Abstract
The nearly universal recurrence of glioblastoma (GBM) is driven in part by a treatment-resistant subpopulation of GBM stem cells (GSCs). To identify improved therapeutic possibilities, we combined the EGFR/HER2 inhibitor lapatinib with a novel small molecule, CBL0137, which inhibits FACT (Facilitates Chromatin Transcription), a histone chaperone complex predominantly expressed in undifferentiated cells. Lapatinib and CBL0137 synergistically inhibited the proliferation of patient-derived GBM cells. Compared to non-stem tumor cells (NSTCs) enriched from the same specimens, the GSCs were extremely sensitive to CBL0137 monotherapy or FACT knockdown. FACT expression was elevated in GSCs compared to matched NSTCs, and decreased in GSCs upon differentiation. Acute exposure of GSCs to CBL0137 increased asymmetric cell division, decreased GSC marker expression, and decreased the capacity of GSCs to form tumor spheres in vitro and to initiate tumors in vivo. Oral administration of CBL0137 to mice bearing orthotopic GBM prolonged their survival. Knockdown of FACT reduced the expression of genes encoding several core stem cell transcription factors (SOX2, OCT4, NANOG and OLIG2), and FACT occupied the promoters of these genes. FACT expression was elevated in GBM tumors compared to non-neoplastic brain tissues, portended a worse prognosis, and positively correlated with GSC markers and stem cell gene expression signatures. Preferential targeting of GSCs by CBL0137 and synergy with EGFR inhibitors support the development of clinical trials combining these two agents in GBM.
Keywords: Glioblastoma stem cells, SSRP1, curaxin, CBL0137, lapatinib
Introduction
Despite the use of aggressive, multimodal treatments for glioblastoma (GBM), the median time to tumor recurrence is 6.9 months, and the median patient survival is 14 months (1, 2). Therefore, there is an urgent need for new, effective therapies. In the majority of GBMs, aberrant amplification, an in frame deletion (e.g., EGFRvIII), or mutation of the EGF receptor (EGFR) has been identified (3, 4). The high frequency of EGFR aberrations has generated considerable interest in using EGFR tyrosine kinase inhibitors (TKIs), which have provided significantly increased progression-free survival in other cancers, such as non-small cell lung cancer (NSCLC) (5). Nevertheless, the promising results in preclinical studies have failed to translate into improved outcomes in the clinic (6–10). One of the proposed mechanisms of resistance to EGFR TKIs is the compensatory up-regulation of related ERBB family members in GBM cells with stem-like properties (11).
Numerous studies provided evidence of a hierarchal organization within GBMs, commonly termed the cancer stem cell (CSC) hypothesis. CSCs are less susceptible to radiotherapy and chemotherapy and are more capable of initiating secondary tumor formation in transplantation assays when compared to non-stem tumor cells (NSTCs) from the same tumors (12–14). Similarly to normal stem cell populations, CSCs self-renew, differentiate, and express many of the same core transcription factors (14). For GBM, several cell-surface markers, including CD133, CD44, CD15, EGFR, L1CAM, and integrin α6, are used to enrich for stem-cell populations, known as GBM stem cells (GSCs) (14, 15). The contribution of GSCs to tumor recurrence following therapeutic intervention necessitates new strategies to target and eradicate this cell population in GBM.
FACT (facilitates chromatin transcription) is a histone chaperone that aids transcriptional elongation by the disassembly and reassembly of nucleosomes, thereby allowing the passage of RNA polymerase II through chromatin during transcription (16). Recently, FACT was found to play a role in maintaining cells in an undifferentiated state by being part of a stem cell-like gene expression signature (17). FACT is also highly expressed in high grade, poorly differentiated cancers and portends worse survival in these tumors (18). Targeting FACT with a novel class of small molecule inhibitors, termed curaxins (19, 20), has been tested in a number of cancers, including NSCLC (21), pancreatic cancer (22), breast cancer (23), and, neuroblastoma (24). Curaxins comprise a novel class of carbazole-based anticancer compounds, discovered as functional analogs of quinacrine, a 9-aminoacridine derivative used originally as an anti-malarial drug (19). More recently, both quinacrine and curaxins were found to harbor anti-tumor activity by targeting FACT, which subsequently leads to inhibition of NF-κB-dependent transcription (19, 20). We have previously shown that quinacrine treatment can overcome resistance to the EGFR TKI erlotinib in NSCLC by targeting FACT and inhibiting NF-κB (21). We now investigate the use of CBL0137, a curaxin currently undergoing multicenter phase I clinical trials in metastatic or unresectable advanced solid neoplasms or refractory lymphomas (NCT01905228), in GBM. We treated patient-derived GBM specimens, maintained as xenografts, with the combination of CBL0137 and lapatinib. In particular, we tested the impact of CBL0137 on GSCs, defined as CD133-positive (CD133+) cells, in comparison to NSTCs, or CD133-negative (CD133−) cells, and the potential role of FACT in maintaining the stem cell phenotype of GSCs.
Materials and Methods
Reagents
Lapatinib was obtained from Selleck Chemicals (# S2111). CBL0137 was provided by Incuron, LLC. For immunoblotting, mouse β-actin antibody (# A5316) was from Sigma; rabbit GFAP antibody (# Z0334) was from Dako; rabbit histone H3 (#4499) was from Cell Signaling; mouse OLIG2 (# ab56643) antibody was from Abcam; rabbit SOX2 antibody (# A301-741) was from Bethyl Laboratories; mouse SSRP1 antibody (# 609701) was from BioLegend. For immunofluorescence assays, mouse CD133/1 (#W6B3C1) antibody was from Miltenyi Biotec.
Isolation and culture of CD133-positive GSCs
Primary patient-derived brain tumor patient specimens were obtained in compliance with Institutional Review Board approved protocol. Known tumor grade and available cytogenetic information (relevant to pathologic classification) for each specimen has been previously published (13, 25, 26). Cells were authenticated by short tandem repeat (STR) analysis by the Duke University DNA Analysis Facility. EGFR amplification status (% cells positive for polysomy) is as follows: 3691 (83%), 08-387 (WT), 3565 (unknown), 4121 (WT, 48% of cells have loss of EGFR locus), CW718 (unknown), 4302 (62%). The specimens were maintained as subcutaneous xenografts in the flanks of NOD scid gamma (NSG) mice. Tumors cells were dissociated using a papain dissociation system (Worthington Biochemical). CD133+ cells were enriched by magnetic-activated cell sorting (MACS; Miltenyi Biotech). All cells were cultured at 37 °C at 5% CO2 and utilized in under 5 passages. CD133+ were cultured in Neurobasal media (Invitrogen) supplemented with B27 (Invitrogen), basic FGF (10 ng/ml; R&D Systems), EGF (10 ng/mL; R&D Systems), l-glutamine (2 mM; Invitrogen), and sodium pyruvate (1 mM; Invitrogen). CD133− cells were cultured in DMEM with 10% FBS. CD133+ and CD133− cells in cell proliferation assays (IncuCyte) were maintained in growth factor-free Neurobasal medium (supplemented with the above components except for growth factors). Unsorted bulk tumor cells were cultured in supplemented Neurobasal media mixed with DMEM with 10% FBS at a 1:1 ratio.
Cell viability assay
Cell viability was determined using the CellTiter-Glo Luminescent Assay (Promega). The combination index (CI) was assessed by CompuSyn software (ComboSyn, Inc) (27). For apoptosis, at 48 hours caspase 3/7 activity was measured by CaspaseGlo 3/7 Assay (Promega). Alternatively, cell confluence was monitored every hour for up to 4 days using the IncuCyte ZOOM live cell imaging system (Essen BioScience), which enables non-invasive, continuous, and direct evaluation of cell numbers and growth using live cell time-lapse microscopy.
Flow cytometric analysis and sorting
Flow cytometry was performed using a FACS Aria II Cell Sorter (BD Biosciences). To enrich for CD133+ cells, single cells were labeled with a phycoerythin-conjugated monoclonal antibody against CD133 (CD133/1, AC133; Miltenyi Biotec). Dead cells were eliminated by DAPI staining (1 μg/mL, added immediately prior to sorting).
Limiting dilution assay and sphere formation
GSCs were plated using FACS-sorting into 96-well plates at 1, 5, 10, 20, or 50 cells/well in supplemented Neurobasal medium. After 14 days, wells were scored for the presence of at least one tumor sphere. Estimated stem cell frequency was calculated using extreme limiting dilution analysis (28).
Mitotic cell collection and analysis
GSCs were treated with vehicle, 300 nM CBL0137, or 3 μM lapatinib for 9 hours. Collection of mitotic pairs and quantification of asymmetry was performed as in (29) and detailed in supplementary materials.
Lentiviral transduction
Lentiviral plasmids encode shRNAs targeting GFP, SSRP1 (TRCN0000019270, “#1″; TRCN0000019272, “#2″) or SUPT16H (SPT16) (TRCN0000001258250, “#2″, TRCN00000001260250, “#4″).
Quantitative RT-PCR
Total mRNA was isolated using RNeasy kit (Qiagen) and reversed transcribed into cDNA using the SuperScript Reverse Transcription Kit (Life Technologies). Real-time PCR was performed on the LightCycler 480 Instrument II using the SYBR Green Master Mix (Roche). Gene-specific primers are in supplementary materials.
Chromatin immunoprecipitation
SSRP1-bound chromatin from GSCs was immunoprecipitated using mouse SSRP1 antibody (# 609701) or mouse IgG (sc-2025). Details are in supplementary materials.
ChIP-qPCR
Two μL of DNA from each ChIP sample was used for qPCR reaction Primers and cycling conditions are in supplementary materials.
In vivo studies
All animal studies were approved by the Cleveland Clinic Foundation IACUC and conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. For intracranial implantation studies, 10 × 103 viable GSCs were implanted into the left striate nucleus of four to six months old NSG mice. Mice were monitored daily for neurological impairment at which time they were sacrificed. For in vivo treatment, 7 d after intracranial implantation of GSCs, DMSO or CBL0137 (0.5 mg/mL) was added to drinking water and replaced every 7 d.
Bioinformatics and Statistical analysis
Statistical analyses were conducted using Graphpad Prism 5 unless otherwise stated. Results are represented by means ± SD. The Gene Set Enrichment Analysis (GSEA) tool (30) was used to analyze single-cell RNA-seq data in primary glioblastoma from GSE57872 (n=430) (31) using stem cell-related gene sets (n=56) from the Molecular Signature Databases at the Broad Institute (MSigDB) (32–35).
Results
GBM cells are resistant to lapatinib but sensitive to CBL0137 alone and CBL0137 plus lapatinib
Previously, we reported that the combination of erlotinib with quinacrine, a curaxin predecessor, is synergistic in NSCLC (21). CBL0137 is a second-generation FACT inhibitor and more potent than quinacrine (20). Importantly, in orthotopic xenograft models of GBM, CBL0137 achieves effective CNS penetration, as evidenced by its high concentration in normal brain tissues in mice (383 μM at 0.5 h after IV injection of 70 mg/kg CBL0137 and steadily decreases with time to 9 μM at 24 h, which is still 30 times above its in vitro IC50 of 300 nM against GBM cells (Supplementary Fig. S1); further, its concentration in tumors was elevated in comparison to normal brain tissues (Barone T, et al., Manuscript under review, February 2016). In these models, CBL0137 has also been shown to significantly prolong the survival of tumor-bearing mice (36).
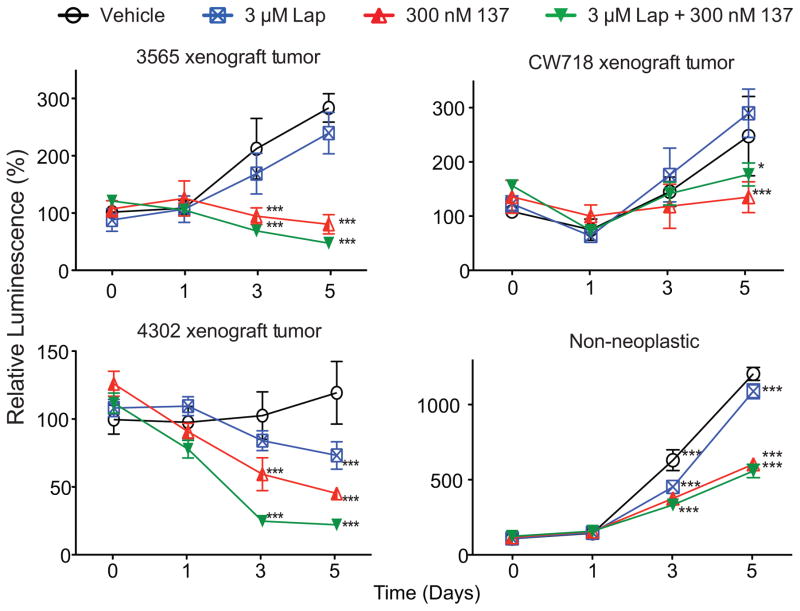
We performed a longitudinal drug treatment experiment with lapatinib alone, CBL0137 alone, or both in unsorted cells from several patient-derived GBM tumor specimens and a non-neoplastic brain specimen (Fig. 1). There was little or no effect of lapatinib in inhibiting tumor cell growth. Importantly, treatment with CBL0137 alone or in combination with lapatinib was highly effective in inhibiting cell proliferation in all three tumor specimens.
Figure 1. GBM cells are resistant to lapatinib but sensitive to CBL0137 and CBL0137 plus lapatinib.
Cells from xenografted GBM patient-derived tumor specimens, or a nonneoplastic brain specimen were treated with the indicated concentrations of lapatinib (Lap) and/or CBL0137 (137). Cell proliferation at the indicated time-points was normalized to the values from Day 0 [*, P<0.05; ***, P<0.001 with two-way ANOVA followed by Bonferroni’s posttest compared to Day 0; all values represent means ± SD].
Prolonged exposure to CBL0137 preferentially kills GSCs over NSTCs and synergizes with lapatinib
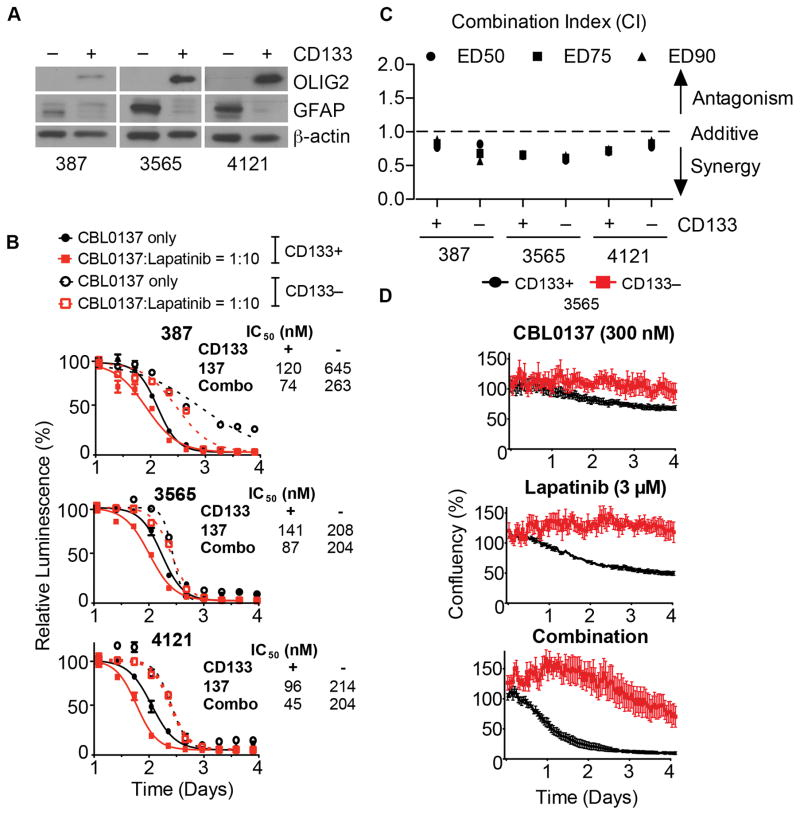
We sorted GBM patient tumor specimens into GSCs and NSTCs based on CD133 (13, 25, 26) and then tested for differences in sensitivity to CBL0137 between GSCs and NSTCs. Sorting purity was confirmed by immunoblotting for the independent stem cell marker OLIG2 (14), present only in the CD133+ cells, or glial fibrillary acidic protein (GFAP), a marker for astrocytes and more differentiated cells (15), expressed only in the CD133− cells (Fig. 2A).
Figure 2. Prolonged exposure to CBL0137 preferentially kills GSCs over NSTCs and synergizes with lapatinib.
A, Purity of CD133+ and CD133− cell sorting was validated by immunoblotting OLIG2 and GFAP, with β-actin as the loading control. B, GSCs (CD133+, solid lines) and NSTCs (CD133−, dotted lines) cells treated with the indicated concentrations of lapatinib, CBL0137 (black lines) or CBL0137 plus lapatinib at a 1:10 ratio (red lines). After 72 h, cell viability was determined and normalized to untreated controls. IC50s in response to single and combination drug treatments are shown. C, Synergy was determined by calculating combination indices (CI) based on the dose-response data. A CI < 0 indicates synergy. D, GSCs (CD133+, black) and NSTCs (CD133−, red) cells were seeded in growth factor-free (GFF) Neurobasal media precoated with Geltrex. Thereafter, 300 nM CBL0137, 3 μM lapatinib, 300 nM CBL0137 plus 3 μM lapatinib, or vehicle (DMSO) were added. Cell confluence was monitored by the IncuCyte ZOOM system every hour thereafter and normalized to vehicle-treated wells.
In all three patient-derived tumor specimens, both GSCs and NSTCs were highly sensitive to CBL0137, with IC50s (half maximal inhibitory concentrations) ranging between 100–600 nM (Fig. 2B); whereas IC50s for lapatinib were between 1–6 μM, approximately 10 fold higher (Supplementary Fig. S2A). EGFR expression and amplification status in the CD133+ and CD133− population are indicated in the method section and Supplementary Fig. S2B respectively.
To evaluate synergy between lapatinib and CBL0137, we combined them at a molar ratio of 10 to 1, based on their IC50s, and repeated the dose-response experiments. GSCs were more sensitive to the combination compared to NSTCs; in fact, the combination was able to reduce the IC50 of CBL0137 for GSCs to values below 100 nM (Fig. 2B). The degrees of synergism were quantified using Chou and Talalay’s median-drug effect analysis method (27). The combination of lapatinib and CBL0137 was synergistic in all three patient-derived tumor specimens, regardless of their CD133 status, when combined at 10:1 ratios, as indicated by combination indices (CIs) below 1.0 (Fig. 2C).
To confirm the above findings, we treated GSCs or NSTCs with 300 nM CBL0137, 3 μM lapatinib, or CBL0137 plus lapatinib, and monitored cell confluence over time using live-cell microscopy (IncuCyte Zoom). CBL0137 or lapatinib significantly decreased cell proliferation of GSCs compared to that of NSTCs, and combination treatment completely inhibited cell proliferation in GSCs but not NSTCs (Fig. 2D). Consistently, significantly more apoptosis was detected in GSCs compared to NSTCs treated with the same doses of CBL0137 (Fig. 3A).
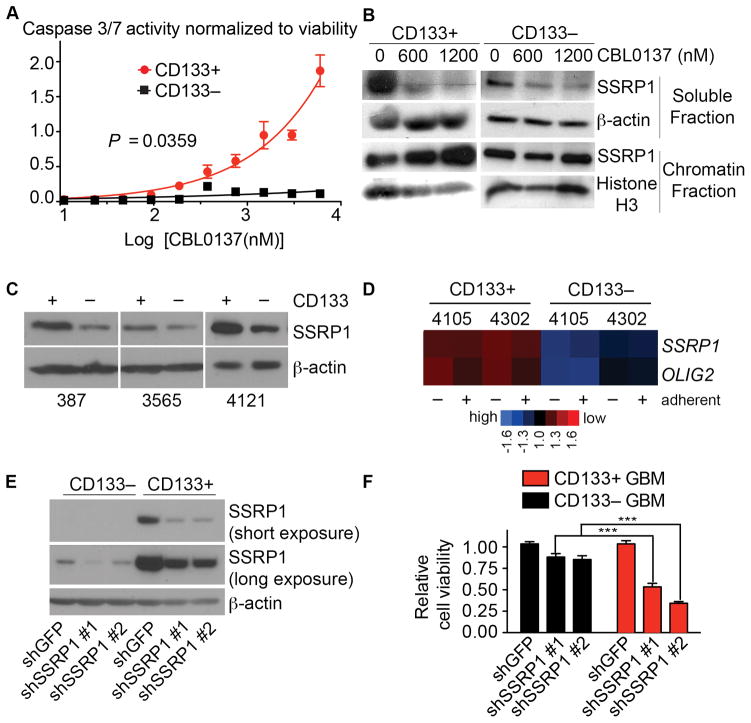
Figure 3. CBL0137 preferentially kills GSCs over NSTCs by targeting FACT.
A, GSCs (CD133+, red circles) and NSTCs (CD133−, black squares) were treated with CBL0137 for 48 h. Caspase 3/7 activity was measured by CaspaseGlo and normalized to viability as determined by CellTiter Glo. B, GSCs or NSTCs cells were treated with 0, 600 or 1200 nM CBL0137 for 5 h. Levels of SSRP1 in soluble and chromatin-bound fractions were detected by immunoblotting, using β-actin or histone H3 as loading controls. C, SSRP1 in GSCs and NSTCs were probed by immunoblotting in whole cell extracts. D, SSRP1 and OLIG2 expression levels were obtained from an independent microarray profiling (GSE24716) of GSCs and NSTCs cells from two human glioma xenograft tumors (4105, 4302) cultured with (adherent) or without (in suspension) laminin (33). E, CD133+ or CD133− cells were transduced with lentiviral vectors bearing puromycin resistance gene and two different shRNAs against SSRP1 or shGFP. Knockdown efficiency was evaluated by immunoblotting of SSRP1. F, Following puromycin selection, equal numbers of GSCs and NSTCs transduced with shRNAs against SSRP1 or shGFP were plated and assayed for cell viability normalized to the values for shGFP.
CBL0137 preferentially kills GSCs over NSTCs by targeting FACT
Curaxins were shown to induce chromatin trapping of FACT, a dimer of SSRP1 and SPT16 (20). Consistently, we observed that CBL0137 depleted FACT from the soluble nucleoplasmic fraction and led to its accumulation in the insoluble chromatin fraction (Fig. 3B), which indicates FACT inhibition (20, 22). Importantly, we observed significantly higher protein levels of the FACT subunits in GSCs compared to NSTCs in untreated cells, which was validated in three independent specimens (Fig. 3C). This was supported by higher SSRP1 mRNA expression in independent specimens from a transcriptome profiling of CD133+ versus CD133− GBM cells (Fig. 3D) (37).
To test whether the increased sensitivity to CBL0137 might be due to a higher expression and reliance on FACT in GSCs than in NSTCs, we transduced them with two different shRNAs to SSRP1 or scrambled shRNA and assayed cell viability. Consistent with our hypothesis, depletion of SSRP1 significantly decreased cell viability in the GSCs compared to the NSTCs (Fig. 3E, F).
Acute exposure to CBL0137 attenuates GSCs self-renewal and tumor initiation
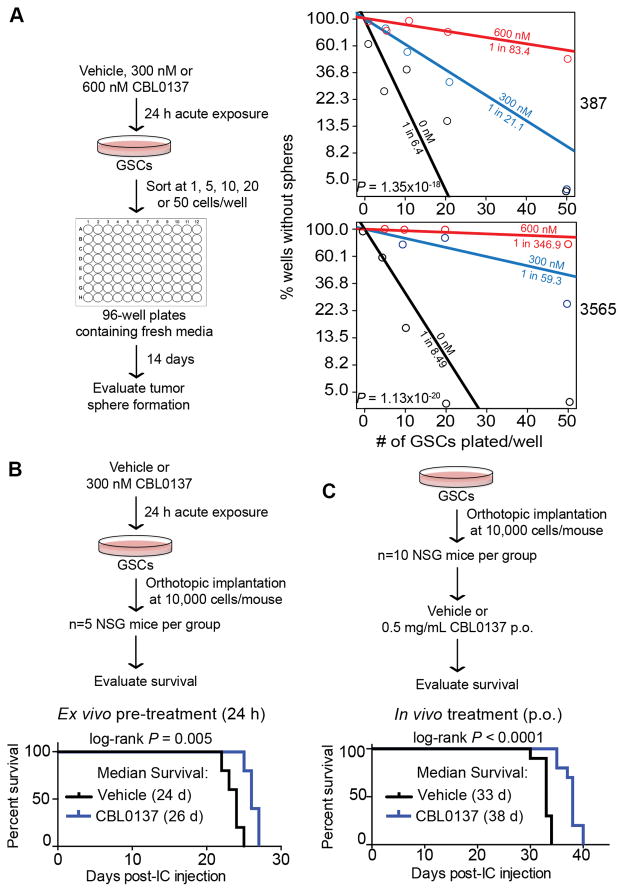
A hallmark of tumor-initiating cells is their ability to form a tumorsphere from a single cell in vitro (14). We assessed whether CBL0137 impacts the ability of GSCs to form tumorspheres in an in vitro extreme limiting dilution assay (eLDA), which permits quantified estimation of stem-like cell frequencies (28). We exposed GSCs to vehicle, 300 nM, or 600 nM of CBL0137 for 24 h. Cells were then washed and plated at densities of 1, 5, 10, 20, or 50 cells per well. CBL0137 markedly decreased self-renewal of the GSCs based on evaluation of tumorsphere formation 14 days later (Fig. 4A).
Figure 4. Acute exposure to CBL0137 attenuates GSCs self-renewal and tumor initiation.
A, For in vitro extreme limiting dilution assays (eLDA), GSCs were pretreated with vehicle (DMSO), 300 nM or 600 nM CBL0137 for 24 h before plating. Fourteen days later, each well was evaluated for the presence or absence of tumor spheres. Estimated stem cell frequencies for each condition are indicated. P-values indicate significance of differences in stem cell frequencies between groups (chi-square test). B–C, CBL0137 reduces tumor-initiating capacity of GSCs in vivo. 10 x 103 of viable cells were intracranially (IC) implanted. The Kaplan–Meier survival curves post-intracranial implantation of GSCs are shown [P=0.005 with log-rank analysis]. B, For ex vivo treatment, GSCs (specimen 387) were pre-treated for 24 h with vehicle (DMSO) or 300 nM CBL0137 prior to IC implantation (n=5 per group). C, For in vivo treatment, 7 d after IC implantation (specimen 3691), mice receive vehicle or CBL0137 (0.5 mg/mL) orally ad libitum by addition of drug to drinking water, which was replaced every 7 d (n=10 per group).
To examine whether CBL0137 treatment has an impact on tumor initiation and survival of tumor-bearing animals in vivo, we pretreated GSCs with vehicle or CBL0137 and intracranially implanted viable cells into immunocompromised mice. Exposure of GSCs to low dose CBL0137 (300 nM) for 24 h ex vivo prior to xenografting was sufficient to increase latency in tumor initiation and prolonged survival (Fig. 4B). Of note, whenever we examined the impact of CBL0137 on stem cell phenotypes, we chose a time-point of 24 h or less for treatment with 300 nM CBL0137 based on our analysis of cell viability in CD133+ cells, which showed no immediate or delayed cell death compared to vehicle (Supplementary Fig. S3), indicating the decrease in overall stemness was not due to a decrease in viability.
To evaluate in vivo efficacy, mice with intracranially implanted GSCs were given DMSO or CBL0137 in their drinking water (at libitum, 0.5 mg/ml). Treatment with CBL0137 significantly prolonged survival (Fig. 4C).
Acute exposure to CBL0137 decreases CD133 cell-surface expression and increases asymmetric cell division in GSCs
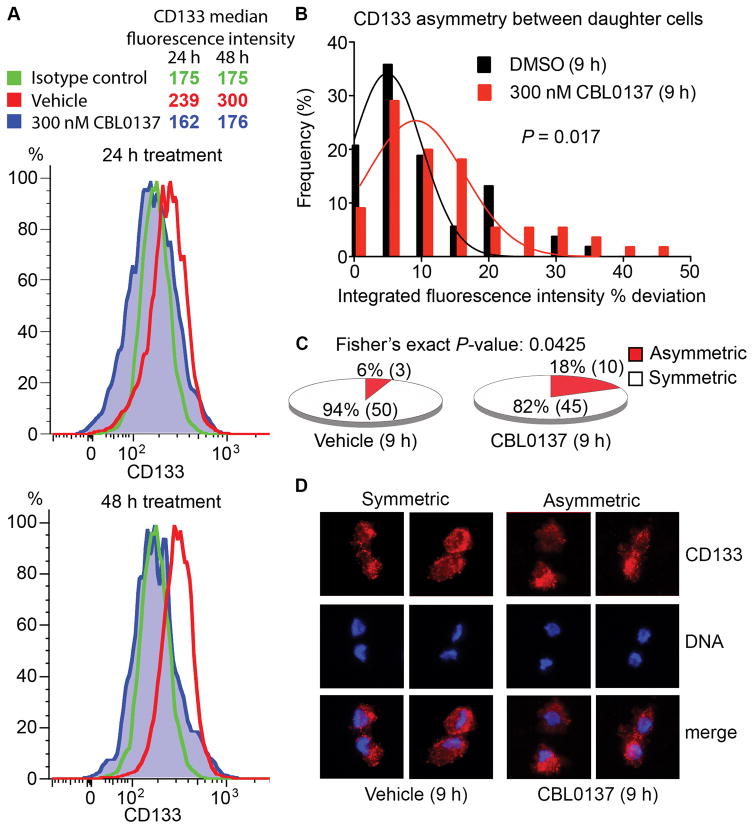
To further explore the impact of CBL0137 on GSC phenotypes, we treated GSCs with 300 nM CBL0137 and analyzed the expression of CD133 in the viable population. We observed a decline in CD133 levels with drug treatment compared to vehicle (Fig. 5A). Recently, GSCs have been shown to undergo predominantly symmetric rather than asymmetric cell division at the single cell level (29). Asymmetric cell division is an important mechanism by which normal stem cells maintain a constant stem cell population and also generate differentiated progeny, whereas symmetric cell division has been proposed to occur at a higher frequency in tumors to amplify the fraction of CSCs (38). We hypothesized that a switch from symmetric to asymmetric cell division could account for the decrease in CD133+ cells. To this end, we exposed GSCs to vehicle or CBL0137 (300 nM) for 9 h, collected mitotic cells, and then measured the frequencies of symmetric and asymmetric cell division by quantifying the degree of polarization of CD133 fluorescence signals between the dividing daughter cells (% deviation). Similarly to previous reports (29), asymmetric cell division was a rare event in the GSCs we examined (6% in the vehicle control). CBL0137 treatment significantly increased the frequency of asymmetric cell division in GSCs to 18% (Fig. 5B–D). As a positive control, we observed a similar but less significant effect when we exposed the cells to 10% fetal bovine serum, an agent commonly used to differentiate GSCs (Supplementary Fig. S4). Our finding suggests that CBL0137 can shift the balance from symmetric cell division to asymmetric cell division, implying increased differentiation, leading to a decrease in the frequency of CD133+ cells among the GSCs.
Figure 5. Acute exposure to CBL0137 decreases CD133 cell-surface expression and increases asymmetric cell division in GSCs.
A, GSCs in suspension were treated with DMSO or 300 nM CBL0137 for 24 or 48 h, and then stained with DAPI, isotype control or an anti-CD133-PE antibody. CD133 expression was subsequently analyzed by FACS within the DAPI-negative populations. B, Following release from 15 h of thymidine block, GSCs were treated with DMSO or 300 nM CBL0137 for 9 h. The histograms of the frequency distribution of the % deviation of CD133 staining (Integrated fluorescence intensities, IFI) between daughter cells in mitotic pairs in vehicle- (n=53) and drug-treated (n=55) are shown [one-tailed P-value with student’s t-test]. C, Fractions of symmetric vs. asymmetric distribution in the vehicle (50 vs. 3) and CBL0137 (45 vs. 10) group are represented as pie charts [frequency comparison with Fisher’s exact test]. Values above or below mean ± 2SD of the % deviation of CD133 fluorescence signals from the vehicle group (i.e., −24 to 25.2%) were used as the cutoff for asymmetric distribution. D, Representative fluorescence micrographs showing increased asymmetric distribution of CD133 staining in the CBL0137-treated cells compared to vehicle controls.
FACT is involved in the expression of regulatory stem cell transcription factors
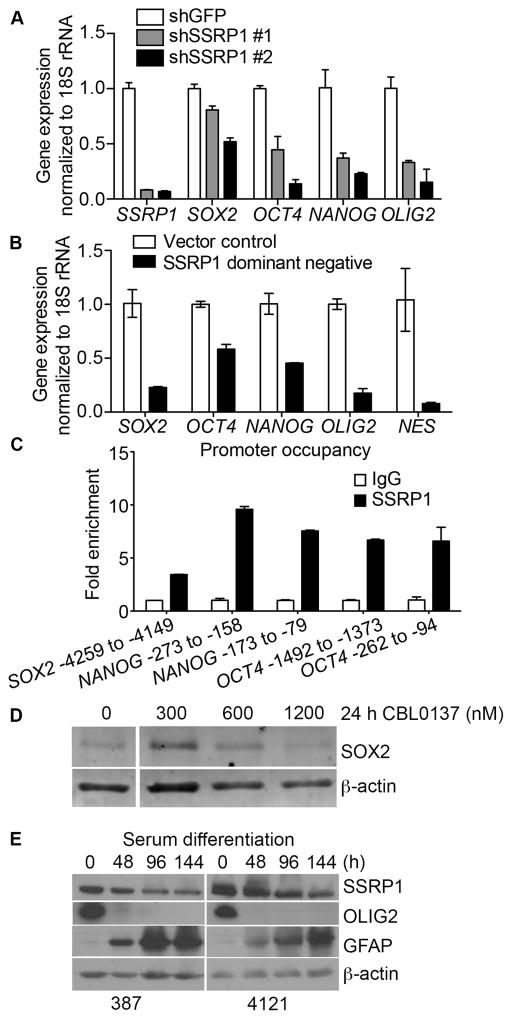
We measured the expression of the stem cell markers SOX2, OCT4, NANOG and OLIG2 following suppression of FACT expression in GSCs (12–15). Knockdown of SSRP1 significantly reduced the levels of these mRNAs (Fig. 6A). This was corroborated when we overexpressed the essential dimerization domain of SSRP1 (SSRP1 DD), a dominant negative derivative of FACT that prevents the dimerization of endogenous SSRP1 and SPT16 (39), and observed a reduction in the expression of SOX2, OCT4, NANOG, OLIG2 and NES (Fig. 6B). Further, using chromatin immunoprecipitation we found that FACT binds to the promoters of the SOX2, OCT4 and NANOG genes (Fig. 6C) in GSCs, further implicating its potential role in regulating the expression of these genes. Consistently, using SOX2 as a surrogate stemness marker, we detected a dose-dependent reduction of SOX2 protein levels in GSCs treated with CBL0137 (Fig. 6D).
Figure 6. FACT is involved in the expression of regulatory stem cell transcription factors.
A–B, The expression of SSRP1, SOX2, OCT4, NANOG, OLIG2, and NES mRNA, normalized to 18S rRNA, in GSCs transduced with shGFP or two different shRNAs against SSRP1 (A), or SSRP1 DD (SSRP1 dimerization domain) vs. vector control (B), was measured using qPCR. C, ChIP was performed in GSCs by using an antibody against SSRP1 or mouse IgG and subsequent qPCR analysis of the promoters of the SOX2, NANOG and OCT4 genes. D, GSCs were plated onto Geltrex-coated plates, and treated with 0, 300, 600 or 1200 nM CBL0137 for 24 h. Changes in the levels of SOX2 were detected by immunoblotting. E, GSCs were plated into Geltrex-precoated plates and exposed to 10% FBS-containing media for 0, 2, 4, or 6 days. Changes in the levels of SSRP1, OLIG2, and GFAP over the course of six days of serum differentiation were evaluated by immunoblotting.
To explore whether FACT expression correlates with the differentiation status of GSCs, we treated them with 10% FBS to induce differentiation. In response to serum, OLIG2 decreased rapidly while GFAP increased (Fig. 6E). Importantly, SSRP1 expression also decreased with differentiation.
FACT is elevated in GBMs compared to normal brain tissues and its level correlates with survival and GSC gene signatures
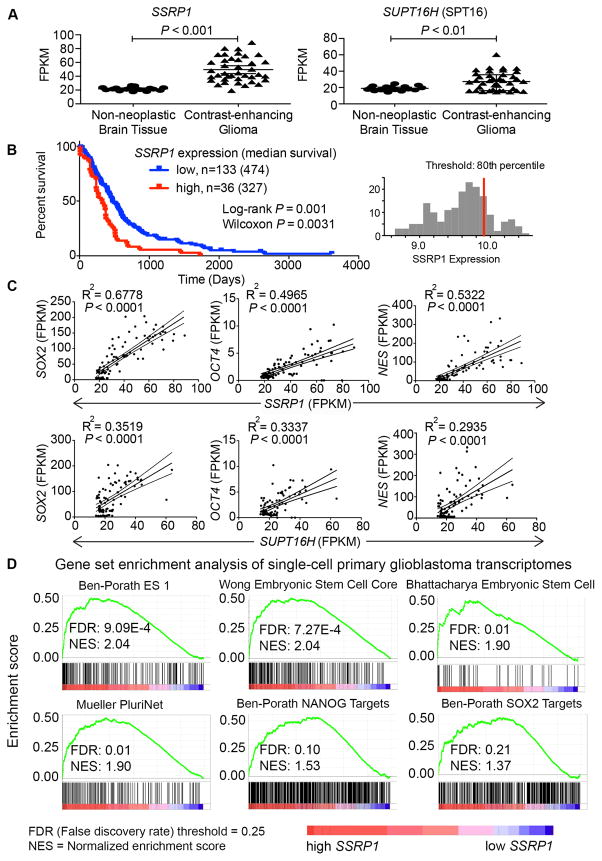
Gill et al (40) recently performed an RNA-seq analysis of GBM samples from radiographically-guided biopsies (n=39) versus non-neoplastic brain (n=17) (GSE9612). Comparing SSRP1 and SPT16 gene expression between these two groups, we found a markedly elevated FACT expression in GBM compared to nonneoplastic brain tissues (Fig. 7A). Moreover, analysis of GBM patient survival data from NIH’s REpository for Molecular BRAin Neoplastic DaTa (REMBRANDT) (41) showed that upregulation of SSRP1 expression (> 80th percentile, n=36), compared to low expression (n=133), is significantly associated with worse survival (Fig. 7B). Additionally, SSRP1 and SPT16 expression is positively correlated with well-established cancer stem cell markers: SOX2, OCT4, and NESTIN in the contrast-enhanced gliomas (Fig. 7C). Interestingly, there is no positive correlation between the expression of FACT and the GSC markers in the non-neoplastic brain tissues (Supplementary Fig. S5).
Figure 7. FACT is elevated in GBMs compared to normal brain tissues and its level correlates with survival and GSC gene signatures.
A, Normalized RNA expression levels in fragments per kilobase of transcript per million mapped reads (FPKM) of SSRP1 and SUPT16H were retrieved from GSE59612 (40) comparing their expression levels between contrast-enhancing glioma (n=39) versus non-neoplastic brain tissues (n=17) [unpaired Student’s t-test]. B, Kaplan-Meier survival plot of GBM patients with high (n=36) and low (n=133) expression of SSRP1 was downloaded from REMBRANDT (41). C, Expression levels (FPKM) of SOX2, OCT4, and NES were retrieved from RNA-seq data of contrast-enhancing glioma tissues (n=39) from GSE59612 and plotted against those of SSRP1 and SUPT16H [R2, regression coefficients; P values for linear regression]. D, Enrichment plots from GSEA for stem cell-related gene sets (false discovery rate, FDR ≤ 25%) in the primary single cell glioblastoma transcriptomes (GSE57872) expressing high SSRP1 (> 75 percentile, n=107) versus low SSRP1 (< 25 percentile, n=227).
Furthermore, since the existence of GSCs in GBMs reflects intratumoral heterogeneity, we examined RNA-seq data from 430 primary single cell glioblastoma transcriptomes (GSE57872) (31). Data from samples with high SSRP1 (> 75 percentile, n=107) or low SSRP1 expression (< 25 percentile, n=227) were input into the GSEA tool (30) to analyze for enrichment of gene sets that are associated with stem cell identities (32–35). A positive normalized enrichment score reflects the degree to which genes positively correlated with the high SSRP1 phenotype are overrepresented in the gene sets analyzed, and is normalized to the size of the gene sets. Genes positively correlated with high SSRP1 expression were significantly enriched in gene signatures associated with stemness (Fig. 7D, GSE57872). None of the 56 stem cell-related gene sets analyzed were significantly enriched in samples with the low SSRP1 phenotype.
Taken together, our data support the efficacy of CBL0137 against GSCs and strongly suggest the importance of FACT in maintaining a stem cell phenotype in GBM, offering preclinical insight into targeting the stem cell-like neoplastic population and possibly reducing tumor recurrence.
Discussion
To date, no targeted therapy against GBM has been successfully translated into improved survival. For instance, anti-EGFR therapies such as erlotinib and lapatinib have failed to reproduce in GBM the success seen in the treatment of NSCLC (8–10). The standard-of-care treatment in GBM, extensive surgical resection followed by radiotherapy and chemotherapy (mostly temozolomide), may allow treatment-resistant GSCs to survive. Hence, tumor recurrence after initial eradication has been attributed to GSCs due to their inherent or acquired resistance to therapy, e.g., elevated DNA damage repair activity and aberrations in apoptotic pathways (12–15, 42). To identify a drug combination to overcome resistance to lapatinib, we examined CBL0137, previously shown to have anticancer activity in multiple tumor types (18–24). CBL0137 was also shown to accumulate to high levels in normal brain tissues and even more so in brain tumors (Supplementary Fig. S1 and Barone T, et al. Manuscript under review, February 2016), and to significantly increase survival in orthotopic GBM preclinical models (36).
We discovered a biphasic impact of CBL0137 on GSC: prolonged treatment with CBL0137 preferentially impairs the viability of GSCs over NSTCs by targeting FACT, whereas acute exposure compromises crucial stem cell phenotypes, including expression of stem cell markers, capacity for self-renewal, and in vivo tumor initiation. We also showed higher FACT expression in GSCs compared to NSTCs, both in our own panel of patient specimens and in external, independent datasets, and that FACT expression correlates with stem cell gene signatures in patient samples. We are aware of the limitations and controversies surrounding the use of CD133 as a GSC marker. Nevertheless, for the particular models that we use, we have validated the functional segregation of tumor sphere formation and in vivo tumor initiation using CD133 sorting (12, 25, 26). In addition, we showed that sorting with CD133 was sufficient to segregate the GBM cells into those predominantly expressing GSC but not differentiation markers, or vice versa.
Our observation that FACT depletion reduces expression of the core stem cell transcription factors SOX2, OCT4, NANOG and OLIG2 points to its possible regulatory role in stem cell transcriptional programming (43). A recent ChIP-seq experiment performed in human fibrosarcoma cells showed an enrichment of SSRP1 at transcription factor genes, e.g., OCT4, which are involved in embryonic development or highly expressed in poorly differentiated cancers (18).
Our finding that exposure to CBL0137 switches GSCs from a symmetric to an asymmetric mode of cell division and reduces CD133 expression suggests that CBL0137 reduces the CD133+ GSC population by promoting a switch towards asymmetric cell division Certain oncogenes in cancer stem cells disrupt the tightly regulated asymmetric mode of cell division in normal stem cells by promoting symmetric cell division and thereby uncontrolled expansion of stem cells (38). Therefore, FACT could play a role in sustaining the generation of the GSC population within the tumor and thus permitting the progressive development of cancer.
A recent pancreatic cancer study using CD24Hi/CD44Hi as CSC markers showed that CBL0137 decreased their numbers in vitro and that SSRP1 was more highly expressed in this population compared to the CD24Lo/CD44Lo population (22). Our study, in addition to demonstrating increased FACT expression in CD133+ GSCs compared to CD133− NSTCs, demonstrate not only the ability of CBL0137 to kill GSCs, but also how this effect was more marked compared to that in matched NSTCs. Furthermore, the novelty and significance of our study lie in our findings that CBL0137 treatment or FACT depletion affects several defining characteristics of cancer stem cells (14–15): in vitro self-renewal capacity, in vivo tumor-initiating capacity, expression of core stem cell transcription factors, and propensity to undergo asymmetric cell division, which in turn potentially drives differentiation into NSTCs. In addition, we showed synergy between CBL0137 and lapatinib, illustrating the potential of combining FACT targeting agents and EGFR TKIs to improve treatment outcome of the latter. We investigated EGFR protein levels between the CD133+ and CD133− populations (Supplementary Fig. S2B). It is difficult to make a conclusion regarding drug synergies based on EGFR immunoblot data alone especially in light of the dual targeting of lapatinib to HER2 and the reported elevation of HER2 in GSCs (11). Follow-up studies could include manipulation of these receptors in both GSCs and NSTCs to better understand the difference in drug sensitivity between the GSCs and NSTCs as well as elucidation of the mechanism behind the synergy. Nonetheless, our studies highlight the novel indication of CBL0137 for GBM.
Therefore, this is the first comprehensive study that not only holds promising translational potential by showing the novel, preferential targeting of cancer stem cells by CBL0137, but also elucidates FACT’s role in regulating crucial cancer stem cell-defining functions by detailed mechanistic investigations.
CBL0137 is currently undergoing multicenter phase I clinical trials in advanced or metastatic solid tumors and lymphomas (NCT01905228), and has been shown to significantly prolong survival alone or in combination with temozolomide, the current standard-of-care treatment in GBM, in orthotopic preclinical models (36). One promising approach would be to use GSC-targeting therapy prior to or simultaneously with tumor-eradicating radiotherapy and chemotherapy, which effectively kills NSTCs but to which GSCs are largely resistant (12, 13, 44), to prevent tumor recurrence by also eradicating GSCs. Another important avenue to explore is combination treatment with CBL0137 and single agent pathway inhibitors, e.g., PDGF, mTOR, VEGF, which showed promising preclinical efficacy but modest survival benefits in patients (45). Our novel finding of preferential targeting of GSCs through FACT and the ready availability of a drug that effectively sustains its inhibition presents a highly translatable and targeted therapeutic approach that can be exploited clinically in GBM.
Supplementary Material
Acknowledgments
Financial Support: This research was supported by the NCI Grant P01 CA062220 to G.R.S; NIH grants CA154130, CA169117, CA171652, NS087913, and NS089272 to J.N.R.; NIH K99/R00 CA157948 Pathway Independence Award and a Distinguished Scientist Award from the Sontag Foundation to J.D.L.; and Incuron LLC to KVG.
We thank the Flow Cytometry Core for excellent technical support and Dr. Catherine Burkhart for critical reading of the manuscript.
Footnotes
Conflict of Interest: K.V.G and A.A.P. are among the co-inventors on a patent application that covers composition of matter and use of curaxins (CBL0137), #61/102,913, “Carbazole compounds and therapeutic uses of the compounds”. K.V.G. has received an honorarium from and A.A.P. is the Vice President of Chemistry in Incuron LLC., which holds patents on and develops curaxins. The other authors declare no competing interests.
Author contributions: J.D., M.H., D.S., Q.W., A.P. and M.V. performed experiments. J.D., M.H., J.L., K.G., G.S. and M.V. conceived and designed experiments. J.R. and A.S. provided patient-derived cell specimens and mice for in vivo studies. K.G. and P.S. provided CBL0137 and plasmids for SSRP1 knockdown and dominant negative FACT. M.V. and G.R.S. supervised the studies. J.D. and M.V. wrote the manuscript. J.D., M.H., J.L., K.G., G.S., and M.V. reviewed and edited the manuscript.
References
- 1.Stupp R, Brada M, van den Bent M, Tonn J, Pentheroudakis G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25:93–101. [Google Scholar]
- 2.Stupp R, Hegi M, Mason W, van den Bent M, Taphoorn M, Janzer R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Brennan C, Verhaak R, McKenna A, Campos B, Noushmehr H, Salama S, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelloski C, Ballman K, Furth A, Zhang L, Lin E, Sulman E, et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J Clin Oncol. 2007;25:2288–94. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Yang K, Kuang K. The efficacy and safety of EGFR inhibitor monotherapy in non-small cell lung cancer: a systematic review. Curr Oncol Rep. 2014;16:390. doi: 10.1007/s11912-014-0390-4. [DOI] [PubMed] [Google Scholar]
- 6.Emlet D, Gupta P, Holgado-Madruga M, Del Vecchio C, Mitra S, Han S, et al. Targeting a glioblastoma cancer stem-cell population defined by EGF receptor variant III. Cancer Res. 2014;74:1238–49. doi: 10.1158/0008-5472.CAN-13-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazzoleni S, Politi L, Pala M, Cominelli M, Franzin A, Sergi Sergi L, et al. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70:7500–13. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- 8.Padfield E, Ellis H, Kurian K. Current therapeutic advances targeting EGFR and EGFRvIII in glioblastoma. Front Oncol. 2015;5:5. doi: 10.3389/fonc.2015.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peereboom D, Shepard D, Ahluwalia M, Brewer C, Agarwal N, Stevens G, et al. Phase II trial of erlotinib with temozolomide and radiation in patients with newly diagnosed glioblastoma multiforme. J Neurooncol. 2010;98:93–9. doi: 10.1007/s11060-009-0067-2. [DOI] [PubMed] [Google Scholar]
- 10.Thiessen B, Stewart C, Tsao M, Kamel-Reid S, Schaiquevich P, Mason W, et al. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother Pharmacol. 2010;65:353–61. doi: 10.1007/s00280-009-1041-6. [DOI] [PubMed] [Google Scholar]
- 11.Clark P, Iida M, Treisman D, Kalluri H, Ezhilan S, Zorniak M, et al. Activation of multiple ERBB family receptors mediates glioblastoma cancer stem-like cell resistance to EGFR-targeted inhibition. Neoplasia. 2012;14:420–8. doi: 10.1596/neo.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Li Y, Yu T, McKay R, Burns D, Kernie S, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao S, Wu Q, McLendon R, Hao Y, Shi Q, Hjelmeland A, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 14.Lathia J, Mack S, Mulkearns-Hubert E, Valentim C, Rich J. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–17. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venere M, Fine H, Dirks P, Rich J. Cancer stem cells in gliomas: Identifying and understanding the apex cell in cancer’s hierarchy. Glia. 2011;59:1148–54. doi: 10.1002/glia.21185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatesh S, Workman J. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–89. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 17.Garcia H, Fleyshman D, Kolesnikova K, Safina A, Commane M, Paszkiewicz G, et al. Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget. 2011;2:783–96. doi: 10.18632/oncotarget.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia H, Miecznikowski J, Safina A, Commane M, Ruusulehto A, Kilpinen S, et al. Facilitates chromatin transcription complex is an “accelerator” of tumor transformation and potential marker and target of aggressive cancers. Cell Rep. 2013;4:159–73. doi: 10.1016/j.celrep.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurova K, Hill J, Guo C, Prokvolit A, Burdelya L, Samoylova E, et al. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-κB-dependent mechanism of p53 suppression in tumors. Proc Natl Acad Sci U S A. 2005;102:17448–53. doi: 10.1073/pnas.0508888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasparian A, Burkhart C, Purmal A, Brodsky L, Pal M, Saranadasa M, et al. Curaxins: anticancer compounds that simultaneously suppress NF-κB and activate p53 by targeting FACT. Sci Transl Med. 2011;3:1–12. doi: 10.1126/scitranslmed.3002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dermawan J, Gurova K, Pink J, Dowlati A, De S, Narla G, et al. Quinacrine overcomes resistance to erlotinib by inhibiting FACT, NF-κB, and cell-cycle progression in non-small cell lung cancer. Mol Cancer Ther. 2014;13:2203–14. doi: 10.1158/1535-7163.MCT-14-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burkhart C, Fleyshman D, Kohrn R, Commane M, Garrigan J, Kurbatov V, et al. Curaxin CBL0137 eradicates drug resistant cancer stem cells and potentiates efficacy of gemcitabine in preclinical models of pancreatic cancer. Oncotarget. 2014;30:11038–53. doi: 10.18632/oncotarget.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koman I, Commane M, Paszkiewicz G, Hoonjan B, Pal S, Safina A, et al. Targeting FACT complex suppresses mammary tumorigenesis in HER2/neu transgenic mice. Cancer Prev Res. 2012;5:1025–35. doi: 10.1158/1940-6207.CAPR-11-0529. [DOI] [PubMed] [Google Scholar]
- 24.Carter D, Murray J, Cheung B, Gamble L, Koach J, Tsang J, et al. Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci Transl Med. 2015;7:312ra176. doi: 10.1126/scitranslmed.aab1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eyler C, Wu Q, Yan K, MacSwords J, Chandler-Militello D, Misuraca K, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146:53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schonberg D, Miller T, Wu Q, Flavahan W, Das N, Hale J, et al. Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell. 2015;28:441–55. doi: 10.1016/j.ccell.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou T. Drug Combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, Smyth G. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Lathia J, Hitomi M, Gallagher J, Gadani S, Adkins J, Vasanji A, et al. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel A, Tirosh I, Trombetta J, Shalek A, Gillespie S, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Porath I, Thomson M, Carey V, Ge R, Bell G, Regev A, et al. An embryonic stem cell–like gene expression signature in poorly differentiated aggressive human tumors. Nat Gen. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong D, Liu H, Ridky T, Cassarino D, Segal E, Chang H. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–44. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharya B, Miura T, Brandenberger R, Mejido J, Luo Y, Yang A, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–64. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 35.Müller F, Laurent L, Kostka D, Ulitsky I, Williams R, Lu C, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–5. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barone T, Burkhart C, Purmal A, Gudkov A, Gurova K, Plunkett R. ET-006. Curaxin (CBL0137) significantly increases survival in orthotopic models of glioblastoma multiforme alone and in combination with temozolomide. Neuro Oncol; Paper presented at the 4th Quadrennial Meeting of the World Federation of Neuro-Oncology held in conjunction with the 18th Annual Meeting of the Society for Neuro-Oncology; San Francisco, CA. 21 to 24 November 2013.2013. pp. iii37–iii61. [Google Scholar]
- 37.Shats I, Gatza M, Chang J, Mori S, Wang J, Rich J, et al. Using a stem cell-based signature to guide therapeutic selection in cancer. Cancer Res. 2010;71:1772–80. doi: 10.1158/0008-5472.CAN-10-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison S, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–74. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 39.Winkler D, Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem. 2011;286:18369–74. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gill B, Pisapia D, Malone H, Goldstein H, Lei L, Sonabend A, et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc Natl Acad Sci U S A. 2014;26:12550–5. doi: 10.1073/pnas.1405839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.National Cancer Institute. [Accessed 2015 December 17];REMBRANDT home page. 2005 < http://www.betastasis.com/glioma/rembrandt/kaplan-meier_survival_curve>.
- 42.Venere M, Hamerlik P, Wu Q, Rasmussen R, Song L, Vasanji A, et al. Therapeutic targeting of constitutive PARP activation compromises stem cell phenotype and survival of glioblastoma-initiating cells. Cell Death Differ. 2013;21:258–69. doi: 10.1038/cdd.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyer L, Lee T, Cole M, Johnstone S, Levine S, Zucker J, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beier D, Schulz J, Beier C. Chemoresistance of glioblastoma cancer stem cells – much more complex than expected. Mol Cancer. 2011;10:128. doi: 10.1186/1476-4598-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polivka J, Polivka J, Rohan V, Topolcan O, Ferda J. New molecularly targeted therapies for glioblastoma multiforme. Anticancer Res. 2012;32:2935–46. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.