Abstract
Genetic association mapping in structured populations of model organisms can offer a fruitful complement to human genetic studies by generating new biological hypotheses about complex traits. Here we investigated prepulse inhibition (PPI), a measure of sensorimotor gating that is disrupted in a number of psychiatric disorders. To identify genes that influence PPI, we constructed a panel of half-sibs by crossing 30 females from common inbred mouse strains with inbred C57BL/6J males to create male and female F1 offspring. We used publicly available single nucleotide polymorphism (SNP) genotype data from these inbred strains to perform a genome-wide association scan using a dense panel of over 150 000 SNPs in a combined sample of 604 mice representing 30 distinct F1 genotypes. We identified two independent PPI-associated loci on Chromosomes 2 and 7, each of which explained 12 – 14% of the variance in PPI. Searches of available databases did not identify any plausible causative coding polymorphisms within these loci. However, previously collected expression quantitative trait locus (eQTL) data from hippocampus and striatum indicated that the SNPs on Chromosomes 2 and 7 that showed the strongest association with PPI were also strongly associated with expression of several transcripts, some of which have been implicated in human psychiatric disorders. This integrative approach successfully identified a focused set of genes which can be prioritized for follow-up studies. More broadly, our results show that F1 crosses among common inbred strains can be used in combination with other informatics and expression datasets to identify candidate genes for complex behavioral traits.
Keywords: Acp2, Arfgap2, expression QTL, Fam171b, Gtf2h1, Htatip2, prepulse inhibition, Prkd3, Rbm45, schizophrenia, Smarcad1, Timm10
Impairments in the ability to filter out unnecessary sensory information are a common characteristic of patients with a number of neuropsychiatric disorders including schizophrenia, bipolar disorder, obsessive – compulsive disorder, Tourette’s syndrome, panic disorder and some social disorders (Braff et al. 1978, 2001; Larsen et al. 2002; Perry et al. 2001, 2007; Swerdlow et al. 1993). Sensorimotor gating is often measured as the relative reduction in startle reflex that occurs when a weak prepulse is given before a stronger stimulus, which is referred to as prepulse inhibition (PPI). While genome-wide association studies (GWASs) in humans have begun to identify dozens of loci implicated in schizophrenia (Ripke et al. 2013, 2014), the function of those genes has yet to be delineated. Furthermore, the genes that influence endophenotypes for psychiatric disease and whether they overlap with schizophrenia risk genes remains unknown. We and others have used endophenotypes such as PPI to study the aspects of psychiatric disease using animal models (Geyer et al. 2001). Numerous studies have identified specific chromosomal regions that are associated with heritable differences in PPI in rodents. These studies have used a variety of crosses including backcrosses (Palmer et al. 2003), chromosome substitution strains (Leussis et al. 2009; Petryshen et al. 2005), recombinant inbred strains (Loos et al. 2012), panels of inbred mice (Webb et al. 2009), heterogeneous stock mice (Hitzemann et al. 2008) and advanced intercross lines (Samocha et al. 2010). These approaches have various strengths and weaknesses; some permit finer scale mapping resolution and higher power to detect variants, while others incorporate greater amounts of genetic diversity, which may better approximate the diversity of human populations (Flint & Eskin 2012; Mott & Flint 2013).
Large panels of inbred strains can be an efficient choice for mapping quantitative trait loci (QTL). One major advantage of mapping using inbred strains is that because all individuals of a given strain are genetically identical, genotype data only need to be collected once. An additional advantage is that the resolution of mapped intervals is better than classic F2 crosses because of the much greater number of ancestral recombinations captured by the inbred strains. Third, there are many more alleles segregating among inbred strains. Finally, the results can be integrated with other accumulated data about the same inbred strains. Early attempts to use this approach used modest numbers of inbred strains to map the genetic basis of various quantitative traits (Berndt et al. 2011; Grupe et al. 2001; Liao et al. 2004; Liu et al. 2006) and did not always correctly account for unequal relatedness among the inbred strains (Chesler 2001). More recently, larger panels of inbred strains have been developed and densely genotyped to further improve precision and power. For example, the hybrid mouse diversity panel (HMDP), which includes 29 inbred as well as 71 recombinant inbred lines, has been used to map the genetic basis of various physiological traits including gene expression networks in brain, bone mineral density and other metabolic traits (Bennett et al. 2010; Farber et al. 2011; Park et al. 2011). The HMDP can map QTL to regions smaller than 1 Mb (Ghazalpour et al. 2012).
We conducted a GWAS of PPI using a panel of 30 inbred mouse strains. Rather than phenotyping inbred strains from a vendor or breeding 30 inbred lines in house, we used a more efficient breeding scheme in which we produced a panel of 30 F1 offspring from crosses of C57BL/6J males and inbred strain females. Because both parents of F1 offspring are inbred, the F1 genotypes can be inferred directly from publicly available genotype data. We used two independent cohorts of mice. We then conducted a combined analysis of the data from both cohorts. A critical aspect of GWAS in populations used for high-resolution mapping is to account for the unequal relatedness among individuals; we accomplished this by using mixed models, similar to our (Cheng et al. 2010) and other prior publications (Bennett et al. 2010; Farber et al. 2011; Kang et al. 2008; Park et al. 2011). In order to follow-up on the implicated regions, we used expression QTL (eQTL) derived from the HMDP in hippocampus and striatum. This strategy identified a number of candidate transcripts that underlie the effects of the identified loci on PPI.
Materials and methods
Inbred mice and structured panel of F1 offspring
All animals used in this study were housed in a single, pathogen-free barrier facility. All procedures were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC). Mice had access to food and water ad libitum. Lights were on a 12 h on/12 h off cycle with lights on at 0600 h. We bred C57BL/6J males with females from a range of 30 commonly used inbred lab strains, creating an F1 generation of mice to serve as the study’s test population. C57BL/6J was used as the common strain to all crosses because of its inclusion in mouse genome resequencing projects. The creation of F1 s rather than using purely inbred strains avoids several detrimental effects of homozygosity including small litter sizes, some vision and hearing problems and extreme phenotypic differences. We attempted to produce 10 male and 10 female mice for each of the 30 possible F1 crosses; however, the actual numbers varied somewhat, because of variable breeding success and other practical considerations. The exact numbers of F1 mice are listed in Table S1, Supporting Information.
The 30 strains used to produce F1 s were adapted from the Mouse Phenome Database list of ‘priority strains’. We excluded wild-derived strains from consideration because of the severe population structure they introduce and because of the low power to map QTL to rare wild-specific variants (Ghazalpour et al. 2012). We favored inbred strains for which genome-wide dense genotype data were available. We eliminated several strains that failed to breed in our colony (CE/J, BPL/1J, C57Br/cdJ). We also eliminated pairs of strains that were extremely genetically similar to one another (e.g. BALB/cByJ vs. BALB/cJ; Kirby et al. 2010; Petkov et al. 2004). The different maternal strains and their respective abbreviations are as follows: 129S1/SvImJ (129), A/J (AJ), AKR/J (AKR), BALB/cByJ (BALB), BTBR T+tf/J (BTBR), BUB/BnJ (BUB), C3H/HeJ (C3H), C57BL/6J (B6), C57BLKS/J (BKS), C57L/J (57L), C58/J (C58), CBA/J (CBA), DBA/2J (DBA), DDY/JclSidSeyFrkJ (DDY), FVB/NJ (FVB), I/LnJ (ILN), KK/HlJ (KK), LP/J (LP), LG/J (LG), MRL/MpJ (MRL), NOD/ShiLtJ (NOD), NON/ShiLtJ (NON), NZB/BlNJ (NZB), NZW/LacJ (NZW), PL/J (PL), RIIIS/J (RII), SEA/GnJ (SEA), SJL/J (SJL), SM/J (SM), and SWR/J (SWR). We generated two cohorts of F1 mice using this same design; the second cohort was produced to provide replication for the results obtained from the initial study and to increase sample size. Production and testing of each cohort occurred over a period of 8 and 6 months, respectively. For each cohort, females were obtained from the Jackson Laboratory (2 – 6 per strain depending on predicted fecundity) and were bred to C57BL/6J males using harem breeding. Individual females were removed from harems and single housed when visibly pregnant. Four or more litters per F1 cross were used to minimize possible litter-specific effects. Male and female F1 offsprings were weaned and ear tagged at 21 – 23 days of age and housed in same-sex cages containing two to five littermates. Mice were tested between 64 and 99 days of age. The sample size used for genetic mapping was 604 mice (see Table S1).
Prepulse inhibition
Testing was conducted between 0900 and 1600 h. Five PPI testing apparati were used, which were 5-cm cylindrical Plexiglass containers on a platform contained within a lighted, ventilated chamber (San Diego Instruments, San Diego, CA, USA). The cylinder was connected to a piezoelectric accelerometer that measured the mouse’s startle response. The system was calibrated according to manufacturer’s specifications before each day of testing began.
Mice were placed in a clean holding cage with bedding where they awaited testing for 20 min. An experimenter blind to genotype then placed them in the testing apparatus. The protocol consisted of a 5-min acclimation period of 70-db white noise, followed by 62 trials occurring with 70 db noise in the background (Dulawa & Geyer 1996). Trials included pulse-alone trials (40 ms, 120 db burst); no stimulus trials and three types of prepulse trials (20 ms prepulse; 3, 6 or 12 db above background noise) followed 100 ms later by a 40-ms, 120-db pulse. Prepulse trials were arranged into four blocks. Blocks 1 and 4 were pulse-alone trials; blocks 2 and 3 contained pseudo-random combinations of pulse alone, no stimulus and each type of prepulse trial (3, 6 and 12 db). Responses were recorded for 65 ms after the beginning of 120-db stimulus. The intertrial interval was 9 – 20 s (average 15 s) throughout the test.
Prepulse inhibition was quantified as previously reported (Samocha et al. 2010). The PPI phenotype was defined as the difference of the average startle amplitude during the 3-, 6- and 12-db prepulse trials from the average startle amplitude during the pulse-alone trials, normalized by the pulse-alone amplitude: PPI = (SApulse – SAprepulse )/SApulse . As expected, PPI increased as the prepulse intensity increased; PPI 3 db < PPI 6 db < PPI 12 db; see Fig. S1. This study focuses on PPI 12 db because it had the highest heritability of the three PPI phenotypes (Appendix S1).
PPI cannot exceed a value of 1; in rare cases, mice startled more during the prepulse trials, which produced negative PPI values. We transformed the PPI measurements using the logit function, allowing the empirical distribution of phenotypes to closely resemble a normal distribution. To avoid extremely small or extremely large values after the transformation, small (or negative) PPI values less than 0.01 were set to 0.01 and any values greater than 0.99 were fixed at 0.99.
Genome-wide SNP data and informatics
Mouse diversity array SNP panel
An advantage of working with inbred lab strains is that genotype data are readily available. A total of 198 mouse strains have been genotyped using the AffyMetrix (Santa Clara, CA) mouse diversity array (MDA). This array was specifically designed to capture genetic diversity in commonly used laboratory mouse strains (Yang et al. 2009, 2011). The MDA single nucleotide polymorphism (SNP) data, after discarding low-quality SNPs, consist of 155 283 SNPs on Chromosomes 1 through 19, X, Y and the mitochondria (Wang et al. 2012). Single nucleotide polymorphism genotype data for a large number of inbred strains have also been released as a part of the mouse HapMap project (Kirby et al. 2010). These data were used to impute genotypes for the recombinant inbred strains in the HMDP (Bennett et al. 2010). We used MDA genotype data instead because it includes genotypes for a larger number of SNPs ascertained in a more comprehensive collection of strains. Mouse diversity array genotype data from the Jackson Laboratory Center for Genome Dynamics webpage were retrieved on 28 January 2014 (http://cgd.jax.org/datasets/popgen/diversityarray/yang2011.shtml). Mitochondrial and sex-linked markers as well as SNPs with missing genotypes were removed. We also filtered out SNPs polymorphic in 5 or fewer of the 30 strains because power to detect significant associations at such loci is low. For the same reason, we removed any SNPs with missing genotypes. We did not consider the X chromosome for mapping QTL because the X chromosome introduces several additional complications to the analysis (Broman et al. 2006; Wise et al. 2013), and a treatment that properly addresses confounding because of relatedness for SNPs on the X chromosome is beyond the scope of this study. After these filtering steps, we included 155 283 SNPs on Chromosomes 1 – 19 in the association analysis (the list of SNPs is available upon request). All physical base-pair positions are based on NCBI release 37 of the Mouse Genome Assembly (mm9). Mapped loci are also presented in the text using GRCm38/mm10.
Imputed SNP panel
The MDA SNP data together with the full DNA sequences of 11 lab strains from the Wellcome Trust Sanger mouse genomes project (Keane et al. 2011; Wong et al. 2012), plus the mouse reference genome (Church et al. 2009; Waterston et al. 2002), have been previously used to impute genotypes in 88 MDA lab strains in a higher density panel of 1 181 330 SNPs (Wang et al. 2012). We used this resource to ‘fine-map’ the QTL initially identified using the lower density MDA SNP panel. We downloaded imputed SNP data for 11.8 million markers on Chromosomes 1 – 19 (http://csbio.unc.edu/imputation) on 16 September 2014. Heterozygous genotypes and low-confidence genotypes (confidence score = 0) were set to missing, any SNPs with missing genotypes were discarded, SNPs that were not polymorphic in our inbred strains were removed and a small number of SNPs that had more than two alleles were also removed. We found that a large number of SNPs from the MDA SNP panel (329 210) were not included in the imputed SNP panel. Presumably, this was because they were highly correlated with SNPs that were already included. We combined these SNPs with the UNC SNPs to create a final SNP panel for fine-mapping. We also found that 53 211 SNPs were common to both MDA and UNC SNP panels. We compared the MDA genotypes against the UNC imputed genotypes for this overlapping set of SNPs and found only a very small discrepancy in these genotypes, in line with the error rate reported previously (Wang et al. 2012); only 142 out of 53 211 SNPs (0.3%) had one or more genotype discrepancies between the two panels. The discrepant SNPs were not used for the analysis.
QTL mapping
We took several steps to prepare the phenotype data for QTL mapping. We computed a linear model with terms representing each potential covariate, including sex, body weight, chamber number and cohort, to evaluate their effect(s) on the PPI phenotype. Cohort explained 0.6% of the variance; sex explained 0.2% and body weight explained 0.3%. Because cohort and sex had negligible effects on PPI, data were combined across these factors. Covariates explaining less than 2% of the variance were not used in the final linear model used for genetic mapping. Two of the five PPI testing chambers (boxes 3 and 5) explained ~3% of the variance in PPI phenotype, so we included binary indicators for these testing chambers as a covariate for subsequent analyses. Next, we inspected the phenotype residuals obtained by removing linear effects of covariates. We removed outlying data points that were more than two standard deviations away from the mean of each strain (N = 17). This ensures that statistics calculated in tests for phenotype –genotype correlations were not overly sensitive to unusually large or unusually small phenotype measurements. The residuals of our linear model had empirical quantiles that closely match expected quantiles under the normal distribution, suggesting that a normal distribution was a good fit. Figure S2 contains genome-wide associations from cohorts 1 and 2.
As the amount of the genome that is shared can vary considerably among the inbred mouse strains, these varying levels of relatedness are likely to confound our tests for phenotype –genotype correlations, leading to inflation of spurious associations (Astle & Balding 2009; Kang et al. 2008; Marchini et al. 2004; Newman et al. 2001; Price et al. 2010). To correct for confounding because of hidden relatedness in our genome-wide association analysis, we used a linear-mixed model approach implemented in the program GEMMA (v. 0.95 alpha) (Zhou & Stephens 2012). Linear mixed model (LMM)-based approaches for QTL mapping have emerged as a robust strategy to account for confounding because of the population structure (Astle & Balding 2009; Cheng et al. 2010, 2011; Kang et al. 2010; Lippert et al. 2011; Listgarten et al. 2012; Price et al. 2010; Yang et al. 2014; Yu et al. 2006).
We have at least three reasons for using GEMMA over other LMM-based approaches: (1) the numerical computations in GEMMA scale well to large numbers of markers and samples; (2) unlike other implementations of LMMs for genome-wide mapping [e.g. EMMAX (Kang et al. 2010), GRAMMAR (Aulchenko et al. 2007)], GEMMA avoids making approximations which lead to a reduction of power to detect QTL in certain circumstances; and (3) GEMMA only models additive effects of genetic markers (but not dominance), which is appropriate for our study design because our F1 mice only have two possible genotypes (B6/B6 or B6/non-B6). GEMMA yields calculations that are equivalent to exact test statistics [e.g. as implemented in EMMA (Kang et al. 2008)] but computes these statistics much faster. We expect that FaST-LMM would have been a good alternative (Lippert et al. 2011; Listgarten et al. 2012). We used the genotypes of the genetic markers to specify the genetic relationship matrix. A critical consideration for LMM-based association mapping is that including a genetic marker in the genetic similarity matrix K can deflate the test statistic for this marker, leading to a loss of power to detect a QTL. This phenomenon has been called ‘proximal contamination’ (Cheng et al. 2013; Listgarten et al. 2012). The basic intuition for this loss of power is that including the candidate SNP in the genetic relationship matrix makes the null log-likelihood higher when the SNP helps to explain variance in the trait. In human GWASs with smaller sample sizes, this loss in power is expected to be small (Yang et al. 2014). In our study, we expect that proximal contamination will have a larger impact on QTL detection because of the extended patterns of linkage disequilibrium and because we anticipate alleles with relatively large effect sizes. To avoid proximal contamination, we computed 19 different matrices, each one excluding one of the 19 autosomes [we have previously taken this approach in Parker et al. (2014)]. To scan markers on a given chromosome, we used the version of K that did not include that chromosome. This leave-one-chromosome-out (LOCO) approach provides a simple solution for avoiding the problem of proximal contamination (Cheng et al. 2013).
We report the evidence of a QTL at each SNP using the P value calculated from the likelihood-ratio test with 1 df (Zhou & Stephens 2012). Although other association tests are also implemented in GEMMA, we use the likelihood-ratio test because it is ‘well-behaved’ in that it controls for type 1 error; by contrast, the Wald and score test statistics for the LMM are not well-calibrated when the sample size (the number of genetically unrelated samples) is very small (Zhou & Stephens 2014). To calculate significance thresholds for the P values, we must first obtain the distribution of this test statistic under the null distribution. This distribution is commonly estimated by permuting the phenotype values relative to the genotypes (Broman & Sen 2009). However, standard permutation tests are based on the assumption that the samples are exchangeable and do not preserve the covariance structure in the samples because of population structure. Therefore, the permutation-based estimates may lead to inadequately stringent significance thresholds and inflated type 1 error rates (Abney et al. 2002; Aulchenko et al. 2007; Cheng & Palmer 2013; Cheng et al. 2010; Zou et al. 2005). Alternative permutation test procedures have been developed that preserve the relationship between the phenotype and polygenic effect (Abney 2015). However, we opted not to use this approach to assess significance because (1) these procedures are computationally intensive for the dense SNP sets in this study and (2) the accuracy of these permutation tests hinge on how well the polygenic covariance matrix captures the true covariance structure in the phenotype, and this accuracy may not be high given that the effect sample size (the number of inbred strains) is small. A simple alternative to this approach is to use a Bonferroni correction of the P values obtained from GEMMA, which controls for the probability of making at least one false discovery (‘family-wise error rate’). Bonferroni correction typically leads to overly stringent significance thresholds in GWASs because it ignores correlations between markers (i.e. it ignores the fact that the association tests are not independent). In our data, this threshold is easily improved by observing that we often have blocks of SNPs in complete or near-complete linkage disequilibrium. Therefore, we reduced the effective number of tests in the Bonferroni correction by pruning the set of markers so that no pair of markers has a (Pearson’s) correlation coefficient greater than 0.98. This reduced number of tests from 155 000 to about 35 000. Using this as our effective number of independent tests, P values of 1.4 × 10−6 , or −log10 P values of 5.86, yield Bonferroni-adjusted P values of 0.05; P values of 2.8 × 10−6 , yield Bonferroni-adjusted P values of 0.1. The full code and data for reproducing the analysis are available for download at http://palmerlab.org/protocols-data/.
Bioinformatics
The Mouse Genome Database at the Mouse Genome Informatics website (http://www.informatics.jax.org) was used to search for disease associations and mouse phenotypes for genes in mapped regions (Eppig et al. 2015). Coding polymorphisms, indels and structural variants in mapped regions were searched using Wellcome Trust mouse genomes project (Keane et al. 2011). Sorting intolerant from tolerant (SIFT) (http://sift.jcvi.org) was used to predict the impact of nonsynonymous coding variants (Kumar et al. 2009). The Allen Mouse Brain Atlas was used to identify evidence for brain gene expression (Lein et al. 2007).
eQTL data
We used gene expression data from the Hybrid Mouse Diversity Panel, a panel of 29 classical inbred strains (including 24 of the 30 strains used in our study) and 71 recombinant inbred strains. Expression data included 25 697 transcripts measured on the Illumina MouseRef-8 v2.0 expression beadchip for both hippocampus and striatum in male mice (Park et al. 2011). The hippocampus and striatum have key roles in the neural regulation of PPI of the startle response in rodents (Swerdlow et al. 2001). For each region, eQTL were defined using a genome-wide significance threshold corresponding to a false discovery rate/Q value <5% (Park et al. 2011). Transcript abundance values were normalized using the neqc() function from the LIMMA R package. A non-normalized version of expression array data is available from NCBI-GEO under Dataset record number GDS3900. Strains included in the eQTL data are given in Table S1.
Human phenotypic associations
We used the genome-wide repository of associations between SNPs and phenotypes (GRASP) to explore genes implicated in our study. The NHLBI GRASP catalog contains human genetic association results reported in primary manuscripts, their supplemental information and web-based content. Included studies use 25 000 or more markers to study one or more traits (Eicher et al. 2015; Leslie et al. 2014). Genome-wide repository of associations between SNPs and phenotypes v 2.0 contains searchable data from 2082 studies totaling 8.87 million association results extracted with a P < 0.05 threshold. Genome-wide repository of associations between SNPs and phenotypes v 2.0 searches were conducted via the web interface.
Results
Heritability and genome-wide association
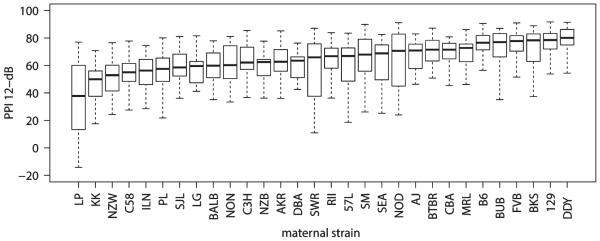
Prepulse inhibition was evaluated in 604 mice from 30 F1 strains (Fig. 1 and Fig. S1). In order to estimate the heritability (h2) of the PPI trait (12 db prepulse), we compared between- and within-strain variations from a one-way analysis of variance (ANOVA) of PPI given strain, resulting in an estimated heritability h2 = 0.32 (Hegmann & Possidente 1981) (Appendix S1). In addition, we also estimated h2 using all of the SNPs from the GWAS scan; this approach is often called ‘SNP heritability’ or ‘chip heritability’. The chip heritability was estimated at 0.43 with 95% posterior interval from 0.29 to 0.57. We suspect that the chip heritability may be higher because it can better account for the degree of divergence between each pair of strains in the study, whereas the other method implicitly assumes that all strains are equally unrelated.
Figure 1. Percent prepulse inhibition at 12 db across 30 F1 mouse strains.
Box-and-whisker plots indicate median (black bar), boxes indicate the upper and lower quartiles and whiskers indicate high and low values; N = 604 mice balanced across strain and sex.
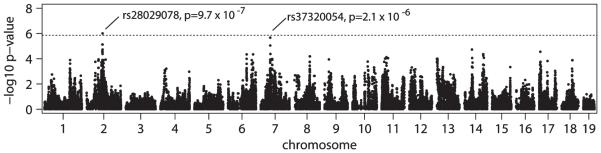
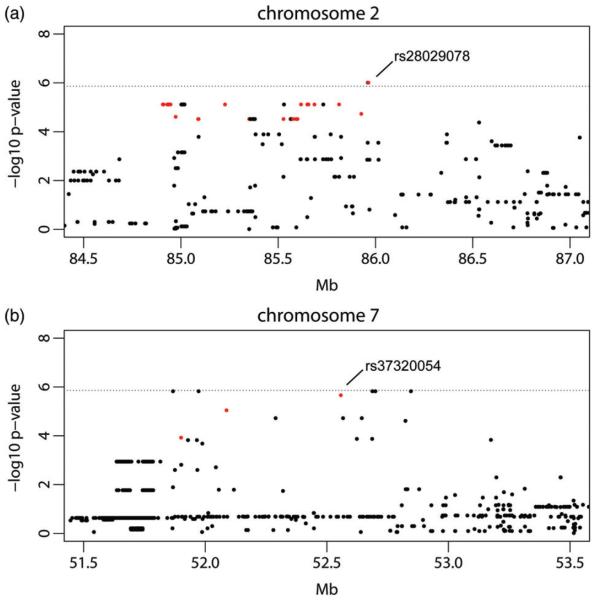
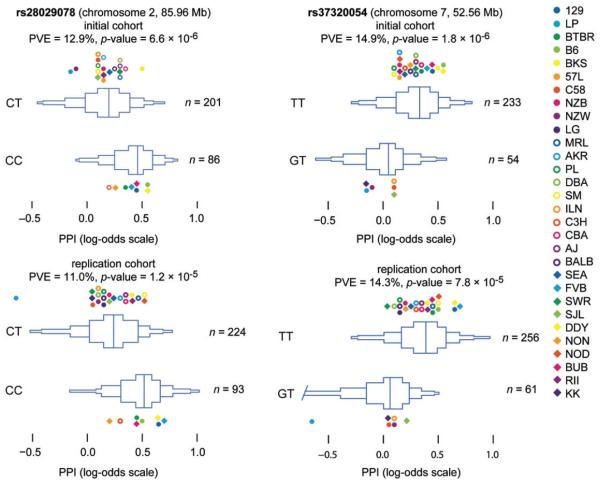
We performed a GWAS for PPI (12 db prepulse) using the set of 155 283 SNPs. We identified loci on Chromosomes 2 and 7 (Fig. 2). Using a conservative Bonferroni-adjusted correction for multiple testing, the locus on Chromosome 2 (P = 9.7 × 10−7) met the α< 0.05 level for genome-wide significance and the association on Chromosome 7 (P = 2.1 × 10−6) met the α< 0.1 threshold. To better establish the location of genetic variants underlying these QTL, we assessed support for PPI associations in a higher density panel of imputed SNPs. Several imputed SNPs within the Chromosome 7 QTL had lower P values than that obtained in the MDA mapping (imputed P = 1.5 × 10−6), and thus just exceeded the α< 0.05 threshold (P = 1.4 × 10−6). The implicated region, as defined by SNPs with log P values within two of the highest P value were Chr 2: 84.82 – 86.53 Mb (mm10: 84.97 – 86.69 Mb) and Chr 7: 51.82 – 52.85 Mb (mm10: 44.56 – 45.59 Mb) (Fig. 3). We examined the distribution of PPI phenotype across the 30 strains after removing the linear effects of covariates on phenotype (Fig. 4). The proportion of variance in PPI explained by genotype was estimated to be 12.0% for the Chromosome 2 locus and 14.6% for the Chromosome 7 locus.
Figure 2. Genome-wide scan for prepulse inhibition in F1 mice from 30 inbred strains.
Both the initial and the replication cohort data were used to calculate P values using a linear-mixed model that accounts for differences in genetic sharing among the F1 mice (N = 604). P values are shown on the log-scale for 155 283 SNPs on autosomes. The dotted line gives the α= 0.05 significance threshold after applying Bonferroni correction.
Figure 3. Region plots showing fine-mapped genetic loci for PPI.
Regions were identified on Chromosome 2 (a) and Chromosome 7 (b). Red SNPs are eQTL in hippocampus and striatum (see Table 1 and Table S2). A higher density panel of SNPs with imputed genotypes was used to further evaluate support for association at the loci identified in the genome-wide scan. Both initial and replication cohort data were used for fine-mapping (N = 604). The dotted line gives the α= 0.05 significance threshold after applying Bonferroni correction.
Figure 4. Distribution of (logit-transformed) PPI in F1 crosses.
Data from the initial (a) and replication (b) cohorts, stratified by genotype for SNP rs28029078 (left-hand panels) and SNP rs37320054 (right-hand panels). Box-percentile plots show the distribution of PPI across all the individual F1 samples. Dots give strain means over genetically identical F1 crosses, colored by strain. Proportion of variance explained (PVE) by genotype is estimated for each SNP after removing linear effects of covariates on phenotype.
Informatics
The locus on Chromosome 2 contains 7 genes plus a large cluster of olfactory receptor genes, and the Chromosome 7 contains approximately 100 coding genes. Although many of these are expressed in the brain, none of these genes have been previously reported to influence the PPI phenotype. Mice with a null allele of Slc17a7, which is within the Chromosome 7 locus, have an altered startle response but also exhibit severe neurological deflcits that we did not observe in this study (Fremeau 2004). There are no reported coding polymorphisms in Slc17a7 among the panel of strains we used. Several other genes within the Chromosome 7 locus have been previously implicated in behavioral phenotypes in mouse models: Shank1, Emc10, IL4i1 and Cpt1c. Of these, only Shank1 has a missense variant predicted to have a ‘moderate’ impact, but the allelic distribution of this variant within our panel of strains does not match that of the peak PPI-associated variant (data not shown).
We searched the Wellcome Trust mouse genomes project for additional potentially deleterious coding variants with strain distribution patterns similar to our PPI-associated SNPs. We found that Olfr1014 and Olfr1015 each contained one predicted deleterious variant. Olfactory receptor (Olfr -) genes represent a large family of more than a thousand genes that are believed to mediate the detection of odorants (Buck & Axel 1991); these genes are unlikely to underlie our QTL for PPI. There was one predicted deleterious coding variant each in Zfp473 and Hrc, but according to the Allen Brain Atlas, these genes do not appear to be expressed in the mouse brain. Known indels within the mapped regions were all limited to introns. We did not identify evidence suggesting that the homologous human genes/regions have been strongly associated with any psychiatric diseases.
eQTL at PPI-associated markers
As described in the previous section, there were no coding polymorphisms likely to underlie the loci on Chromosomes 2 and 7. Another possibility is that these loci influence gene expression, rather than protein coding. Expression polymorphisms are enriched among complex trait-associated polymorphisms (Nicolae et al. 2010), so we considered the possible role for regulatory variation affecting gene expression. We used public eQTL data from the HMDP to determine whether the SNPs associated with PPI were eQTL in hippocampus and/or striatum. The SNP on Chromosome 2 that was most strongly associated with PPI (rs28029078) was strongly associated with the expression of five transcripts in both the hippocampus and the striatum, Fam171b, Timm10, Arfgap2, Rbm45 and Acp2 (Table 1). Within the confidence intervals of the association on Chromosome 2, there were 31 additional SNPs with more modest P values that were also eQTL for these same five transcripts (Fig. 3 and Table S2). The SNP on Chromosome 7 that was most strongly associated with PPI (rs37320054) was associated with hippocampal and/or striatal expression of four transcripts: Prkd3, Smarcad1, Htatip2 and Gtf2h1 (Table 1).
Table 1.
Brain expression QTL at peak prepulse inhibition-associated markers
| Marker | eQTL P value, region |
Location | Gene | Psychiatric GWAS (human) |
GWAS P value (human) |
|---|---|---|---|---|---|
| rs28029078 (Chr 2) | 1.3E–14 HIP 1.9E–19 STR |
cis | Fam171b | – | – |
| 6.0E–12 HIP 2.2E–16 STR |
cis | Timm10 | – | – | |
| 2.9E–09 HIP 9.2E–07 STR |
trans | Acp2 | – | – | |
| 2.3E–09 HIP 3.5E–11 STR |
trans | Arfgap2 | – | – | |
| 1.2E–09 HIP 8.8E–12 STR |
trans | Rbm45 | – | – | |
| rs37320054 (Chr 7) | 3.7E–08 HIP 1.9E –08 STR |
trans | Prkd3 | Schizophrenia (Ripke et al. 2013) |
6.7 E–09 |
| 7.4E–07 STR | trans | Smarcad1 | Bipolar disorder and schizophrenia (Wang et al. 2010) |
3.7E–06 | |
| 6.9E–08 STR | cis | Htatip2 | – | – | |
| 3.5E–09 STR | trans | Gtf2h1 | – | – |
Hybrid mouse diversity panel eQTL were determined in hippocampus (HIP) and striatum (STR). Genes with start position ±3Mb from eQTL were called cis-eQTL. Genome-wide repository of associations between SNPs and phenotypes (GRASP) was used to determine associations with psychiatric phenotypes.
Phenotypic associations of genes with eQTL at PPI-associated markers
Next, we asked whether genes with eQTL at the PPI-associated SNPs had been previously implicated in human disease. Variants in two of the eQTL genes have been associated with psychiatric phenotypes in humans: Prkd3 and Smarcad1. Prkd3 resides within a linkage disequilibrium block in humans that have been significantly associated with schizophrenia (Ripke et al. 2013). Smarcad1 was suggestively associated with bipolar disorder and schizophrenia in a European-American sample (P = 3.7 × 10−6; Wang et al. 2010), and it is one of a groups of Wnt-signaling genes implicated in bipolar disorder (Pandey et al. 2012).
Discussion
We conducted genome-wide association in F1 mice to generate new hypotheses about the genetic basis of PPI, which is an endophenotype for multiple psychiatric diseases. The mice were derived from 30 common inbred strains and were produced and tested in two independent cohorts. We identified loci on Chromosomes 2 and 7 that explained 12% and 14% of the variance in PPI, respectively. Given the absence of coding polymorphisms that could explain these associations, we examined eQTL data from the HMDP. The most strongly associated SNP on Chromosome 2 is also associated with the expression of five transcripts in both hippocampus and striatum. The most strongly associated SNP on Chromosome 7 is associated with the expression of four transcripts, one of which has a human homologue that is implicated in schizophrenia and another which has been implicated in bipolar disorder. The results of this study generate new biological hypotheses about the genes underlying PPI and thus identify genes which might be involved in the etiology of human psychiatric diseases for which successful human GWAS are nascent or have met technical limitations. More broadly, these results show that panels of inbred F1 mice can be used to efficiently map other complex behavioral traits.
Our effective sample size of 30 different inbred genotypes is small and has limited power to detect QTL (Kirby et al., 2010; Bennett et al., 2010); as a consequence the genetic associations we identified had large effect sizes (12 – 14%). Moreover, the Bonferroni-corrected genome-wide significance threshold used here was conservatively chosen such that no pair of markers had a correlation coefficient greater than 0.98; this further reduced our power. The association on Chromosome 2 met the α< 0.05 significance criterion, while the evidence for association at the Chromosome 7 locus met the α< 0.1 criterion at our conservative significance threshold. It is likely that adding more strains to the panel would increase power, which has been part of the motivation behind other experimental designs such as the BXD recombinant inbred strains (Andreux et al. 2012; Peirce et al. 2004), and the HMDP, which is composed of more than 100 strains (Bennett et al. 2010; Ghazalpour et al. 2012; Park et al. 2011).
This study is the first to use a panel of F1 mice to map complex phenotypes. This approach differs in several ways from using a panel of traditional inbred mice. We were able to produce these F1 mice more easily because we only had to obtain females from the 30 inbred strains, and we obtained larger litters, presumably because of hybrid vigor. Similar to a panel of fully inbred strains, genotypes could be inferred from publicly available genotype data. The results were significant and identified small regions (approximately 1 – 2 Mb), even when using conservative confidence intervals. The size of our mapped regions is consistent with the size of ancestral haplotype/linkage disequilibrium blocks across inbred mouse strains (Kirby et al. 2010). Given that the mice in this population are F1s, phenotypic differences across strains could be because of direct genetic effects, maternal factors (which are indirect genetic effects) and/or parent-of-origin effects.
The loci that we mapped in this study have not been previously implicated in PPI in other QTL studies in mice or rats (Hitzemann et al. 2008; Leussis et al. 2009; Loos et al. 2012; Palmer et al. 2003; Petryshen et al. 2005; Samocha et al. 2010; Webb et al. 2009). This could be because of minor differences in PPI testing protocols, different statistical methods, a lack of power to detect true positive associations and the use of different mouse strains/crosses across studies, including the new population of F1s used here. The most similar study to our own mapped PPI in silico in a large panel of inbred strains, 25 of which were also used here (Webb et al. 2009). That study used publicly available PPI data for 37 strains and ~17k SNPs from various sources (Willott et al. 2003) and identified QTL on Chromosomes 1 and 13, neither of which show evidence for association here. Differences between these two studies include our use of F1 mice rather than fully inbred individuals, the SNPs used for phenotypic association, the statistical procedures and the thresholds for significance.
The most comprehensive available databases did not show coding variants likely to explain our observed associations. We found instead that eQTL for multiple brain transcripts overlap with the behavioral QTL. Most eQTLs have an additive genetic architecture, suggesting that the use of inbred strain expression data was appropriate although the phenotypic data were derived from F1s rather than fully inbred strains. The peak PPI-associated SNP on Chromosome 7 is an eQTL for Prkd3 and Smarcad1, which are within regions previously implicated in schizophrenia and bipolar disorder by human GWASs (Ripke et al. 2013; Wang et al. 2010). Genes that have eQTL at our Chromosome 2 locus included Fam171b, Timm10, Arfgap2, Rbm45 and Acp2. Acp2 is involved in the development of cerebellum (Bailey et al. 2014). Rbm45 is found in neuronal and glial nuclear inclusions in patients with several neurodegenerative disorders (Collins et al. 2012). Arfgap2 is involved in vesicle formation and trafficking (Frigerio et al. 2007; Kartberg et al. 2010) and is a protein binding partner of secretagogin, a calcium-binding protein within the calmodulin superfamily (Bauer et al. 2011). Of all the eQTL co-localizing with the top PPI-associated SNPs, Fam171b was the strongest (see Table 1, Table S2). A member of a family of secreted proteins, Fam171b exhibits intense and selective expression levels in brain but its function has not yet been studied. As a result of these characteristics, Fam171b is one of a set of 106 genes that has been described as the ‘core brain ignorome’ (Pandey et al. 2014). Because of its putative association with PPI and its potential impact on brain functions, Fam171b may be an especially interesting candidate for knockout and other biological studies.
In summary, we used high-density SNP data downloaded from public databases to conduct genome-wide association in F1 inbred mice, leading to the identification of several small (<2 Mb) loci associated with PPI. We then used extant eQTL data collected in the same inbred strains that formed our F1 panel to efficiently narrow down possible candidate genes within each locus. This study shows two advantages of using F1 mice. First, breeding is less time-consuming and expensive because female but not male inbred mice from 30 strains were purchased. Second, one may use existing genotype and expression data to inform the genetic analysis. This same approach can be applied to other complex traits as a complement to human GWAS and as a means of generating new biological hypotheses.
Supplementary Material
Acknowledgments
The authors acknowledge the helpful statistical advice obtained from Shyam Gopalakrishnan, Xiang Zhou, Mark Abney, Riyan Cheng, Eleazar Eskin, Matthew Stephens, Heejung Shim, Robert Williams and Karl Broman. We thank John Zekos for maintaining the compute cluster. We thank Alexander Ho and Philip Stephens for technical assistance. This work was funded by the National Institutes of Health (F32 MH102931 to L.J.S.) and (R21 MH102728 to A.A.P.).
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s web-site:
Appendix S1: Methods for Heritability estimation.
Figure S1: (a) Startle, (b) PPI 3 db and (c) PPI 6 db in 30 F1 strains. Box-and-whisker plots indicate median (black bar), upper and lower quartiles (boxes) and high and low values (whiskers); N = 604 mice balanced across strain and sex. (d) Mean PPI ± standard error of the mean for each prepulse intensity (3, 6 and 12 db).
Figure S2: Genome-wide scans of prepulse inhibition in F1 mice from the initial cohort (N = 287) and the replication cohort (N = 317). The dotted lines give the significance threshold after applying Bonferroni correction for multiple testing (α= 0.05).
Table S1: Numbers of F1 mice. Our goal was to have five male and five female mice per strain per cohort.
Table S2: Genes with expression QTL within confidence intervals of PPI associations.
The authors declare that they have no competing interests.
References
- Abney M. Permutation testing in the presence of polygenic variation. Genet Epidemiol. 2015;39:249–258. doi: 10.1002/gepi.21893. DOI:10.1002/gepi.21893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abney M, Ober C, McPeek MS. Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites. Am J Hum Genet. 2002;70:920–934. doi: 10.1086/339705. DOI:10.1086/339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy M-F, Henry H, Schoonjans K, Williams RW, Auwerx J. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. DOI:10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astle W, Balding DJ. Population structure and cryptic relatedness in genetic association studies. Stat Sci. 2009;24:451–471. [Google Scholar]
- Aulchenko YS, de Koning D-J, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177:577–585. doi: 10.1534/genetics.107.075614. DOI:10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey K, Rahimi Balaei M, Mannan A, Del Bigio MR, Marzban H. Purkinje cell compartmentation in the cerebellum of the lysosomal acid phosphatase 2 mutant mouse (nax – naked-ataxia mutant mouse) PLoS One. 2014;9:e94327. doi: 10.1371/journal.pone.0094327. DOI:10.1371/journal.pone.0094327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MC, O’Connell DJ, Maj M, Wagner L, Cahill DJ, Linse S. Identification of a high-affinity network of secretagogin-binding proteins involved in vesicle secretion. Mol Biosyst. 2011;7:2196–2204. doi: 10.1039/c0mb00349b. DOI:10.1039/c0mb00349b. [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang W, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. DOI:10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A, Cario CL, Silva KA, Kennedy VE, Harrison DE, Paigen B, Sundberg JP. Identification of fat4 and tsc22d1 as novel candidate genes for spontaneous pulmonary adenomas. Cancer Res. 2011;71:5779–5791. doi: 10.1158/0008-5472.CAN-11-1418. DOI:10.1158/0008-5472.CAN-11-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff D, Stone C, Callaway E, Geyer M, Glick I, Bali L. Prestimulus effects on human startle reflex in normals and schizophrenics. Psychophysiology. 1978;15:339–343. doi: 10.1111/j.1469-8986.1978.tb01390.x. [DOI] [PubMed] [Google Scholar]
- Braff DL, Geyer MA, Swerdlow NR. Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology (Berl) 2001;156:234–258. doi: 10.1007/s002130100810. [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. Springer; New York: 2009. [Google Scholar]
- Broman KW, Sen S, Owens SE, Manichaikul A, Southard-Smith EM, Churchill GA. The X chromosome in quantitative trait locus mapping. Genetics. 2006;174:2151–2158. doi: 10.1534/genetics.106.061176. DOI:10.1534/genetics.106.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. DOI:10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- Cheng R, Palmer AA. A simulation study of permutation, bootstrap, and gene dropping for assessing statistical significance in the case of unequal relatedness. Genetics. 2013;193:1015–1018. doi: 10.1534/genetics.112.146332. DOI:10.1534/genetics.112.146332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics. 2010;185:1033–1044. doi: 10.1534/genetics.110.116863. DOI:10.1534/genetics.110.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Abney M, Palmer AA, Skol AD. QTLRel: an R package for genome-wide association studies in which relatedness is a concern. BMC Genet. 2011;12 doi: 10.1186/1471-2156-12-66. DOI:10.1186/1471-2156-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Parker CC, Abney M, Palmer AA. Practical considerations regarding the use of genotype and pedigree data to model relatedness in the context of genome-wide association studies. G3 (Bethesda) 2013;3:1861–1867. doi: 10.1534/g3.113.007948. DOI:10.1534/g3.113.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ. In silico mapping of mouse quantitative trait loci. Science. 2001;294:2423. doi: 10.1126/science.294.5551.2423a. DOI:10.1126/science.294.5551.2423a. [DOI] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. DOI:10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M, Riascos D, Kovalik T, An J, Krupa K, Krupa K, Hood BL, Conrads TP, Renton AE, Traynor BJ, Bowser R. The RNA-binding motif 45 (RBM45) protein accumulates in inclusion bodies in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) patients. Acta Neuropathol. 2012;124:717–732. doi: 10.1007/s00401-012-1045-x. DOI:10.1007/s00401-012-1045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulawa SC, Geyer MA. Psychopharmacology of prepulse inhibition in mice. Chin J Physiol. 1996;39:139–146. [PubMed] [Google Scholar]
- Eicher JD, Landowski C, Stackhouse B, Sloan A, Chen W, Jensen N, Lien J-P, Leslie R, Johnson AD. GRASP v2.0: an update on the genome-wide repository of associations between SNPs and phenotypes. Nucleic Acids Res. 2015;43:D799–D804. doi: 10.1093/nar/gku1202. DOI:10.1093/nar/gku1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The mouse genome database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–D736. doi: 10.1093/nar/gku967. DOI:10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber CR, Bennett BJ, Orozco L, Zou W, Lira A, Kostem E, Kang HM, Furlotte N, Berberyan A, Ghazalpour A, Suwanwela J, Drake TA, Eskin E, Wang QT, Teitelbaum SL, Lusis AJ. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 2011;7:e1002038. doi: 10.1371/journal.pgen.1002038. DOI:10.1371/journal.pgen.1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Eskin E. Genome-wide association studies in mice. Nat Rev Genet. 2012;13:807–817. doi: 10.1038/nrg3335. DOI:10.1038/nrg3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremeau RT. Vesicular glutamate transporters 1 and 2 target to functionally distinct synaptic release sites. Science. 2004;304:1815–1819. doi: 10.1126/science.1097468. DOI:10.1126/science.1097468. [DOI] [PubMed] [Google Scholar]
- Frigerio G, Grimsey N, Dale M, Majoul I, Duden R. Two human ARFGAPs associated with COP-I-coated vesicles. Traffic. 2007;8:1644–1655. doi: 10.1111/j.1600-0854.2007.00631.x. DOI:10.1111/j.1600-0854.2007.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer MA, Krebs-Thomson K, Braff DL, Swerdlow NR. Pharmacological studies of prepulse inhibition models of sensorimotor gating deficits in schizophrenia: a decade in review. Psychopharmacology (Berl) 2001;156:117–154. doi: 10.1007/s002130100811. [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Rau CD, Farber CR, et al. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm Genome. 2012;23:680–692. doi: 10.1007/s00335-012-9411-5. DOI:10.1007/s00335-012-9411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe A, Germer S, Usuka J, Aud D, Belknap JK, Klein RF, Ahluwalia MK, Higuchi R, Peltz G. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–1918. doi: 10.1126/science.1058889. DOI:10.1126/science.1058889. [DOI] [PubMed] [Google Scholar]
- Hegmann JP, Possidente B. Estimating genetic correlations from inbred strains. Behav Genet. 1981;11:103–114. doi: 10.1007/BF01065621. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Malmanger B, Belknap J, Darakjian P, McWeeney S. Short-term selective breeding for high and low prepulse inhibition of the acoustic startle response; pharmacological characterization and QTL mapping in the selected lines. Pharmacol Biochem Behav. 2008;90:525–533. doi: 10.1016/j.pbb.2008.04.004. DOI:10.1016/j.pbb.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. DOI:10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S-Y, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. DOI:10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartberg F, Asp L, Dejgaard SY, Smedh M, Fernandez-Rodriguez J, Nilsson T, Presley JF. ARFGAP2 and ARFGAP3 are essential for COPI coat assembly on the Golgi membrane of living cells. J Biol Chem. 2010;285:36709–36720. doi: 10.1074/jbc.M110.180380. DOI:10.1074/jbc.M110.180380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. DOI:10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A, Kang HM, Wade CM, Cotsapas C, Kostem E, Han B, Furlotte N, Kang EY, Rivas M, Bogue MA, Frazer KA, Johnson FM, Beilharz EJ, Cox DR, Eskin E, Daly MJ. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics. 2010;185:1081–1095. doi: 10.1534/genetics.110.115014. DOI:10.1534/genetics.110.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. DOI:10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Larsen DK, Norton GR, Walker JR, Stein MB. Analysis of startle responses in patients with panic disorder and social phobia. Cogn Behav Ther. 2002;31:156–169. DOI:10.1080/165060702321138555. [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. DOI:10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Leslie R, O’Donnell CJ, Johnson AD. GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics. 2014;30:i185–i194. doi: 10.1093/bioinformatics/btu273. DOI:10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leussis MP, Frayne ML, Saito M, Berry EM, Aldinger KA, Rockwell GN, Hammer RP, Baskin-Hill AE, Singer JB, Nadeau JH, Sklar P, Petryshen TL. Genomic survey of prepulse inhibition in mouse chromosome substitution strains. Genes Brain Behav. 2009;8:806–816. doi: 10.1111/j.1601-183X.2009.00526.x. DOI:10.1111/j.1601-183X.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Wang J, Guo J, Allard J, Cheng J, Ng A, Shafer S, Puech A, McPherson JD, Foernzler D, Peltz G, Usuka J. In silico genetics: identification of a functional element regulating H2-Ealpha gene expression. Science. 2004;306:690–695. doi: 10.1126/science.1100636. DOI:10.1126/science.1100636. [DOI] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. DOI:10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- Listgarten J, Lippert C, Kadie CM, Davidson RI, Eskin E, Heckerman D. Improved linear mixed models for genome-wide association studies. Nat Methods. 2012;9:525–526. doi: 10.1038/nmeth.2037. DOI:10.1038/nmeth.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y, Liu Y, You M. Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet. 2006;38:888–895. doi: 10.1038/ng1849. DOI:10.1038/ng1849. [DOI] [PubMed] [Google Scholar]
- Loos M, Staal J, Pattij T, Smit AB, Spijker S. Independent genetic loci for sensorimotor gating and attentional performance in BXD recombinant inbred strains. Genes Brain Behav. 2012;11:147–156. doi: 10.1111/j.1601-183X.2011.00754.x. DOI:10.1111/j.1601-183X.2011.00754.x. [DOI] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. DOI:10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- Mott R, Flint J. Dissecting quantitative traits in mice. Annu Rev Genomics Hum Genet. 2013;14:421–439. doi: 10.1146/annurev-genom-091212-153419. DOI:10.1146/annurevgenom-091212-153419. [DOI] [PubMed] [Google Scholar]
- Newman DL, Abney M, McPeek MS, Ober C, Cox NJ. The importance of genealogy in determining genetic associations with complex traits. Am J Hum Genet. 2001;69:1146–1148. doi: 10.1086/323659. DOI:10.1086/323659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000888. DOI:10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AA, Breen LL, Flodman P, Conti LH, Spence MA, Printz MP. Identification of quantitative trait loci for prepulse inhibition in rats. Psychopharmacology (Berl) 2003;165:270–279. doi: 10.1007/s00213-002-1258-0. DOI:10.1007/s00213-002-1258-0. [DOI] [PubMed] [Google Scholar]
- Pandey A, Davis NA, White BC, Pajewski NM, Savitz J, Drevets WC, McKinney BA. Epistasis network centrality analysis yields pathway replication across two GWAS cohorts for bipolar disorder. Transl Psychiatry. 2012;2:e154. doi: 10.1038/tp.2012.80. DOI:10.1038/tp.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, Lu L, Wang X, Homayouni R, Williams RW. Functionally enigmatic genes: a case study of the brain ignorome. PLoS One. 2014;9:e88889. doi: 10.1371/journal.pone.0088889. DOI:10.1371/journal.pone.0088889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CC, Gale GD, de Jong S, Ghazalpour A, Bennett BJ, Farber CR, Langfelder P, Lin A, Khan AH, Eskin E, Horvath S, Lusis AJ, Ophoff RA, Smith DJ. Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC Syst Biol. 2011;5:43. doi: 10.1186/1752-0509-5-43. DOI:10.1186/1752-0509-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Carbonetto P, Sokoloff G, Park YJ, Abney M, Palmer AA. High-resolution genetic mapping of complex traits from a combined analysis of F2 and advanced intercross mice. Genetics. 2014;198:103–116. doi: 10.1534/genetics.114.167056. DOI:10.1534/genetics.114.167056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. DOI:10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry W, Minassian A, Feifel D, Braff DL. Sensorimotor gating deficits in bipolar disorder patients with acute psychoticmania. Biol Psychiatry. 2001;50:418–424. doi: 10.1016/s0006-3223(01)01184-2. [DOI] [PubMed] [Google Scholar]
- Perry W, Minassian A, Feifel D, Braff DL. Sensorimotor gating deficits in adults with autism. Biol Psychiatry. 2007;61:482–486. doi: 10.1016/j.biopsych.2005.09.025. DOI:10.1016/j.biopsych.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. DOI:10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryshen TL, Kirby A, Hammer RP, Purcell S, O’Leary SB, Singer JB, Hill AE, Nadeau JH, Daly MJ, Sklar P. Two quantitative trait loci for prepulse inhibition of startle identified on mouse chromosome 16 using chromosome substitution strains. Genetics. 2005;171:1895–1904. doi: 10.1534/genetics.105.045658. DOI:10.1534/genetics.105.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. DOI:10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. DOI:10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. DOI:10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Lim JE, Cheng R, Sokoloff G, Palmer AA. Fine mapping of QTL for prepulse inhibition in LG/J and SM/J mice using F2 and advanced intercross lines. Genes Brain Behav. 2010;9:759–767. doi: 10.1111/j.1601-183X.2010.00613.x. DOI:10.1111/j.1601-183X.2010.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow NR, Geyer MA, Braff DL. A preliminary assessment of sensorimotor gating in patients with obsessive compulsive disorder. Biol Psychiatry. 1993;33:298–301. doi: 10.1016/0006-3223(93)90300-3. [DOI] [PubMed] [Google Scholar]
- Swerdlow NR, Geyer MA, Braff DL. Neural circuit regulation of prepulse inhibition of startle in the rat: current knowledge and future challenges. Psychopharmacology (Berl) 2001;156:194–215. doi: 10.1007/s002130100799. [DOI] [PubMed] [Google Scholar]
- Wang K-S, Liu X-F, Aragam N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res. 2010;124:192–199. doi: 10.1016/j.schres.2010.09.002. DOI:10.1016/j.schres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Wang JR, de Villena FP-M, Lawson HA, Cheverud JM, Churchill GA, McMillan L. Imputation of singlenucleotide polymorphisms in inbred mice using local phylogeny. Genetics. 2012;190:449–458. doi: 10.1534/genetics.111.132381. DOI:10.1534/genetics.111.132381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. DOI:10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Webb BT, McClay JL, Vargas-Irwin C, York TP, van den Oord EJCG. In silico whole genome association scan for murine prepulse inhibition. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005246. DOI:10.1371/journal.pone.0005246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willott JF, Tanner L, O’Steen J, Johnson KR, Bogue MA, Gagnon L. Acoustic startle and prepulse inhibition in 40 inbred strains of mice. Behav Neurosci. 2003;117:716–727. doi: 10.1037/0735-7044.117.4.716. [DOI] [PubMed] [Google Scholar]
- Wise AL, Gyi L, Manolio TA. eXclusion: toward integrating the X chromosome in genome-wide association analyses. Am J Hum Genet. 2013;92:643–647. doi: 10.1016/j.ajhg.2013.03.017. DOI:10.1016/j.ajhg.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Bumpstead S, Weyden L, Reinholdt LG, Wilming LG, Adams DJ, Keane TM. Sequencing and characterization of the FVB/NJ mouse genome. Genome Biol. 2012;13:R72. doi: 10.1186/gb-2012-13-8-r72. DOI:10.1186/gb-2012-13-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell TA, Paigen BJ, Graber JH, Villena FP, Churchill GA. A customized and versatile high-density genotyping array for the mouse. Nat Methods. 2009;6:663–666. doi: 10.1038/nmeth.1359. DOI:10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH-T, Nachman MW, Pialek J, Tucker P, Boursot P, McMillan L, Churchill GA, de Villena FP-M. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43:648–655. doi: 10.1038/ng.847. DOI:10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet. 2014;46:100–106. doi: 10.1038/ng.2876. DOI:10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. DOI:10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–824. doi: 10.1038/ng.2310. DOI:10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Methods. 2014;11:407–409. doi: 10.1038/nmeth.2848. DOI:10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F, Gelfond JA, Airey DC, Lu L, Manly KF, Williams RW, Threadgill DW. Quantitative trait locus analysis using recombinant inbred intercrosses: theoretical and empirical considerations. Genetics. 2005;170:1299–1311. doi: 10.1534/genetics.104.035709. DOI:10.1534/genetics.104.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.