supercoiling [76], with a demonstrated influence on fitness [77, 78], deleting Fis, one of the NAPs with which H-NS controls supercoiling, has only marginal effects, depending on the conditions under which the bacterium grows [78]. NAPs may thus also only modulate the more fundamental patterns of co-expression imposed by the relationships of proximities between genes.
As evidence for the evolutionary prevalence of regulation by facilitated co-transcription, we showed that pairs of genes with conserved strong co-expression in distant species correspond typically to genes that remained proximal or that belong to segments that share housekeeping SFs, not to genes that are directly controlled by common TFs or common SFs (Fig 6C). Together with the observation that distant segments can be further coupled by specific TFs in E. coli (Fig 4C) or by alternative SFs in B. subtilis, i.e., SFs other than the housekeeping SFs (Fig 4F and S13 Fig), this strongly suggests that the division of task between TFs and alternative SFs is evolutionarily more recent than the co-regulation of adjacent genes by facilitated co-transcription.
Our hypothesis that facilitated co-transcription of co-directional and divergent genes is at the evolutionary origin of gene clustering also disposes of the paradoxes usually associated with the evolution of operons. Although controversial [79–81], the selfish operon scenario has indeed challenged the commonly-held assumption that selection for co-regulation drove the evolution of operons [82]. In particular, it has questioned the selective advantage of evolutionary intermediates when forming a new operon by bringing together several genes and an operator. Under our hypothesis, the clustering of transcriptionally independent genes may enhance their co-expression, independent of the presence of operators. This may confer an adaptive benefit to a bacterium even before an operon is formed. Consistent with this scenario, gene clustering, just as gene gain [83], appears to be under positive selection [52]. Co-expression may, however, not be the main selective pressure behind the clustering of genes: as proposed for eukaryotic genomes, another selective pressure may come from the need to reduce fluctuations in gene expression [84].
Finally, data from S. cerevisiae suggests that facilitated co-transcription may be relevant beyond bacteria. Genes that are proximal in S. cerevisiae and whose orthologs are in synteny in bacteria show indeed a much stronger co-expression than those that are not in synteny in bacteria (S14 Fig). Moreover, whereas micro-array data associates virtually every gene of S. cerevisiae with at least one TF, ChIP-seq data suggests that only a small fraction of these associations stem from direct physical interactions [85], consistent with the presence of a form of seg-regulation. Higher than expected levels of co-expression between proximal genes have thus far been attributed to chromatin remodeling [35]. Facilitated co-transcription offers an alternative explanation, without, nevertheless, excluding other contributions. Beyond S. cerevisiae, clusters of co-expressed genes are a common feature of eukaryotic genomes [30]. As these genomes do not contain operons and have regulatory mechanisms significantly different from those of bacteria, the presence and conservation of gene clustering support the hypothesis of generic mechanisms behind the co-transcription of proximal genes. Transcriptional read-through and divergent promoters have, in fact, also been proposed to account for the conservation of gene cluster in mammals [34], and supercoiling, one mechanism that we propose for facilitated co-transcription, is also recognized as a crucial factor for the local properties of gene regulation in eukaryotes [86].
Conclusion
Our identification by synteny of transcriptional units beyond the usual scale of operons does not call into question the existence of well-established operons as much as it challenges the very notion of “transcriptional unit”: at different times, the same genes may indeed be co-transcribed either as short operons or as longer segments. The extent of co-transcription may depend on internal and external conditions and, given these conditions, be partly stochastic. Given their evolutionary conservation, we can conclude, however, that the larger units are as much, if not more, functionally meaningful than the smaller ones.
From an evolutionary standpoint, our hypothesis that facilitated co-transcription is both historically primitive and currently primary shifts the challenge from explaining how the expressions of adjacent genes became coupled to the challenge of explaining how they became partially uncoupled [87]. This perspective confers an important role to the regulation of termination. While this aspect of the problem is beyond the scope of the present work, we note that transcriptional termination is as regulated as initiation [18], can be strongly conserved [88], and is at the heart of the hierarchical properties of co-expression in TF-depleted bacteria [25].
Materials & Methods
Micro-array data and transcriptional co-expression analyses
Our analysis of co-expression in E. coli is based on transcription profiles generated from the M3D database, which concerns the expression of 4320 genes across 466 conditions normalized altogether using a quantile normalization procedure [8]. Our analysis in B. subtilis is based on the dataset produced by the BaSysBio consortium using 22 bases tiling resolution micro-arrays and concerns the expression of 4162 genes in 104 different conditions (for a total of 269 different experiments given the various replicates) [9]; for consistency with E. coli, we quantile normalized the data. For each dataset, given a gene i in condition s, we define its expression level, or activity asi, as the average of the values associated with the micro-array probes overlapping with the gene—a quantity already computed in the original data of B. subtilis. We quantify the co-expression of a pair i, j of genes by the Pearson correlation of their activities across all conditions: where with Nc the number of conditions. Patterns of co-expression are visualized by representing the matrix Cij with the genes ordered as along the chromosome (Fig 1B); to visualize large-scale patterns in Fig 1D, we apply a Gaussian filter with a standard deviation of 10 genes. Finally, we quantify the distance-dependence of the correlations by defining an autocorrelation function Γ(d) as the average value of Cij over the pairs ij of genes at a given distance d ± Δd, with Δd = 0.5 kb. This autocorrelation can be computed using all pairs of genes, or only the pairs satisfying a given criterion, such as belonging to different operons, or comprising no gene annotated as regulated by a TF or/and by a common SF.
Construction of synteny segments
Genomes
The synteny segments are defined from a systematic comparison of the relative positions of orthologous genes across multiple genomes. We downloaded all the complete and COG-annotated bacterial genomes available in the NCBI databases as of March 2015 (ftp.ncbi.nih.gov), representing a data-set of 1445 genomes. COGs are Clusters of Orthologous Genes [50], which we use to map the genes to orthology classes. COGs are defined on the principle that any group of at least three genes from distant genomes that are more similar in sequence to each other than to any other genes from the same genomes should belong to the same COG [50]. As a result, a genome may contain one, several or no gene associated with any given COG, and a gene may be associated with one or no COG. Our analysis is based on the most recent update of this approach [89], which includes 4764 different COGs.
A synteny segment in a given genome is defined as a set of consecutive genes that are also proximal in a significant number of other genomes. To identify these segments, we first define an inter-gene distance and then a criterion to assess whether two genes are proximal in a significant number of genomes. To take into account the phylogeny of the genomes when counting, we use weights that reduce the contribution of genomes with a large number of closely phylogenetically related genomes in the data-set. Finding two genes nearby in a large number of closely related genomes can thus be less significant than finding them nearby in a smaller number of more distantly related genomes.
Inter-gene distances
We measure the distance between two genes in base pairs, from the mid-point of their nucleotide sequences. To account for the fact that genomes may comprise several chromosomes, which may be non-circular and of different lengths, we formally circularize linear chromosomes and normalize them to a common length of L = 500 kb, by setting all distances exceeding L/2 = 250 kb to 250 kb: if d is the actual distance in base pairs, we thus define a normalized distance x by x = min(1, 2d/L). The normalized distance between genes on distinct chromosomes is also set to x = 1. As L = 500 kb is by far larger than the typical extension of the synteny segments that we find, the exact value of this cut-off is not determining.
Genome weights for counting statistics
The number Mij(x) of genomes in which genes i and j are at normalized distance xij ≤ x is computed as Mij(x) = ∑g ωg1(xij ≤ x) (we consider genomes where at least one gene is present and we set xij = 1 if one of the two genes is missing), with genome weights defined by ωg = 1/|{h: Dgh < δ}|, where |{h: Dgh < δ}| denotes the number of genomes h at phylogenetic distance at most δ from g. Here, we fix δ to δ = 0.25, which is large enough to treat as equivalent the different strains of a same species (larger values δ may reveal more conserved syntenic relations [52]). This weighting procedure defines an effective number of genomes as M′ = ∑g ωg with here M′ = 500—for the pair ij, we define the corresponding effective number of genomes, , by considering only the genomes where i and j are present (). We use a simple definition of evolutionary distance based on the sequence similarity of a few representative conserved genes (quantifying the phylogenetic distance between bacterial genomes is a notoriously difficult task, given that different genes in a same genome often have different histories [90]). Specifically, we selected the 10 genes associated with the COGs 126G, 173J, 202K, 2255L, 481M, 497L, 541U, 544O, 556L, 1158K. These genes were taken from a list of genes shown to reflect phylogenetic distances between bacterial strains [91], with the additional constraint that they comprise a single copy in most of the genomes of our dataset. We aligned the amino sequences of these genes with MAFFT [92] and defined the similarity between any two genes by their fraction of common amino acids in the resulting multiple sequence alignment, excluding positions with gaps in the two genes. The evolutionary similarity Sgh between two strains g and h was obtained by averaging these similarities over the representative genes, taking only into account those genes present in single copy in the two strains. We then defined an evolutionary distance between strains as Dgh = 1 − Sgh. We checked that this procedure yields a robust estimation of evolutionary distance by repeating the analysis with subsets of only 5 of the 10 genes and verifying that it leads to equivalent results (S14 Fig).
Significance of proximity
Assuming a uniform distribution of genes along a circular genome of length L, the probability of observing a distance less than xL/2 between 2 given genes is just x. In this null model, the number Mij(x) of genomes with normalized distance xij ≤ x thus follows a binomial law , where is the effective number of genomes. The probability πij(x) of observing Mij(x) events is therefore , where Ix(m, n) is the regularized incomplete beta function. The least likely and therefore most significant normalized distance between a given pair of genes ij, is the one minimizing πij(x), which defines and an associated p-value .
To treat pairs of COGs ij with multiple copies (genes), we fix a gene gi in i, count the number n of genes in j at normalized distance less than x to i, and compute the probability of the event as p(x) = 1 − (1 − x)n. The analysis is then performed as for a single gene (n = 1) but with πgij(x) now standing for πgij(p(x)), thus defining . We then define as the most significant observation when considering successively each gene gi in i, i.e., . As different numbers of genes in i and j may imply , we finally define a symmetrical measure of significance by .
Threshold of significance
Under the null model, the distribution of is found to have an exponential tail [52], ψ0(y) ∼ e−ay, with here an exponent a ≃ 3.25 (S15 Fig). Given a threshold of significance π*, we compute the fraction σs of significant pairs, with , and estimate the fraction of false positive pairs as . Following [93], we set a threshold of significance π* by imposing a given false discovery rate FDR = σfp/σs, which we take to be 0.005. This leads us to a threshold π* ≃ 4.10−4.
Synteny segments
For a given genome, we call synteny segment a maximal set of consecutive genes that are all proximal between each other in a significant number of other genomes. More formally, a synteny segment is defined as a set of consecutive genes such that any two genes i, j in the segment verify , and where none of the two genes k1, k2 at the external border of the segment verifies with all genes i in the segment—we skip genes that are not COG-annotated. The later criterion ensures that the segments are maximal, with no larger segment containing them. Note that this definition allows for overlapping segments; as a consequence, a given gene may belong to several segments, but also to no segment at all.
Analysis of structural properties of synteny segments
Inclusion of operons inside segments
To relate the synteny segments to operons, we count the fraction of operons shared between two or more segments, and compare the result with counts obtained from randomized operon maps (see the legend of Fig 3 for details).
Profiles of NAPs with respect to the segment and operon borders
To compute the average binding profile of each NAP (here H-NS and Fis taken separately) with respect to the synteny segments of E. coli, we first compute a binding profile ρk(x) for each segment k. To this end, we define the two borders of every segment, , as the positions of the TSS(s) and/or stop codon(s) located at the extremities of the segment. We then define the profiles with respect to these borders as , with δ(x) = 1 if position x is bound by the NAP and 0 otherwise and with a normalization factor ensuring that ∑x ρk(x) = 1. Denoting Nseg the number of segments, an average profile is finally defined by ρseg(x) = Σk ρk(x)/Nseg. For comparison, we compute for each NAP the average binding profile ρop(x) of the NAP with respect to the 1649 operons that are not located at the border of a segment.
Directionality of operons in a segment
To analyze the relative orientations of operons inside a segment, we consider segments made of 2 operons and make a statistics of the following three configurations: co-directional (on the leading or lagging strand), divergent or convergent. To compute a p-value and a z-score (number of standard deviations) for each configuration, we use a null model where the operon map is translated by an arbitrary number of operons, while the segment map is fixed (as in Fig 3). We then compute for all possible translations the resulting distributions for the numbers of co-directional, divergent and convergent orientations in the segments, and consider these distributions to be Gaussian. We analyze similarly segments made of 3 operons, in which case 4 configurations must be considered, which are represented in Fig 5A. To analyze more generally the relative orientation of genes in segments for genomes that are not annotated in operons, we consider, for a given set of genes, the number of consecutive genes in that are divergent, the number of consecutive genes in that are convergent and the number of consecutive genes in that are co-directional. For consisting of an entire circular chromosome, the ratio Dir(chrom)/Conv(chrom) must be 1. To quantify the particular orientation of genes inside segments, we define a divergence index as DI = Dir(in)/Conv(in) − Dir(out)/Conv(out), where (respectively, ) is the set of genes inside (respectively, outside) a segment. This index is computed for every genome. We also compute for every genome a co-directionality index defined as the difference of the ratio of co-directional over divergent or convergent orientations: CoDI = Codir(in)/[Conv(in) + Dir(in)] − Codir(out)/[Conv(out) + Dir(out)].
Transcriptional read-through analysis
To analyze transcription in non-coding, inter-operon regions of E. coli, we use RNA-seq data from [24], which we retrieved in the form of.sra files. RNA reads were mapped to the genome of E. coli K12 MG1655 using bowtie2. The number of reads per bp was then computed as the genomic coverage of the data (using genomeCoverageBed and the flags “-d -split”), with the final expression levels equal to the log-value of the mean number of reads found in the regions of interest. We considered datasets for which more than ∼ 90% of the reads were uniquely mapped. Our results are averaged over 7 different conditions corresponding to the following GEO Accession Number: GSM1104381 (sgrS- with vector), GSM1104384 (sgrS- with sgrS+ plasmid), GSM1104387 (WT in LB +αMG), GSM1104401 (WT in defined medium with glycerol +αMG), GSM1104402 (WT in defined medium with glycerol −αMG), GSM1104405 (sgrS- in defined medium with glycerol +αMG) and GSM1104408 (sgrS- in defined medium with glycerol −αMG). Analyzing inter-operonic transcription also requires identifying transcription start sites (TSS). We retrieved TSS datasets from the most recent update of RegulonDB (Morett dataset [46]) and from the recent dataset of Palsson’s group [5]. We combined these two datasets into a single list of TSSs, and considered operons for which the first gene had an associated TSS in the immediate upstream inter-operonic region. For genes with several potential TSSs in the inter-operonic region, we considered the closest upstream start sites. To assess whether synteny segments display any specific inter-operon transcriptional activity between co-directional consecutive operons, we further limited biases from mis-annotations by considering only inter-operon regions of size larger than 100 bp, which corresponds in E. coli to 243 cases of co-directional consecutive operons (29 pairs are intra-segment pairs). Considering the 7 different RNA-seq conditions of E. coli, we thus analyzed 203 (29 × 7) situations inside a same segment and 1498 (214 × 7) situations outside segments.
To investigate the phenomenon of transcriptional read-through in B. subtilis, we analyzed the tendency of adjacent genes from different operons to belong to one of the transcriptional units identified by the BaSysBio consortium. These transcriptional units represent blocks of contiguous expression that often extend the known operons of B. subtilis [9].
Supporting Information
(TXT)
(TXT)
(PDF)
A. As in Fig 1A for E. coli, micro-array data reporting the expression levels of 4320 genes (rows) in 466 conditions (columns) with high expression in red and low expression in green. B. Applying a singular value decomposition to the micro-array data yields two principal components, V1 along the genes and U1 along the conditions. The co-expression matrix of Fig 1B is shown here with, above the diagonal, the genes sorted by V1: this component classifies the genes according to their contribution to one of the two anti-correlated clusters visible in Fig 1D. C. Same expression data as in A, but with the conditions sorted by U1 and the genes sorted by V1, thus revealing the main pattern of variation. D. Distribution of the conditions along the principal component U1, with different colors for the different phases of growth at which the measurements of transcriptional activity were made, showing that U1 correlates with the growth rate. E. Fraction of genes controlled by σ70 (gray squares) and with a binding site for the NAP Fis (red triangles) as a function of V1, showing that genes that are transcribed in growing phases (negative values of V1) are more likely to be regulated by σ70 and bound by Fis.
(PDF)
A. Transcriptional co-expression between the 1231 genes of E. coli having σ70 as unique SF. Genes are reordered along the first component V1 from the SVD decomposition of the data as in S1B Fig. B. In E. coli, fraction of pairs of genes belonging to different operons that share a TF, a SF or one of the two, showing that, except at very high level of co-expression (Cij > 0.85), the majority (∼ 75%) of correlated pairs of genes do not share a common TF or SF. C. Same analysis in B. subtilis.
(PDF)
Synteny as a proxy for high co-expression. Taken two genes within 10 kb along the chromosome of a reference genome, what is the probability that these genes have orthologs within the same distance in the chromosome of another bacterium? We obtain an answer from a statistics over > 1000 bacterial genomes (left panel). This answer depends not only on the phylogenetic divergence between the query and reference genomes, but also very strongly on the level of co-expression of the two genes in the reference genome (plots): the more co-expressed are the two genes in E. coli (top) or in B. subtilis (bottom), the more likely they are to remain proximal in the chromosome of distant bacteria. The curves in the graph represent the fraction of pairs of genes within 10 kb in the reference genome (E. coli or B. subtilis) that are also within 10 kb in another genome as a function of the phylogenetic divergence between the two genomes (this divergence is measured by sequence divergence, see Materials and methods). Different colors correspond to pairs of genes with different levels of co-expression in the reference genome: proximity between highly co-expressed pairs, in red, is thus much more conserved than between weakly co-expressed pairs, in yellow. The plain lines are based on pairs of genes that do not belong to the same operon, and the dotted lines on pairs of operonic genes: this shows that the relation between co-expression and synteny extends beyond operons.
(PDF)
Genomic distribution of segments in E. coli (top) and in B. subtilis (bottom): the histograms of the location of the segments along the chromosome reveal a fairly uniform distribution (bin size of 65 kb). The vertical dashed lines indicate the origin (oriC) and terminus (ter) of replication. In B. subtilis, the depletion close to ter is mainly due to a poor gene annotation in this region.
(PDF)
Size distributions of synteny segments (solid circles) in three phylogenetically distant bacteria and of polycistronic operons in E. coli and in B. subtilis (crosses), showing a similar exponential decrease up to ∼ 10 kb.
(PDF)
Binding profile of tsEPODs [53] with respect to synteny segments and operons, showing, as in the case of H-NS (Fig 2D), a strikingly high density of tsEPODs at the external boundaries of segments together with a depletion inside segments (in red). In agreement with their role in transcription silencing [94], we also observe an enrichment around the promoter region, and over the first gene for operons not at the border (in black).
(PDF)
Co-expression analysis for two additional bacteria: A. Mycoplasma pneumoniae (classified as close to Gram-positive) and B. Dickeya dadantii (formerly Erwinia chrysanthemi, Gram-negative). These two bacterial strains have very different genome lengths (they contain respectively ca. 650 and 4500 protein coding genes) and lifestyles (M. pneumoniae is a human parasit living in the respiratory tract, D. dadantii is a plant pathogen); they are also phylogenetically distant from both E. coli and B. subtilis (analyzed in Fig 4). M. pneumoniae is known to have a tiny repertoire of TFs and a single major SF, while the regulatory network of D. dadantii is mostly unknown (as is the case for most bacteria). The graphs compare co-expression inside synteny segments (red triangles) to co-expression outside segments (gray squares). In both cases, only genes belonging to different operons are considered (operon map from [25] for M. pneumoniae and from the ProOpDB database [95] for D. dadantii). Co-expression levels are computed from rRNA normalized RNA-seq data obtained in 151 different conditions for M. pneumoniae [25] and from rRNA normalized micro-array data obtained in 32 different conditions for D. dadantii [96]. Although global levels of co-expression differ between strains (see [25] for a detailed analysis of co-expression properties in M. pneumoniae), a systematic enhancement of co-expression is observed inside synteny segments, which is nearly independent of the distance separating the genes.
(PDF)
A. The red triangles correspond to those of Fig 4B (E. coli), and the gray squares and cyan points show that restricting to co-directional or divergent pairs has little incidence. B. Similar to A, but considering the smallest segments (< 4 kb) instead of the largest ones (> 10 kb): the overall level of correlation is lower for shorter segments.
(PDF)
Average number of operons directly controlled by at least one TF (upper panels) or by at least one SF (lower panels) as a function of the number of operons in the segment. Results show that both in E. coli (left panels) and in B. subtilis (right panels) there is roughly a constant number (close to 1) of operons directly regulated by a TF. In contrast, most operons are directly regulated by a SF in E. coli (left lower panel). In B. subtilis, not all operons of the segment are regulated by a SF, but at least one. The dashed lines in the lower panels indicate the bisectors y = x.
(PDF)
Co-expression between E. coli genes in different operons that are not regulated by any TF and that do not share the same SF (gray squares). Pairs in synteny, independently of whether they are proximal in the chromosome of E. coli, are on average more co-expressed than those not in synteny (red triangles). The phenomenon appears to be specific since replacing the first gene in these pairs by its nearest neighbor not in synteny (while keeping the second gene) systematically decreases the mean level of co-expression at all distances.
(PDF)
Fraction of adjacent genes that belong to a same transcriptional unit (TU) in B. subtilis [9]. Two types of TUs are considered as proposed in [9]: “short TUs” (left panel), which are minimal TUs found in most conditions, and “long TUs” (right panel), which are maximal TUs found in at least one condition. The fraction is computed for genes inside synteny segments (red bars) and for genes outside synteny segments (gray bars). In each panel, the two bars on the left are based on all pairs of genes in different operons and those on the right on pairs of co-directional genes in different operons.
(PDF)
A. Extension of the results of Fig 6C, showing that conserved high co-expression is mostly due to a seg-regulation by housekeeping SFs (σ70 in E. coli and SigA in B. subtilis). B. Contribution of the seg-regulation by housekeeping SFs in each organism. C. Same as in B but considering only genes that belong to synteny segments, showing a strong relationship in both bacteria between gene co-expression and seg-regulation by a housekeeping SF. In B and C, the drop at high co-expression level for B. subtilis may either come from a too partial annotation of SF binding sites, or from the imperfect match between our synteny segments and the actual relevant co-expression unit of B. subtilis.
(PDF)
Distribution in B. subtilis of the co-expression Cij between pairs of genes that are not directly regulated by a TF or a SF and that belong to different synteny segments. Gray distribution: pairs in segments with different sets of SFs. Red distribution: pairs in segments that have one single seg-SF, the housekeeping SigA. Cyan distribution: pairs in segments that have exactly the same seg-SFs, excluding SigA.
(PDF)
Co-expression for pairs of genes in synteny (red triangles) or not (gray squares) in S. cerevisiae. Synteny is defined from our dataset of bacterial genomes, which does not include any yeast genome. Co-expression is computed from micro-array data retrieved from the M3D database [8]. Pairs of genes in synteny in bacteria are in average more co-expressed in S. cerevisiae than pairs that are not in synteny in bacteria.
(PDF)
Robustness of the calculation of evolutionary distances. We compare two evolutionary distances that were computed using two disjoint groups of 5 genes that reflect phylogenetic distances between bacterial strains (Materials and methods). One can observe a linear relationship (in red) for almost the full range of similarities, except at very low similarities. All genome pairs formed from the 1445 genomes of our dataset are reported. The dashed black line indicates the bisector y = x.
(PDF)
Probability density of for the empirical data (red triangles) obtained for an effective number of genomes M′ = 500. For small enough values of , the density decays exponentially with (black line). The deviation from an exponential at large values (gray area) indicates the conservation of co-localization. For the null model (gray points), for which we consider the same effective number of genomes but where gene positions are randomized, the exponential decay extends to larger values of . Here, we consider a false discovery rate FDR = 0.005, leading to a threshold π* ≃ 4.10−4 (vertical blue line).
(PDF)
Acknowledgments
We thank Luis Serrano’s and William Nasser’s groups for facilitating access to their transcriptomic data and for valuable discussions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by ATIP-Avenir CNRS to I.J. and Agence Nationale de la Recherche (ANR-10-PDOC-004-01) to O.R. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Browning DF, Busby SJW. The regulation of bacterial transcription initiation. Nature Reviews Microbiology. 2004;2:57–65. 10.1038/nrmicro787 [DOI] [PubMed] [Google Scholar]
- 2. Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature genetics. 2006;38(6):626–635. 10.1038/ng1789 [DOI] [PubMed] [Google Scholar]
- 3. David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, et al. A high-resolution map of transcription in the yeast genome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(14):5320–5325. 10.1073/pnas.0601091103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, et al. The transcription unit architecture of the Escherichia coli genome. Nature Biotechnology. 2009;27:1043–1049. 10.1038/nbt.1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cho BK, Kim D, Knight EM, Zengler K, Palsson BØ. Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states. BMC biology. 2014;12(1):4 10.1186/1741-7007-12-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic expression programs in the response of yeast cells to environmental changes. Molecular biology of the cell. 2000;11(12):4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome research. 2004;14(6):1085–1094. 10.1101/gr.1910904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faith JJ, Driscoll ME, Fusaro VA, Cosgrove EJ, Hayete B, Juhn FS, et al. Many Microbe Microarrays Database: uniformly normalized Affymetrix compendia with structured experimental metadata. Nucleic acids research. 2007;36(Database):D866–D870. 10.1093/nar/gkm815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- 10. Ma Q, Yin Y, Schell MA, Zhang H, Li G, Xu Y. Computational analyses of transcriptomic data reveal the dynamic organization of the Escherichia coli chromosome under different conditions. Nucleic acids research. 2013;41:5594–5603. 10.1093/nar/gkt261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature genetics. 2003;34(2):166–176. 10.1038/ng1165 [DOI] [PubMed] [Google Scholar]
- 12. Bonneau R. Learning biological networks: from modules to dynamics. Nature Chemical Biology. 2008;4(11):658–664. 10.1038/nchembio.122 [DOI] [PubMed] [Google Scholar]
- 13. Brooks AN, Reiss DJ, Allard A, Wu WJ, Salvanha DM, Plaisier CL, et al. A system-level model for the microbial regulatory genome. Molecular Systems Biology. 2014;10:740 10.15252/msb.20145160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goelzer A, Bekkal Brikci F, Martin-Verstraete I, Noirot P, Bessières P, Aymerich S, et al. Reconstruction and analysis of the genetic and metabolic regulatory networks of the central metabolism of Bacillus subtilis. BMC systems biology. 2008;2:20 10.1186/1752-0509-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nature Reviews Microbiology. 2014;12(5):327–340. 10.1038/nrmicro3238 [DOI] [PubMed] [Google Scholar]
- 16. Waters LS, Storz G. Regulatory RNAs in Bacteria. Cell. 2009;136:615–628. 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morris KV, Mattick JS. The rise of regulatory RNA. Nature Reviews Genetics. 2014;15(6):423–437. 10.1038/nrg3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. BioEssays. 2002;24:700–707. 10.1002/bies.10125 [DOI] [PubMed] [Google Scholar]
- 19. Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. 10.1016/j.cell.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berthoumieux S, de Jong H, Baptist G, Pinel C, Ranquet C, Ropers D, et al. Shared control of gene expression in bacteria by transcription factors and global physiology of the cell. Molecular Systems Biology. 2013;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dorman CJ. DNA topology and the global control of bacterial gene expression: implications for the regulation of virulence gene expression. Microbiology. 1995;141(6):1271–1280. 10.1099/13500872-141-6-1271 [DOI] [PubMed] [Google Scholar]
- 22. Hatfield GW, Benham CJ. DNA topology-mediated control of global gene expression in Escherichia coli. Annual Review of Genetics. 2002;36:175–203. 10.1146/annurev.genet.36.032902.111815 [DOI] [PubMed] [Google Scholar]
- 23. Travers A, Muskhelishvili G. DNA supercoiling—a global transcriptional regulator for enterobacterial growth? Nature Reviews Microbiology. 2005;3:157–169. 10.1038/nrmicro1088 [DOI] [PubMed] [Google Scholar]
- 24. McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, et al. Computational analysis of bacterial RNA-Seq data. Nucleic acids research. 2013;41:e140–e140. 10.1093/nar/gkt444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Junier I, Besray Unal E, Yus E, Llorens V, Serrano L. Insights into the mechanisms of basal coordination of transcription using a genome-reduced bacterium. Cell Systems, in press, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mering Cv. STRING: a database of predicted functional associations between proteins. Nucleic acids research. 2003;31:258–261. 10.1093/nar/gkg034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bergmann S, Ihmels J, Barkai N. Similarities and differences in genome-wide expression data of six organisms. PLoS biology. 2004;2(1):E9 10.1371/journal.pbio.0020009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(47):17973–17978. 10.1073/pnas.0605938103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsaparas P, Mariño-Ramírez L, Bodenreider O, Koonin EV, Jordan IK. Global similarity and local divergence in human and mouse gene co-expression networks. BMC Evol Biol. 2006;6:70 10.1186/1471-2148-6-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008;91:243–248. 10.1016/j.ygeno.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 31. Lee JM, Sonnhammer ELL. Genomic gene clustering analysis of pathways in eukaryotes. Genome research. 2003;13(5):875–882. 10.1101/gr.737703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hurst LD, Pál C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nature Reviews Genetics. 2004;5(4):299–310. 10.1038/nrg1319 [DOI] [PubMed] [Google Scholar]
- 33. Singer GAC, Lloyd AT, Huminiecki LB, Wolfe KH. Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Molecular biology and evolution. 2005;22(3):767–775. 10.1093/molbev/msi062 [DOI] [PubMed] [Google Scholar]
- 34. Semon M, Duret L. Evolutionary origin and maintenance of coexpressed gene clusters in mammals. Molecular Biology and Evolution. 2006;23:1715–1723. 10.1093/molbev/msl034 [DOI] [PubMed] [Google Scholar]
- 35. Batada NN, Urrutia AO, Hurst LD. Chromatin remodelling is a major source of coexpression of linked genes in yeast. Trends in genetics. 2007;23:480–484. 10.1016/j.tig.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 36. Engström PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome research. 2007;17(12):1898–1908. 10.1101/gr.6669607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(6):2896–2901. 10.1073/pnas.96.6.2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lathe WC III, Snel B, Bork P. Gene context conservation of a higher order than operons. Trends in biochemical sciences. 2000;25:474–479. 10.1016/S0968-0004(00)01663-7 [DOI] [PubMed] [Google Scholar]
- 39. Tamames J. Evolution of gene order conservation in prokaryotes. Genome biology. 2001;2:RESEARCH0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rogozin IB, Makarova KS, Murvai J, Czabarka E, Wolf YI, Tatusov RL, et al. Connected gene neighborhoods in prokaryotic genomes. Nucleic acids research. 2002;30:2212–2223. 10.1093/nar/30.10.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang G, Rocha EP, Danchin A. Persistence drives gene clustering in bacterial genomes. BMC genomics. 2008;9:4 10.1186/1471-2164-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ermolaeva MD, White O, Salzberg SL. Prediction of operons in microbial genomes. Nucleic acids research. 2001;29(5):1216–1221. 10.1093/nar/29.5.1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Korbel JO, Jensen LJ, Von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nature Biotechnology. 2004;22:911–917. 10.1038/nbt988 [DOI] [PubMed] [Google Scholar]
- 44. Janga SC, Collado-Vides J, Moreno-Hagelsieb G. Nebulon: a system for the inference of functional relationships of gene products from the rearrangement of predicted operons. Nucleic acids research. 2005;33(8):2521–2530. 10.1093/nar/gki545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carpentier AS, Torrésani B, Grossmann A, Hénaut A. Decoding the nucleoid organisation of Bacillus subtilis and Escherichia coli through gene expression data. BMC genomics. 2005;6:84 10.1186/1471-2164-6-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Salgado H, Peralta-Gil M, Gama-Castro S, Santos-Zavaleta A, Muniz-Rascado L, Garcia-Sotelo JS, et al. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic acids research. 2012;41(D1):D203–D213. 10.1093/nar/gks1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Research. 2008;36(Database issue):D93–6. 10.1093/nar/gkm910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeong KS, Ahn J, Khodursky AB. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome biology. 2004;5:R86 10.1186/gb-2004-5-11-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sobetzko P, Travers A, Muskhelishvili G. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E42–E50. 10.1073/pnas.1108229109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic acids research. 2000;28:33–36. 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1293–301. 10.1073/pnas.1111471108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Junier I, Rivoire O. Synteny in bacterial genomes: inference, organization and evolution; 2013. [Google Scholar]
- 53. Vora T, Hottes AK, Tavazoie S. Protein occupancy landscape of a bacterial genome. Molecular Cell. 2009;35:247–253. 10.1016/j.molcel.2009.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kahramanoglou C, Seshasayee ASN, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, et al. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic acids research. 2011;39:2073–2091. 10.1093/nar/gkq934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martínez-Antonio A, Janga SC, Thieffry D. Functional organisation of Escherichia coli transcriptional regulatory network. Journal of molecular biology. 2008;381(1):238–247. 10.1016/j.jmb.2008.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang GZ, Chen WH, Lercher MJ. Coexpression of linked gene pairs persists long after their separation. Genome Biology and Evolution. 2011;3:565–570. 10.1093/gbe/evr049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hershberg R, Yeger-Lotem E, Margalit H. Chromosomal organization is shaped by the transcription regulatory network. Trends in genetics. 2005;21:138–142. 10.1016/j.tig.2005.01.003 [DOI] [PubMed] [Google Scholar]
- 58. Babu MM, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. Journal of Molecular Biology. 2006;358:614–633. 10.1016/j.jmb.2006.02.019 [DOI] [PubMed] [Google Scholar]
- 59. Gruber TM, Gross CA. Multiple sigma subunits and the partioning of bacterial transcription space. Annual Review of Microbiology. 2003;57:441–466. 10.1146/annurev.micro.57.030502.090913 [DOI] [PubMed] [Google Scholar]
- 60. Yin Y, Zhang H, Olman V, Xu Y. Genomic arrangement of bacterial operons is constrained by biological pathways encoded in the genome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6310–6315. 10.1073/pnas.0911237107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Junier I, Hérisson J, Képès F. Genomic organization of evolutionarily correlated genes in bacteria: limits and strategies. Journal of Molecular Biology. 2012;419:369–386. 10.1016/j.jmb.2012.03.009 [DOI] [PubMed] [Google Scholar]
- 62. Wade JT, Grainger DC. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nature Reviews Microbiology. 2014;12:647–653. 10.1038/nrmicro3316 [DOI] [PubMed] [Google Scholar]
- 63. Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:7024–7027. 10.1073/pnas.84.20.7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meyer S, Beslon G. Torsion-mediated interaction between adjacent genes. PLoS Computational Biology. 2014;10:e1003785 10.1371/journal.pcbi.1003785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dornenburg JE, Devita AM, Palumbo MJ, Wade JT. Widespread antisense transcription in Escherichia coli. mBio. 2010;1 10.1128/mBio.00024-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20172–20177. 10.1073/pnas.1113521108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes & Development. 2004;18:1766–1779. 10.1101/gad.1207504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Le TBK, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. 10.1126/science.1242059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome biology. 2004;5(11):R87 10.1186/gb-2004-5-11-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lozada-Chávez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Research. 2006;34(12):3434–3445. 10.1093/nar/gkl423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Price MN, Dehal PS, Arkin AP. Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli. Genome biology. 2008;9(1):R4 10.1186/gb-2008-9-1-r4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shou C, Bhardwaj N, Lam HYK, Yan KK, Kim PM, Snyder M, et al. Measuring the evolutionary rewiring of biological networks. PLoS Computational Biology. 2011;7:e1001050 10.1371/journal.pcbi.1001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Isalan M, Lemerle C, Michalodimitrakis K, Horn C, Beltrao P, Raineri E, et al. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–845. 10.1038/nature06847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kolesov G, Wunderlich Z, Laikova ON, Gelfand MS, Mirny LA. How gene order is influenced by the biophysics of transcription regulation. Proceedings of the National Academy of Sciences. 2007;104(35):13948–13953. 10.1073/pnas.0700672104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Janga SC, Salgado H, Martínez-Antonio A. Transcriptional regulation shapes the organization of genes on bacterial chromosomes. Nucleic acids research. 2009; p. gkp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO reports. 2006;7:710–715. 10.1038/sj.embor.7400729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Crozat E, Philippe N, Lenski RE, Geiselmann J, Schneider D. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics. 2005;169:523–532. 10.1534/genetics.104.035717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Crozat E, Winkworth C, Gaffe J, Hallin PF, Riley MA, Lenski RE, et al. Parallel genetic and phenotypic evolution of DNA superhelicity in experimental populations of Escherichia coli. Molecular Biology and Evolution. 2010;27:2113–2128. 10.1093/molbev/msq099 [DOI] [PubMed] [Google Scholar]
- 79. Pál C, Hurst LD. Evidence against the selfish operon theory. Trends in genetics. 2004;20:232–234. 10.1016/j.tig.2004.04.001 [DOI] [PubMed] [Google Scholar]
- 80. Price MN, Huang KH, Arkin AP, Alm EJ. Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome Research. 2005;15:809–819. 10.1101/gr.3368805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ballouz S, Francis AR, Lan R, Tanaka MM. Conditions for the evolution of gene clusters in bacterial genomes. PLoS computational biology. 2010;6(2):e1000672 10.1371/journal.pcbi.1000672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lawrence JG, Roth JR. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 2002;143:1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Snel B, Bork P, Huynen MA. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Research. 2002;12:17–25. 10.1101/gr.176501 [DOI] [PubMed] [Google Scholar]
- 84. Batada NN, Hurst LD. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nature genetics. 2007;39(8):945–949. 10.1038/ng2071 [DOI] [PubMed] [Google Scholar]
- 85. Geistlinger L, Csaba G, Dirmeier S, Küffner R, Zimmer R. A comprehensive gene regulatory network for the diauxic shift in Saccharomyces cerevisiae. Nucleic acids research. 2013;41:8452–8463. 10.1093/nar/gkt631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Frontiers in bioscience: a journal and virtual library. 2007;12:4409–4423. 10.2741/2398 [DOI] [PubMed] [Google Scholar]
- 87. Singh SS, Singh N, Bonocora RP, Fitzgerald DM, Wade JT, Grainger DC. Widespread suppression of intragenic transcription initiation by H-NS. Genes & Development. 2014;28:214–219. 10.1101/gad.234336.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends in genetics. 2005;21:260–264. 10.1016/j.tig.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 89. Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic acids research. 2014; p. gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gabaldón T, Koonin EV. Functional and evolutionary implications of gene orthology. Nature Reviews Genetics. 2013;14:360–366. 10.1038/nrg3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zeigler DR. Gene sequences useful for predicting relatedness of whole genomes in bacteria. International Journal of Systematic and Evolutionary Biology. 2003;53:1893–1900. [DOI] [PubMed] [Google Scholar]
- 92. Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic acids research. 2005;33:511–518. 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 94. Dorman CJ. H-NS, the genome sentinel. Nature Reviews Microbiology. 2007;5(2):157–161. 10.1038/nrmicro1598 [DOI] [PubMed] [Google Scholar]
- 95. Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. ProOpDB: prokaryotic operon database. Nucleic acids research. 2012;40(D1):D627–D631. 10.1093/nar/gkr1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jiang X, Sobetzko P, Nasser W, Reverchon S, Muskhelishvili G. Chromosomal Stress-Response Domains Govern the Spatiotemporal Expression of the Bacterial Virulence Program. MBio. 2015;6(3):e00353–15. 10.1128/mBio.00353-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TXT)
(TXT)
(PDF)
A. As in Fig 1A for E. coli, micro-array data reporting the expression levels of 4320 genes (rows) in 466 conditions (columns) with high expression in red and low expression in green. B. Applying a singular value decomposition to the micro-array data yields two principal components, V1 along the genes and U1 along the conditions. The co-expression matrix of Fig 1B is shown here with, above the diagonal, the genes sorted by V1: this component classifies the genes according to their contribution to one of the two anti-correlated clusters visible in Fig 1D. C. Same expression data as in A, but with the conditions sorted by U1 and the genes sorted by V1, thus revealing the main pattern of variation. D. Distribution of the conditions along the principal component U1, with different colors for the different phases of growth at which the measurements of transcriptional activity were made, showing that U1 correlates with the growth rate. E. Fraction of genes controlled by σ70 (gray squares) and with a binding site for the NAP Fis (red triangles) as a function of V1, showing that genes that are transcribed in growing phases (negative values of V1) are more likely to be regulated by σ70 and bound by Fis.
(PDF)
A. Transcriptional co-expression between the 1231 genes of E. coli having σ70 as unique SF. Genes are reordered along the first component V1 from the SVD decomposition of the data as in S1B Fig. B. In E. coli, fraction of pairs of genes belonging to different operons that share a TF, a SF or one of the two, showing that, except at very high level of co-expression (Cij > 0.85), the majority (∼ 75%) of correlated pairs of genes do not share a common TF or SF. C. Same analysis in B. subtilis.
(PDF)
Synteny as a proxy for high co-expression. Taken two genes within 10 kb along the chromosome of a reference genome, what is the probability that these genes have orthologs within the same distance in the chromosome of another bacterium? We obtain an answer from a statistics over > 1000 bacterial genomes (left panel). This answer depends not only on the phylogenetic divergence between the query and reference genomes, but also very strongly on the level of co-expression of the two genes in the reference genome (plots): the more co-expressed are the two genes in E. coli (top) or in B. subtilis (bottom), the more likely they are to remain proximal in the chromosome of distant bacteria. The curves in the graph represent the fraction of pairs of genes within 10 kb in the reference genome (E. coli or B. subtilis) that are also within 10 kb in another genome as a function of the phylogenetic divergence between the two genomes (this divergence is measured by sequence divergence, see Materials and methods). Different colors correspond to pairs of genes with different levels of co-expression in the reference genome: proximity between highly co-expressed pairs, in red, is thus much more conserved than between weakly co-expressed pairs, in yellow. The plain lines are based on pairs of genes that do not belong to the same operon, and the dotted lines on pairs of operonic genes: this shows that the relation between co-expression and synteny extends beyond operons.
(PDF)
Genomic distribution of segments in E. coli (top) and in B. subtilis (bottom): the histograms of the location of the segments along the chromosome reveal a fairly uniform distribution (bin size of 65 kb). The vertical dashed lines indicate the origin (oriC) and terminus (ter) of replication. In B. subtilis, the depletion close to ter is mainly due to a poor gene annotation in this region.
(PDF)
Size distributions of synteny segments (solid circles) in three phylogenetically distant bacteria and of polycistronic operons in E. coli and in B. subtilis (crosses), showing a similar exponential decrease up to ∼ 10 kb.
(PDF)
Binding profile of tsEPODs [53] with respect to synteny segments and operons, showing, as in the case of H-NS (Fig 2D), a strikingly high density of tsEPODs at the external boundaries of segments together with a depletion inside segments (in red). In agreement with their role in transcription silencing [94], we also observe an enrichment around the promoter region, and over the first gene for operons not at the border (in black).
(PDF)
Co-expression analysis for two additional bacteria: A. Mycoplasma pneumoniae (classified as close to Gram-positive) and B. Dickeya dadantii (formerly Erwinia chrysanthemi, Gram-negative). These two bacterial strains have very different genome lengths (they contain respectively ca. 650 and 4500 protein coding genes) and lifestyles (M. pneumoniae is a human parasit living in the respiratory tract, D. dadantii is a plant pathogen); they are also phylogenetically distant from both E. coli and B. subtilis (analyzed in Fig 4). M. pneumoniae is known to have a tiny repertoire of TFs and a single major SF, while the regulatory network of D. dadantii is mostly unknown (as is the case for most bacteria). The graphs compare co-expression inside synteny segments (red triangles) to co-expression outside segments (gray squares). In both cases, only genes belonging to different operons are considered (operon map from [25] for M. pneumoniae and from the ProOpDB database [95] for D. dadantii). Co-expression levels are computed from rRNA normalized RNA-seq data obtained in 151 different conditions for M. pneumoniae [25] and from rRNA normalized micro-array data obtained in 32 different conditions for D. dadantii [96]. Although global levels of co-expression differ between strains (see [25] for a detailed analysis of co-expression properties in M. pneumoniae), a systematic enhancement of co-expression is observed inside synteny segments, which is nearly independent of the distance separating the genes.
(PDF)
A. The red triangles correspond to those of Fig 4B (E. coli), and the gray squares and cyan points show that restricting to co-directional or divergent pairs has little incidence. B. Similar to A, but considering the smallest segments (< 4 kb) instead of the largest ones (> 10 kb): the overall level of correlation is lower for shorter segments.
(PDF)
Average number of operons directly controlled by at least one TF (upper panels) or by at least one SF (lower panels) as a function of the number of operons in the segment. Results show that both in E. coli (left panels) and in B. subtilis (right panels) there is roughly a constant number (close to 1) of operons directly regulated by a TF. In contrast, most operons are directly regulated by a SF in E. coli (left lower panel). In B. subtilis, not all operons of the segment are regulated by a SF, but at least one. The dashed lines in the lower panels indicate the bisectors y = x.
(PDF)
Co-expression between E. coli genes in different operons that are not regulated by any TF and that do not share the same SF (gray squares). Pairs in synteny, independently of whether they are proximal in the chromosome of E. coli, are on average more co-expressed than those not in synteny (red triangles). The phenomenon appears to be specific since replacing the first gene in these pairs by its nearest neighbor not in synteny (while keeping the second gene) systematically decreases the mean level of co-expression at all distances.
(PDF)
Fraction of adjacent genes that belong to a same transcriptional unit (TU) in B. subtilis [9]. Two types of TUs are considered as proposed in [9]: “short TUs” (left panel), which are minimal TUs found in most conditions, and “long TUs” (right panel), which are maximal TUs found in at least one condition. The fraction is computed for genes inside synteny segments (red bars) and for genes outside synteny segments (gray bars). In each panel, the two bars on the left are based on all pairs of genes in different operons and those on the right on pairs of co-directional genes in different operons.
(PDF)
A. Extension of the results of Fig 6C, showing that conserved high co-expression is mostly due to a seg-regulation by housekeeping SFs (σ70 in E. coli and SigA in B. subtilis). B. Contribution of the seg-regulation by housekeeping SFs in each organism. C. Same as in B but considering only genes that belong to synteny segments, showing a strong relationship in both bacteria between gene co-expression and seg-regulation by a housekeeping SF. In B and C, the drop at high co-expression level for B. subtilis may either come from a too partial annotation of SF binding sites, or from the imperfect match between our synteny segments and the actual relevant co-expression unit of B. subtilis.
(PDF)
Distribution in B. subtilis of the co-expression Cij between pairs of genes that are not directly regulated by a TF or a SF and that belong to different synteny segments. Gray distribution: pairs in segments with different sets of SFs. Red distribution: pairs in segments that have one single seg-SF, the housekeeping SigA. Cyan distribution: pairs in segments that have exactly the same seg-SFs, excluding SigA.
(PDF)
Co-expression for pairs of genes in synteny (red triangles) or not (gray squares) in S. cerevisiae. Synteny is defined from our dataset of bacterial genomes, which does not include any yeast genome. Co-expression is computed from micro-array data retrieved from the M3D database [8]. Pairs of genes in synteny in bacteria are in average more co-expressed in S. cerevisiae than pairs that are not in synteny in bacteria.
(PDF)
Robustness of the calculation of evolutionary distances. We compare two evolutionary distances that were computed using two disjoint groups of 5 genes that reflect phylogenetic distances between bacterial strains (Materials and methods). One can observe a linear relationship (in red) for almost the full range of similarities, except at very low similarities. All genome pairs formed from the 1445 genomes of our dataset are reported. The dashed black line indicates the bisector y = x.
(PDF)
Probability density of for the empirical data (red triangles) obtained for an effective number of genomes M′ = 500. For small enough values of , the density decays exponentially with (black line). The deviation from an exponential at large values (gray area) indicates the conservation of co-localization. For the null model (gray points), for which we consider the same effective number of genomes but where gene positions are randomized, the exponential decay extends to larger values of . Here, we consider a false discovery rate FDR = 0.005, leading to a threshold π* ≃ 4.10−4 (vertical blue line).
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.