Abstract
A lack of experimental models of tumor heterogeneity limits our knowledge of the complex subpopulation dynamics within the tumor ecosystem. In high grade gliomas (HGG), distinct hierarchical cell populations arise from different glioma stem-like cell (GSC) subpopulations. Extracellular vesicles (EV) shed by cells may serve as conduits of genetic and signaling communications, however, little is known about how HGG heterogeneity may impact EV content and activity. In this study, we performed a proteomic analysis of EV isolated from patient-derived GSC of either proneural or mesenchymal subtypes. EV signatures were heterogeneous, but reflected the molecular make-up of the GSC and consistently clustered into the two subtypes. EV-borne protein cargoes transferred between proneural and mesenchymal GSC increased pro-tumorigenic behaviors in vitro and in vivo. Clinically, analyses of HGG patient data from the TCGA database revealed that proneural tumors with mesenchymal EV signatures or mesenchymal tumors with proneural EV signatures were both associated with worse outcomes, suggesting influences by the proportion of tumor cells of varying subtypes in tumors. Collectively, our findings illuminate the heterogeneity among tumor EV and the complexity of HGG heterogeneity which these EV help maintain.
Keywords: exosome, extracellular vesicles, glioblastoma, heterogeneity
Introduction
Characterization of the genome (1, 2) and RNA transcriptome (3-6) provided an increasingly high-resolution picture of the heterogeneity of the HGG landscape. Single cell RNA sequencing of HGG reveals that tumor cell subpopulations with different transcriptome subtypes coexist and have developmentally evolved over a period of time from a distantly related precursor, likely a cancer stem-like cell (7, 8). This intratumoral heterogeneity with redundant signaling network aberrancies underlies the ineffectiveness of conventional and targeted therapies (9-11).
Recently there is increasing appreciation for the role that EVs play in cell- cell communication in cancers. However, it is not known whether the profound genetic and phenotypic diversity of cells in a HGG is related to the intercellular transfer of functional molecules contained within EVs (12-14). Here, we show that EVs released by GSCs retain subtype characteristics and that their transfer between GSC subtypes leads to pro-tumorigenic changes in the target GSC. Experimentally, in mouse brains, there is a hierarchical clustering of different GSC subtypes with visible transfer of EVs. Coupled with the finding that the EV proteome is predictive of HGG patient outcome, this study shows that EV communication may be a critical linchpin in HGG GSC subtype diversity.
Methods
Human specimens and primary cells
Tumor samples were obtained as approved by IRB at and The Harvard Medical School (HMS). Surgery was performed by E.A.C. and I.N. GSCs were obtained by dissociation of tumor samples and cultivated in stem cells enriching conditions using Glutamine, B27, FGF, EGF supplemented Neurobasal medium (14, 15). The unique identity of cultured patient-derived cells was confirmed by short tandem repeats analysis (16).
Purification of EVs
The conditioned media were collected and EVs were isolated by differential centrifugation and analyzed using a NanoSight as previously described (14).
Mass spectrometry and immunoblotting
All mass spectra were acquired at the Bioproximity LLC and data were validated by Western blotting (14).
In vitro assays
For EV transfer: 3 × 105 GSCs/ml were maintained overnight in un-supplemented medium followed by 24h treatment with EV derived from mesenchymal (M) GSCs (EVM) or form proneural (P) GSCs EVP (5 μg protein/ml).
For spheroid formation: GSCs were dissociated to single cells and plated at 200 (M GSCs) or 1000 (P GSCs) cells/well and in 96-well plate in un-supplemented media and treated with EVs (5μg of protein/ml) on 0h and 48h and analyzed after 96h.
For co-culture assay: single-cell suspensions of M GSCs, P GSCs and M/P GSC co-culture (at 1:4 ratio) were cultured for 48h in neurosphere medium.
In vivo studies
Female athymic mice were purchased from Envigo. Mice were housed in HMS animal facility in accordance with NIH regulations. Protocols were approved by the HMS IACUC. Intracranial tumor injection was performed as described (14). For GSC co-implantation experiments, either 1×103 M GSCs or 5×105 P GSCs, or both combined were used. For GSC/EV co-implantation cells were pre-treated with EVs for 24h, pelleted and admixed with EVs (at 5μg of protein/ml).
Microscopy
EV release was visualized by embedding either PALM-Tomato M GSC or PALM-GFP P GSC spheroids in collagen. All fluorescent and bright field microscopy assays were observed using a Nikon Eclipse Ti.
Data Analysis
Experimental and clinical data were analyzed for molecular profiles of glioblastoma using the GBM-BioDP (17). Data are expressed as mean ± SD. The unpaired two-tail t-test was used to compare between 2 groups. One-way ANOVA, followed by Bonferroni's test, was conducted to test for significance among multiple groups. P < 0.05 was considered significant.
Results and Discussion
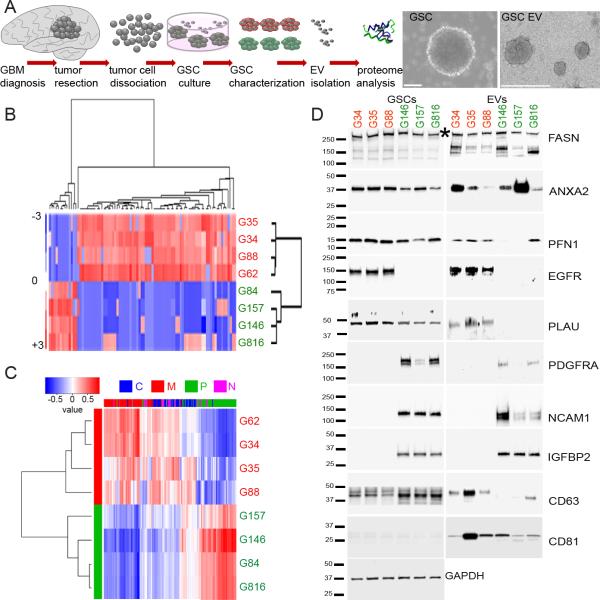
Recent data have shown that gene expression datasets may be used to classify GSCs from different subtypes (15). To investigate whether our collection of GSCs (Fig. 1A) display distinct expression profiles, an analysis of gene signature was performed (15) followed by principal component analysis (PCA). This defined two distinct subpopulations of GSCs, belonging to the P or the M subtype. As expected, there were also several GSCs that did not cluster into either subtype (Supplementary Fig. 1A). To validate whether the observed cellular transcriptional subtype diversity was reflected by the protein composition of EVs released by these GSCs, EVs were isolated and quantified followed by global mass spectrometry (MS) - based analysis of their proteome. Interestingly, there was significant physical heterogeneity of EVs released by both subtypes: larger and homogenously sized EVM, and smaller but with broader diversity in size EVP. There was no significant difference in the number of EVs shed by either subtype in culture (Supplementary Fig. 1B). Comprehensive MS analysis revealed over 1400 proteins within the EV proteome. Over 90% of them were detected in both subtypes (Supplementary Fig. 1C). For subtype signature profiling, a differential expression analysis of EVP vs. EVM was carried out and 98 proteins were found to be significantly differentially released in a subtype-dependent manner (Fig. 1B, Supplementary Table 1), indicating that the proteome of GSC EVs recapitulated subtype clustering. In fact, when the GSC EV proteome data was queried with known molecular subtypes from TCGA (4), the EV signature clustered into the two same subgroups (P and M) as those from their respective GSCs (Fig. 1C, Supplementary Table 2). In silico analysis revealed that proteins identified in EVs from each subtype regulate different biological processes and molecular functions. EVP proteins primarily control processes related to neurons/nervous system development, and EVM proteins function as regulators of receptor binding, protein synthesis and gene expression (Supplementary Fig. 1C). To provide validation of the proteome analysis by Western blots, we selected: 1- specific EV proteins that were most predictive of a subtype by TCGA classification (for example NCAM1, PLAU for P vs. M GSCs, see Supplementary Fig. 1D), 2- proteins that were most differentially expressed between the two subtypes (for example, IGFPB2 or PNF1 for P vs. M GSCs, see Supplementary Fig. 1E), and 3- proteins that are most commonly present in HGG EVs (ANXA2, FASN) (14) (Supplementary Fig. 1E). The analysis showed a cellular and EV subtype-specific pattern of distribution (Fig. 1D), supporting the global analysis (Fig. 1B).
Figure 1.
Heterogeneity of GSCs is mirrored by the diversity of EV protein composition.
A. Workflow depicting isolation of GSCs from primary tumors for GSC culture and proteomic analysis of EVs (left). Representative micrographs of GSCs (middle) and GSC EVs (right).
B. The GSC EV proteome profile distinguishes P (green) from M (red) GSC subtypes. Proteins sets that vary coherently between subtypes were identified by clustering.
C. The GSC EV proteome profile separates P and M glioblastoma subtypes. Genes coding for proteins sets that vary coherently between GSC subtypes (classical – C, blue; mesenchymal – M, red; proneural – P, green; neural – N, magenta) from 89- proteins signature were retrieved from TCGA GBM dataset and identified by clustering: power of prediction of top 6 genes is shown.
D. The GSC EV proteome content partially mimic cellular expression. Selected protein sets were validated by Western blotting using indicated antibodies, * indicate specific band on FASN blot.
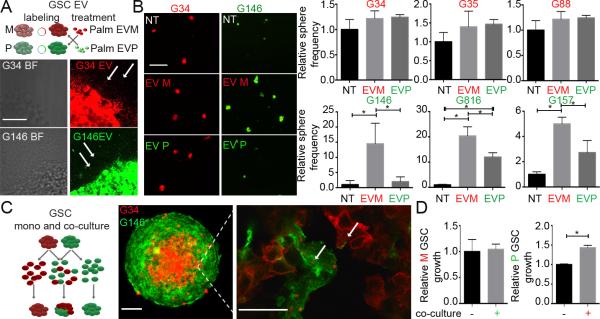
To evaluate whether there were functional consequences after EVM and EVP exchange, GSCs of each subtype were exposed to EVs isolated from the other subtype, using membrane-tagged GSCs for visualization of EV internalization (18) (Fig. 2A). When P GSCs were exposed to EVM there was a significant increase in GSC sphere frequency, number, viability and volume (Fig. 2B, Supplementary Fig. 2A-D). Exposure of P GSCs to their own EVs had no effect. When M GSCs were exposed to EVP there was no significant change in M GSC frequency (Fig. 2B) and viability (Supplementary Fig. 2C). However, the frequency of larger spheroids was moderately increased upon exposure to either EV (Supplementary Fig. 2B). Next, to model coexistence of P and M GSCs within tumors (6), single cell GSCs from either M or P subtype were co-cultured together to form spheroids (Fig. 2C left). All of the spheroids ended up growing as mosaics instead of growing as homogenous cultures of M or P GSCs only (Fig. 2C right). This exclusivity of mosaic growth indicated a selective pressure for GSCs to grow in a heterogenous rather than a homogenous subtype environment (Supplementary movie). Interestingly, these mosaic spheroids formed a visibly distinct architecture with the M GSCs clustered together in the inner core of the spheroid, while P GSCs localized to the outer layers. Importantly, frequent exchange of EVs between cells from both subtypes was observed, suggesting dynamic and ongoing EV-mediated communication between cell types. To verify whether growth of each subtype benefited from the observed coexistence, growth of GSCs was assessed in co-culture vs. mono-culture conditions. Similar to the EV treatment experiments, growth of M GSCs was not affected in co-culture vs. mono-culture, but there was a significant increase in P GSCs growth when co-cultured with M GSCs (Fig. 2D). To determine which signaling pathways may be involved in GSC subtype cross-talk, the expression of key pro-oncogenic kinases was analyzed: AKT was more phosphorylated in P GSCs, while ERK, SRC and P65 were more phosphorylated in M GSCs (Supplementary Fig. 2E). Although these kinases were more phosphorylated in EVM-treated P GSCs, suggesting a possible mechanism for the observed increase in P GSC growth, the involvement of other signaling pathways cannot be excluded. The analysis of the GSC/EV proteome showed that EGFR was M GSC-enriched with little immuno-positivity in P GSCs (Fig. 1D) which was further validated by in situ staining and qPCR (Supplementary Fig. 2F). When P GSCs were exposed to EVM, EGFR was detected in P GSCs, demonstrating that EGFR could be transferred from M GSCs to P GSCs by EVs. Such transfer, coupled with the previously observed kinase phosphorylation, could explain the significant growth enhancement observed in P GSCs exposed to EVM.
Figure 2.
Differential in vitro effects of GSC co-culture and exposure to EVs.
A. Workflow depicting labelling and co-treatment of GSCs and GSC EVs (top). Representative micrographs of GSC spheroids releasing labelled EVs (bottom). Arrows indicate EVs shed by P and M GSCs. Scale bar 50μm.
B. EVM promote growth of P GSCs. Representative micrographs of GSC spheroids co-cultured with EVs (left) and quantification of spheroid frequency (right). * P < 0.05. Scale bar 100μm.
C. Workflow depicting co-culture of labelled GSCs (left). Representative micrographs of GSC spheroids and transfer of EVs between GSCs in the spheroid (right). Arrows indicates EV internalization. Scale bars 100μm (left) and 25μm (right).
D. M GSCs promote growth of P GSCs. Quantification of cell growth in mono- and co-culture. * P < 0.05.
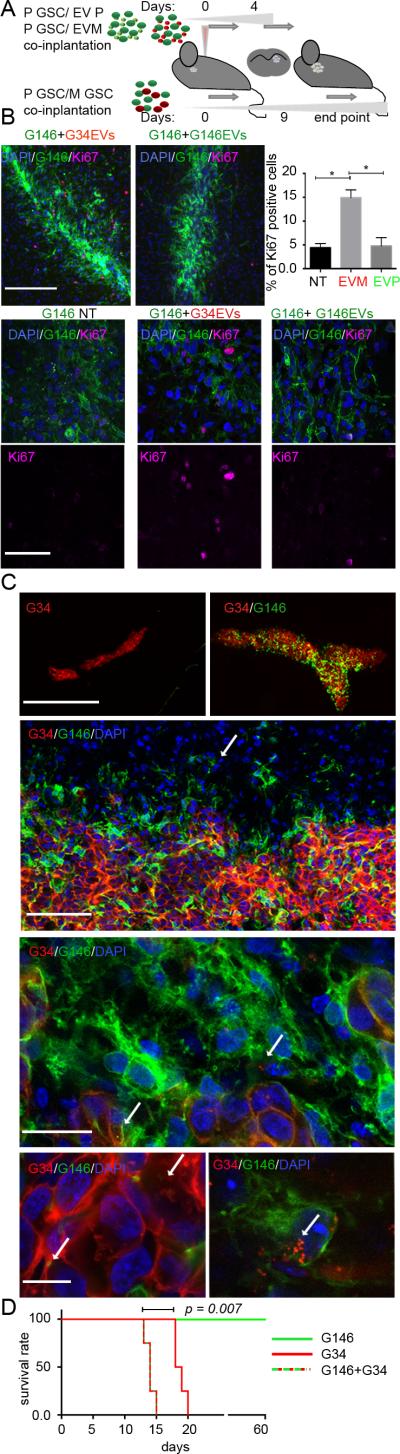
To determine whether the in vivo microenvironment influenced the EV-mediated cross-talk between GSCs observed in vitro, we performed set of experiments using an intracranial GSC xenograft model. First, P GSCs were co-injected with either EVP or EVM. Second, P GSCs were co-injected with M GSCs (Fig. 3A). For the first experiment, animals were sacrificed four days after GSC implantation: a ~3-fold higher percentage of Ki-67-positive P GSCs upon co-injection with EVM was observed when compared to P GSCs alone. However, this increase was not observed upon co-injection with EVP (Fig. 3B). Because M GSCs grow more aggressively than do P GSCs (15), P GSCs and M GSCs were admixed in a 500:1 ratio in the second co-implantation experiment. Nine days after implantation, tumors formed by mixed GSCs were significantly larger than those comprised of M GSCs only (Fig. 3C (upper panels)), perhaps unsurprisingly as the latter were initially formed by much fewer cells. However, volume of tumors originated by M GSCs implanted with P GSCs was ~4-fold greater than that of M GSCs alone (Supplementary Fig. 3A), suggesting that the heterogeneous environment provided a mitotic advantage to M GSCs. This differed somewhat from the in vitro data where EVPs did not provide a growth advantage to M GSCs, possibly suggesting additional influences to growth in the in vivo model. Analysis of tumor architecture revealed that M GSCs tended to form the non-invasive tumor core, while P GSCs located preferentially to the peripheral zone with single cells invading into surrounding tissue, similarly to in vitro observation of spheroids’ architecture. Both cell types were found with numerous internalized EVs from the other cell type (Fig. 3C (lower panels), Supplementary Fig. 3B) suggesting that EV-mediated communication is in fact frequent between these cell types in vivo. Finally, a cohort of animals from the co-implantation experiment was studied for survival, showing a significant survival disadvantage for tumors composed of two cell subtypes (Fig. 3D), when compared to those formed by M GSCs or by P GSCs alone. Analysis of tumor composition revealed that despite the initial dominance of P GSCs, by day 9 most of the tumor was composed of M GSCs and by the terminal phase almost the entire tumor was composed of M GSCs (Supplementary Fig. 3C-D). Likely, M GSCs benefited from the P GSCs co-implantation at the beginning and overgrew to take over the entire neoplasm, suggesting that heterogeneity coexisting within one tumor ecosystem drives tumor progression.
Figure 3.
Heterogeneous GSCs exchange EVs in vivo and promote tumorigenicity.
A. Workflow depicting in vivo model.
B. EVM promote proliferation of P GSCs in vivo. Representative micrographs of P GSCs 4 days after co-implantation with EVM with Ki-67 immunostaining (upper left), higher magnification insets (bottom) and Ki-67 quantification (upper right). * P < 0.05. Scale bars 200μm (upper) and 50μm (bottom).
C. EVs are transferred intra-tumorally in vivo. Representative micrographs of co-implanted tumors (top row). Distinct distribution of P and M GSCs (second row, arrow indicates infiltrating P GSCs). Intratumoral EV transfer between P and M GSCs (third and fourth row, arrows indicate internalized EVs). Scale bars from top to bottom: 1mm, 100μm, 25μm, 10 μm.
D. Heterogenous GSC-originated tumors are associated with worsened survival. Kaplan-Meier curves are shown. N=4; P = 0.007. Average survival: G34 – 18.5 days, G146+G34 – 14 days.
Finally, we examined whether the subtype-specific heterogeneity of the GSC EV proteome was relevant to HGG patients’ outcome according to TCGA (4, 19). Genes encoding for all GSC EV proteins were associated with poorer outcome in a full cohort of HGG patients (Fig. 4A, Supplementary Fig. 4A). However, when we examined the EV protein profiles separately, the EVM signature was associated with a significantly worse outcome in P but not M tumors (Fig. 4B, Supplementary Fig. 4B). Conversely, the EVP signature was associated with a significantly worse outcome in M but not P tumor sub-classes (Fig. 4C, Supplementary Fig. 4C). These datasets indicated that heterogeneity within tumors, which may be propagated throughout the tumor via EV communication, was associated with decreased survival. This is in agreement with the scenario proposed by Patel et al. (6) where the clinical outcome of a glioblastoma subclass is influenced by the proportion of tumor cells of alternate subtypes and emphasizes the clinical importance of intratumoral heterogeneity.
Figure 4.
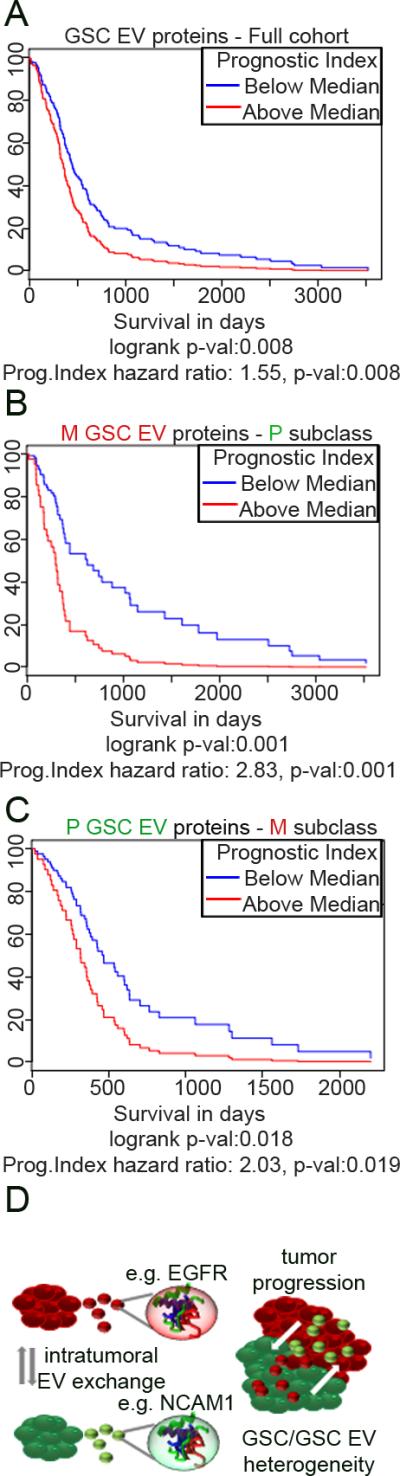
GSC EV proteome predicts patients’ outcome.
A-C. Survival analysis based on the impact of the prognostic index of multiple protein-coding genes from all GSC EVs (A), EVM (B) and EVP (C) based on retrospective data extrapolated from the TCGA.
D. Summarizing cartoon: the heterogeneity of EV cargo contributes to the diverse complexity and enhanced progression of HGG.
The combination of tumor EVs and cell-specific secreted molecules that can be taken up by neighboring or distant recipient cells leading to changes in gene expression, suggests a cell-specialized role in physiological and pathological conditions (20). We have leveraged the analysis of GSC EV proteomics to characterize heterogeneous extracellular programs within HGG tumors interrelating their release, uptake and function which have fundamental implications for biology and therapeutic strategies of HGG, as EVs provide two-way influence through release/uptake between tumors’ cells in vivo (Fig. 4D). Analysis of single-cell transcriptome demonstrated transcriptional diversity within an individual tumor (6) and our analysis reveals that transfer of EV protein cargo between tumor cells subpopulations offers a means and opportunity for dynamic transitions.
Supplementary Material
Acknowledgments
The authors are grateful to Hannes Ricklefs for helping in the data analysis by writing a python program.
Financial Support: NCI P01 CA69246 (EAC, RW, XOB) and by NCI P01 CA163205 (EAC), NCI 1R01 CA176203-01A1 (JG) and DFG RI 2616/1-1 (FR)
Footnotes
Authors declare no conflict of interest
References
- 1.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proceedings of the National Academy of Sciences. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2008;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan Cameron W, Verhaak Roel GW, McKenna A, Campos B, Noushmehr H, Salama Sofie R, et al. The Somatic Genomic Landscape of Glioblastoma. Cell. 2014;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hale JS, Sinyuk M, Rich JN, Lathia JD. Decoding the cancer stem cell hypothesis in glioblastoma. CNS Oncology. 2013;2:319–30. doi: 10.2217/cns.13.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes & Development. 2015;29:1203–17. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schonberg DL, Lubelski D, Miller TE, Rich JN. Brain tumor stem cells: Molecular characteristics and their impact on therapy. Molecular Aspects of Medicine. 2014;39:82–101. doi: 10.1016/j.mam.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of Receptor Tyrosine Kinases Affects the Response of Tumor Cells to Targeted Therapies. Science. 2007;318:287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 11.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, et al. Targeted Therapy Resistance Mediated by Dynamic Regulation of Extrachromosomal Mutant EGFR DNA. Science. 2014;343:72–6. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Curry WT, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godlewski J, Krichevsky AM, Johnson MD, Chiocca EA, Bronisz A. Belonging to a network— microRNAs, extracellular vesicles, and the glioblastoma microenvironment. Neuro-Oncology. 2015;17:652–62. doi: 10.1093/neuonc/nou292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bronisz A, Wang Y, Nowicki MO, Peruzzi P, Ansari KI, Ogawa D, et al. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer research. 2014;74:738–50. doi: 10.1158/0008-5472.CAN-13-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8644–9. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SH, Ezhilarasan R, Phillips E, Gallego-Perez D, Sparks A, Taylor D, et al. Serine/Threonine Kinase MLK4 Determines Mesenchymal Identity in Glioma Stem Cells in an NF-kappaB-dependent Manner. Cancer cell. 2016;29:201–13. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Celiku O, Johnson S, Zhao S, Camphausen K, Shankavaram U. Visualizing molecular profiles of glioblastoma with GBM-BioDP. PloS one. 2014;9:e101239. doi: 10.1371/journal.pone.0101239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nature communications. 2015;6:7029. doi: 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015 doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.