Abstract
Multiple myeloma (MM) remains incurable and the majority of patients die within 5 years of diagnosis. Reolysin, the infusible form of human reovirus (RV), is a novel viral oncolytic therapy associated with antitumor activity likely resulting from direct oncolysis and a virus-mediated antitumor immune response. Results from our phase I clinical trial investigating single agent Reolysin in patients with relapsed MM confirmed tolerability, but no objective responses were evident, likely because the virus selectively entered the MM cells but did not actively replicate. To date, the precise mechanisms underlying the RV infectious life cycle and its ability to induce oncolysis in patients with MM remain unknown. Here we report that Junctional Adhesion Molecule 1 (JAM-1), the cellular receptor for RV, is epigenetically regulated in MM cells. Treatment of MM cells with clinically relevant histone deacetylase inhibitors (HDACi) results in increased JAM-1 expression as well as increased histone acetylation and RNA polymerase II recruitment to its promoter. Further, our data indicate that the combination of Reolysin with HDACi, potentiates RV killing activity of MM cells in vitro and in vivo. This study provides the molecular basis to use these agents as therapeutic tools to increase the efficacy of RV therapy in MM.
Keywords: RV, HDACi, multiple myeloma, apoptosis, JAM-1
INTRODUCTION
Reovirus (RV) Serotype 3 – Dearing Strain is a non-enveloped double-stranded RNA oncolytic virus (OV) that is widespread in nature (1). RV is capable of selectively replicating in transformed cells leading to selective anti-tumor activity without harming normal tissues (2, 3). The surface protein Junctional Adhesion Molecule 1 (JAM-1) and in the central nervous system Negative Growth Regulatory Protein 1 (NGR1) are the cellular receptors responsible for RV entry in human cells (4, 5). The virus mediates its antitumor activity via induction of direct cytolysis, and activation of a tumor-directed immune response (6–8).
Multiple myeloma (MM) is an incurable blood cancer affecting approximately 83,000 people in the United States of America. Over 50% of these patients die within 5 years of diagnosis (9), and those with high risk disease (1p deletion, 1q amplification, 17p deletion) have more dismal prognoses associated with an estimated 24 month survival (10). In most patients, relapse remains inevitable due to the presence of resistant residual disease (9).
Preclinical evidence has demonstrated that Reolysin induces MM cell oncolysis alone, but that it is more effective in combination with the proteasome inhibitor (PI) by increasing the expression of endoplasmic reticulum (ER) stress-related genes (11–13). We recently reported that single agent Reolysin is safe and well tolerated in patients with relapsed MM (14). Our correlative analyses revealed that viral RNA, but not active viral replication, was evident in the patient’s MM cells approximately one week following initial RV treatment.
Correlative studies from our phase I trial revealed that patient’s MM cells displayed minimal, or no expression of the RV receptor JAM-1 (4); findings consistent with RV resistance and with the importance of viral cell entry to affect RV-mediated tumor cell killing in cancer cells (15).
Histone acetyltransferases and histone deacetylases (HDACs) affect a broad-array of genes involved in cell cycle, apoptosis, and protein folding by regulating the acetylation status of their promoters, or through post-translational modifications (16).
In MM, HDACi’s have shown only minimal activity as a single agent (17). That being said, two phase 1B trials showed that some patients were able to be salvaged by a combination of the HDACi, vorinostat (SAHA), or panobinostat (LBH) with proteasome inhibitor (18, 19). Based on the recent FDA approval for the use of LBH in the treatment of MM patients (20), HDACi are now relevant therapeutic options that require further investigation to identify ways to best optimize their clinical use.
Here we show that clinically relevant pan-HDACi upregulate the expression of the RV receptor JAM-1 (4), an effect that increases the sensitivity of MM cells to the oncolytic effects of RV in vitro and in vivo. These data support that combining RV with HDACi is a promising therapeutic strategy in patients with MM.
METHODS
Tissue culture
RPMI-8226 (CRM-CCL-155), U266 (TIB-196), NCI-H929 (CRL-9068), and MM.1S (CRL-2974) MM cell lines were purchased from American Type Cell Culture Collection (ATCC, Manassas, VA, USA). L363 (ACC-49) MM cells were purchased from DSMZ German Collection of Microrganisms and Cell Cultures. Cell lines were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated FBS and penicillin and streptomycin antibiotics. Karyotype analysis was used to validate MM cell lines used. Primary MM patient cells were obtained from bone marrow aspirates, following informed consent according to an Institutional Review Board (IRB)-approved protocol, and CD138-positive cells were isolated using EasySep CD138 positive selection kit (Stem Cell Technologies, Vancouver, BC, Canada cat# 18357).
Reagents
Reolysin used for preclinical studies was a gift from Dr. Matt Coffey (Oncolytics Biotech Inc.). AR-42 was provided by Arno Therapeutics (Flemington, NJ), JQ12 was a gift from the laboratory of Dr. Baiocchi, and 5-Aza-2′-deoxycytidine was obtained from Sigma Aldrich (St. Louis, MO, USA cat# A3656). SAHA (cat# S1047), LBH-589 (cat# S1030), RGFP966 (cat# S7229), and Entinostat (cat# S1053) were purchased from Selleckchem (Boston, MA, USA).
In vitro assessment of RV and HDACi treatment of MM cells
MM cells were plated in triplicate on 96-well plates with DMSO and treated with RV alone, or in combination with different HDACi at the concentrations indicated. To calculate the effectiveness of the different HDACi (IC50) at 48hrs, the drugs were added every 24hrs for a total of two times. For RV+HDACi combination experiments, MM cells were treated with RV and HDACi in combination or as single agents for 48hrs, without addition of extra drug (HDACi) at 24hrs. To exclude different HDACi half-lives could affect the read out of our experiments, MM cells were pre-treated for 10hrs with HDACi, and following drug washout, were infected for 48hrs with RV. BD Pharmingen™ FITC Annexin V - BD Biosciences (cat#556419) staining kit was used to assess MM cell viability. We examined the ability of HDACi to synergize with RV using the Chou-Talalay synergy analysis, a mathematical way of evaluating interactions between two drugs (21–24). This analysis is frequently utilized to investigate synergy between anti-cancer agents (24). Data shown are a standard synergy analysis. Briefly, the 50% effective dose (ED50) of HDACi and RV were each defined as the dosage yielding 50% cell viability 48hrs following treatment, as compared to untreated controls (RV MOI5, AR-42 0.2μM, SAHA 1.00μM, LBH 0.01 μM and Entinostat 2 μM). To evaluate if the combination resulted in synergistic cell killing, the cells were treated with each HDACi alone, RV alone, or HDACi+RV in combination. Concentrations of HDACi and RV were serially diluted at fixed ratios of 0.0625, 0.125, 0.25, 0.5, 1, 2 and 4 times their ED50.
The viability data were utilized to calculate the Combination Index (CI) via the Compusyn program in which CI<1 indicates synergistic interaction, CI>1 is antagonistic, and CI=1 is additive. Cell proliferation was assessed with Aqueous Non-radioactive Cell Proliferation Assay (cat# G3582, CellTiter 96®, Promega, Madison, WI) according to the manufacturer’s instructions.
To determine if the combination of HDACi and RV resulted in enhanced productive infection and generation of new functional viral particles, H929 and L363 cells were treated with DMSO, RV at MOI 5 alone, or RV with 0.2 μM AR-42, and 1.2×106 cells were seeded in 3 mL per well on 24-well plates. After 48hrs, cells were collected, washed three times in cold PBS, and then viral RNA and viral capsid protein (σ3) was analyzed using qRT-PCR and western blot, respectively. Supernatants were also harvested and used to treat additional MM cells at a 1:100 dilution, the effect of which was determined after 24hrs using a cell proliferation assay (CellTiter 96®, Promega, Madison, WI) according to the manufacturer’s instructions.
Immunohistochemistry (IHC) and RNA in situ hybridization (ISH)
The following antibodies were used in this study: antibody to RVRV capsid protein (compliments of Dr. Matt Coffey of Oncolytics Biotech, Inc.), caspase-3 (1:33, antigen retrieval, cat#Ab4051, Abcam, Cambridge, MA), p38 (1:250, antigen retrieval, cat#Ab7952, Abcam, Cambridge, MA), p-p38 (pospho-T180/Y180; cat#Ab38238, Abcam, Cambridge, MA) and Junctional Adhesion Molecule 1 (JAM-1, 1:150 with antigen retrieval; cat#Ab52647, Abcam, Cambridge, MA), as previously described (14). Heat Mediated Antigen Retrieval was performed using Solution Antigen retrival pH 6.0 (cat#ab973, Abcam, Cambridge, MA). The viral RNA in situ hybridization (ISH) protocol has also been previously published (14). In brief, after digestion with protease, the tissue and reoviral RNA probes (locked nucleic acid modified 5′ digoxigenin tagged, Exiqon, Woburn, MA) were co-incubated at 60°C for 5 minutes, then hybridized from 2 to 15 hrs at 37°C. After a wash in 0.1% of Saline Sodium Citrate Buffer pH7.0 (20X stock solution: 3M NaCl/0.3M Sodium citrate) and 2% bovine serum albumin (BSA) at 50°C for 10 minutes, the reoviral RNA–probe complexes were visualized via NBT/BCIP (La Roche Inc, Basel, Switzerland) by alkaline phosphatase conjugation to antidigoxigenin antibody. Negative controls included myeloma cells not exposed to RV and omission of the probe.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted with the Trizol reagent (Invitrogen, Grand Island, NY, USA, cat# 15596018), according to the manufacturers protocol. For the detection of RV genome, reverse transcription reactions were done using 100ng of RNA in a 10μl reaction using the iScript cDNA synthesis kit (BioRad, Hercules, CA, USA cat# 1708891). For F11R, cDNA synthesis was preformed using 500ng of RNA in a 20μl reaction with the high capacity cDNA Reverse Transcription Kit (Applied Biosystems, Grand Island, NY, USA, cat# 4368814). Reverse transcription reactions were run using a Mastercycler pro. For the quantification of RV genome qRT-PCR reactions were conducted using the iQ SYBR Green Super Mix (Bio Rad, Hercules, CA, USA, cat# 1708880) and the following primers: Reo9: 5′-TGCGCAAGAGGCAGCAATCG-3′ and Reo10: 5′-TTCGCGGGCCTCGCACATTC-3′. F11R qRT-PCR reactions were performed using TaqMan gene expression assay (cat#4331182, Applied Biosystems, Grand Island, NY, USA). RV genome and F11R expression were normalized against GAPDH expression using the GAPDH PrimePCR SYBR Green assay (Bio Rad, Hercules, CA, USA, cat# qHsaCED0038674) and GAPDH TaqMAN gene expression assay (Applied Biosystems, Grand Island, NY, USA, cat# 4331182), respectively. All qRT-PCR reactions were run on the Eppendorf Realplex4.
Immunoblotting
Cells were washed in PBS and lysed in RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA cat# R0278) supplemented with 10 % phosphatase (Thermo Fisher, cat# 1862495) and protease inhibitors (Thermo Fisher, cat# 186178).
Immunoblotting
Cells were washed in PBS and lysed in RIPA buffer (St. Louis, MO, USA cat# R0278) supplemented with 10 % phosphatase (Thermo Fisher cat# 1862495) and protease inhibitors (Thermo Fisher cat# 186178). Lysates were then clarified by spinning at 14,000 rpm at 4°C and protein concentration quantified by Bradford assay. 50 μg of protein was boiled in loading buffer for three minutes, electrophoresed in Criterion TGX 4–20% gels (Bio Rad, Hercules, CA, USA cat# 5671095), and transferred to nitrocellulose membranes. Blots were blocked in 5% milk/TBST (20mM Tris pH7.5, 150 mM NaCl, 0.5% Tween 20) solution and stained at 4°C overnight with primary antibodies diluted in 2% BSA (bovine serum albumin)/TBST. The antibodies used were anti-JAM-1 (1:1000; cat#Ab52647 Abcam Cambridge, MA), GAPDH (1:1000; cat#Ab9485 Abcam Cambridge, MA), acetylated histone 3 lysine 27 (Ac-H3-K27; 1:1000; cat#Ab729 Abcam Cambridge, MA), and anti-RV σ-NS (1:1000; provided by Oncolytics Biotech Inc.). Blots were washed three times for 15 minutes with TBST, and stained with horseradish peroxidase (HRP)-conjugated secondary antibodies (diluted 1:4000) for 2 hrs at room temperature. Following two 10-minute washes in TBST, signals were detected with Super Signal HRP substrate (Thermo Fisher cat# 34080).
Flow cytometry
Cells were stained with PE-conjugated mouse anti-JAM-1 (cat# bs-3651R-PE; BD Biosciences, San Jose, CA), or PE-conjugated mouse IgG1/λ isotype control (cat#550339; BD Biosciences, San Jose, CA, USA). Data were collected using either a Beckman-Coulter Fc500, or Beckman Coulter Gallios Flow Cytometer and analyzed using the Beckman-Coulter Kaluza software package. In addition, near-IR Live/Dead cell-staining kit (Invitrogen, Grand Island, NY, USA, cat# L10119) was used to ensure data were collected from live cells.
RV Tropism
Cell lines were treated with RV at a multiplicity of infection (MOI) of 500, for either 10, 20, or 45 minutes. Cells were then rinsed three times using cold PBS, incubated for 5 minutes in Trypsin-EDTA (0.25%) to remove un-internalized virus, and washed in PBS again. RV genome was then quantified as described above. To determine if HDACi mediated increased JAM-1 expression resulted in increased association of RV with MM cells, H929 and L-363 cell lines were first treated with DMSO (0.001%) or 0.2 μM AR-42 for 24 hours. Cells were then treated with RV at MOI of 500 for 10, 20, or 30 minutes. Cells were collected and RV genome quantified as indicated.
JAM-1 knockdown
Commercially available FlexiTube siRNA (FlexiTube GeneSolution cat#GS50848 for F11R, Qiagen, Valencia, CA, USA) was used to target mRNA from the F11R gene that encodes the JAM-1 protein. Cells were transfected using a Lonza Nucleofector® 4D electroporation system (Basel, Switzerland), according to the manufacturer’s instructions. Briefly, 5×106 cells were suspended in 100 μl solution SF containing 300nM siRNA from the FlexiTube kit. H929 and L363 cells were then electroporated using the DS150 and CM138 programs, respectively, and then allowed to rest for 12hrs prior to treatment.
Chromatin Immunoprecipitation
L-363 cells were incubated with DMSO, or 0.2 μM AR-42 for 24hrs. ChIP experiments were then performed using the EZ ChIP chromatin immunoprecipitation kit (Millipore, Billerica, MA, USA, cat# 17371) according to the manufacturer’s instructions. Anti-acetylated H3K27 (cat#Ab4729, Abcam, Cambridge, MA), anti-total acetylated histone 3 (cat#06-599, Millipore, Billerica, MA), anti-acetylated histone 4 (Cat#06-866, Millipore, Billerica, MA), and anti-RNA polymerase II (cat#05-623, Millipore, Billerica, MA) antibodies were used for immunoprecipitation. Chromatin antibody complexes were captured with salmon sperm DNA/protein G agarose. Following washes, the DNA-protein crosslinks were reversed at 65°C overnight. Immunoprecipitated DNA was then treated with RNase A and proteinase K, and purified by phenol-chloroform extraction and ethanol precipitation. Input and immunoprecipitated DNA were then analyzed by qRT-PCR using SYBR green, as indicated above. The following primers for the F11R promoter region were used: F11RproF: 5′-GAGGTTGGAGGAAGGCTCTC-3′; and F11RproR: 5′-CCAGTGAGGACGAGGAGTGT-3′. Negative control forward primer: 5′-GGTGTGGGATTTACGGAGAC-3′ and negative control reverse primer 5′-CCCAGCGTAGCTCATCTCTC-3′, were designed away of the promoter region (exon 3 in JAM-1 gene).
Animal experiments
All animal experiments were performed in compliance with federal laws and in strict accordance with the Animal Care and Use Committee (IACUC) policies and guidelines of the Ohio State University. A total of 5 × 106 MM.1S GFP+/Luc+ cells (25) were injected into 30 NOD-SCID mice (Foxn1nu/Fox1nu; Charles River). Two weeks later mice with similar levels of tumor burden, as determined by bio-luminescence using the In Vivo Imaging System (IVIS), were divided into four groups containing 5 mice per group. The groups were treated with AR-42 (25mg/kg) alone, RV alone (5 × 108 TCID50), AR-42 combined with RV, and vehicle controls (8% DMSO in PBS for AR-42 control, or PBS for RV control). The AR-42 treatment schedule was Mon-Tue-Wed, 25mg/kg via intraperitoneal injection, and RV or PBS (for control) were delivered by single tail vein injections. AR-42 treatments continued for 3 weeks. At the end of treatment bone marrow cells were isolated from the femurs of mice and infiltration of MM cells was analyzed by detection of GFP+ cells by flow cytometry.
Statistics
Two sample t-test were used to analyze the experiment with two groups involved. For experiments with multiple groups, data were analyzed by analysis of variance (ANOVA). Trend analysis was used to evaluate the correlation between the cell viability and intracellular level of RV genome. We classified cells into five groups based on the levels of virus genome, calculated as 2e-DCT: control (<10), Low (< 5000), Median (< 100,000), High (<2,000,000), and Super high (>2,000,000). Since we classified the cells into groups, we used slope test to analyze the association between cell viability and viral genome levels instead of using the Pearson correlation test (for continuous variables) Negative slope and significant p-value indicated an inverse correlation.
RESULTS
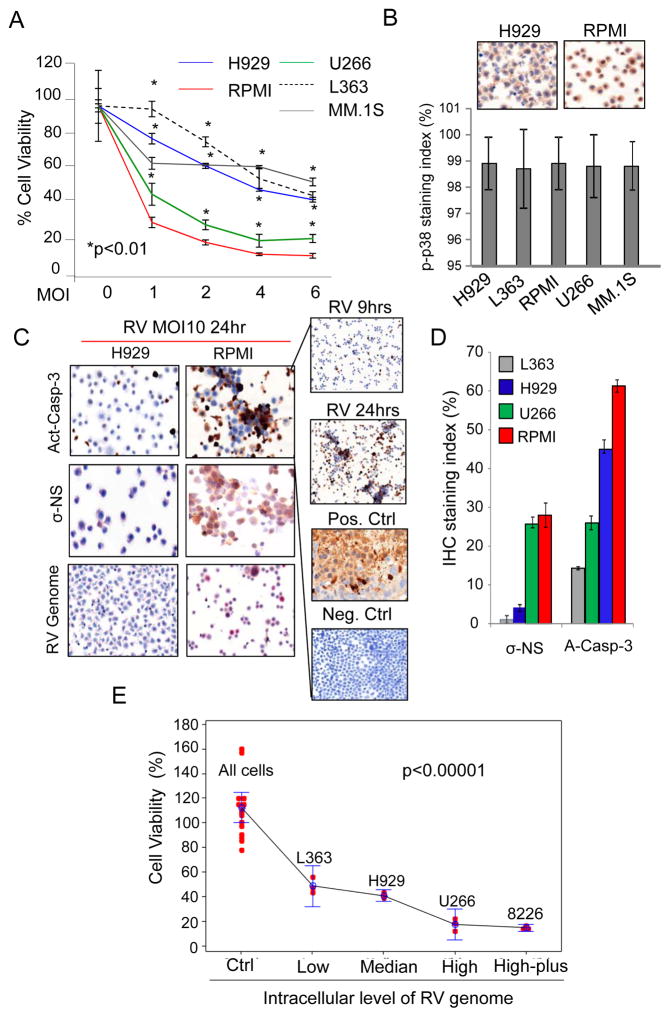
MM cell lines show differential sensitivities to RV
To investigate the molecular mechanism(s) responsible for the lack of RV replication in our phase 1 study (14), we initially infected multiple MM cell lines with a clinically achievable multiplicity of infection (MOI 1-6) of RV. Cell proliferation assays at 48hrs showed that some cell lines, such as RPMI-8226 (RPMI) and U226 were highly sensitive to RV, while others, including H929, L-363 and MM.1S, were inherently less susceptible to RV induced growth arrest (Figure 1A). Since it has been previously reported that the selective replication of RV in cancer cells relies on the presence of an activated Ras/RalGEF/p38 pathway (3, 26), we initially screened for the presence of phospho-p38 MAPK (p-p38) by immunohistochemistry (IHC). We found that over 98% of cells in all cell lines were p-P38 positive upon RV infection (Figure 1B), and thus, no correlations were found to explain the differential sensitivity of the cell lines to RV infection. The lower sensitivity of semi-adherent MM.1S cells to RV infection compared to the more sensitive suspension cells (RPMI, U266) could be explained by the fact that these cells require trypsinization that may affect the viral entry, but it could not explain why suspension H929 and L363 cells displayed increased RV resistance (27, 28). Hence, our comparisons focused only on suspension cells, including those that were sensitive (RPMI and U266) and resistant (H929 and L363) to RV. Cell lines were treated with RV and analyzed for the presence of RV genome using RNA in situ hybridization (ISH), and sigma non-structural viral capsid protein (σ-NS), as well as activated caspase-3 by immunohistochemical analysis (IHC) (Figure 1C). We saw increased caspase-3 activation (A-Casp-3) in the sensitive cell lines (RPMI and U266), which also displayed higher levels of σ-NS capsid protein expression (Figure 1D) compared to the resistant cell lines, findings that were further supported by viability assay at 24hrs after treatment with RV. Additionally, based on AnnexinV-PI staining following 48hrs of infection, viability of the different MM cell lines was inversely correlated with the amount of virus genome present during the first hours of infection (3–9hrs) as detected by quantitative real-time PCR (qRT-PCR) (p < 0.0001, trend analysis) (Figure 1E).
Figure 1.
MM cell lines display differential sensitivities to RV. A) MM cell lines were treated for 48 hrs with RV at a multiplicity of infection (MOI) of 1, 2, 4, and 6, or PBS as a control (0). Proliferation was assessed by MTS assay. Graph represents results from three independent experiments with each treatment performed in triplicate. B) p-p38 MAPK expression in MM cell lines was determined by IHC. Images of H929 and RPMI are representative of other cell lines, which all have equivalent p-p38 activation upon RV infection as evaluated by the p-p38 staining index shown in the graph. C) MM cell lines were treated with RV at MOI 10 for 24 hours. The presence of the RV genome was detected by in ISH (bottom), while the levels of the RV protein σ-NS (middle) and activated caspase-3 (Act-Casp-3; top) were measured by IHC. Images of H929 and RPMI are representative for all MM cell lines tested. D) Caspase 3 activation RV σ-NS protein expression and RV genome following treatment of MM cell lines with RV at MOI 10 for 24 hours, as detected by IHC and ISH analysis. On the right panel activated Caspase-3 after 9hrs and 24hrs of RV infection. RPMI xenograft tumors treated (+Ctrl) with proteosome inhibitor (Velcade) or stained with not-selective IgG (-Ctrl) were used as positive and negative control respectively. E) MM cell lines (RPMI, U266, H929 and L-363) were treated for 3 and 9 hrs with RV at MOI10. Untreated cells were used as control (Ctrl). RV genome expression levels were assessed by q-RT-PCR and correlated with cell viability as assessed by Annexin V and propidium iodide staining after 48hrs of RV treatment. RV untreated MM cells are defined as All Cells. Data were divided into 5 groups based on the virus genome levels detected by q-RT-PCR (2−DCT): Low (< 5000 2−DCT); Median (< 100,000 2−DCT); High (<2,000,000 2−DCT); High-plus (>2,000,000 2−DCT). Data were presented by dots with 95% confidence interval of mean for each group, and each dot represents an observation. Slope analysis showed as the virus genome decreased, the percent alive cells decreases (slope=−2.26, p-value<0.0001).
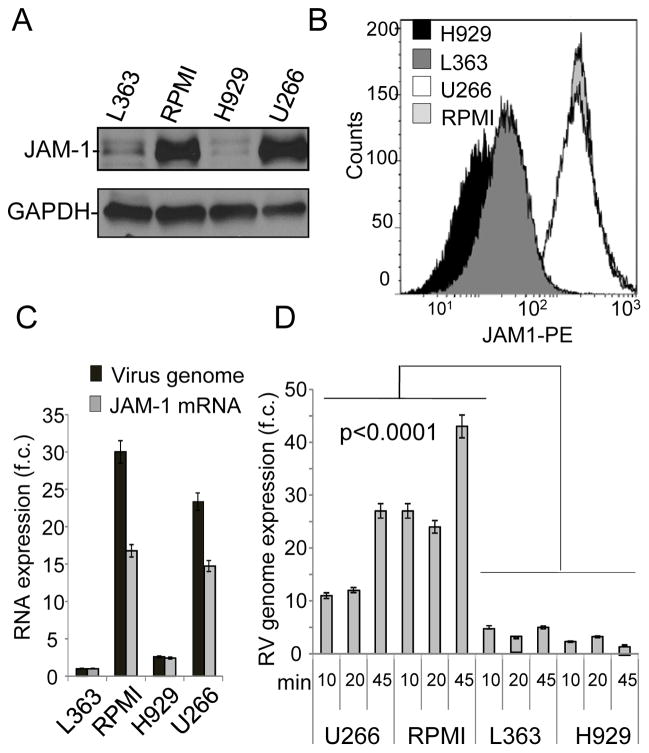
Expression of RV receptor JAM-1 in MM cell lines
Investigation of the RV receptor expression (JAM-1) in MM cell lines by western blot (Fig. 2A) and flow cytometry (Fig. 2B) showed that RV sensitive cell lines have higher JAM-1 expression levels compared to the resistant ones. We also found higher levels of RV genome in those cell lines that had higher surface JAM-1 expression (RPMI and U266), than those with lower surface JAM-1 expression (H929 and L-363) (Figure 2C). To assess whether differences in viral genome levels among the cell lines were associated with different tropism and replication of the virus, we treated MM cell lines with RV at a high MOI (500). Cells were collected at early time points (10, 20, and 45 minutes after infection), washed with trypsin to remove non-internalized virus from the cell surface, and the RV genome was then quantified. qRT-PCR indicated that even at early time points, sensitive MM cells had greater intracellular viral genome levels when compared to the resistant cell lines (Figure 2D). Correlations between JAM-1 surface expression and RV infection (Pearson correlation coefficient=−0.68, p-value=0.002) were also identified in mantle cell lymphoma cells (MCL) (Supplementary Fig. S1B), another B cell malignancy. In conclusion, our data support the idea that RV tropism correlates with JAM-1 receptor expression, and this may play a role in affecting RV tropism in B cell malignancies.
Figure 2.
MM cell lines variably express the high affinity RV protein receptor JAM-1. A) Western blot showing total protein expression of JAM-1 in MM cell lines L-363, RPMI, H929, and U266. GAPDH expression was used as a loading control. The data presented are representative of 3 biological replicates. B) Cell surface expression of the RV receptor JAM-1 in MM cell lines H929, L-363, U266, and RPMI was determined by flow cytometry using PE-conjugated JAM-1 antibody. C) JAM-1 mRNA expression affects the efficiency of RV infection. L363 cells were treated with RV at MOI of 10 for 9 hrs and viral genome expression was determined by qRT-PCR and normalized to GAPDH. Data are expressed in relative fold change (f.c.) compared to the controls. D) MM cell lines more susceptible to RV infection (RPMI and U266) show higher levels of the RV genome at early time-points of infection (10, 20 and 45min), compared to more resistant L-363 and H929 cells. Cells were washed in PBS to remove unbound virus and in trypsin to remove bound un-internalized virus. RV genome expression was determined by q-RT-PCR and normalized against GAPDH.
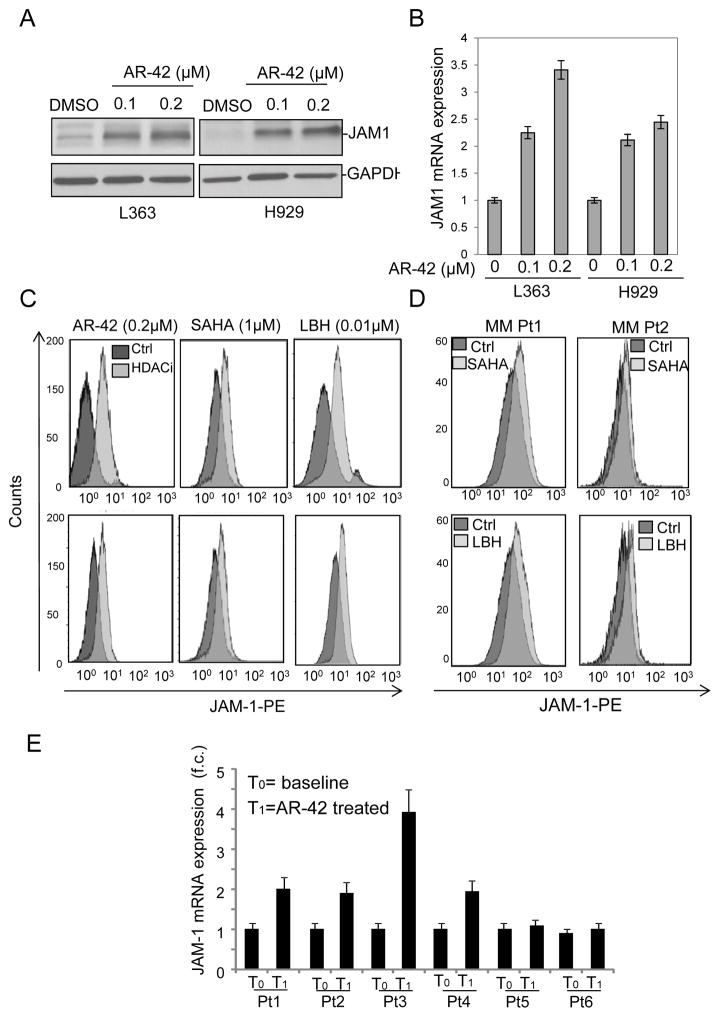
JAM-1 expression is epigenetically regulated in MM cells
The JAM-1 gene (also known as F11R) is located at 1q21.1-1q21.3, a chromosomal region frequently amplified and epigenetically regulated in MM (29). The reported 1q21 copy number in the cell lines used in this study had no correlation with the observed level of JAM-1 protein expression (29), which led us to hypothesize that JAM-1 expression could be epigenetically regulated in MM. We then tested whether treatment with various HDACi’s in clinical use such as AR-42, SAHA, and LBH changed JAM-1 protein expression in MM cells that were less sensitive to RV infection. In MM cell lines the drugs were used at comparable IC50 concentrations (AR-42 0.2μM, SAHA 1.0μM and LBH 0.01μM), and their effect was assessed at 48hrs (Supplementary Fig. S2A). We observed a massive dose dependent increase of JAM-1 protein and mRNA levels after 24hrs of treatment with all 3 HDACi’s (Figure 3A–C). An increase in JAM-1 mRNA and surface expression upon HDACi treatment was also evident in MM.1S cells (Supplementary Fig. S2B) and to a lesser extend in U266 and RPMI that already display high surface JAM-1 expression levels (data not shown). Importantly, we show that treatment with HDACi increased surface JAM-1 expression in primary MM patient samples (Figure 3D). We analyzed RNA samples isolated from the CD138+ cell fractions of the BM aspirates obtained from refractory MM patients enrolled in our single agent AR-42 trial (NCT 01798901). RNA was isolated before and 24hrs after the second cycle of AR-42 treatment (15 days), and qRT-PCR analysis demonstrated increased JAM-1 mRNA expression in 4 of 6 patients (Figure 3E) (p=0.03). Collectively, our data supports that JAM-1 expression is regulated by HDACi in vitro and in MM patients.
Figure 3.
HDACi increase JAM-1 expression in MM cells. A) L363 and H929 cell lines were treated with 0.01% DMSO as control, 0.1μM AR-42, or 0.2μM AR-42 for 24 hrs and JAM-1 protein was measured by immunoblot with GAPDH as a loading control. Immunoblots displayed are representative of 3 independent experiments. B) L363 and H929 cells were treated as in (A) and JAM-1 mRNA expression was measured by qRT-PCR, using GAPDH for normalization. C) Increased cell surface expression of JAM-1 in H929 (top) and L-363 (bottom) MM cell lines following 24-hr treatments with 0.2μM AR-42, 1μM SAHA, or 0.01μM LBH, as compared to control (0.5% DMSO), measured by flow cytometry using PE-conjugated anti-JAM-1 antibody. The LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit was used to ensure data were collected from live cells. D) HDACi treatment of primary plasma cells isolated from MM patients increases JAM-1 cell surface expression. CD138+ cells were isolated by positive selection from bone marrow aspirates of 2 MM patients (MM Pt1 and MM Pt2) and treated for 24 hrs with 0.5μM SAHA, 0.008μM LBH, or control (0.01% DMSO). Cell surface JAM-1 expression was then determined by flow cytometry with PE-conjugated anti-JAM-1 antibody. E) AR-42 increases JAM-1 mRNA expression in CD138+ cells from MM patients. As part of a phase I clinical trial, patients received 40mg AR-42 orally Mon-Wed-Fri, in cycles of 28 days, followed by a 7-day break. RNA was isolated from CD138+ cells purified by positive selection from bone marrow aspirates before AR-42 treatment and 15 days after receiving the last dose of AR-42. JAM-1 mRNA expression was determined by qRT-PCR and normalized for GAPDH.
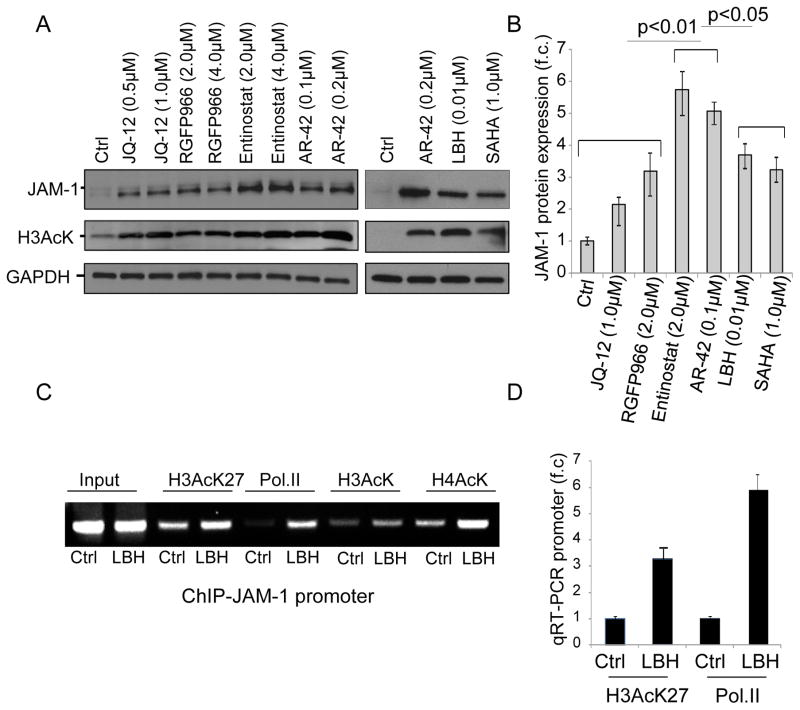
Investigation of HDACi mediated JAM-1 upregulation
To determine whether increased promoter acetylation could be associated with JAM-1 upregulation upon non-selective HDACi treatment, we assessed whether the selective inhibition of Class 1 histone deacetylases enzymes (HDAC1,2 and 3), which are found primarily in the nucleus and play an important role in the regulation of gene expression through direct histone deacetylation (30) could also modulate JAM-1 expression in MM cells. Hence, we compared the effects of the HDAC 1- and 2-selective JQ12, HDAC 3-selective RGFP966, HDAC 1- and 3-selective Entinostat, and the pan-HDACi’s AR-42, LBH and SAHA on JAM-1 expression upon 24hrs treatment. The efficacy of the treatments was assessed using lysine acetylation of histone 3 (H3AcK) (Figure 4A) and for drugs in clinical use by using comparable IC50 (Fig. S2A). Our data show that AR-42 and Entinostat increased JAM-1 protein expression to higher degree than the other selective and non-selective HDACi we tested (figure 4A,B). Further, using a chromatin immunoprecipitation (ChIP) experiment incorporating the JAM-1 promoter before and after 24hrs of treatment of RV-refractory L363 cells with pan-HDACi, we found increased acetylation of lysine 27 on histone 3 (H3AcK27) and increased association of RNA polymerase II (Pol II) with the JAM-1 promoter (Figure 4C,D), supporting the idea that increased in promoter acetylation is one of the primary mechanisms underlying the upregulation of JAM-1 following HDACi treatments.
Figure 4.
JAM-1 expression is epigenetically regulated in MM cells. A) L363 cells were treated for 24 hrs with indicated concentrations of JQ-12, RGFP966, Etinostat, AR-42, SAHA and LBH or DMSO as control. Whole cell extracts were analyzed by western blot using JAM-1 antibody (top). Staining with anti-Acetylated histone 3 antibody (H3AcK) (middle) was used to confirm activity of all drugs and GAPDH (bottom) served as a loading control. B) JAM-1 mRNA fold change ±SD in L363 cells after treatment with HDACi. GADPH was used for normalization proposes. C) L363 cells were treated with 0.01μM LBH or 0.01% DMSO for 24 hrs and analyzed by ChIP assay using H3AcK27, anti-RNA Polymerase II (PolII), anti-H3AcK, and anti-H4AcK antibodies. The association between acetylated histones and RNA polymerase II with the JAM-1 promoter were visualized by agarose gel electrophoresis. D) Immunoprecipitated chromatin obtained in the ChIP experiment in (C) was also tested by qRT-PCR and demonstrated increased levels of the JAM-1 promoter in association with acetylated H3AcK27 and Pol II. Control primers for JAM-1 exon 3 and for GAPDH were used for normalization purposes.
Pan-HDACi sensitize MM cell to RV treatment in vitro
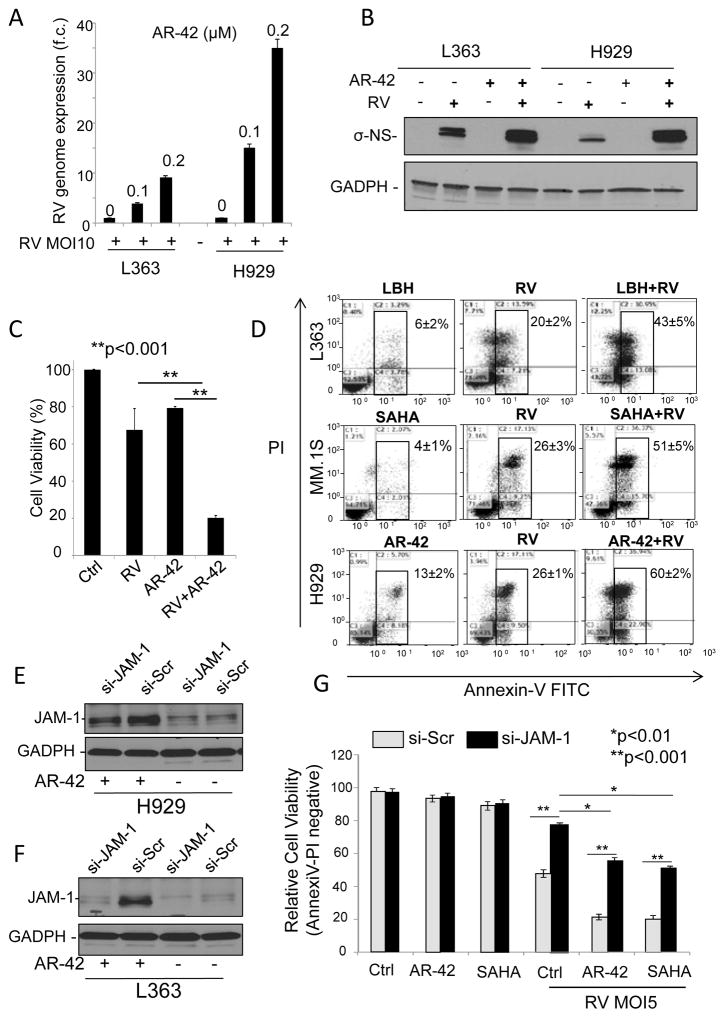
Based on our observations that MM cell RV sensitivity is associated with expression of the RV receptor JAM-1 and that treatment with HDACi increases JAM-1 expression, we hypothesized that combining HDACi with RV would enhance viral induced oncolysis. To test this, we treated RV-refractory L-363 and H929 cells with single-agent RV and RV in combination with AR-42. Q-RT-PCR showed that the amount of intracellular RV viral genome increased after AR-42 was added (Figure 5A). Additionally, western blot indicated that the combination treatment increased σ-NS viral capsid protein production (Figure 5B). To this end, we found that there was a greater than 3-fold increase in viral particles evident in the supernatant obtained from MM cells incubated for 24hrs with both RV and AR-42 when compared to either single agent alone (Figure 5C).
Figure 5.
Combination treatment of MM cells with RV and HDACi’s results in increased levels of RV genome, RV protein production, and enhanced MM cell death compared to single agent treatments. A) L-363 and H929 MM cell lines were treated with RV at MOI of 10 (+) in combination with 0.01% DMSO (0), 0.1μM AR-42 (0.1), or 0.2μM AR-42 (0.2) for 48 hours. RNA was isolated and expression of the RV genome was determined by q-RT-PCR and normalized to GAPDH. B) L-363 and H929 cells were treated with 0.01% DMSO and PBS for controls (-), or where indicated by “+”, with RV at MOI of 5, 0.2μM AR-42, or the combinations of RV and AR-42 for 48 hours. Protein lysates were probed for expression of the viral σ-NS capsid protein. GAPDH antibody staining served as a loading control. C) Supernatants of H929 cells harvested after 48 hrs from the treatments outlined in 5B were used to treat additional MM cells at a 1:100 dilution. Cell proliferation was then assessed by MTS assay 48hrs after cells were exposed to the supernatants. D) L-363 (top), MM.1S (middle) and H929 (bottom) cell lines were treated for 48 hrs with 0.5% DMSO and PBS as controls, or HDACi: 0.2μM AR-42, 0.01μM LBH, 1μM SAHA, RV at MOI of 5, or the combinations of pan-HDACi’s and RV. Cells were then stained with Annexin V and PI to detect apoptosis by flow cytometry. Flow cytometry plots are representative of each cell line and for each drug combination. E) siRNA targeting JAM-1 reduces JAM-1 protein expression upon AR-42 treatment. H929 and L363 cells were transfected with siRNA targeting JAM-1 (si-JAM-1), or scramble oligo (si-Scr) for control and allowed to rest for 12 hours. Cells were then treated with 0.5% DMSO (−), or where indicated by “+” with 0.2μM AR-42 for 24hours. JAM-1 protein expression was determined by Western blot. F) Twelve hrs after transfection with si-JAM-1 or si-Scr H929 and L363 cells were treated with vehicle control (Ctrl; 0.01% DMSO and PBS), 0.2μM AR-42, or 1μM SAHA in the absence, or presence of RV at MOI of 5. Cell viability was determined by flow cytometry using AnnexinV/PI staining 48 hrs later ±SD. Each experiment was performed in triplicate.
Further, after 48hrs, increased cell killing activity was observed in all MM cells (L-363, H929, MM.1S) treated for 48hrs with the combination of RV+HDACi compared to the single agents (Figure 5D and data not shown).
Since different HDACi have different half-lives and because by q-RT-PCR JAM-1 mRNA upregulation is already evident after 1hr of treatment reaching the highest level at 8hrs (data not shown), MM cells were pretreated for 10hrs with the different HDACi (SAHA, LBH, AR-42, Entinostat and JQ12) followed by drug washout and then treatment with RV for 48hrs. As shown in supplementary Fig. S3A, reduced drug exposure did not affect the increased anti-MM activity of the different HDACi in combination with RV. Using Chou-Talalay synergy analyses, we examined the ability of each HDACi (LBH, SAHA, AR-42 and Entinostat) to synergize with RV (21–24). Viability data after 48hrs of treatment were utilized to calculate the Combination Index (CI) via the Compusyn program, in which CI<1 indicates synergistic interaction, CI>1 is antagonistic, and CI=1 is additive. Our data show that the combination of all HDACi and RV synergistically killed the tumor cells, even at the lowest drug concentrations (Supplementary Fig. S3B). Increased cell killing was also observed in MCL cell lines when RV was used in combination with AR-42 compared to the single agents (Supplementary Fig. S4A,B).
To further support the functional role of JAM-1 in promoting RV induced MM cell death, we performed knockdown of JAM-1 expression using small interfering RNA (siRNA) followed by treatment with HDACi’s (AR-42, SAHA), or vehicle control in both H929 and L363 cells (Figure 5E). JAM-1 knockdown in MM cells was able to prevent the majority of the apoptotic effect of cells treated with single agent RV and more than 60% of the apoptotic rate of cells those treated with combination RV+HDACi (Figure 5F).
HDACi sensitizes MM cell to RV treatment in vivo
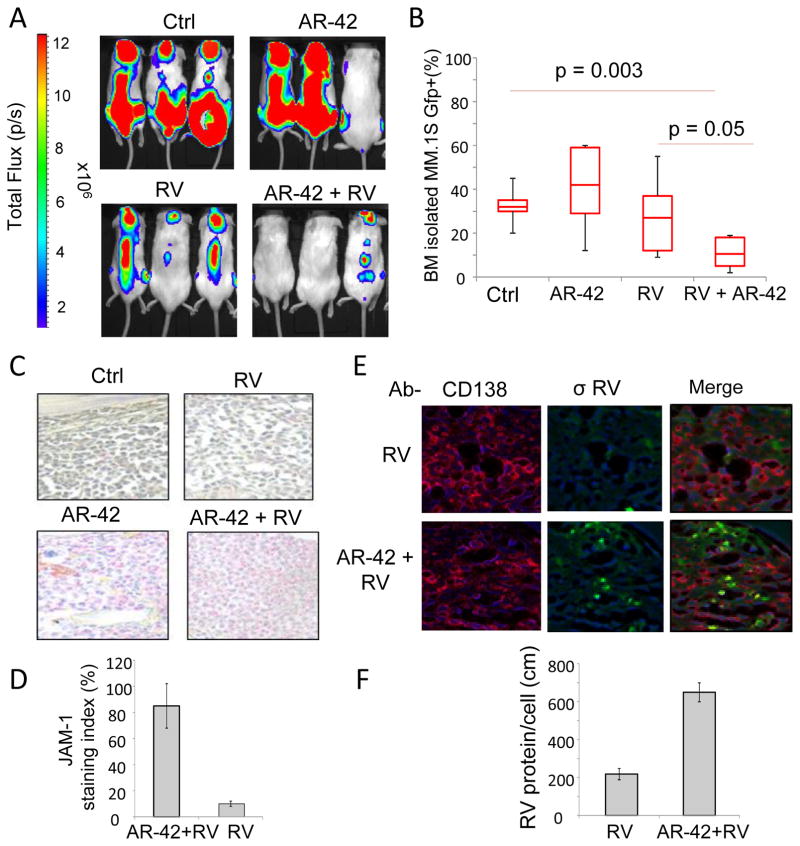
We used our previously described human MM cell homing model to investigate whether the combination of RV and HDACi could be a successful therapeutic strategy for treating MM (25, 31). Forty NOD-SCID mice were injected intravenously with 5×106 GFP+/Luc+ MM.1S cells. Three weeks later, mice with similar tumor burden, as calculated by luminescence, were divided into 4 treatment groups (6 mice per group): AR-42 alone plus RV control vehicle (PBS), RV plus AR-42 control vehicle (8% DMSO in PBS), AR-42 plus RV, and the combination of both vehicle controls. We used the combination of intraperitoneal injection (IP) of AR-42 (25 mg/kg on Monday, Wednesday, and Friday), single IV treatment of Reolysin (5×108 TCID50), and vehicle controls for a total of 3 weeks (Figure 6A). At the end of the treatment, the extent of bone marrow engraftment was determined by detecting GFP+ cells using flow cytometry. We found that neither single agents AR-42 or RV showed significant effects on tumor burden (Fig. 6B). However, the combination of AR-42 and RV significantly decreased MM cell bone marrow infiltration when compared to either RV (p=0.05) or AR-42 (p=0.003) alone (Figure 6B). Additionally, IHC staining showed increased expression of JAM-1 by MM cells in the bone marrow of mice treated with AR-42 or the combination of AR-42 and RV compared to control and RV treated mice (Fig. 6C and data not shown). Using RNA ISH and IHC, we also observed increased levels of RV genome (green) in the MM cells (red, CD138+) in the BM of mice treated with the combination (Figure 6D). These findings collectively support that treatment with HDACi increases RV replication in MM cells.
Figure 6.
The combination of RV and AR-42 reduces tumor burden in vivo. Forty male NOD-SCID mice were injected with 5×106 MM.1S GFP+/Luc+ cells, and two weeks later 5 mice with similar tumor burden determined by IVIS bioluminescent imaging were allocated to each treatment group: control (8% DMSO in PBS; Ctrl), AR-42 (25mg/kg i.p. Mon-Wed-Fri), single intravenous injection of RV (RV; 5×108 TCID50), or the combination of AR-42 and RV (AR-42+RV). Representative IVIS bioluminescent images of mice from each experimental group at the conclusion of three weeks of the treatments are shown. B) After three weeks of treatments mice were sacrificed and bone marrow cells were harvested from both femurs of each mouse. The degree of MM cell bone marrow infiltration was then determined by flow cytometry to detect GFP+ cells (n= at least 4 mice per group). C) IHC detection of JAM-1 expression in bone marrow sections from mice in different treatment groups (pink staining indicates JAM-1 positive cells). Representative images show increased JAM-1 expression with the addition of AR-42, compared to the treatment with RV alone. D) Immunofluorescence staining for CD138+ MM cells (CD138; red) and capsid σ-NS (green) protein expression. In merged images, MM cells with active RV protein production are indicated by yellow fluorescence, showing co-localization of CD138 and the σ-NS protein. Nuclei of cells are stained with DAPI blue (blue). Quantification of the immunofluorescent staining (bar graph below) showed that the addition of AR-42 resulted in increased RV protein production within the bone marrow of treated mice.
DISCUSSION
Our group is the first to investigate the use of RV in patients with hematologic malignancies. Our single agent RV phase 1 trial confirmed tolerability of the virus in patients with relapsed MM, but the best response was a stable disease in 3 different patients for 4, 5, and 8 cycles (14). Correlative analyses from this study revealed only minimal RV proliferation associated with the lack of expression of the RV receptor JAM-1 in MM cells, and indicated that further exploration of the basic mechanisms underlying RV infection in myeloma cells was needed.
The present study aimed at defining the role of the RV receptor, JAM-1, on viral cell entry, induction of productive infection, and MM cell killing, but we also set out to define strategies to exploit the oncolytic potential of RV in myeloma cells. To begin to address these questions, we treated MM cells with clinically achievable doses of virus and found that RV resistant MM cell lines have lower expression of JAM-1 compared to sensitive cell lines. We then showed that JAM-1 expression is epigenetically regulated in MM cells and can thus be modulated by HDACi. Further, we report that HDACi’s are capable of increasing the expression of JAM-1 by epigenetically regulating its promoter through histone acetylation, and that the combination of HDACi and RV synergistically increases MM cell killing. The role of increased JAM-1 expression in producing this effect was evaluated by siRNA experiments to knockdown JAM-1 expression. While decreased JAM-1 expression by siRNA greatly reduced the amount of cell death produced by RV alone, JAM-1 knockdown could not completely rescue the killing effect of the HDACi and RV combination. Several possible explanations exist for this effect. First, siRNA significantly reduced JAM-1 levels, but did not completely silence its expression. Second, it has been reported that activity levels of the cysteine proteases cathepsins B and L are correlated with the efficiency of RV-mediated cell killing of cancer cells (15) and because HDACi’s can increase the activity of cytoplasmic cathepsin B in MM cells (32), we cannot exclude that this alternative mechanism also supports RV replication following combination treatment. Further investigations are needed to clarify the contribution of HDACi’s in supporting RV replication in MM cells.
HDACi strongly enhance the killing ability of RV in MM cells and this treatment combination could potentially play an important role in improving the clinical therapeutic efficacy of systemic RV treatment in patients with MM. Here we show that Entinostat and AR-42, display a greater ability to upregulate JAM-1 protein expression and greater synergistic activity compared to other selective and non-selective HDACi, though in general, synergistic activity was observed with all HDACi tested. That being said, further preclinical analyses are necessary to assess the tolerability and efficacy of RV in combination with clinically relevant HDACi. In fact, although AR-42 as a single agent in our trial (NCT 01798901) was well tolerated in 26 patients with hematopoietic diseases, including MM, it displayed a shorter half-life (4–56hrs, Cmax 0.3–1.2μM) compared to Entinostat (33). On the other hand, LBH has been recently approved for the treatment of MM and may result in the most valuable companion drug for RV therapy in relapse patients.
Our data also showed that HDACi strongly enhances the killing ability of RV not only in MM cells, but also in MCL cells, and this seems to be dependent on JAM-1 expression, supporting the idea that the use of HDACi to potentiate the RV killing activity could be used not only in MM but also in other B cell malignancies.
In conclusion, HDACi strongly enhance the killing ability of RV in MM cells via upregulation of the RV receptor, JAM-1, and this treatment combination could potentially play an important role in enhancing the clinical therapeutic efficacy of systemic RV in patients with MM. Collectively, our data provides the scientific foundation for future phase 2 combination treatment studies.
Supplementary Material
Acknowledgments
We thank Deborah Parris for her scientific advice, Laura Hanley for her administrative support, and Nita Williams and Jones Desiree for collecting clinical trials samples and clinical trial data. We thank Dr. Hanna S. Radomska for the scientific editing of the manuscript and Robert Weingart and Joseph Liu for the technical support.
A. Stiff was supported by The Ohio State University Pelotonia research fellowship, D.W. Sborov was supported under Award Number T32CA165998. C.C. Hofmeister and F. Pichiorri were supported under NIH R21CA156222. Research was also supported from Multiple Myeloma Opportunities for Research & Education (MMORE) and NIH R21CA156222 (C.C. Hofmeister).
Footnotes
Financial disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict.
References
- 1.Tyler KL, Fields BN. RVes. In: Fields BN, Knipe DM, Chanock RM, et al., editors. Virology. 2. New York: Raven Press, Ltd; 1990. pp. 1307–28. [Google Scholar]
- 2.Coffey MC, Strong JE, Forsyth PA, Lee PW. RV therapy of tumors with activated Ras pathway. Science. 1998;282:1332–4. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- 3.Strong JE, Coffey MC, Tang D, Sabinin P, Lee PW. The molecular basis of viral oncolysis: usurpation of the Ras signaling pathway by RV. The EMBO journal. 1998;17:3351–62. doi: 10.1093/emboj/17.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guglielmi KM, Kirchner E, Holm GH, Stehle T, Dermody TS. RV binding determinants in junctional adhesion molecule-A. The Journal of biological chemistry. 2007;282:17930–40. doi: 10.1074/jbc.M702180200. [DOI] [PubMed] [Google Scholar]
- 5.Konopka-Anstadt JL, Mainou BA, Sutherland DM, Sekine Y, Strittmatter SM, Dermody TS. The Nogo receptor NgR1 mediates infection by mammalian RV. Cell host & microbe. 2014;15:681–91. doi: 10.1016/j.chom.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall K, Scott KJ, Rose A, Desborough M, Harrington K, Pandha H, et al. RV-mediated cytotoxicity and enhancement of innate immune responses against acute myeloid leukemia. BioResearch open access. 2012;1:3–15. doi: 10.1089/biores.2012.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steele L, Errington F, Prestwich R, Ilett E, Harrington K, Pandha H, et al. Pro-inflammatory cytokine/chemokine production by RV treated melanoma cells is PKR/NF-kappaB mediated and supports innate and adaptive anti-tumour immune priming. Molecular cancer. 2011;10:20. doi: 10.1186/1476-4598-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prestwich RJ, Errington F, Steele LP, Ilett EJ, Morgan RS, Harrington KJ, et al. Reciprocal human dendritic cell-natural killer cell interactions induce antitumor activity following tumor cell infection by oncolytic RV. Journal of immunology. 2009;183:4312–21. doi: 10.4049/jimmunol.0901074. [DOI] [PubMed] [Google Scholar]
- 9.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 10.Kapoor P, Kumar S, Fonseca R, Lacy MQ, Witzig TE, Hayman SR, et al. Impact of risk stratification on outcome among patients with multiple myeloma receiving initial therapy with lenalidomide and dexamethasone. Blood. 2009;114:518–21. doi: 10.1182/blood-2009-01-202010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly KR, Espitia CM, Mahalingam D, Oyajobi BO, Coffey M, Giles FJ, et al. RV therapy stimulates endoplasmic reticular stress, NOXA induction, and augments bortezomib-mediated apoptosis in multiple myeloma. Oncogene. 2012;31:3023–38. doi: 10.1038/onc.2011.478. [DOI] [PubMed] [Google Scholar]
- 12.Thirukkumaran CM, Shi ZQ, Neri P, Bahlis N, Morris D. RV synergy with proteosome inhibitor carfilzomib and Akt inhibitor perifisone overcomes therapy resistance of multiple myeloma: promising preclinical activity [abstract]. Proceedings of the 105th Annual Meeting of the American Association of Cancer Research; 2014 Apr 5–9; San Diego, CA. Philadelphia (PA): AACR; 2014. p. Abstract nr 1709. [Google Scholar]
- 13.Thirukkumaran CM, Shi ZQ, Luider J, Kopciuk K, Gao H, Bahlis N, et al. RV modulates autophagy during oncolysis of multiple myeloma. Autophagy. 2013;9:413–4. doi: 10.4161/auto.22867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sborov DW, Nuovo GJ, Stiff A, Mace T, Lesinski GB, Benson DM, Jr, et al. A phase I trial of single-agent reolysin in patients with relapsed multiple myeloma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:5946–55. doi: 10.1158/1078-0432.CCR-14-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katayama Y, Terasawa Y, Tachibana M, Mizuguchi H, Sakurai F. Proteolytic disassembly of viral outer capsid proteins is crucial for RV-mediated type-I interferon induction in both RV-susceptible and RV-refractory tumor cells. BioMed research international. 2015;2015:468457. doi: 10.1155/2015/468457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimopoulos K, Gimsing P, Gronbaek K. The role of epigenetics in the biology of multiple myeloma. Blood cancer journal. 2014;4:e207. doi: 10.1038/bcj.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson P, Mitsiades C, Colson K, Reilly E, McBride L, Chiao J, et al. Phase I trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) in patients with advanced multiple myeloma. Leukemia & lymphoma. 2008;49:502–7. doi: 10.1080/10428190701817258. [DOI] [PubMed] [Google Scholar]
- 18.Dimopoulos M, Siegel DS, Lonial S, Qi J, Hajek R, Facon T, et al. Vorinostat or placebo in combination with bortezomib in patients with multiple myeloma (VANTAGE 088): a multicentre, randomised, double-blind study. The lancet oncology. 2013;14:1129–40. doi: 10.1016/S1470-2045(13)70398-X. [DOI] [PubMed] [Google Scholar]
- 19.San-Miguel JF, Richardson PG, Gunther A, Sezer O, Siegel D, Blade J, et al. Phase Ib study of panobinostat and bortezomib in relapsed or relapsed and refractory multiple myeloma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:3696–703. doi: 10.1200/JCO.2012.46.7068. [DOI] [PubMed] [Google Scholar]
- 20.San-Miguel JF, Hungria VTM, Yoon S-S, Beksac M, Dimopoulos MA, Elghandour A, et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. The Lancet Oncology. 2014;15:1195–206. doi: 10.1016/S1470-2045(14)70440-1. [DOI] [PubMed] [Google Scholar]
- 21.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer research. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 22.Aghi M, Rabkin S, Martuza RL. Effect of chemotherapy-induced DNA repair on oncolytic herpes simplex viral replication. Journal of the National Cancer Institute. 2006;98:38–50. doi: 10.1093/jnci/djj003. [DOI] [PubMed] [Google Scholar]
- 23.Cheema TA, Kanai R, Kim GW, Wakimoto H, Passer B, Rabkin SD, et al. Enhanced antitumor efficacy of low-dose Etoposide with oncolytic herpes simplex virus in human glioblastoma stem cell xenografts. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7383–93. doi: 10.1158/1078-0432.CCR-11-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo JY, Hurwitz BS, Bolyard C, Yu JG, Zhang J, Selvendiran K, et al. Bortezomib-induced unfolded protein response increases oncolytic HSV-1 replication resulting in synergistic antitumor effects. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:3787–98. doi: 10.1158/1078-0432.CCR-14-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer cell. 2010;18:367–81. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Bodkin DK, Fields BN. Growth and survival of RV in intestinal tissue: role of the L2 and S1 genes. Journal of virology. 1989;63:1188–93. doi: 10.1128/jvi.63.3.1188-1193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarone G, Galetto G, Prat M, Comoglio PM. Cell surface molecules and fibronectin-mediated cell adhesion: effect of proteolytic digestion of membrane proteins. The Journal of cell biology. 1982;94:179–86. doi: 10.1083/jcb.94.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. CD46 is a cellular receptor for human herpesvirus 6. Cell. 1999;99:817–27. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- 29.Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–32. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.New M, Olzscha H, La Thangue NB. HDAC inhibitor-based therapies: can we interpret the code? Molecular oncology. 2012;6:637–56. doi: 10.1016/j.molonc.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–80. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheriyath V, Kuhns MA, Kalaycio ME, Borden EC. Potentiation of apoptosis by histone deacetylase inhibitors and doxorubicin combination: cytoplasmic cathepsin B as a mediator of apoptosis in multiple myeloma. British journal of cancer. 2011;104:957–67. doi: 10.1038/bjc.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frys S, Simons Z, Hu Q, Barth MJ, Gu JJ, Mavis C, et al. Entinostat, a novel histone deacetylase inhibitor is active in B-cell lymphoma and enhances the anti-tumour activity of rituximab and chemotherapy agents. British journal of haematology. 2015;169:506–19. doi: 10.1111/bjh.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.