Abstract
Persimmon fruit are unique in accumulating proanthocyanidins (tannins) during development, which cause astringency in mature fruit. In ‘Mopanshi’ persimmon, astringency can be removed by treatment with 95% CO2, which increases the concentrations of ethanol and acetaldehyde by glycolysis, and precipitates the soluble tannin. A TGA transcription factor, DkTGA1, belonging to the bZIP super family, was isolated from an RNA-seq database and real-time quantitative PCR indicated that DkTGA1 was up-regulated by CO2 treatment, in concert with the removal of astringency from persimmon fruit. Dual-luciferase assay revealed that DkTGA1 had a small (less than 2-fold), but significant effect on the promoters of de-astringency-related genes DkADH1, DkPDC2 and DkPDC3, which encode enzymes catalyzing formation of acetaldehyde and ethanol. A combination of DkTGA1 and a second transcription factor, DkERF9, shown previously to be related to de-astringency, showed additive effects on the activation of the DkPDC2 promoter. Yeast one-hybrid assay showed that DkERF9, but not DkTGA1, could bind to the DkPDC2 promoter. Thus, although DkTGA1 expression is positively associated with persimmon fruit de-astringency, trans-activation analyses with DkPDC2 indicates it is likely to act by binding indirectly DkPDC2 promoter, might with helps of DkERF9.
Introduction
Persimmon (Diospyros kaki L.) is a crop of high economic importance in China. There are two main types, either non-astringent or astringent [1]. However, most persimmon fruit have astringency at the commercial mature stage [2], caused by accumulation of soluble condensed tannins (proanthocyanidins, SCTs). Artificial postharvest treatments have been developed to remove astringency, including high CO2 (usually >90%), ethylene and ethanol [3–7]. CO2 is the most effective treatment and depends on the production of acetaldehyde [6,8,9].
The biochemical basis of postharvest de-astringency in persimmon has been comprehensively investigated. The activities of pyruvate decarboxylase (PDC, EC 4.1.1.1) and alcohol dehydrogenase (ADH, EC 1.1.1.1) have been shown to increase during CO2 treatment [6,10,11] leading to the formation of acetaldehyde and ethanol from pyruvate [12–14] and the acetaldehyde produced precipitates the soluble tannins. More recently, the genes involved in this process have been investigated. Five PDC genes and three ADH genes have been isolated from persimmon and their expression shown to be induced by the application of CO2 [6]. Transient over-expression of DkPDC2 rapidly decreased SCT in persimmon leaves [6], thus, DkPDC2 is an important structural gene for persimmon fruit de-astringency. Five transcription factors were reported to be involved in persimmon de-astringency regulation, including four ethylene responsive factor genes (DkERF9/10/19/22) and a MYB transcription factor (DkMYB6) [6,7,15]. All five transcription factor could transcriptionally activated de-astringency related target genes, meanwhile DkMYB6 could also enhance activity of the DkERF19 promoter, which suggested possible transcriptional regulatory cascades involving different transcription factors. Thus, de-astringency is based on the induction under anaerobic conditions of genes leading to acetaldehyde production, but more information about the transcriptional regulation of these processes is still limited.
In Arabidopsis, at least four ERF genes, including HRE1, HRE2, RAP2.2 and RAP2.12 have been identified as the main plant oxygen-sensing regulators. HRE1 and RAP2.2 could transcriptionally regulate the anaerobic fermentation-related ADH and PDC genes [16,17], and lines containing T-DNA knockouts of RAP2.2 had lower survival rates than the wild type in Arabidopsis [16]. Moreover, some other transcription factors have also been reported as hypoxia responsive, such as AtMYB2, which is induced by hypoxia, can bind to the promoter of AtADH1 [18], and increased the expression of AtADH1 when overexpressed [19]. Also, in wheat, TaMYB1has been shown to be responsive to low oxygen [20] and recent omics-based studies have highlighted more hypoxia responsive genes, and RNA-seq has been used to identify more than 180 transcription factors differentially expressed in response to low oxygen [21]. Thus, it is likely that more transcription factors would be expected to be involved in persimmon fruit postharvest de-astringency.
In the present research, a TGA-like transcription factor, DkTGA1, was identified, using data from the RNA-seq analysis performed by Min et al. [7]. The expression pattern of DkTGA1 in response to the application of 95% CO2 was studied by real-time PCR. The potential role(s) of DkTGA1 in regulating previously characterized deastringency-related target genes were investigated, using dual-luciferase assay, testing synergistic interactions between DkTGA1 and DkERF genes and yeast one hybrid analysis.
Materials and Methods
Plant materials and treatment
Commercial mature persimmon (Diospyros kaki L. cv. Mopanshi) fruit were harvested and bought from a commercial orchard (Yangjingliang’s farm) at Fangshan (Beijing, China) in 2010. We confirm that the field studies did not involve endangered or protected species. Fruit were transported to the laboratory at Zhejiang University (Hangzhou, China) and then were selected with uniform size, free from visible mechanical wounds for postharvest treatments. Selected fruit were divided into two batches, one treated with 95% CO2 for the de-astringency and the other (control) placed in air, with both batches held in air-tight plastic containers for 1 d. After treatments, the fruit were held in air at 20°C for 4 d. All of the treatments were performed with three biological replicates. At each sampling point, fruit flesh samples were separately taken from three replicates of four fruit each. The samples were frozen in liquid nitrogen and stored at -80°C until further use.
Fruit physiology evaluation
In order to reflect fruit astringency, SCTs were measured using Folin-Ciocalteu reagent (Sigma) according to the method described by Yin et al. [5]. The results were expressed as tannin acids equivalents per g-1 fresh weight.
Acetaldehyde and ethanol production were measured with a gas chromatograph (Agilent 6890N, USA), fitted with a FID column (HP-INNOWAX, 0.25mm, 30m, 0.25μm, Agilent J&W, CA, USA). Measurements were conducted according to our previous report [6]. In brief, 2 g frozen flesh sample was ground in liquid nitrogen and added to 5 ml saturated NaCl. Three ml of the mixture were transferred to 10 ml air-tight vials with crimp-top caps, and were incubated at 60°C in a water-bath for 1 h. Then, 0.2 ml head-space gas was removed for gas chromatography. The injector, detector and oven temperatures were set at 150, 160 and 100°C, respectively. Sec-butyl alcohol (Sigma) was used as an internal standard. The results were calculated using standard curves for acetaldehyde and ethanol, respectively.
RNA extraction and cDNA synthesis
Total RNA was extracted from frozen fruit flesh (2.0 g) by the method developed by Chang et al. [22]. The trace amount of genomic DNA in total RNA was digested with TURBO DNA free kit (Ambion). First strand cDNA was synthesized from 1.0 μg DNA-free RNA, using iScript cDNA Synthesis Kit (Bio-Rad). For each sampling point, three biological replicates were used for RNA extraction and subsequent cDNA synthesis.
Gene Isolation and Sequence Analysis
RNA-seq analysis was conducted with CO2-treated persimmon fruit, which were described in Min et al. [7]. A TGA-like de-astringency responsive unigene was indicated by RNA-seq. The full-length TGA gene was isolated with a SMART RACE cDNA Amplification Kit (Clontech). The sequences of primers used for RACE and full-length amplification of DkTGA1 are described in Table 1. Based on the deduced amino acid sequences, a phylogenetic tree of TGA genes was generated using Clustal X (v 1.81) and MEGA (v 6.0). The deduced of amino acid sequences of homologous genes in Arabidopsis were obtained from NCBI.
Table 1. Sequences of the primers used for RACE, full-length amplification, real time PCR and vector construction.
| Genes | Methods used | Primers (5’-3’) |
|---|---|---|
| DkTGA1 | 3’RACE (Primary PCR) | CAACAGGGTCTGTATATAGGTGGTGG |
| DkTGA1 | 3’RACE (Secondary PCR) | GGTCTGGATAAACTCCAGCAAACT |
| DkTGA1 | 5’RACE (Primary PCR) | GTAAGAGCATCTTCCAATTGCTGACA |
| DkTGA1 | 5’RACE (Secondary PCR) | TTGGTCAAACTTTTTTGAAGGTAGT |
| DkTGA1 | Full-length clone (FP) | CTCAAAGGTCATATTGGAAACTCA |
| DkTGA1 | Full-length clone (RP) | AGTTCCATCTAAAGGCACTCT |
| DkTGA1 | Q-PCR (FP) | TCATCTTCGCAAGGAAACGC |
| DkTGA1 | Q-PCR (RP) | CGACTAGGCAGGCTCGTGAA |
| DkTGA1 | SK vector construction (FP) | CGCGTCGACATGACCTCTCCAACTGC |
| DkTGA1 | SK vector construction (RP) | GACGGTACCCTAGGCAGGCTCGTGA |
| DkTGA1 | Y1H constructs (FP) | CGCCATATGATGACCTCTCCAACTGC |
| DkTGA1 | Y1H constructs (RP) | GACGGATCCCTAGGCAGGCTCGTGA |
| DkERF9 | Y1H constructs (FP) | CATGAATTCATGGTTGGCTTTGTGAA |
| DkERF9 | Y1H constructs (RP) | GCAGGATCCTCATCCAGCAGCTTCA |
Note: underlined sequences show cutting sites for restriction enzymes.
Real-time PCR analysis
Oligonucleotide primers for real-time PCR analysis were designed with primer premier 5.0. The specificity of primers was determined by melting curves and PCR products resequencing. The sequences of oligonucleotide primers are in Table 1. Primers used for DkADH, DkPDC and DkERF genes were the same as in previous reports [7].
Real-time PCR was carried out with LightCycler 480 SYBR Green I Master (Roche) and LightCycler 480 II instrument (Roche) for gene expression studies. The PCR reaction mixture (20 μl total volume) comprised 10 μl 2 × real-time PCR mix (Roche), 1 μl of each primer (10 μM), 2 μl diluted cDNA, and 6 μl Diethy pyrocarbonate (DEPC) H2O. The PCR program was initiated for 3 min at 95°C, followed by 50 cycles of 95°C for 10 s, 55°C for 30 s, and completed with a melting curve analysis program. The relative expression of each gene was calibrated with values for day 0 fruit set as 1. The housekeeping gene, Actin, was used as the internal control [1].
Dual Luciferase assay
The trans-activation ability of DkTGA1 on de-astringency related genes was investigated by dual luciferase assay. Full-length DkTGA1 was inserted into pGreen II 002962-SK vector, using the primers listed in Table 1. Full-length DkERF and DkADH and DkPDC promoter constructs were obtained from our previous studies [6,7]. All constructs were electroporated into Agrobacterium tumefaciens GV3101. The dual luciferase assay was performed with Nicotiana benthamiana leaves, using the protocol described by Min et al. [6,7]. Dual luciferase assays were carried out in three independent experiments, with five biological replicates for each experiment.
Yeast One-hybrid Assay (Y1H)
The yeast one-hybrid assays were performed, using the Matchmaker Gold Yeast One-hybrid Library Screening System (Clontech, USA). As described in Min et al. [7], DkADH1 and DkPDC3 showed auto-activation activities, thus only DkPDC2 promoter was included for yeast one-hybrid analysis. The full-lengths of DkERF9 and DkTGA1 were subcloned into pGADT7 AD vector (AD) (primers are listed in Table 1). Transformed Y1HGold were cultured on SD/-Leu containing 200 ng/ml aureobasidin A at 30°C for 3 d to test interaction. pGADT7-Rec (AD-Rec-P53) was co-transformed with the p53-promoter fragment in Y1HGold as positive control.
Statistical Analysis
Statistical significance of differences was calculated using least significant difference (LSD) with DPS software (v.3.11).
Results
Gene isolation and sequence analysis
One hypoxia responsive TGA-like unigene, which showed a 25-fold induction by CO2 treatment, was identified by RPKM (Reads per kb per million reads) from RNA-seq (data not shown) and the full-length sequence was obtained using RACE. Sequence analysis indicated that this differentially expressed gene had a YCQRLRALSMLW domain (Fig 1a), which is a characteristic of TGA genes [23], and was designated as DkTGA1 (KU589287).
Fig 1. Phylogenetic tree of TGA.
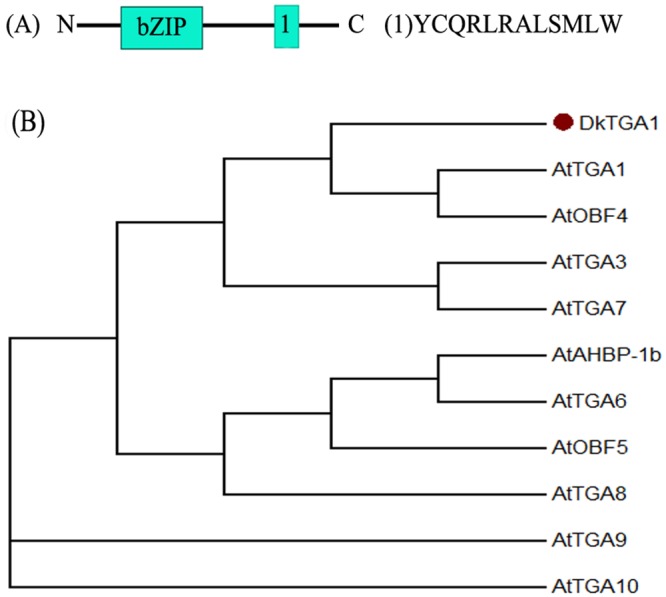
(A) Schematic analysis of DkTGA1. The bZIP domain and TGA related motif are indicated in blue. (B) Persimmon DkTGA is indicated with a red dot. The amino acid sequences of AtTGA transcription factors were obtained from The Arabidopsis Information Resource or National Center for Biotechnology Information. The phylogenetic tree was constructed with MEGA (v 6.0)
A phylogenetic tree was generated from the deduced amino acid sequences of the DkTGA1 and 10 TGA genes from Arabidopsis thaliana, which have been divided into five clades [24,25]. DkTGA1 showed the highest homology to clade I that comprises AtTGA1 (At5g65210) and AtTGA4 (At5g10030) (Fig 1b).
Association between DkTGA1 mRNA accumulation and persimmon fruit postharvest de-astringency
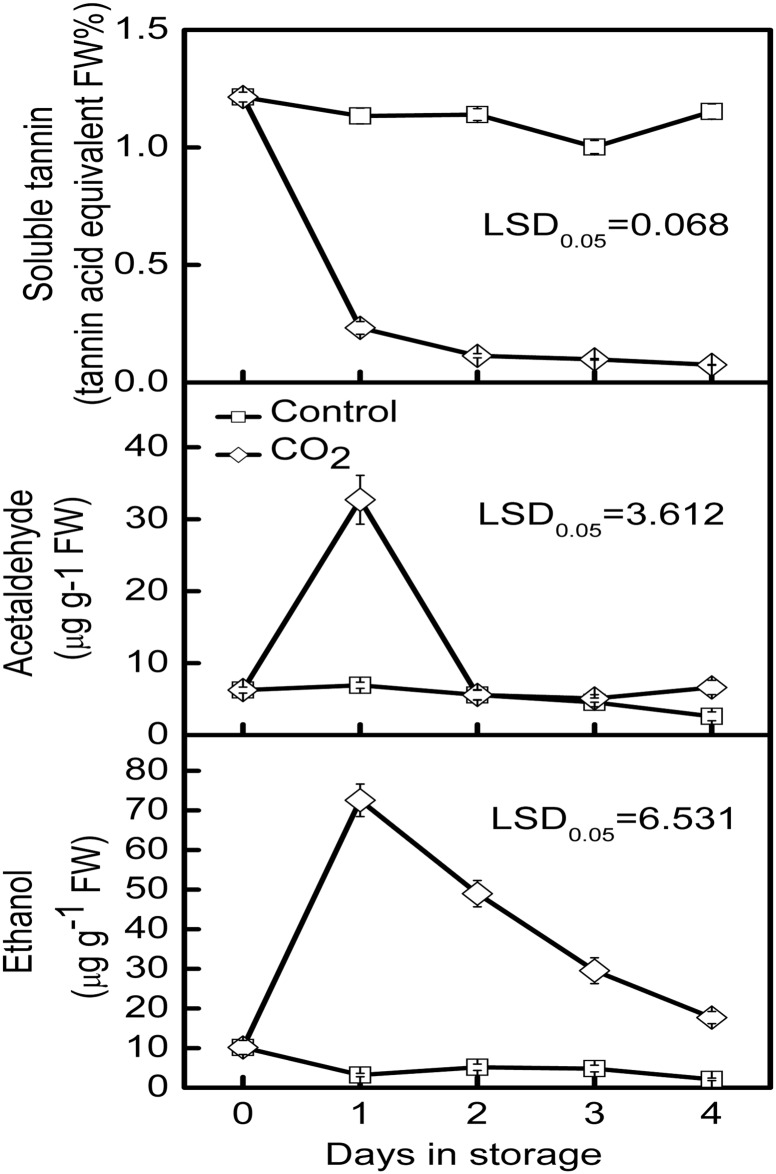
CO2 treatment (95%) promoted persimmon fruit de-astringency and caused a rapid decrease in the concentration of soluble tannin from 1.214% at 0 d to 0.232% at 1 d while in the control fruit the soluble tannin content was almost constant during storage (Fig 2). Accumulation of acetaldehyde and ethanol, the products of anaerobic respiration, were observed in CO2-treated fruit, concomitantly with loss of astringency-related SCT. Acetaldehyde increased from 6.26 μg/g at 0 d to 32.70 μg/g after CO2 treatment while the ethanol increased from 10.18 μg/g to 72.56 μg/g (Fig 2). Following removal of CO2 treatment, acetaldehyde content rapidly declined to the basal level observed at 0 d, while the decrease in ethanol content occurred more slowly than acetaldehyde, and remained higher than that in control fruit (Fig 2).
Fig 2. Changes of soluble tannins, acetaldehyde and ethanol in ‘Mopanshi’ persimmon fruit in response to treatment with 95% CO2 for 1 day.
Error bars represent ±SE from three replicates. LSDs represent least significant difference at p = 0.05.
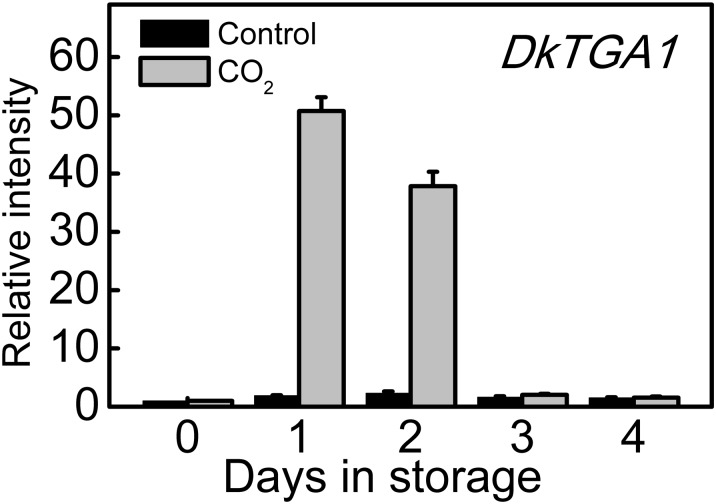
Transcripts of DkTGA1 was transiently induced during CO2 treatment, peaking after 1 d, with an approximate 50-fold induction at 1 d, and then decreased to similar level of that in control fruit following removal of CO2 (Fig 3). All of the previously isolated de-astringency related genes, DkADH1, DkPDC2, DkPDC3, DkERF9, DkERF10, DkERF19 and DkERF22, were up-regulated by CO2 treatment (S1 Fig), which confirmed previous observations [6,7].
Fig 3. Expression of DkTGA1 in response to CO2 treatment (95%, 1 day).
Relative mRNA abundance was evaluated by real-time PCR. Day 0 fruit values were set as 1. Error bars represent ±SE from three replicates. LSDs represent least significant differences at p = 0.05.
Trans-activation of persimmon DkADH1, DkPDC2 and DkPDC3 promoters by DkTGA1
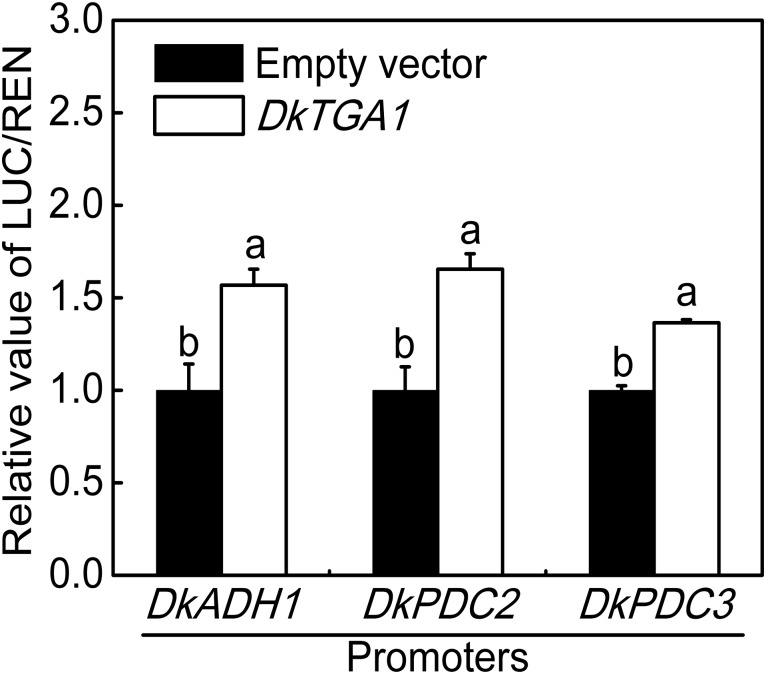
A dual luciferase assay was carried out to investigate the possible transcriptional regulatory linkage between DkTGA1 and target genes known to be involved in astringency removal. The results indicated that DkTGA1 had a small (but statistically significant) effect on genes induced by anaerobic conditions and involved in the de-astringency process, including DkADH1, DkPDC2 and DkPDC3, with an approximately 1.5-fold activation of the promoters of these genes (Fig 4).
Fig 4. Trans-activation effects of DkTGA1 on the promoters of deastringency-related genes (DkADH1, DkPDC2, DkPDC3) using the dual-luciferase assay.
The value of LUC/REN for the empty vector (SK) was set as 1. Error bars indicate S.E.s from five replicates. Different letters above the columns indicate significant differences (P < 0.05)
Synergistic effect of DkTGA1 and DkERF on trans-activation of promoters of de-astringency-related target genes
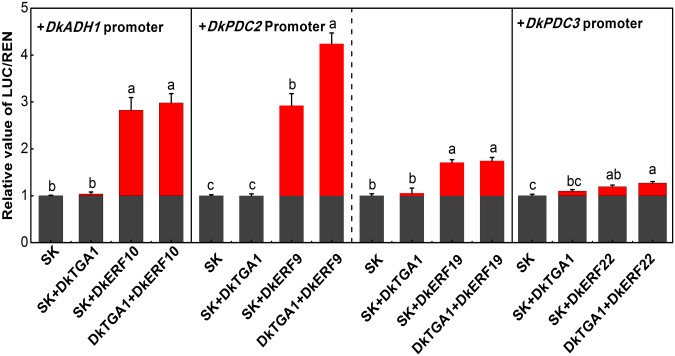
It was shown previously that DkERF9 and DkERF19 could activate the DkPDC2 promoter while DkERF10 activated the DkADH1 promoter [6,7]. As shown in Fig 4, DkTGA1 could trans-activate the DkADH1, DkPDC2 and DkPDC3 promoters. Thus, further investigations were conducted using DkERF9, DkERF10 and DkERF19 promoters, however, the results indicated that DkTGA1 also had very limited effects on promoters of these DkERF genes (S2 Fig).
Subsequently, the effect of combining DkTGA1 and DkERF transcription factors was analyzed. DkTGA1 and DkERF9 together significantly enhanced promoter activation, compared with the effects of DkTGA1 or DkERF9 singly (Fig 5). However, no such enhancement was found when DkTGA1 was tested in combination with DkERF10, DkERF19 or DkERF22 (Fig 5).
Fig 5. Synergistic effect of DkTGA1 and DkERF on trans-activation of the promoters of deastringency-related genes (DkADH1, DkPDC2, DkPDC3) using the dual-luciferase assay.
Yeast one/two-hybrid assays were performed in view of the results obtained from the dual luciferase assay. Motif analysis indicated the existing of the potential ERF and TGA binding sites (Fig 6A). Yeast one-hybrid results indicated that DkERF9 could bind to DkPDC2 promoter, whereas no physical interaction was found between DkTGA1 and the DkPDC2 promoter (Fig 6B). However, yeast two-hybrid studies indicated that there was no direct protein-protein interaction between DkTGA1 and DkERF9 (S3 Fig), despite the additive effects observed between DkTGA1 and DkERF9 on expression from the DkPDC2 promoter.
Fig 6. Yeast one-hybrid analysis of DkTGA1 and DkERF9 binding to promoter of DkPDC2.
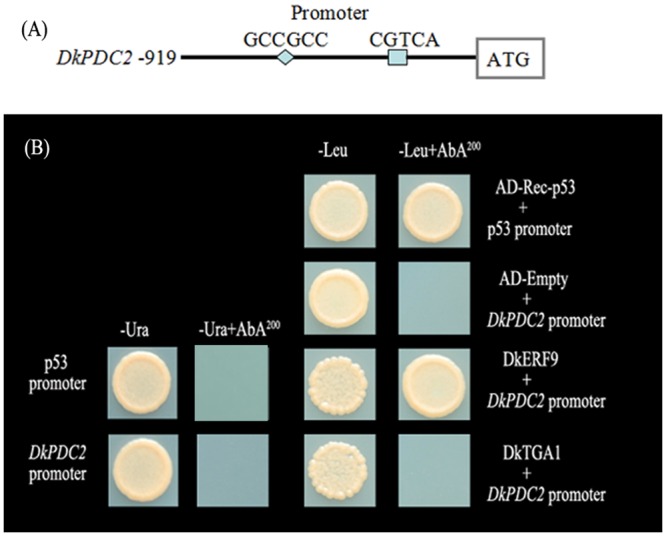
(A) Schematic representation of DkERF (GCCGCC) and DkTGA (CGTCA) possible binding sites in DkPDC2 promoter. (B) Auto-activation of promoters were tested on SD medium lacking Ura in presence of aureobasidin A (AbA). Interaction was determined on SD medium lacking Leu in presence of AbA.
Discussion
Persimmon astringency removal had been widely studied and several technologies had been applied to reduce the SCTs in persimmon fruit, including treatment with high concentrations of CO2 or N2 [4,6,7,12,26], C2H4 treatment [5], dipping in hot water [27], all of which can lead to the persimmon fruit astringency removal. The effectiveness of CO2 treatment is widely considered to be due to the hypoxia response, which stimulates acetaldehyde production in persimmon fruit by anaerobic respiration, which precipitates the insoluble tannins, thereby removing astringency [6,15,28]. The results obtained in the present research are entirely consistent with this, as a rapid decrease in SCTs and transient stimulation of acetaldehyde and ethanol were observed in ‘Mopanshi’ persimmon in response to 95% CO2 treatment.
A few transcription factors have been suggested to be involved in the de-astringency response in persimmon fruit, however, only five transcription factors have been verified as having trans-activation ability on promoters of de-astringent related target genes (eg. DkADH1, DkPDC2 and DkPDC3). These include four DkERF genes (DkERF9/10/19/22) [6,7] and a MYB transcription factor (DkMYB6) [15]. Here, a de-astringency responsive bZip transcription factor, detected in an RNA-seq database and designated as DkTGA1, based on phylogenetic comparisons with related genes from other plants. TGA is a member of a sub-class of bZIP transcription factors that have been widely reported to be involved in stress responses, for instance, AtTGA1-7 has been characterized with respect to their interaction with pathogenesis-related gene1 (NPR1) [29]; AtTGA1 and AtTGA4 also played important roles in Arabidopsis root developmental responses to nitrate [30]. DkTGA1 was clustered with AtTGA1 and AtTGA4, suggesting DkTGA1 may also participate in response to stress in persimmon fruit. Although no TGA genes have been reported as being responsive to low oxygen in other plants, the present results indicate a significant (50-fold) induction of DkTGA1 expression in response to high CO2/low O2 treatment (Fig 3). This suggests that the involvement of TGA genes in the anoxia response in other plants should be investigated. DkTGA1 had a small transactivation effect on the promoters of DkADH1, DkPDC2 and DkPDC3, which further indicated its possible involvement in persimmon de-astringency and the hypoxia response. However, the effects of DkTGA1 on de-astringency related target genes were weaker than our previously reported results for other transcription factors such as DkERF9/10/19/22 and DkMYB6 [6,7,15]. It is notable that increasing expression of DkTGA1 only occurred at 1 d and 2 d, which is similar to DkERF9/10/19/22 [6,7] and differ to DkMYB6 [15]. As deastringent process for ‘Mopanshi’ fruit in response to high CO2 treatment, generally happened on 1 d or 2 d, thus expression patterns of DkTGA1, as well as DkERF9/10/19/22, suggested that these transcription factors might be more specific for deastringency.
It is worth emphasizing that the synergistic effects of DkTGA1 and DkERF9 were observed on DkPDC2 promoter. Synergistic effects of different transcription factors have been widely reported, for example MaNAC1 is involved in cold tolerance of banana fruits, and interacts with MaCBF1 [31] and EjAP2-1 interacts with EjMYB1/2 to form a complex involved in loquat fruit lignification [32]. Moreover, other TGAs and ERFs have been shown to interact with each other, for instance, AtTGA4 interacts with AtEBP (ethylene-responsive element binding protein, the previous name for ERF), which binds the ethylene response promoter elements and may therefore be important in regulating gene expression during the plant defense response [33]. However, additive effects of DkTGA1 and DkERF9 were not due to direct protein-protein interaction between, as Y2H experiments indicated no interaction between two transcription factors. Thus, synergistic effects of DkTGA1 and DkERF9 not only raised a possible involvement of a new transcription factor(s) in persimmon fruit de-astringency, but also provided a new evidence for relationships, with unknown mechanisms, between members of the TGA and ERF families, different from those previously reported. It is clear that DkERF9 can physically interact with the DkPDC2 promoter, while DkTGA1 is not able to bind directly to the promoter. These results suggest that DkTGA1 is a novel and indirect regulator of persimmon fruit postharvest de-astringency, and may function via indirect interaction with DkERF9, a direct regulator on DkPDC2. As persimmon fruit deastringency related transcription factors are still limited, future researches might bridge the gap between DkTGA1 and deastringency.
Conclusion
In conclusion, CO2 (95%) treatment is effective in promoting and accelerating persimmon fruit postharvest de-astringency. The present study isolated a de-astringency inducible DkTGA1, which is an indirect activator of anaerobic response target genes. Furthermore, DkTGA1 and DkERF9 were shown to have an additive effect on activation of DkPDC2. As CO2-driven persimmon fruit postharvest de-astringency is mainly based on fermentation under anoxia, the activity of DkTGA1 suggests the likely involvement of TGA transcription factors in hypoxia responses in other plants.
Supporting Information
(TIF)
(TIF)
(TIF)
Abbreviations
- ADH
alcohol dehydrogenase
- ERF
ethylene response factor
- PDC
pyruvate decarboxylase
- SCT
soluble condensed tannins
- AD
pGADT7
- BD
pGBKT7
- AbA
aureobasidin A
- cv
Cultivar
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the Special Fund for Agro-scientific Research in the Public Interest [no. 201203047], National Natural Science Foundation of China [no. 31372114], Program of International Science and Technology Cooperation [2011DFB31580] and the Natural Science Foundation of Zhejiang Province, China [LR16C150001]. There was no additional external funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, et al. DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 2009a; 4, 2028–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akagi T, Ikegami A, Suzuki Y, Yoshida J, Yamada M, Sato A, et al. Expression balances of structural genes in shikimate and flavonoid biosynthesis cause a difference in proanthocyanidin accumulation in persimmon (Diospyros kaki Thunb.) fruit. Planta 2009b; 230, 899–915. [DOI] [PubMed] [Google Scholar]
- 3.Ikegami A, Eguchi S, Kitajima A, Inoue K, Yonemori K. Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit. Plant Sci. 2007; 172, 1037–1047. [Google Scholar]
- 4.Salvador A, Arnal L, Besada C, Larrea V, Quiles A, Pérez-Munuera I. Physiological and structural changes during ripening and deastringency treatment of persimmon fruit cv. ‘Rojo Brillante’. Postharvest Biol Technol. 2007; 46, 181–188. [Google Scholar]
- 5.Yin XR, Shi YN, Min T, Luo ZR, Yao YC, Xu Q, et al. Expression of ethylene response genes during persimmon fruit astringency removal. Planta 2012; 235, 895–906. 10.1007/s00425-011-1553-2 [DOI] [PubMed] [Google Scholar]
- 6.Min T, Yin XR, Shi YN, Luo ZR, Yao YC, Grierson D, et al. Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon (Diospyros kaki) fruit de-astringency. J Exp Bot. 2012; 63, 6393–6405. 10.1093/jxb/ers296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Min T, Fang F, Ge H, Shi YN, Luo ZR, Yao YC, et al. Two novel anoxia-induced ethylene response factors that interact with promoters of deastringency-related genes from persimmon. PLOS One 2014; 9, 97043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taira S, Ikeda K, Ohkawa K. Comparison of insolubility of tannins induced by acetaldehyde vapor in fruit of three types of astringent persimmon. J Jpn Soc Hortic Sci. 2001; 48, 684–687. [Google Scholar]
- 9.Pesis E, Ben-Arie R. Involvement of acetaldehyde and ethanol accumulation during induced deastringency of persimmon fruits. J Food Sci. 2006; 49, 896–899. [Google Scholar]
- 10.Tamura F, Tanabe K, Itai A, Hasegawa M. Characteristics of acetaldehyde accumulation and removal of astringency with ethanol and carbon dioxide treatments in ‘Saijo’ persimmon fruit. J Jpn Soc Hortic Sci. 1999; 68, 1178–1183. [Google Scholar]
- 11.Liu CP, Zheng ZM, Liang YN, Li B. Relationship between activity of alcohol dehydrogenase and soluble tannin in persimmon. Acta Hortic Sin. 2008; 35, 741–746. (In Chinese with English abstract) [Google Scholar]
- 12.Yamada M, Taira S, Ohtsuki M, Sato A, Iwanami H, Yakushiji H, et al. Varietal differences in the ease of astringency removal by carbon dioxide gas and ethanol vapor treatments among Oriental astringent persimmons of Japanese and Chinese origin. Sci Hortic. 2002; 94: 63–72. [Google Scholar]
- 13.Pesis E. The role of the anaerobic metabolites, acetaldehyde and ethanol, in fruit ripening, enhancement of fruit quality and fruit deterioration. Postharvest Biol Technol. 2005; 37, 1–19. [Google Scholar]
- 14.Botondi R, Russo V, Mencarelli F. Anaerobic metabolism during short and long term storage of kiwifruit. Postharvest Biol Technol. 2012; 64, 83–90. [Google Scholar]
- 15.Fang F, Wang MM, Zhu QG, Min T, Grierson D, Yin XR, et al. DkMYB6 is involved in persimmon fruit deastringency, via transcriptional activation on both DkPDC and DkERF. Postharvest Biol Technol. 2016; 111, 161–167. [Google Scholar]
- 16.Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, et al. Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010; 153, 757–772. 10.1104/pp.110.155077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang CY, Hsu FC, Li JP, Wang NN, Shih MC. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011; 156, 202–212. 10.1104/pp.111.172486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoeren FU, Dolferus R, Wu YR, Peacock WJ, Dennis ES. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 1998; 149, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe H, Urao T, Ito T, Seki M. Arabidopsis AtMYB2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003; 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee TG, Jang CS, Kim JY, Kim DS, Park JH, Kim DY, et al. A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol Plantarum 2007; 129, 375–385. [Google Scholar]
- 21.Licausi F. Regulation of the molecular response to oxygen limitations in plants. New Phytol. 2011; 190, 550–555. [DOI] [PubMed] [Google Scholar]
- 22.Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993; 11, 113–116. [Google Scholar]
- 23.Schindler U, Beckmann H, Cashmore AR. TGA1 and G-box binding factors: two distinct classes of arabidopsis leucine zipper proteins compete for the G-box like element TGACGTGG. Plant Cell 1992; 4, 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002; 7, 106–111. [DOI] [PubMed] [Google Scholar]
- 25.Gatz C. From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol Plant Microbe In. 2013; 26, 151–159. [DOI] [PubMed] [Google Scholar]
- 26.Arnal L, Del Río MA. Removing astringency by carbon dioxide and nitrogen-enriched atmospheres in persimmon fruit cv.’Rojo Brillante’. J Food Sci. 2003; 68, 1516–1518. [Google Scholar]
- 27.Kitagawa H. Studies on the removal of the astringency and storage of Kaki (Oriental persimmons). 5. A relation between removal of the astringency and acetaldehyde formation during the warm water treatment for removal of the astringency. J Jpn Soc Hortic Sci. 1969; 37, 379–382. (in Japanese with English abstract). [Google Scholar]
- 28.Del Bubba M, Giordani E, Pippucci L, Cincinelli A, Checchini L, Galvan P. Changes in tannins, ascorbic acid and sugar content in astringent persimmons during on-tree growth and ripening and in response to different postharvest treatments. J Food Compos Anal. 2009; 22, 668–677. [Google Scholar]
- 29.Kesarwani M, Yoo J, Dong XN. Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 2007; 144, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez JM, Riveras E, Vidal EA, Gras DE, Contreras-Lopez O, Tamayo KP, et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014; 80, 1–13. [DOI] [PubMed] [Google Scholar]
- 31.Shan W, Kuang JF, Lu WJ, Chen JY. Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ. 2014; 37, 2116–2127. [DOI] [PubMed] [Google Scholar]
- 32.Zeng JK, Li X, Xu Q, Chen JY, Yin XR, Ferguson IB, et al. EjAP2-1, An AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol J. 2015; 13, 1325–1334. [DOI] [PubMed] [Google Scholar]
- 33.Butiner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. P Natl Acad Sci USA 1997; 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.