Abstract
This article presents a reproducible research workow for amplicon-based microbiome studies in personalized medicine created using Bioconductor packages and the knitr markdown interface. We show that sometimes a multiplicity of choices and lack of consistent documentation at each stage of the sequential processing pipeline used for the analysis of microbiome data can lead to spurious results. We propose its replacement with reproducible and documented analysis using R packages dada2, knitr, and phyloseq. This workow implements both key stages of amplicon analysis: the initial filtering and denoising steps needed to construct taxonomic feature tables from error-containing sequencing reads (dada2), and the exploratory and inferential analysis of those feature tables and associated sample metadata (phyloseq). This workow facilitates reproducible interrogation of the full set of choices required in microbiome studies. We present several examples in which we leverage existing packages for analysis in a way that allows easy sharing and modification by others, and give pointers to articles that depend on this reproducible workow for the study of longitudinal and spatial series analyses of the vaginal microbiome in pregnancy and the oral microbiome in humans with healthy dentition and intra-oral tissues.
Keywords: Illumina, amplicon, DADA2, phyloseq, microbiota, microbiome, microbial ecology, longitudinal data, spatial, personalized medicine, random effects models
1. Introduction
High-throughput (HT) DNA sequencing is allowing major advances in microbial studies; our understanding of the presence and abundance of microbial species relies heavily on the observation of their nucleic acids in a “culture independent” manner.1 At present, the most common and cost-effective method for characterizing microbes and their communities is amplicon sequencing: PCR amplification of a small (100–500 bp) fragment of a conserved gene (phylogenetic marker) for which there are taxonomically-informative reference sequences available. The standard phylogenetic marker gene for bacteria is the small subunit ribosomal RNA (16S rRNA) gene,1 for which there are also convenient tools and large reference databases.2–4 16S rRNA amplicon sequencing provides a census of the personalized bacterial communities present in a sampled individual.
After obtaining the amplicon sequences, a standard series of bioinformatic and statistical analyses are used to evaluate these data: filtering out low quality sequences and samples, constructing a taxonomic feature table of observations from each sample, incorporating the sample metadata, transforming and normalizing the feature table, and performing exploratory and inferential analyses. Here we explore the multiplicity of choices made during this process, show examples of their consequences, and motivate the need for better and easier reproducibility of the standard analytic workow on amplicon dataa.
We focus here on two Bioconductor5 packages—dada26 and phyloseq7,8—created specifically to analyze amplicon sequencing data within the R environment, and show how they enable reproducible research in several microbiome studies. We begin by illustrating the need for reproducible research workows in microbiome studies with a typical workow example.
2. A Case study in multiple outcomes: the Enterotypes
A few years ago, Arumugam, M. et al.9 published an article in Nature that concluded that humans could be grouped into intestinal gut types. In fact, some bioinformatic forensics (presented in detail in the supplementary material) shows that during the course of the analysis, the following choices were made.
| • A preliminary choice of data transformation from the original counts to proportions was made, although the authors could have chosen to take logarithms, variance stabilizing transformations10 (here proportions replaced the original counts), choose between | log, rlog, subsample, prop, orig (5) |
| • Nine points were dropped from the study as they were considered outliers; of course the authors could have chosen to | .. leave out 0, 1, 2,..,9, + criteria (100) |
| • Certain taxa were filtered out as they were considered too rare or unlabeled. filter taxa | … remove rare taxa, ie threshold at 0.01%, 1%, 2%,… (10) |
| • A distance was chosen (Jensen-Shannon, JSD) to quantify similarities between samples. Distances | … 40 choices in vegan/phyloseq (40) |
| • An ordination method and number of coordinates has to be chosen. Ordination and axes | … MDS, NMDS, DCA, k=2,3,4,5.. (16) |
| • A clustering method and a number of clusters is chosen | … PAM, KNN, hclust … (16) |
| • The authors chose an underlying continuous variable, an alternative could have been a linear or curved latent variable or group of variables. | latent variable choices… (4) |
According to this rough list, there are more than 200 million possible ways of analyzing these data b. Thus, there is a combinatorial explosion of the number of possible choices that an investigator makes. Some choices can impact conclusions drawn from microbiome studies; it becomes necessary for experimentalists to develop and adopt pipelines documenting choices used in these analyses with the intention of providing an assessment of the robustness and reproducibility of the analyses. In fact, an errata was published to the paper,9 substantially weakening the original conclusions. Figure 1 shows graphical representations made after the Jensen-Shannon distance was computed on the data. The authors made an inappropriate supplementary step using the data based clusters as labels in a supervised classification between group analysis that separated the clusters more than they actually appear in the middle figure. There is unfortunately no way to use multiple hypothesis testing corrections for this number of possible analyses, thus the only way of ensuring robustness of the conclusions is to repeat the analyses with many different settings. In the supplementary material we include the output showing the ordination with 40 different distances. In particular, the clustering is not always as obvious; different choices of distance such as chisquare or Jaccard give very different results.
Fig. 1.
On the left we show the analysis as done in9, in the middle we have done the same analysis with the Jensen Shannon distance but without the extra (invalid) supervised separation and on the right we have the minimum spanning tree exhibiting a clear gradient in the data.
As this re-analysis demonstrates, access to reproducible analysis workows is necessary for the interpretation of modern microbiome studies. In this example, in just one stage of the analysis (clustering of samples based on taxonomic features), the reported outcome was one out of millions of analogous alternatives, many of which differed qualitatively. Other parts of standard amplicon analysis, such as the construction of OTU tables and the evaluation of differential abundances, are accompanied by a similar myriad of choices. For this reason it is crucial that the analysis of amplicon data be made accessible — sharing the data alone is not enough.
3. A reproducible workow in R
Here we present a workow for the analysis of amplicon data within R (Figure 2). This workow takes as input the amplicon sequencing reads and associated sample metadata, and provides as output exploratory and inferential statistical analyses as well as sharable analysis scripts and data files that fully reproduce those analyses. Here we focus on two particular packages developed by our group for the analysis of amplicon data within the R environment: dada2 and phyloseq.
Fig. 2.
Diagram of the new reproducible workow including denoising, data integration and statistical analyses.
3.1. Inferring sample sequences and abundances using DADA2
The DNA sequence errors introduced by PCR and sequencing complicate the interpretation of amplicon data, and present different challenges than the more well known problem of resequencing. When re-sequencing a diploid organism (like a human being) it is known that there exist either 1 or 2 variants at every position in the genome. Thus increasing depth eventually trivializes the problem of making genotype calls by overwhelming the error rate with data. However, when amplicon sequencing microbial communities the number of variants and their associated frequencies are unknown, which fundamentally changes the inference problem. When increasing sampling depth reveals new sequence variants, these might represent rare errors or rare members of the community. In addition, the PCR amplification step introduces chimeras and additional errors with a different structure than sequencing errors.
Most current studies use two methods to deal with amplicon errors, reducing their incidence by filtering out low quality reads, and lumping similar sequences together into Operational Taxonomic Units (OTUs). However, there are a significant number of choices made during this process: the type and stringency of quality filtering, the minimum abundance threshold, the size of the OTUs, the OTU construction method, and more. All of these choices can have significant downstream consequences for later analysis.11
This has led to serious problems for the reproducibility of amplicon-based studies. The methods used to filter sequences, construct OTUs and then perform analysis are often performed in separate environments (e.g., shell scripts vs. python vs. R). This makes the creation of a single coherent record of the analysis from input data to final product difficult and time-consuming. In practice few studies can be reproduced from the original raw data.
We have addressed this shortcoming by developing the dada2 package6 for R which performs the crucial filtering and sample inference steps that turn a set of raw amplicon sequences into a feature table of the types observed in each sample (e.g., an OTU table). Because dada2 shares the R environment with downstream analysis methods already present in R, such as those in the phyloseq package, the publication of reproducible workows encompassing the entirety of the analysis is far easier. One unified R script, and one unified Rdata data object, can provide a complete record of the published analysis, and allows interrogation of the full set of choices made in that process.
3.2. Performing exploratory and inferential analysis with phyloseq
Phyloseq allows the user to import a species by sample contingency table matrix (aka, an OTU Table) and data matrices from metagenomic, metabolomic, and or other –omics type experiments into the R computing environment. Phyloseq is unique in that it allows the user to integrate the OTU Table, the phylogenetic tree, the “representative sequence” fasta file, and the metadata mapping file into a single “phyloseq-class” R object. The microbial ecologist can then harness all the statistical and graphical tools available in R, including knitr, ggplot2 to generate reproducible research reports with beautiful graphics, as detailed in our supplementary .Rmd file and in the case studies below.
Combining this environment with a number of other important R packages (e.g., vegan, ade4, DESeq2, multtest …) allows for powerful and specific analyses to be performed on amplicon-sequenced microbiome data. We share several such examples, along with the data and code necessary to reproduce them.
4. Examples: Longitudinal data analysis
Tackling the challenges involved in longitudinal patient-dependent data requires methods specifically tailored for the human body sites studied. For instance, the vaginal community is the one human body habitat that has been shown to robustly cluster into discrete community state types (CSTs).12 This feature allows the complex information about community composition to be simplified by projecting into a small number of CSTs. Combined with longitudinal sampling information, this simplified projection is then amenable to analysis as a Markov chain.
In a 2015 study12 we used this Markov chain representation to analyze the dynamics of the vaginal community during pregnancy. Transition rates were estimated from a set of 652 pairs of samples collected one week apart during the pregnancies of 40 women, producing an estimation of the dynamics of the vaginal community as illustrated in Figure 3. These results reproduced previous qualitative and semi-quantitative observations,13 such as the high stability of Lactobacillus crispatus communities, but also provided a more detailed quantification of the stability of each CST as well as the connectivity between them.
Fig. 3.
Markov chain modeling of CST states across pregnancy and preterm birth.12 Numbers indicate the one-week self-transition rate for each state. The high-diversity, low Lactobacillus class 4 is the least stable and most connected to the other CSTs. A more complete version of this figure appears in the aforementioned PNAS paper.12
Markov chain analysis is a powerful way to quantify and visualize dynamics, but it can only be applied to systems that are representable by a set of discrete states (a property which is often not trivial to establish as in Section 2). In the context of microbial communities this is a substantial limitation, as few communities can be so represented. A second concern, especially when applied to human-associated communities, is how the estimation of the transition rates should be performed across subjects. If the community dynamics are subject-independent, then an average over the observed transitions in each subject is appropriate, and this was the method used in our analysis of pregnant vaginal communities. However, it is possible that subject-specific factors (eg. host genetics) may inuence transition rates, in which case the set of states should be expanded to include the subject effect.
Finally, the uncertainties that exist in mapping community states to discrete CSTs can also have significant consequences for the estimation of transition rates between those states, as in the case where a rare and unmodeled community state exists intermediate between the centers of two CSTs and is sometimes assigned to one or the other.
Even in the relatively very simple case of the vaginal community, this set of concerns cannot be comprehensively addressed within a single manuscript. Thus the need for access to the analytical workow. In this study, we used the reproducible R workow, and Rmd and Rdata files, to make our analysis easily accessible and modifiable and have deposited the data and code in a permanent repository (permanent url) maintained by the Stanford Digital Repository at http://purl.stanford.edu/hg140kw6221.
5. Example: Spatial data analysis
Patterns of diversity and community composition across human body-sites have been well characterized.14 When comparing human-associated microbial communities across different anatomic sites, skin and gut for instance, dramatic differences in the acquisition, development, and maintenance of microbial community composition are observed. Few studies have yet examined the extent to which microbial communities vary across fine scale spatial gradients on the human body, such as between and across individual tooth surfaces in the oral cavity. Datasets that have attempted to examine the spatial variation of oral microbial communities have shown interpersonal variation to be the strongest effect with secondary effects exerted by tooth position.15,16 We have extended the current exploratory approaches through the use of the statistical packages available in R specifically tailored to analyze spatial or longitudinal data.
In this study, we demonstrate the usability of the phyloseq7 package for applied spatial analysis of microbial communities in the oral cavity. As a test case, we generated data for this demonstration, collecting 186 independent samples from the facial (cheek-facing) and lingual (tongue-facing) surfaces of every tooth (excluding the third molars) of one adult female on each of two non-consecutive days. We extracted DNA from each sample, amplified the V3-V5 region of the 16S rRNA gene using golay-barcoded primers, and sequenced the amplicons using the 454-Titanium platform, generating 216,965 sequences with a median sequencing depth of 2,479. We use two methods to examine the spatial variation of oral microbial communities: a Between Class Analysis and a Principal Components Analysis with respect to Instrumental Variables.
5.1. Between Class Analysis
When dealing with a priori classes in which we know teeth communities segregate, we want to highlight differences in a supervised analysis. The segregation of supragingival communities might arise because teeth are situated along an ecological gradient. As a first examination of the spatial relationship between the oral communities, after filtering and preprocessing the data we used ade4 to perform a Between Class Analysis (BCA) in which tooth class was used as the spatial partition.
Using teeth groupings, we found that 12% of the total variance could be explained by Tooth-Class with the first component accounting for 56.5% of the explained variance. The BCA revealed that not only can communities be distinguished from one another based on tooth class, but that these communities may exist along an ecological gradient. Molar and Premolar communities appeared to be associated with positive scores along the first BCA axis while the Central Incisors and Lateral Incisors appeared to be associated with more negative scores along the first BCA axis, regardless of whether we examine communities in the top (Maxillary) or bottom (Mandibular) jaws or whether we examined the buccal or the lingual aspect of teeth. The distribution of teeth along the first and second components suggested that supragingival plaque varies along a gradient in the mouth. Interestingly, the variance of community scores along CS1 varied according to tooth class (Figure 4) with Central Incisor communities appearing to be the most variable community class especially if communities were sampled from the lingual aspect of the teeth or from the buccal aspect of teeth in the lower jaw. This is interesting given the relative proximity of these sites to the secretions of the submandibular/sublingual glands, which may give rise to the observed structure.
Fig. 4.
Comparison of Buccal and Lingual sides of five different teeth types.
5.2. Principal Components with Instrumental Variables
It can be challenging to incorporate covariates such as spatial variation into studies of microbial communities. Here, we explicitly model the spatial variation of microbial communities in the human mouth using a Principal Components Analysis with respect to Instrumental Variables (PCA-IV)17 where a third-order polynomial function of the geographic coordinates was used as the constraint.
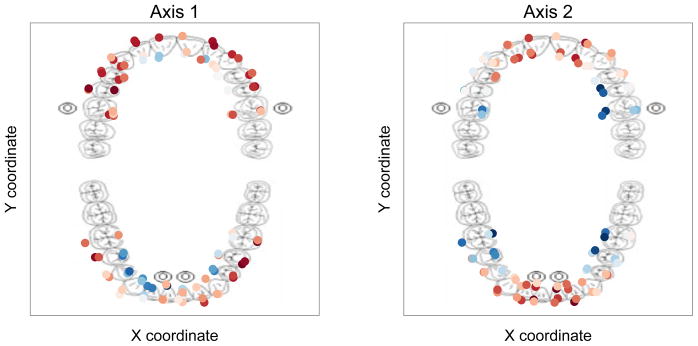
The PCA-IV accounts for 27% of the variance. The first principal coordinate separates buccal from lingual communities with lingual communities being associated with more positive scores along Axis 1 compared to buccal communities, especially for communities in the bottom jaw. Examining multiple individuals should confirm whether this pattern is consistent across multiple subjects and may reveal the sum of factors that structure the spatial variation of microbial communities. For now, we speculate that in this subject the relative proximity of the the submandibular glands to the upper anterior sites, the lower anterior, and posterior sites are important factors contributing to variation along Axis 1.
Axis 2, on the other hand, accounts for 23.4% of the total variance and appears to separate molar from incisor communities with molar communities being associated with positive scores along Axis 1 and incisors being associated with negative values along Axis 1.
While the data presented here pertain to just a single subject, and therefore our ability to make population level inferences limited, in this analysis we generated and provided an .Rmd script that can be used by other expeirmentalists to test hypotheses related to the spatial structure of host-associated communities in the oral cavity or at other body sites.
6. Relevance to Precision Medicine
The vaginal and oral community examples provided above both have relevant applications to personalized medicine. In the first example, the state of the vaginal community during pregnancy has been shown to be related to the likelihood of preterm birth outcomes in some women, with higher relative abundances of particular taxa such as Gardnerella vaginalis and Ureaplasma implicated as specific risk factors.12 Furthermore, the duration of time spent in the high-risk states further stratified preterm risk. This suggests that longitudinal monitoring of the vaginal community during pregnancy, concomitant with the standard schedule of prenatal care, might provide a biomarker for preterm birth risks early in pregnancy allowing for pre-emptive intervention and education.
In the second example, we show an unpublished example of a spatial analysis of the oral microbiome which will have broad applicability in the dental clinic. Dentists have long known that most people experience greater difficulty brushing the lingual aspects of posterior teeth as compared to the buccal aspects of those same teeth, and that whether an individual is right or left handed impacts brushing efficacy. Differences in brushing technique may therefore give rise to differences in microbial community composition in different areas of the mouth, and these differences may be highly specific to individual patients and may relate to the incidence of dental disease. One possible application of this type of analysis in the dental clinic would be to provide patients with customized diagrams of the microbial inhabitants of their oral cavities in order to help them to better understand the impact of brushing technique on their oral health. If analyses were conducted at every semi-annual dental exam the relative impact that different dental interventions have on the spatial structure of communities could be elucidated on a per-patient basis. By wrapping up this type of metadata with phylogenetic sequence analysis and depositing the information into public repositories, we increase our ability to make this type of inference. Analysis of spatial and temporal series may also have relevance to dental disease, which is patchy. Cavities (which are not generalized) are known to form on discrete, localized surfaces of dental enamel, and in the healthy individual a single cavity typically takes years to develop. Our ability to decompose the spatial and temporal components of health-associated supragingival communities would enable clinicians to develop models that detect deviations from health, such as incipient caries, which are reversible. These types of models might then allow clinicians to detect and reverse early carious lesions before the need for costly and invasive dental restorations arises.
These examples demonstrate the need for reproducibility in microbiome research and the relevance of the reproducibility problem to the precision medicine initiative.
7. Difficulties encountered in the production of Reproducible Research
There are many biological and technical choices, which are challenging to standardize across projects, institutions, and investigators, that make experimental and analytical findings difficult to replicate. Studies that examine the ecological structure of human-associated microbial communities are tough to compare when data are generated using different sequencing technologies (e.g., 454 vs. Illumina), or even when investigators simply sequence different regions of the 16S rRNA gene. The rapid increase in the number of sequencing technologies makes it difficult to standardize the highest level technical factors that give rise to irreproducible data. In contrast, how we make use of data that have already been generated is one of the easiest ways to increase the reproducibility of experimental findings. The difficulty that any investigator faces in replicating the steps and choices of another investigator in executing their data analysis, when provided with the same raw data, is a more subtle, often-overlooked problem that is easily addressed. When considering this question in the context of precision medicine it is easy to recognize that a clinician trying to provide an individualized report on, say, oral health must be able to generate a record exactly as intended by the investigators who developed the report. To facilitate the use of these analyses in the clinic, reports could be developed to run with the Shiny Phyloseq8 interface, which places the computational power of R in the hands of individuals less customized to working with scripts. Experimentalists will benefit from considering their data analysis pipelines from this point of view. While most bench scientists use a laboratory notebook to document choices that guide their decisions at the bench, fewer bench scientists have adopted the tools (e.g., LaTex, RMarkdown, etc.) widely used in mathematics and statistics to document the analytic choices used during data analysis. The pipelines presented here provide microbial ecologists with a single platform with which to analyze 16S rRNA gene amplicon data, providing bench scientists with the ability to generate scripts that can be executed by other scientists (or eventually by clinicians in the clinic) and enabling the genesis of figures and findings that are precisely replicable and usable in a variety of other related contexts. Here we have provided examples of three statistical analyses using R and in particular the Rmd format, easily available in RStudio.
7.1. Open Data Access Barriers to Reproducibility
The NIH Open Data Access Policy should dramatically increase our ability to reproduce findings from published datasets in addition to allowing researchers to leverage existing findings in analyses of new experiments. Nonetheless, researchers face several other barriers when trying to access these files. Non-standardized or non-existent mapping files make it difficult to analyze data that have been deposited in public repositories as multiple short read archives.
We advocate the use of platforms (e.g., Github, Bioconductor, CRAN) used by statisticians and bioinformaticians in which experimentalists can deposit not just their sequencing data but also packages containing their complete metadata mapping files, taxonomy files, reference sequence files, which can all be wrapped into a single phyloseq object within R.
7.2. Advantages and weaknesses of the R workow
While rarely achieved in practice, the need for reproducible analysis in amplicon studies has been recognized and there are several existing approaches. The two most common pieces of pipeline software – mothur18 and qiime19 – allow analysis workows to be shared as batch files or IPython notebooks20 respectively. Also, restricted versions of a number of amplicon analysis tools are available through cloud based platforms such as Galaxy.21
However, an amplicon analysis workow in R provides several advantages over these existing approaches. The most compelling advantage is R’s access to a constellation of state-of-the art statistical methods. While common statistical tests have been implemented by pipelines like mothur and QIIME, there is no other platform that has a suite of statistical tools as broad as that implemented in R. In practice, many studies use the popular amplicon pipelines for the initial stages of their analysis, and then transition into R for deeper analysis, with the result being that even when analysis scripts are shared, they only cover part of the full workow. Additionally, R markdown, as implemented by the knitr package, allows the construction of R analysis for which both the underlying code and the full output of that code can be easily shared.
There are also weaknesses in the workow we present. One of those weaknesses is that our current R workow does not include the optimal storage of raw sequence data. A second weakness is in the taxonomic assignment of amplicon sequences or OTUs. While one R package does exist for this purpose (clstutils), it requires parallel computation in an external program (pplacer22), preventing full reproducibility from the R script alone. Taxonomic assignment fully within R is an area of particular interest for future development.
8. Conclusions
We describe reproducible research in the context of studies of the human microbiome. Bioconductor packages, dada2, phyloseq, allow for denoising, handling, filtering, and analyzing high-throughput phylogenetic sequencing data. The phyloseq package provides extensions for leveraging analysis from other ecology-related packages, such as spatstat for spatial data analyses, ade4 for multiway data analyses and lme4 for mixed model analyses. We believe that this workow provides a useful way to document choices in the analyses of phylogenetic sequencing data, its quality control, filtering, processing and its inferential validation.
Fig. 5.
A representation of the output from PCA-IV. The left and right panels show scores along the first and second PCA-IV axes, respectively. Each dot represents a sample, and its position with respect to the image of the teeth shows the area it was sampled from. Color represents whether the sample scored high (blue) or low (red) on the first and second PCA-IV axes. The first PCA-IV axis reveals a complex interaction between tooth aspect, the front vs. the back of the mouth, and jaw. Communities on the lingual aspect of most anterior teeth share negative scores along the first coordinate with a more pronounced difference between tooth aspect in the bottom jaw. The second axis describes a posterior to anterior gradient.
Acknowledgments
We would like to thank the developers of all the open source packages we used including Joey McMurdie for his continuing dedication to phyloseq, Hadley Wickham for ggplot2, Yihui Xie for knitr. We are thankful to the team at RStudio, their IDE and the Shiny platform has made reproducible research much easier. Dan DiGuilio worked with us on the pregnancy study, Elies Bik, Les Dethlefsen, Daniel Sprockett and Jessica Grembi provided useful feedback. This work was partially supported by NSF DMS-1162538 (BC, SPH), NIH R01AI112401(SPH, DAR), NIH R01DE023113 (DAR), NIH R01GM099534 (DAR, SPH) and a Microbiome Seed Grant from Stanford-BioX. DAR is supported by the Thomas C. and Joan M. Merigan Endowment at Stanford University. The supplementary Rmd and html files for this paper can be found at http://statweb.stanford.edu/~susan/papers/PSBRR.html.
Footnotes
Throughout this article we use regular or texttype font for packages/applications with names that are capitalized or uncapitalized, respectively. We use a courier style font for R code, including function and class names. The supplementary material contains Rmd files of the complete R code that enable the reproduction of all the figures in the article.
5 × 100 × 10 × 40 × 16 × 16 × 4 = 204800000
Contributor Information
BENJAMIN CALLAHAN, Statistics Department, Stanford, CA 94305, USA.
DIANA PROCTOR, Departments of Microbiology & Immunology, and Medicine, Stanford University, Stanford, CA 94305 and VA, Palo Alto, CA 94304, USA.
DAVID RELMAN, Departments of Microbiology & Immunology, and Medicine Stanford University, Stanford, CA 94305 and VA, Palo Alto, CA 94304, USA.
JULIA FUKUYAMA, Statistics Department, Stanford University, Stanford, CA 94305, USA.
SUSAN HOLMES, Statistics Department, Stanford University, Stanford, CA 94305, USA.
References
- 1.Pace NR. Science. 1997;276:734. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Applied and Environmental Microbiology. 2006;72:5069. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. Nucleic Acids Research. 2009;37:D141. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. Nucleic Acids Research. 2007;35:7188. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Ole AK, Pages H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Nat Methods. 2015 Feb;12:115. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2015 doi: 10.1038/nmeth.3869. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMurdie PJ, Holmes S. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMurdie PJ, Holmes S. Bioinformatics. 2015;31:282. doi: 10.1093/bioinformatics/btu616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende D, Fernandes G, Tap J, Bruls T, Batto J, et al. Nature. 2011;473:174. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMurdie PJ, Holmes S. PLoS Comput Biol. 2014 Apr;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt TS, Matias Rodrigues JF, Mering C. Environmental microbiology. 2014 doi: 10.1111/1462-2920.12610. [DOI] [PubMed] [Google Scholar]
- 12.DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, Sun CL, Aliaga-Goltsman DS, Wong RJ, Shaw GM, Stevenson DK, Holmes SP, Relman DA. Proceedings of the National Academy of Sciences. 2015;112:11060. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, et al. Science translational medicine. 2012;4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Science. 2009;326:1694. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato Y, Yamagishi J, Yamashita R, Shinozaki N, Ye B, Yamada T, Yamamoto M, Nagasaki M, Tsuboi A. PloS one. 2015;10:e0131607. doi: 10.1371/journal.pone.0131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haffajee A, Teles R, Patel M, Song X, Veiga N, Socransky S. Journal of periodontal research. 2009;44:511. doi: 10.1111/j.1600-0765.2008.01154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes S. Probability and statistics: Essays in honor of David A. Freedman. Institute of Mathematical Statistics; 2008. Multivariate data analysis: the french way; pp. 219–233. [Google Scholar]
- 18.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Applied and environmental microbiology. 2009;75:7537. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. Nature methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragan-Kelley B, Walters WA, McDonald D, Riley J, Granger BE, Gonzalez A, Knight R, Perez F, Caporaso JG. The ISME journal. 2013;7:461. doi: 10.1038/ismej.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova KD, et al. Genome research. 2007;17:960. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsen FA, Kodner RB, Armbrust EV. BMC bioinformatics. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]