Abstract
The CD4+ and CD8+ T cell dichotomy is essential for effective cellular immunity. How the individual T cell identity is established remains poorly understood. Here we show that the high mobility group (HMG) transcription factors Tcf1 and Lef1 are essential for repressing CD4+ lineage-associated genes including Cd4, Foxp3 and Rorc in CD8+ T cells. Tcf1- and Lef1-deficient CD8+ T cells exhibit histone hyperacetylation, which is ascribed to an unexpected intrinsic histone deacetylase (HDAC) activity in Tcf1 and Lef1. Mutating five conserved amino acids in the Tcf1 HDAC domain diminishes the HDAC activity and the ability to suppress CD4+ lineage genes in CD8+ T cells. These findings reveal that sequence-specific transcription factors can utilize intrinsic HDAC activity to guard cell identity by repressing lineage-inappropriate genes.
Cell identity is established by lineage-determining transcription factors, which initiate and sustain expression of cell type-specific genes while repressing those in alternative lineages1, 2, 3, 4. Important insights and extrapolatable paradigms have been derived from hematopoietic cells. Transcription factors (TFs) play instructive roles in lineage determination; for examples, GATA-1 and PU.1 antagonistically control development of erythroid-megakaryocytes and myeloid cells, respectively5, 6. Lineage-committed cells remain dependent on key TFs to guard cell identity. Deletion of Pax5 in mature B cells causes dedifferentiation to uncommitted progenitors, which generate T-lineage cells7. Loss of Bcl11b induces T cells to acquire properties of natural killer cells8. During and after a cell identity is established, TFs are assisted by epigenetic mechanisms, i.e., covalent modification of DNA and/or nucleosomal histones, to ensure heritable expression of lineage-appropriate genes2, 4.
CD4+ and CD8+ T cells are functionally distinct lymphocytes that play essential roles in immune defense against foreign pathogens and transformed cells. Whereas CD8+ T cells are dedicated cytotoxic cells, CD4+ T cells provide versatile help tailored to specific pathogen types. In spite of the functional disparity, naïve CD4+ and CD8+ T cells show remarkably similar transcriptomes, with few genes exhibiting lineage-restricted expression9. Among the CD4+ signature genes, transcription factor ThPOK protects the CD4+-lineage integrity10. Naïve CD4+ T cells lacking ThPOK exhibit increased expression of CD8 coreceptors and Runx3, and abundant upregulation of cytotoxicity-associated genes10. Two histone deacetylase (HDAC) proteins, HDAC1 and HDAC2, although not exclusively expressed in CD4+ T cells, have a similar CD4+-lineage protective effect. Deleting HDAC1-HDAC2 in CD4+ T cells causes induction of a CD8+ effector-like program11. In contrast, it remains unknown how CD8+ T cell identity is regulated.
Tcf1 and Lef1 (encoded by Tcf7 and Lef1, respectively) are HMG transcription factors that are required for early stages of thymocyte maturation12, 13, 14, 15. Induction of Tcf1 in early thymic progenitors by Notch signaling is essential for T cell lineage specification16, 17. In T lineage-committed thymocytes, Tcf1 prevents malignant transformation and promotes β-selection at the CD4−CD8− double negative (DN) 3 stage18, 19. Upon maturation of DN thymocytes to the CD4+CD8+ double positive (DP) stage, the DP cells face a lineage choice decision of becoming either CD4+ or CD8+ T cells20. In DP thymocytes, Tcf1 and Lef1 act upstream of ThPOK and regulate CD4+-lineage decision13, 21. Therefore, Tcf1 and Lef1 have versatile, developmental stage-specific roles during T cell maturation.
Here we report that although not required for DP thymocytes to choose the CD8+ fate, Tcf1 and Lef1 are essential for establishing CD8+ T cell identity, and this function is mediated by intrinsic HDAC activity embedded in these two factors. Our data suggest that Tcf1 and Lef1 directly bridge genetic and epigenetic regulation to establish a gene expression pattern appropriate for CD8+-lineage T cells.
Results
Ablating Tcf1-Lef1 induces CD4+ lineage genes in CD8+ T cells
Tcf1 and Lef1 are required for CD4+-lineage decision21. Compared to WT mice, CD4-Cre+Tcf7FL/FL Lef1FL/FL mice (called Tcf7−/−Lef1−/− hereafter) had very few CD4+ mature thymocytes but increased CD8+ mature thymocytes (Supplementary Fig. 1a)21. The Tcf7−/−Lef1−/− CD8+ T cells retained CD8+ T cell features such as production of interferon-γ and perforin21 and expression of CD8+-specific genes including Runx3d, Nkg7 and Itgae (Supplementary Fig. 1b), but aberrantly expressed the CD4 coreceptor (Supplementary Fig. 1a)21. To assess the global impact of Tcf1 and Lef1 deficiency, we performed RNA-Seq analysis on sort-purified CD69−CD24−TCRβ+CD8+ mature thymocytes from Tcf7−/−, Tcf7−/−Lef1−/−, and their control littermates (CD4-Cre+Tcf7+/+Lef1+/+, CD4-Cre+Tcf7FL/+Lef1FL/+, or CD4-Cre− mice). By a setting of ≥2 fold expression changes and false discovery rate (FDR)<0.01, 718 genes were downregulated and 472 genes were upregulated in Tcf7−/−Lef1−/− compared to control CD8+ mature thymocytes (Fig. 1a). Among the genes markedly upregulated in Tcf7−/−Lef1−/− CD8+ thymocytes were CD8+ T cell effector molecules such as Blimp1 (Prdm1), perforin (Prf1) and Fas ligand (Fasl; Supplementary Fig. 1c). Tcf7−/−Lef1−/− CD8+ T cells were neither aberrantly activated (as determined by the CD44hiCD62Llo phenotype) nor enriched for IL-2Rβ+CD44hi memory-phenotype T cells (Supplementary Fig. 1a, d). We also surveyed the Vβ TCR repertoire in CD8+ mature thymocytes in Tcf7−/−Lef1−/− and control littermates, and no particular Vβ subtype(s) was over-represented due to Tcf1-Lef1 deficiency (Supplementary Fig. 1e,f). Thus, the global transcriptomic changes in Tcf7−/−Lef1−/− CD8+ T cells were not associated with abnormal expansion of a specific TCR chain or functional T cell subset. Our data suggest that Tcf1 and Lef1 restrain effector molecule expression in naïve CD8+ T cells to prevent aberrant cytotoxic activities.
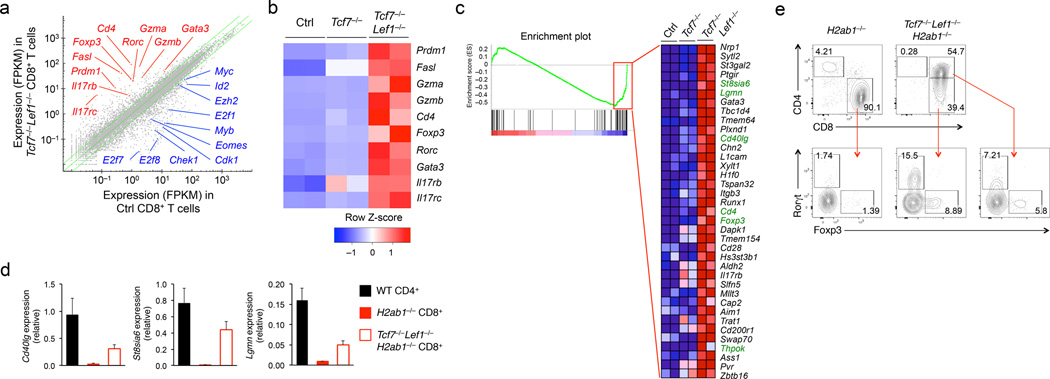
Figure 1. Tcf1 and Lef1 deficiency perturbs CD8+ T cell integrity.
(a) RNA-Seq analysis of genes upregulated (red) or downregulated (blue) in TCRβhiCD24−CD69−CD8+ mature thymocytes sorted from Tcf7−/−Tcf7−/−Lef1−/−, and control littermates, with the average FPKM values of two replicates of control (Ctrl) vs. Tcf7−/−Lef1−/− CD8+ T cells shown in a scatterplot, where the green lines denote gene expression changes of ≥2 fold. (b) Select upregulated genes (right margin) in Tcf7−/−Lef1−/−CD8+ mature thymocytes relative to control and Tcf7−/− CD8+ thymocytes as shown in a heatmap. (c) GSEA showing enriched expression of genes in the CD4+ T cell gene set in Tcf7−/−Lef1−/− CD8+ mature thymocytes, with the enriched genes (red rectangle in the enrichment plot) displayed in a heatmap, where CD4+ signature genes are highlighted in green. (d) Quantitative RT-PCR analysis of Cd40lg, St8sia6 and Lgmn expression (relative the Hprt housekeeping gene) in CD4+ mature thymocytes sorted from wild-type (WT) mice, CD8+ mature thymocytes sorted from H2ab1−/− or Tcf7−/−Lef1−/−H2ab1−/− mice. (e) Intracellular staining for Foxp3 and Rorγt proteins in CD8+ mature thymocytes in H2ab1−/− mice, CD8+CD4− and CD8+CD4+ thymocytes in Tcf7−/−Lef1−/−H2ab1−/− mice. Numbers adjacent to outlined areas indicate percent Foxp3+ and Rorγt+ cells in lower panels. Data are from one experiment measuring duplicate samples (a-c), from 2 experiments (d, means ± s.d., n ≥ 4), or representative of ≥ 5 experiments (e).
Tcf7−/−Lef1−/− CD8+ mature thymocytes also showed increased expression of CD4+ lineage-related genes, including Cd4 and the transcription factors Foxp3, Gata3, and Rorc (Fig. 1b). Based on published data9, we constructed a CD4+ T cell gene set that contained 108 genes expressed ≥ 2 fold in CD4+ compared to CD8+ T cells (Supplementary Table 1). Gene set enrichment analysis (GSEA) revealed that 37 genes in the CD4+ T cell gene set exhibited enriched expression in Tcf7−/−Lef1−/− compared to control CD8+ thymocytes, including all five CD4+ T cell signature genes (Cd4, Thpok, St8sia6, Lgmn, and Cd40lg) that are not expressed in naïve CD8+ T cells (Fig. 1c). The increased expression of CD4+ lineage-associated genes in Tcf7−/−Lef1−/− CD8+ thymocytes were validated with quantitative RT-PCR (for the Cd40lg, St8sia6a, Lgmn, Itgb3 and Gata3 transcripts) or intracellular staining of Foxp3 and Rorγt proteins (Supplementary Fig. 2a,b). Among the CD8+ T cell effector molecules, increased protein expression of FasL was evident in naïve Tcf7−/−Lef1−/− splenic CD8+ T cells, and elevated expression of CD40L and granzyme B proteins was only detected in Tcf7−/−Lef1−/− splenic CD8+ T cells after in vitro priming, compared to control splenic CD8+ T cells (Supplementary Fig. 2c–e).
Because CD4+ T cells are redirected to CD8+ lineage upon loss of Tcf1 and Lef121, the increased expression of CD4+ lineage-associated genes in Tcf7−/−Lef1−/− CD8+ T cells might be derived from their origin as CD4+ T cells. To address this possibility, we crossed Tcf7−/−Lef1−/− mice to MHC-II-deficient H2ab1−/− mice to minimize CD4+ T cell production and hence, their redirection to CD8+ lineage. Compared to CD8+ mature thymocytes in H2ab1−/− mice, Tcf7−/−Lef1−/−H2ab1−/− CD8+ thymocytes showed increased Cd40lg, St8sia6, and Lgmn transcripts and CD4, Foxp3 and Rorγt proteins (Fig. 1d,e), independent of lineage redirection. We also noted that the upregulation of CD4, Foxp3 and Rorγt proteins only occurred in a fraction but not all of Tcf7−/−Lef1−/− CD8+ thymocytes, in both MHC-II-sufficient and –deficient background (Fig. 1e, Supplementary Fig. 2b), indicative of variegated gene expression. These data indicate that Tcf1 and Lef1 are intrinsically required for maintaining the CD8+ T lineage integrity.
Histone hyperacetylation in Tcf1-Lef1-deficient CD8+ T cells
Variegated gene expression, i.e., conversion between silenced and expressed status in a proportion of given cell population, is suggestive of epigenetic regulation22. Cd4 gene silencing in CD8+ T cells is known to be mediated by epigenetic mechanisms23. We thus investigated how Tcf1-Lef1 deficiency affects the epigenome of CD8+ T cells by performing ChIP-Seq analysis of H3K4me3, H3K9Ac, H3K27me3 and H3K27Ac histone marks on wild-type and Tcf7−/−Lef1−/−CD8+ mature thymocytes and wild-type CD4+ mature thymocytes. The loci of 108 genes in CD4+ T cell gene set had stronger H3K4me3 signals in wild-type CD4+ than wild-type CD8+ thymocytes, and there was a moderate increase in both H3K4me3 and H3K27me3 signals at these loci in Tcf7−/−Lef1−/− compared to wild-type CD8+ thymocytes (Supplementary Fig. 3a). On the other hand, the CD4+ T cell gene set showed substantially stronger H3K27Ac signals in both wild-type CD4+ and Tcf7−/−Lef1−/− CD8+ thymocytes than in wild-type CD8+ thymocytes (Fig. 2a). A similar trend was observed for H3K9Ac, albeit at a moderate level compared to H3K27Ac (Fig. 2a). For individual CD4+ T cell signature genes, we found strong H3K27Ac signals in both wild-type CD4+ and Tcf7−/−Lef1−/− CD8+ thymocytes at the transcription start sites (TSSs) and intron regions of Cd4, St8sia6 and Itgb3, and at Cd40lg upstream and Lgmn downstream regulatory regions (Fig. 2b, Supplementary Fig. 3b).
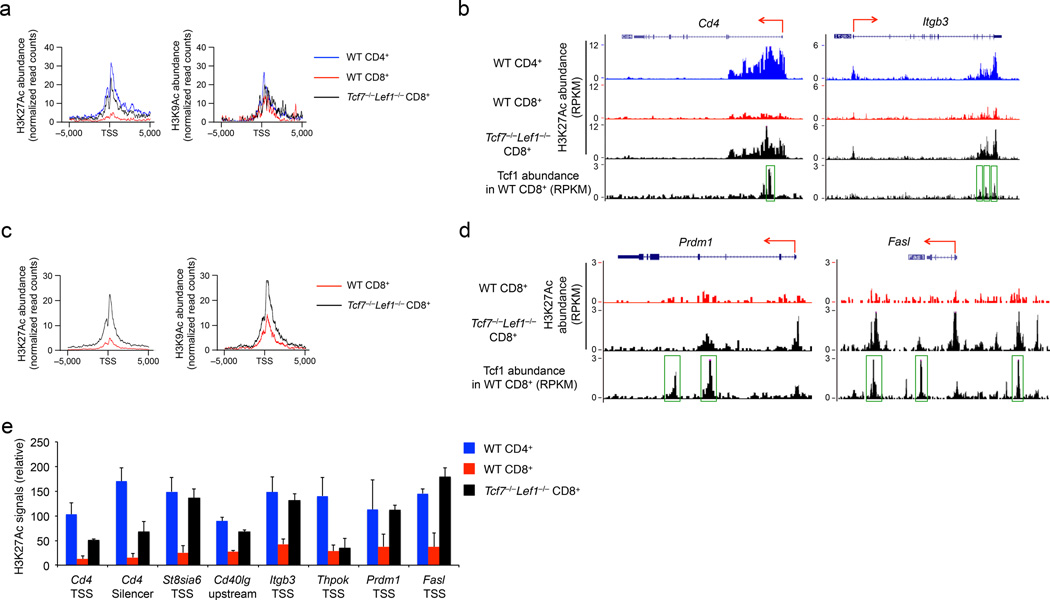
Figure 2. Tcf7−/− Lef1−/− CD8+ T cells exhibit histone hyperacetylation.
(a) ChIP-Seq analysis of H3K27Ac and H3K9Ac histone marks in CD4+ or CD8+ mature thymocytes sorted from wild-type (WT) mice or CD8+ mature thymocytes sorted from Tcf7−/−Lef1−/− mice, with H3K27Ac and H3K9Ac profiles (normalize read counts) shown at the “−5 kb to +5 kb” regions flanking the TSSs of 108 genes in the CD4+ T cell gene set. (b) H3K27Ac ChIP-Seq tracks at the Cd4 and Itgb3 gene loci in WT CD4+, WT or Tcf7−/−Lef1−/− CD8+ mature thymocytes (marked on top of the panels are the gene structures and transcriptional orientations), along with Tcf1 ChIP-Seq tracks in splenic CD8+ T cells, where MACS-called Tcf1 binding peaks are marked by green rectangles. (c) H3K27Ac and H3K9Ac profiles (normalize read counts) at the “−5 kb to +5 kb” regions flanking the TSSs of 472 upregulated genes in Tcf7−/−Lef1−/− CD8+ mature thymocytes. (d) H3K27Ac ChIP-seq tracks at the Prdm1 and Fasl gene loci in WT and Tcf7−/−Lef1−/− CD8+ mature thymocytes, along with Tcf1 ChIP-Seq tracks in splenic CD8+ T cells. (e) ChIP-qPCR analysis of relative H3K27Ac signals (ChIP with H3K27Ac antibody normalized to ChIP with IgG) at the indicated genomic locations in splenic WT CD4+, WT or Tcf7−/−Lef1−/− CD8+ T cells. Data are from one experiment (a-d), or from 2 experiments with each sample measured in duplicate (e, means ± s.d.).
We next analyzed histone modifications of all the 472 genes upregulated in Tcf7−/−Lef1−/− compared to control CD8+ mature thymocytes. In addition to H3K4me3, these gene loci showed markedly elevated H3K27Ac and H3K9Ac signals in Tcf7−/−Lef1−/− compared to wild-type CD8+ thymocytes (Fig. 2c, Supplementary Fig. 3c). Tcf7−/−Lef1−/− CD8+ thymocytes exhibited increased H3K27Ac over wild-type CD8+ thymocytes at the Foxp3 upstream regulatory region and Rorc gene body (Supplementary Fig. 3d), and the TSSs of Prdm1 and Fasl (Fig. 2d). Mature thymocytes and peripheral T cells have virtually identical transcriptomes9, suggesting that transcriptional and epigenetic regulation is preserved in mature T cells during egress from the thymus to peripheral lymphoid tissues. Using ChIP-qPCR, we validated increased H3K27Ac in Tcf7−/−Lef1−/− compared to wild-type splenic CD8+ T cells at the Cd4 TSS and silencer, the TSSs or upstream regulatory regions of other CD4+ signature genes (St8sia6, Cd40lg, and Itgb3) and the TSSs of CD8+ T cell effector genes, such as Prdm1 and Fasl (Fig. 2e). In contrast, an increase in H3K4me3 and/or a decrease in H3K27me3 were only observed at the TSSs of St8sia6 and Fasl in Tcf7−/−Lef1−/− compared to wild-type splenic CD8+ T cells (Supplementary Fig. 4). Tcf1-Lef1 deficiency thus caused increased H3K27Ac and H3K9Ac marks in CD8+ T cells at lineage-inappropriate gene loci, which may account for their aberrant upregulation.
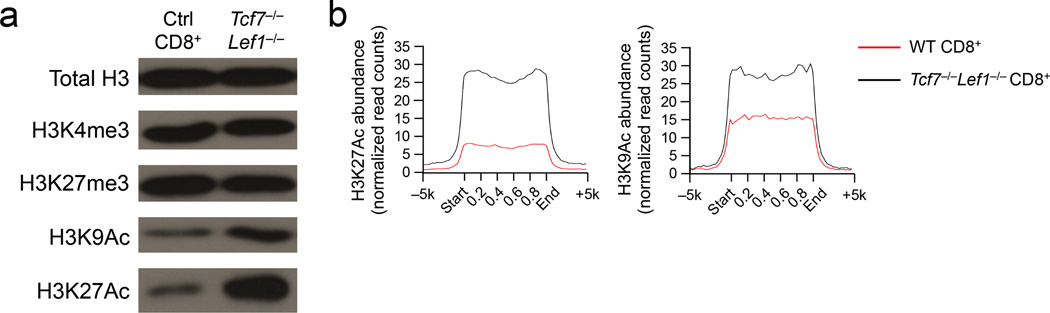
For the overall histone modification status, H3K27Ac and H3K9Ac signals were substantially elevated in Tcf7−/−Lef1−/− compared to control splenic CD8+ T cells, whereas the amount of H3K4me3 and H3K27me3 was similar (Fig. 3a). Re-analysis of the RNA-Seq data comparing Tcf7−/−Lef1−/− and control mature CD8+ thymocytes did not identify major gene expression changes in known HDACs or histone acetyltransferases due to Tcf1-Lef1 deficiency (Supplementary Fig. 5). Next we performed Tcf1 ChIP-Seq using wild-type or Tcf7−/− splenic CD8+ T cells employing a protocol in which sonication intensity and duration were reduced to better preserve protein complexes bound to chromatin. Using a stringent 4-fold enrichment threshold, p<10−5 and FDR<0.05 setting, we identified 7,807 high-confidence Tcf1 binding peaks (more than what we previously identified21). Both H3K27Ac and H3K9Ac signals were relatively weak at the Tcf1 binding sites in wild-type CD8+ mature thymocytes, but were substantially elevated in Tcf7−/−Lef1−/− CD8+ mature thymocytes (Fig. 3b). Among the 472 genes upregulated in Tcf7−/−Lef1−/− CD8+ thymocytes, 202 genes harboured Tcf1 binding peaks within the −5 kb to +5 kb genomic regions flanking the gene loci (Table 1). In addition to Cd421, Tcf1 bound to several other CD4+ lineage-associated genes (Itgb3, St8sia6 and Cd40lg) and CD8+ T cell effector genes (Prdm1 and Fasl) (Fig. 2b, d, Supplementary Fig. 3b). Among the 202 Tcf1-bound genes, over 80% showed increased H3K27Ac and about one third showed increased H3K9Ac signals in Tcf7−/−Lef1−/− compared to control CD8+ thymocytes (Table 1). The same trend was observed when the search of Tcf1 peaks was expanded to −50 kb to +50 kb regulatory regions flanking the gene loci, revealing that Tcf1 was associated with Foxp3 and additional sites in the Fasl gene (Fig. 2d, Supplementary Fig. 3d). These observations suggest that Tcf1 and Lef1 restrain histone acetylation in the CD8+ T cell genome, at their occupancy sites and directly associated genes.
Figure 3. Tcf1 is connected with histone acetylation status in CD8+ T cells.
(a) Immunoblot analysis of total or modified H3 histones in histone protein extracted from splenic CD8+ T cells sorted from Tcf7−/−Lef1−/− or control littermates. (b) H3K27Ac and H3K9Ac profiles (normalize read counts) of wild-type (WT) and Tcf7−/−Lef1−/− CD8+ mature thymocytes within and outside the 7,807 high-confidence Tcf1 binding sites identified by Tcf1 ChIP-Seq analysis in splenic CD8+ T cells. Data are representative of two experiments (a), or from one experiment (b).
Table 1.
Analysis of H3K27Ac and H3K9Ac at the Tcf1 binding peaks
| Genes upregulated in Tcf7−/−Lef1−/−CD8+ mature thymocytes |
472 |
|---|---|
| Tcf1 peaks in −5 to + 5 kb | 202 |
| Increase in H3K27Ac | 165 (81.7%) |
| Increase in H3K9Ac | 73 (36.1%) |
| Tcf1 peaks in −50 to + 50 kb | 354 |
| Increase in H3K27Ac | 267 (75.4%) |
| Increase in H3K9Ac | 97 (27.4%) |
Quantitative analysis of H3K27Ac and H3K9Ac signals (normalized read counts) at the Tcf1 binding peaks identified within the “−5 to +5 kb” or “−50 to +50 kb” genomic regions flanking the gene body of 472 upregulated genes in Tcf7−/−Lef1−/− CD8+ mature thymocytes. The numbers of Tcf1 peaks showing increase in H3K27Ac and H3K9Ac (significance threshold at FDR< 0.05 as determined by the SICER algorithm) in Tcf7−/−Lef1−/− over WT CD8+ mature thymocytes are summarized.
Tcf1 has intrinsic HDAC activity
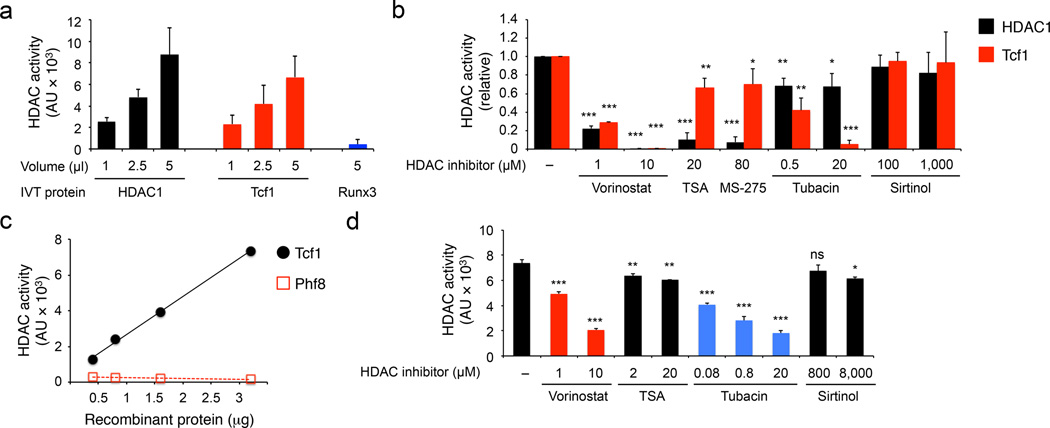
We next investigated if Tcf1 and Lef1 have intrinsic HDAC activities. We employed in vitro translated (IVT) proteins in histone deacetylase assays using a fluorogenic substrate, Boc-Lys(Ac)-AMC. IVT HDAC1 showed dose-dependent deacetylation of the substrate as expected. IVT p45 Tcf1 (45 kDa full-length Tcf1 protein), but not Runx3, exhibited HDAC activity (Fig. 4a). Based on phylogenetic analysis and sequence homology, HDACs are divided into four classes24. With the sirtuin family (including SIRT1−7) constituting class III, the classical HDACs fall into class I (including HDAC1, 2, 3 and 8), class II and class VI (HDAC11). Class II HDACs contains class IIa (HDAC4, 5, 7 and 9) and class IIb (HDAC6 and 10). The HDAC activities of HDAC1 and Tcf1 were both potently inhibited by a pan-HDAC inhibitor Vorinostat and were virtually unaffected by Sirtinol, a sirtuin-specific inhibitor (Fig. 4b). Trichostatin A (TSA) and MS-275, selective inhibitors of class I HDACs25, strongly inhibited HDAC1 but not Tcf1 HDAC activity (Fig. 4b). Tubacin, a selective HDAC6 inhibitor, greatly diminished Tcf1 HDAC activity, but only weakly affected HDAC1 (Fig. 4b). We also purified His-tagged Tcf1 recombinant protein and confirmed dose-dependent HDAC activity in Tcf1 (Fig. 4c). Recombinant Tcf1 was sensitive to Vorinostat and Tubacin, but less responsive to TSA or Sirtinol (Fig. 4d). These data indicate that Tcf1 has intrinsic histone deacetylase activity.
Figure 4. Tcf1 has intrinsic HDAC activity.
(a) Deacetylation assay using IVT HDAC1, Tcf1 or Runx3. AU, arbitrary unit. (b) Deacetylation assay of IVT HDAC1 and Tcf1 in the presence of various HDAC inhibitors at indicated concentrations, with the activity of untreated HDAC1 or Tcf1 set as 1 and the relative activity of inhibitor-treated proteins normalized accordingly. (c) Deacetylation assay using purified His-tagged recombinant Tcf1 or Phf8 segment at indicated protein amounts. (d) Deacetylation assay of purified recombinant Tcf1 (3.2 µg) in the presence of HDAC inhibitors. Data are from ≥ 3 experiments (a, b and d, means ± s.d.) or average from 2 experiments (c). ns, not statistically significant; *, p<0.05; **, p<0.01; and ***, p<0.001 compared with untreated proteins, by Student’s t-test.
Mapping of HDAC activity domain in Tcf1
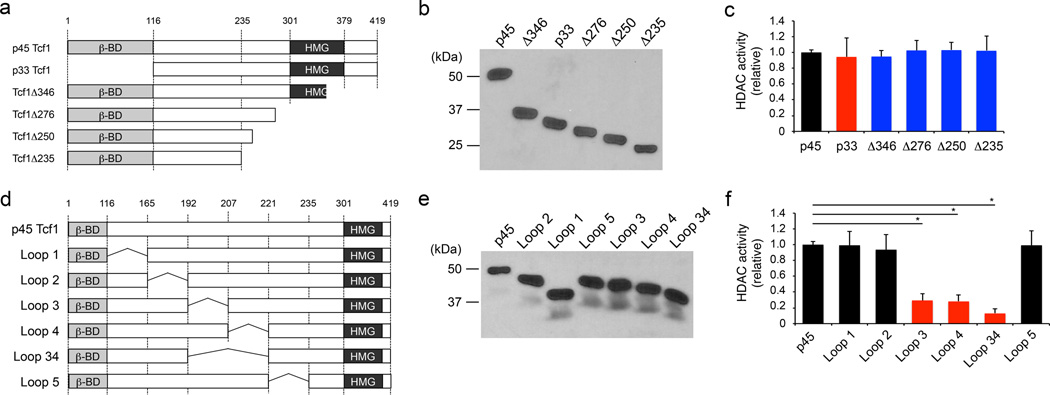
Tcf1 is expressed in multiple isoforms in T cells. Unlike p45 Tcf1, p33 Tcf1 isoform lacks the N-terminal β-catenin-interacting domain. Both isoforms have the same C-terminal HMG DNA-binding domain. A series of C-terminal truncation mutants of p45 Tcf1 that partially or completely lacked the HMG domain as well as p33 Tcf1 maintained HDAC activity (Fig. 5a–c). The HDAC domain is thus located within the 116–235 amino acid (aa) region in Tcf1, independent of the β-catenin-interacting and HMG domains. We then made internal deletions within this 120-aa region (Fig. 5d, e). Deletion of 192–206 aa (Loop 3) or 207–221 aa (Loop 4) segment greatly reduced the Tcf1 HDAC activity, and combined deletion of both segments (Loop 34) further abolished the Tcf1 HDAC activity (Fig. 5f). Thus, a 30-aa domain (Q192-L221) in Tcf1 is essential for its HDAC activity.
Figure 5. Mapping the HDAC activity domain in Tcf1.
(a) Diagram showing N- and C-terminal truncations of p45 Tcf1. (b) Immunoblot analysis of IVT FLAG-tagged Tcf1 truncated proteins with an anti-FLAG antibody. (c) Deacetylation assay using IVT Tcf1 truncated proteins, with the activity of WT p45 Tcf1 set as 1, and the relative activity of Tcf1 truncated proteins normalized accordingly. (d) Diagram showing internal deletions in p45 Tcf1. (e) Immunoblot analysis of IVT FLAG-tagged Tcf1 internal deletion mutant proteins with an anti-FLAG antibody. (f) Deacetylation assay using IVT Tcf1 internal deletion mutant proteins, with relative HDAC activities calculated as in c. Data are representative from 2 experiments (b, e), or from ≥ 3 experiments (c, f, means ± s.d.). *, p<0.001 compared with WT p45 Tcf1, by Student’s t-test.
Tcf1 catalyzes deacetylation of histone substrates
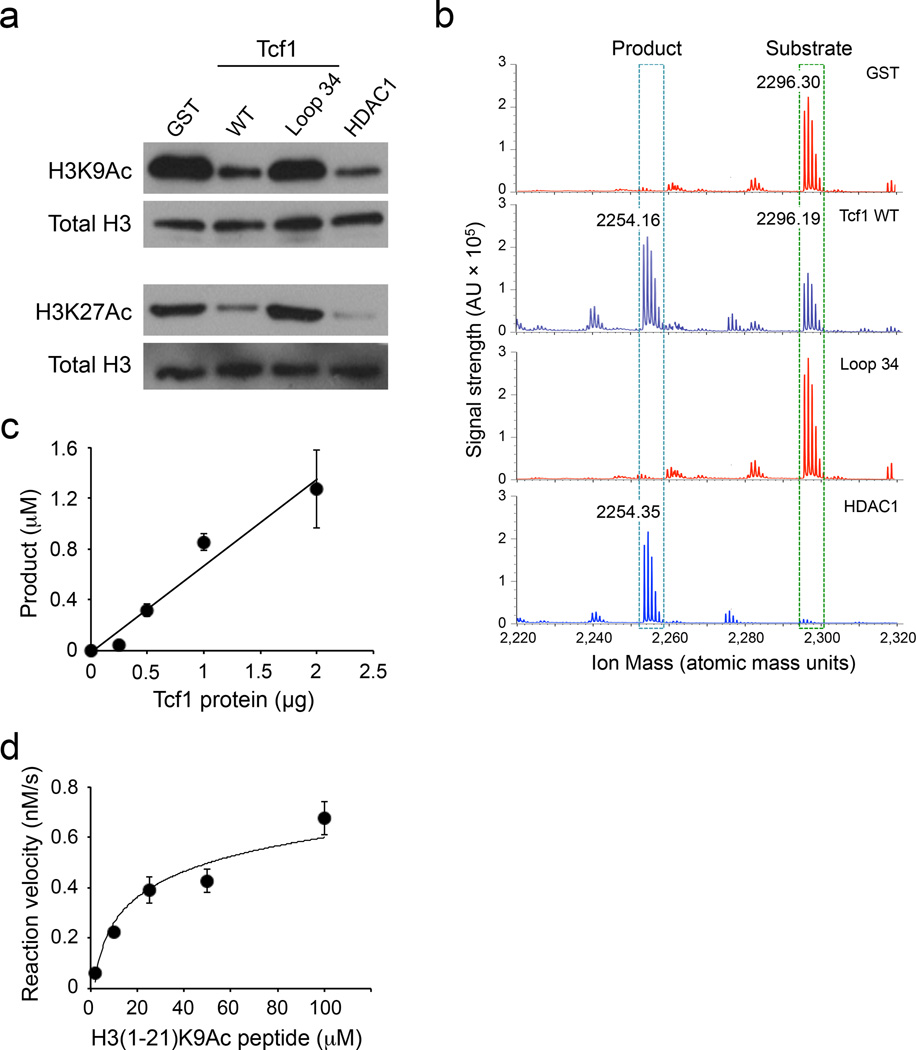
To further analyze the intrinsic HDAC activity of Tcf1, we employed gel filtration-fast protein liquid chromatography (FPLC) to enhance the purity of recombinant Tcf1 protein (Supplementary Fig. 6a, b), and used the FPLC-purified Tcf1 in two newly developed HDAC assays. First, we used acetylated H3 proteins (H3K9Ac and H3K27Ac) as substrates. Similar to a commercially available HDAC1, FPLC-purified Tcf1 caused deacetylation of both H3K9Ac and H3K27Ac proteins (Fig. 6a). In contrast, the Loop 34 Tcf1 mutant had no detectable impact on the acetylation status of H3K9Ac and H3K27Ac substrates, similar to GST purified from the same host bacteria (Fig. 6a).
Figure 6. Tcf1 catalyzes deacetylation of histone protein/peptide substrates.
(a) Deacetylation of H3K9Ac and H3K27Ac protein substrates by GST, FPLC-purified WT p45 Tcf1, Tcf1 Loop34, or HDAC1, as determined by immunoblot analysis of the acetylation levels of the histone substrates. Total H3 protein was detected to show similar amounts of substrates were used in individual assays. (b) Deacetylation of H3(1–21)K9Ac peptide substrate by GST, FPLC-purified WT p45 Tcf1, Tcf1 Loop34, or HDAC1, as determined by MALDI. The acetylated peptide substrate was detected at an m/z of ~2296 Da, and the deacetylated product was detected at an m/z of ~2254 Da on MALDI spectra. (c) Analysis of dose-dependent deacetylation of the H3(1–21)K9Ac peptide by FPLC-purified WT p45 Tcf1, with the amounts of deacetylated products quantified by normalizing to a fixed amount of Angiotensin I peptide (m/z 1296.69 Da) on MALDI. (d) Kinetic analysis of deacetylation of the H3(1–21)K9Ac peptide by FPLC-purified WT p45 Tcf1. Data are representative from 2 experiments (a), or from >3 experiments (b), or from 2 experiments with 8–11 measurements of each assay point (c, d, means ± s.d.).
Second, we used an 1–21 aa segment of H3 protein acetylated at K9, called H3(1–21)K9Ac peptide, as a substrate to determine the kinetic parameters of Tcf1 catalytic activity. Both H3(1–21)K9Ac peptide and its deacetylated form were readily detected on matrix-assisted laser desorption and ionization (MALDI) mass spectrometry, with distinct molecular mass (Supplementary Fig. 6c). Like HDAC1, FPLC-purified Tcf1 effectively converted the H3(1–21)K9Ac peptide to its deacetylated form (Fig. 6b), whose identity was confirmed using laser-induced fragmentation time-of-flight (LIFT) (Supplementary Fig. 6d). The Loop 34 Tcf1 mutant failed to catalyze deacetylation of H3(1–21)K9Ac peptide, similar to GST negative control (Fig. 6b). FPLC-purified Tcf1 showed dose-dependent deacetylation of the H3(1–21)K9Ac peptide (Fig. 6c). By varying the substrate concentrations, the Vmax for Tcf1-catalyzed H3(1–21)K9Ac deacetylation was estimated to be 0.71×10−6 mM/sec with a Km of 0.022 mM (Fig. 6d, Supplementary Fig. 6e). These assays using highly purified Tcf1 protein and physiological substrates further establish the intrinsic HDAC activity in Tcf1.
Structural similarity between Tcf1 and conventional HDACs
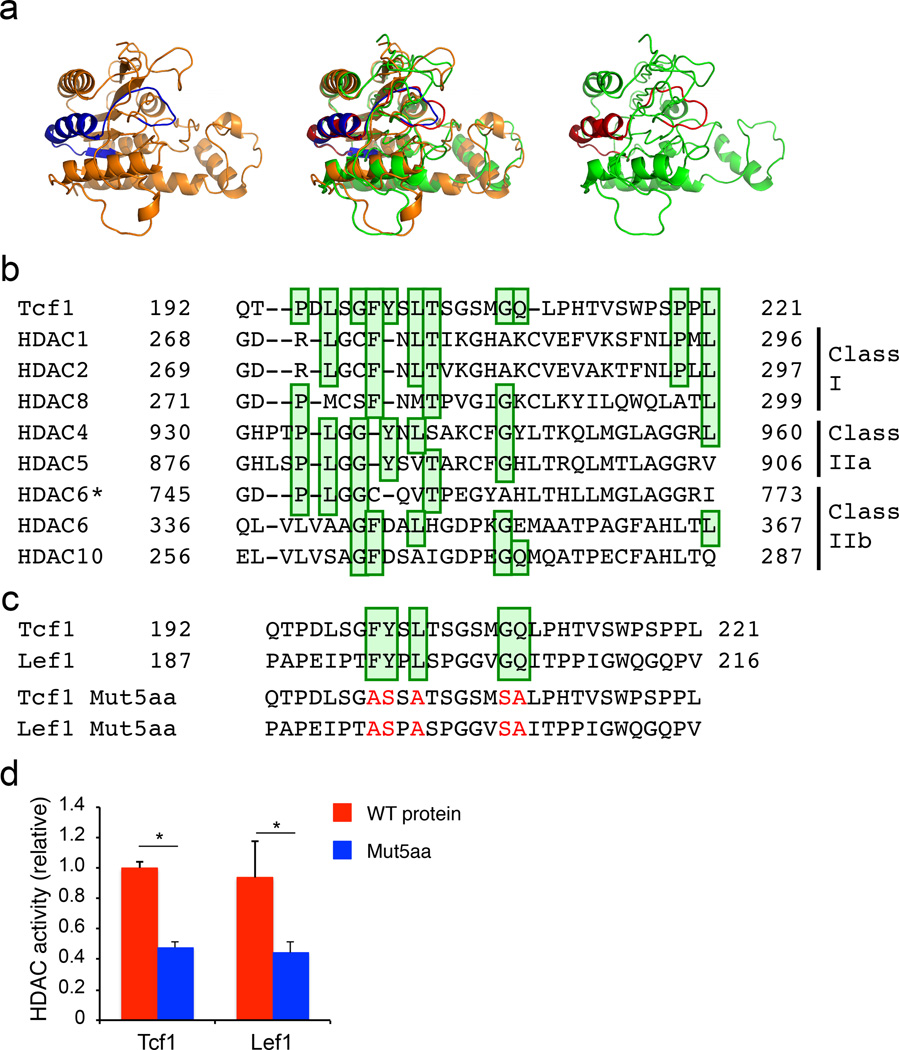
To gain structural insights into Tcf1 HDAC activity, we employed SWISS-MODEL26 to conduct homology modelling. We used HDAC8 as a template, because high-resolution HDAC827, substrate-HDAC828 and ligand-HDAC829 structures are available. The resulting homology model of Tcf1 exhibited a high degree of similarity with HDAC8 (Fig. 7a). In particular, the 30-aa Tcf1 HDAC domain was predicted to form an α-helix with an extended loop and bore strong resemblance to the corresponding region (G271-L299) in HDAC8 (Fig. 7a). The HDAC8 G271-L299 region constitutes an important component in its catalytic pocket29. By extension, we posit that the 30-aa Tcf1 HDAC domain contributes to forming an activity center that catalyzes histone deacetylation.
Figure 7. Homology modelling and sequence conservation predict an HDAC-like structure of Tcf1.
(a) Homology modelling analysis of Tcf1 (D21-N286, right panel in green) with a partial HDAC8 structure (D73-N357, left panel in orange) as a template. The predicted Tcf1 structure and HDAC8 structure were superposed (middle panel) for stereo view. The 30-aa Tcf1 HDAC domain (Q192-L221) is colored in red, and the corresponding HDAC8 domain (G271-L299) is colored in blue. (b) Amino acid sequence alignment of Tcf1 HDAC domain with those in conventional HDACs, with the conserved amino acids highlighted in green boxes. HDAC6 is distinct from other HDACs in that it contains two HDAC domains, and alignments of the first and second domains with Tcf1 are marked as ‘HDAC6’ and ‘HDAC6*’, respectively. (c) Amino acid sequence comparison between Tcf1 and Lef1, with mutations of five conserved amino acids highlighted in red in the Tcf1 Mut5aa and Lef1 Mut5aa sequences. (d) Deacetylation assays of IVT wild-type (WT) Tcf1, Tcf1 Mut5aa, WT Lef1 and Lef1 Mut5aa proteins. Data are from 3 experiments (d, means ± s.d.). *, p<0.001 compared with WT proteins, by Student’s t-test.
To further pinpoint the key residues responsible for the Tcf1 HDAC activity, we constructed multiple sequence alignments comparing HDAC domains in Tcf1 and conventional HDACs based on structural homology, global and local sequence comparisons. The alignments revealed that 11 amino acids in the 30-aa Tcf1 HDAC domain were conserved to various degrees with the HDAC domains in class I, IIa, and IIb HDACs (Fig. 7b). Several of these critical residues were also conserved between Tcf1 and Lef1 (Fig. 7c), and indeed mutating five of these conserved amino acids (Tcf1 Mut5aa) diminished Tcf1 HDAC activity (Fig. 7c, d). IVT Lef1 protein also exhibited similar HDAC activity as Tcf1, and mutating the same five conserved amino acids in Lef1 impaired its HDAC activity as well (Fig. 7c, d). Besides the few conserved residues, the overall sequence conservation between Tcf1 and Lef1 was not high in the putative HDAC domains. Direct coupling analysis (DCA) is a powerful tool to identify residue pairs in protein domains that may have co-evolved together to preserve physical proximity in the three-dimensional conformation of the domain30. DCA on 98 unique primary sequences of Tcf1 and Lef1 orthologs retrieved from 56 species identified several residue pairs that likely co-evolved within their HDAC domains (Supplementary Fig. 7). These co-evolving pairs in Tcf1 and Lef1 suggest that a similar functional structure in the HDAC domains may have been preserved in spite of their sequence divergence. These structure-function analyses further corroborate the notion that Tcf1 and Lef1 are embedded with intrinsic HDAC activity.
The Tcf1 HDAC activity is essential for CD8+-lineage integrity
To further substantiate a requirement for the intrinsic HDAC activity of Tcf1 in vivo, we used retroviruses expressing WT p45 Tcf1 or Tcf1 Mut5aa, along with GFP, to infect lineage-negative bone marrow (BM) cells from CD45.2+ Tcf7−/−Lef1−/− mice, followed by transplantation into wild-type CD45.1+ congenic recipients (Supplementary Fig. 8a). The Tcf1 Mut5aa mutant, over larger internal deletions, was chosen to minimize potential impact on protein structure in vivo. Donor-derived CD45.2+GFP+ T cells were analyzed in the BM chimeras six weeks post-transplantation. Both WT p45 Tcf1 and Tcf1 Mut5aa preserved similar DNA binding capacity based on electrophoretic mobility shift assay (Supplementary Fig. 8b) and the ability to bind to known Tcf1 target genes such as Lef1 and Axin2 in vivo using ChIP (Supplementary Fig. 8c). Retroviral WT p45 Tcf1 and Tcf1 Mut5aa were detected at similar levels in the gene-complemented Tcf7−/−Lef1−/− CD8+ T cells (Supplementary Fig. 8d).
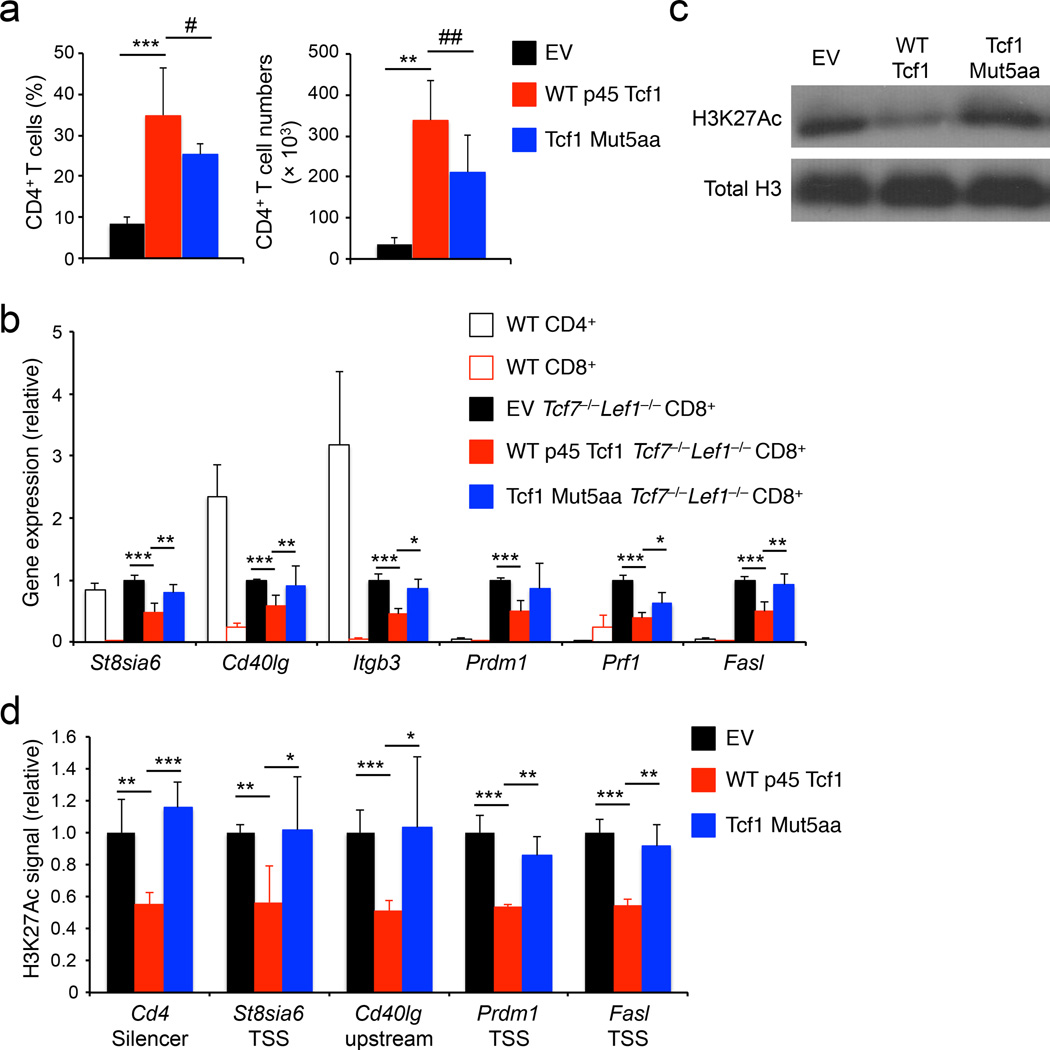
Numbers of CD4+ T cells are greatly diminished in Tcf7−/−Lef1−/− mice21, and this phenotype persisted in empty vector-transduced Tcf7−/−Lef1−/− BM chimeras (Fig. 8a). Forced expression of WT p45 Tcf1 or Tcf1 Mut5aa showed similar capacity to increase CD4+ T cell frequency and numbers in the gene-complemented Tcf7−/−Lef1−/− BM chimeras, but only modestly increased CD8+ T cell output (Fig. 8a, Supplementary Fig. 8e–f). As determined by quantitative RT-PCR, empty vector-transduced Tcf7−/−Lef1−/− CD8+ T cells exhibited derepression of CD4+ lineage-associated genes, such as St8sia6, Cd40lg and Itgb3, and upregulation of CD8+ T cell effector genes Prdm1, Prf1 and Fasl; in contrast, forced expression of WT p45 Tcf1 substantially repressed the expression of these genes (Fig. 8b). Compared to empty vector, retroviral expression of WT p45 Tcf1 decreased the percentages of CD4+ or Foxp3+ Tcf7−/−Lef1−/− CD8+ T cells and repressed FasL expression in Tcf7−/−Lef1−/− CD8+ T cells as measured with flow cytometry (Supplementary Fig. 8g–i). In contrast, all these “rescue” effects by WT p45 Tcf1 were not evidently detected in Tcf1 Mut5aa-transduced Tcf7−/−Lef1−/− CD8+ T cells (Fig. 8b, Supplementary Fig. 8g–i), highlighting an essential requirement for the Tcf1 HDAC activity in suppressing lineage-inappropriate genes in CD8+ T cells. In addition, WT p45 Tcf1-transduced Tcf7−/−Lef1−/− CD8+ T cells showed greatly reduced amount of total H3K27Ac modifications than empty vector-transduced Tcf7−/−Lef1−/− CD8+ T cells, while Tcf1 Mut5aa transduction failed to do so (Fig. 8c). Furthermore, retroviral WT p45 Tcf1, but not Tcf1 Mut5aa, diminished the H3K27Ac signals at the Cd4, St8sia6, Cd40lg, Prdm1, and Fasl gene loci, which showed elevated H3K27Ac in Tcf7−/−Lef1−/− CD8+ T cells (Fig. 8d). These in vivo data indicate that Tcf1 controls histone acetylation to establish an epigenetic landscape suitable for CD8+ T cell identity.
Figure 8. The Tcf1 HDAC activity is essential for establishing CD8+ T cell identity.
(a) Analysis of frequency (left) and numbers (right) of donor-derived CD45.2+GFP+TCRβ+CD4+ T cells in the spleens of BM chimeras that were reconstituted with Tcf7−/−Lef1−/− lineage-negative bond marrow cells retrovirally infected with empty vector (EV), WT p45 Tcf1, or Tcf1 Mut5aa retrovirus. (b) Quantitative RT-PCR analysis of gene expression (relative to Hprt) in wild-type (WT) splenic CD4+ or CD8+ T cells, splenic CD45.2+GFP+TCRβ+CD8+ T cells sorted from the EV-, WT p45 Tcf1-, or Tcf1 Mut5aa-complemented Tcf7−/−Lef1−/− BM chimeras. For each gene, its expression in EV-complemented cells was set as 1, and that in other cell types was normalized accordingly. (c) Immunoblot analysis of H3K27Ac and total H3 histone in histone protein extracted from splenic CD45.2+GFP+TCRβ+ CD8+ T cells sorted from the EV-, WT p45 Tcf1-, or Tcf1 Mut5aa-complemented Tcf7−/−Lef1−/− BM chimeras. (d) ChIP-qPCR analysis of relative H3K27Ac signals at select gene loci in splenic CD45.2+GFP+TCRβ+CD8+ T cells sorted from EV-, WT p45 Tcf1-, or Tcf1 Mut5aa-complemented Tcf7−/−Lef1−/− BM chimeras. For each gene locus, the relative H3K27Ac signal in EV-complemented cells was set as 1, and that in other cell types was normalized accordingly. Data are from 3 experiments (a, b, means ± s.d., n = 4–5), or representative from 3 experiments (c), or from 2 experiments with each sample measured in duplicates (d, means ± s.d.). #, p = 0.11; ##, p = 0.06; *, p<0.05; **, p<0.01; ***, p<0.001 for indicated comparisons, by Student’s t-test.
Discussion
Establishing and maintaining CD4+ and CD8+ T cell dichotomy are essential for effective cellular immunity. The ThPOK transcription factor and HDAC1-HDAC2 are known to maintain the integrity of CD4+ T cells by repressing CD8+ lineage-associated genes10, 11. Here we found Tcf1 and Lef1 transcription factors have a reciprocal role, i.e., establishing CD8+ T cell identity by repressing the CD4+ T cell program. Importantly, this critical function is mediated by HDAC activity that is intrinsically imbedded in Tcf1 and Lef1.
Histone deacetylation is an evolutionarily conserved means for gene repression31. Until now, the HDAC activity is limited to dedicated proteins including HDAC1−11 and SIRT1−724. HDAC proteins were found to regulate key functions in T cells32, such as HDAC1-HDAC2 in DN to DP transition33, HDAC6 in T-cell migration34, and HDAC7 in TCR signaling35. The intrinsic HDAC activity thus adds Tcf1 and Lef1 as new members to the HDAC family. It is noteworthy that although several HDAC or SIRT proteins are expressed in T lineage cells, they cannot compensate for the loss of Tcf1 and Lef1 in regulating histone deacetylation in CD8+ T cells. This consideration highlights the uniqueness of HDAC activity in Tcf1 and Lef1 and/or the requirements for proper positioning of HDAC activity at the target gene loci.
Tcf1 HDAC activity was sensitive to select HDAC inhibitors, suggesting at least partial structural similarity of the Tcf1 HDAC domain with that in conventional HDACs. Homology modelling predicted that the Tcf1 HDAC domain adopts a similar structure as a region in HDAC8 catalytic pocket. The 271G-299L region in HDAC8, which corresponds to the Tcf1 30-aa HDAC domain, constitutes only a portion of the HDAC8 catalytic pocket29. This consideration suggests that Tcf1 HDAC domain, likely together with neighboring residues, may fold to form a catalytic center that has distinct features from conventional HDACs. Partial crystal structures of Tcf-Lef family members have been reported, including the β-catenin-binding domain in XTcf336 and the HMG domain of Lef137. Solving the structure of whole Tcf1 protein or the HDAC domain will provide definitive insights into the molecular basis underlying its intrinsic HDAC activity.
The dual capacity of Tcf1 and Lef1 for direct DNA binding and intrinsic HDAC activity confers unique advantages to these factors in gene regulation. Tcf1-occupied sites in CD8+ T cell genome were associated in hyperacetylation of H3K27 and H3K9 upon loss of Tcf1 and Lef1. On the other hands, among the 472 upregulated genes in Tcf7−/−Lef1−/− CD8+ T cells, not all the gene loci harbor Tcf1 binding sites in the vicinity. One consideration is that the Tcf1 and Lef1 HDAC activity, like the conventional HDACs, may be recruited to proper gene loci by other DNA-binding transcription factors. In fact, direct interaction of Tcf1 or Lef1 with Foxp3, GATA3 and Runx transcription factors has been previously documented21, 38, 39. Histone acetylation is a very dynamic process, with the half-life of the acetyl group on histones in the order of minutes40. Constant addition and removal of the acetyl group to histones may require a more dynamic interaction of Tcf1 or Lef1 with the histone substrates, which may not be readily captured by ChIP-Seq. We postulate that the histone deacetylation capacity of Tcf1 and Lef1 may not necessarily solely depend on their direct contact with DNA and can act in a quite versatile fashion.
Tcf1 binds to the well-characterized Cd4 silencer in the first intron of the Cd4 gene, and Tcf1 directly interacts with Runx3 factor to synergistically suppress Cd4 transcription in CD8+ T cells21. In addition to transcriptional repression, the HDAC activity in Tcf1 and Lef1, brought to the locus by direct binding or recruitment by Runx3, has important contributions to Cd4 gene silencing, as evidence by increased H3K27Ac at the Cd4 silencer in Tcf7−/−Lef1−/− compared to wild-type CD8+ T cells. Beyond the Cd4 gene silencing, we found that Tcf1 and Lef1 have much broader roles in repressing the expression of CD4+ T lineage-associated genes and CD8+ T cell effector genes, and this function of gene repression requires their intrinsic HDAC activity. It is believed that Tcf-Lef-mediated gene repression depends on interaction with known TLE corepressors14. Our findings suggest that Tcf1 and Lef1 HDAC activity may act in synergy with TLE recruitment, or share division of labor with TLE proteins to achieve global and site-specific suppression of gene expression in a given cell type.
Currently, a prevailing view in epigenetic regulation is that the histone modification machinery must be recruited to the target gene loci through sequence-specific transcription factors. The identification of intrinsic HDAC activity in transcription factors thus reveals a flexible regulatory capacity that directly bridges genetic and epigenetic regulation of gene expression. The new dimension of complexity and expanded regulatory capacity in transcription factors may be broadly utilized in cell development, function, and transformation, beyond establishing cell identity.
MATERIALS AND METHODS
Animals
Tcf7fl/fl and Lef1fl/fl mice were described18, 21. B6.SJL mice were from the Jackson Laboratory. H2ab1−/− mice were from Taconic. All mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committee of the University of Iowa. Mice of both genders were analyzed at 6–15 weeks of age, no randomization was used, and no blinding was done.
Flow cytometry and cell sorting
Single cell suspension from the mouse thymus and spleen was prepared as previously described18, 21. The cells were surface-stained with various combinations of the following antibodies: anti-CD4 (M1/69), anti-CD8α (53-6.7), anti-TCRβ (H57–597), anti-CD69 (H1.2F3), anti-CD44 (IM7), anti-CD24 (M1/69), anti-CD62L (MEL-14), anti-CD40L (MR1) and CD45.2 (104) (all from eBiosciences). Anti-IL-2Rβ (TM-β1) and anti-FasL (MFL3) were from BD Biosciences. Anti-granzyme B (GB/2) was from Thermo Fisher Scientific. The Vβ TCR Screening Panel (BD Biosciences) was used to survey the Vβ TCR repertoire. For intranuclear staining, the surface-stained cells were fixed and permeabilized with Foxp3/transcription factor staining buffer set (eBiosciences), and stained with anti-Foxp3 (eBiosciences, FJK-16s), anti-Rorγt (eBiosciences, B2D), or anti-Tcf1 (C63D9, Cell Signaling Technologies). The stained cells were analyzed on FACSVerse (BD Biosciences). All antibodies were used at 1:50 to 1:100 dilution, and validated for mouse and flow cytometry as indicated by the data sheets from the manufacturers. The data were processed using FlowJo software (Version X, TreeStar). For cell sorting, surface-stained thymocytes or splenocytes were sorted on FACSAria (BD Biosciences) from Tcf7−/−Lef1−/− or control mice for RNA-Seq analysis and gene expression validation.
Reverse transcription and quantitative PCR
Total RNA from the sorted cells was extracted and reverse-transcribed using QuantiTech Reverse Transcription Kit (Qiagen). The resulting cDNA was analyzed for expression of different genes using SYBR Advantage qPCR Premix (Clontech) on an ABI 7300 Real Time PCR System (Applied Biosystems). The primers are listed in Supplementary Table 2. The expression of genes of interest was first normalized to Hprt in each sample.
RNA-Seq and data processing
CD8+ thymocytes were sorted from the TCRβhiCD24−CD69− mature thymocyte population in Tcf7−/−, Tcf7−/−Lef1−/− or control animals, and total RNA was extracted as described18. cDNA synthesis and amplification were performed using SMARTer Ultra Low Input RNA Kit (Clontech) starting with 10–100 ng of total RNA per sample following manufacturer’s instructions. cDNA was fragmented with Q800R sonicator (Qsonica) and used as input for NEBNext Ultra DNA Library Preparation Kit (NEB). Libraries were sequenced on Illumina’s HiSeq2000 in single read mode with the read length of 50 nt producing 60–70 million reads per sample. Sequence data in fastq format were generated using CASAVA 1.8.2 processing pipeline from Illumina.
The sequencing quality of RNA-Seq libraries was assessed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, v0.10.1). The RNA-Seq libraries were then processed by RSEM (v1.2.21)41 to estimate expression levels of all genes. The expression of a gene is represented by a gene-level value of Fragments Per Kilobase of transcripts per Million mapped reads (FPKM). The reproducibility of RNA-Seq data was evaluated by computing Pearson’s correlation of FPKM values for all genes between biological replicates. The Pearson’s correlation coefficient between the two biological replicates was 0.999 for both control and Tcf7−/− samples, and 0.996 for the Tcf7−/−Lef1−/− samples, indicating strong reproducibility. EBSeq (v1.5.4), as an integral component of RSEM package, was used to identify differentially expressed genes42. Upregulated or downregulated genes in Tcf7−/−Lef1−/− CD8+ T cells were identified by requiring ≥ 2 fold expression changes and false discovery rate (FDR) < 0.01, as well as FPKM ≥ 1 in Tcf7−/−Lef1−/− CD8+ T cells for upregulated genes or FPKM ≥ 1 in control CD8+ T cells for downregulated genes. UCSC genes from the iGenome mouse mm9 assembly (http://support.illumina.com/sequencing/sequencing_software/igenome.html) were used for gene annotation.
ChIP and quantitative PCR
WT CD4+ or CD8+, Tcf7−/−Lef1−/− CD8+ mature thymocytes or splenic CD8+ T cells were sorted and cross-linked with 1% formaldehyde in medium for 5 minutes, processed using truChIP Chromatin Shearing Reagent Kit (Covaris), and sonicated for 5 minutes on Covaris S2 ultrasonicator. The resulting sheared chromatin fragments were in 200–500 bp range and were immunoprecipitated with anti-H3K4me3 (Millipore, 17–614), H3K9Ac (Abcam, ab4441), H3K27me3 (Millipore, 17–622), H3K27Ac (Abcam, ab4729), or control IgG and washed as previously described18. For calculation of histone mark signals in a given cell type in ChIP-PCR experiments, each histone mark ChIP sample was normalized to corresponding IgG ChIP sample for a target region. In genetic complementation experiments, the H3K27Ac signal was normalized to input DNA from the same sample. The PCR primers for the target regions are listed in Supplementary Table 2.
For Tcf1 ChIP, WT or Tcf7−/− splenic CD8+ T cells were sorted and cross-linked for 10 minutes. The cells were suspended in a nuclear lysis buffer containing 50 mM Tris/HCl (pH 8.1), 10 mM EDTA, 0.1% SDS and 1×protease inhibitor cocktail (Sigma-Aldrich), and sonicated with a Q125 sonicator equipped with an 1/8-inch diameter probe (QSonica) at 20% input amplitude, 20 sec for 8 times. The resulted chromatin fragments were in 300–1,500 bp ranges and were immunoprecipitated with an anti-Tcf1 serum43 followed by proper washing. The immunoprecipitated DNA fragments were used for library construction or PCR quantification.
ChIP-Seq and data processing
DNA segments from ChIP were end-repaired and ligated to indexed Illumina adaptors followed by low-cycle PCR. The resulting libraries were sequenced with the Illumina HiSeq-2000 platform. The sequencing quality of ChIP-Seq libraries was assessed by FastQC. Ends of sequencing reads were trimmed according to the sequencing quality. Bowtie2 v2.2.544 was used to align the sequencing reads to the mouse genome. UCSC genes from the iGenome mouse mm9 assembly were used for gene annotation.
For histone modification ChIP-Seq libraries, SICER v1.145 was used to identify ChIP-enriched islands under the setting of false discovery rate (FDR) < 10−4. The island-filtered histone mark reads were used for downstream analysis. For Tcf1 ChIP-Seq in CD8+ T cells, MACS v1.4.246 was used for peak calling with Tcf1 ChIP-Seq in Tcf7−/− CD8+ T cells as a negative control. We used a stringent setting, i.e., ≥ 4 fold enrichment, p value < 10−5, and FDR < 5%, and identified 7,807 high-confidence Tcf1 binding sites.
Analysis of the distribution of histone marks at specific sets of genomic landmarks
To analyze the profile of a histone mark around transcription start sites (TSSs) of a particular gene set, the TSSs were first aligned, and the island-filtered reads of the histone mark in non-overlapping windows of 100 bases were counted within a +/− 5 kb region flanking all the TSSs of the gene set. The window read counts were then normalized by the number of genes and by the window size. The profile was further normalized by the total number of island-filtered reads in the histone mark library as reads per million.
For the aggregated profile of a histone mark flanking the 7,807 high-confidence Tcf1 binding sites, the island-filtered reads were counted both inside and outside all the sites. Inside the Tcf1 binding sites, the reads were counted in windows equal to 5% of the site length. Outside the Tcf1 binding sites, the reads were counted in non-overlapping windows of 250 bases from −5 kb upstream to the beginning of Tcf1 sites and from the end of Tcf1 sites to +5 kb downstream. All the read counts were normalized by the window size and the number of sites. The profile was further normalized by the total number of island-filtered reads in the histone mark library as reads per million.
GSEA
Microarray data comparing CD4+ and CD8+ single positive thymocytes were downloaded from the Immunological Genome Project (www.immgen.org)9. A total of 108 genes showed ≥ 2 fold more expression in CD4+ than in CD8+ T cells, and was defined as CD4+ lineage-associated gene set (Supplementary Table 1). Gene Set Enrichment Analysis (GSEA) software was downloaded from the Broad Institute47 and used to determine the enrichment of gene sets in Tcf7−/−Lef1−/− and control CD8+ mature thymocytes.
Histone purification and immunoblotting
Tcf7−/−Lef1−/− or control CD8+ mature thymocytes were sort-purified, and histone protein was extracted following a protocol from Abcam. In brief, the cells was washed twice with ice-cold PBS and suspended in Triton Extraction Buffer to isolate the nuclei. The nuclei extract was resuspended in 0.2 N HCl and incubated overnight to extract the histone proteins. The same antibodies for histone marks used in ChIP assays were used for immunoblotting. Total H3 histone was detected with the ab1791 antibody (Abcam).
Generation of Tcf1 mutants and in vitro translation
FLAG-tagged Tcf1 and Lef1 were cloned under the T7-promoter in the pBlueScript vector. To generate C-terminus truncated Tcf1 proteins, stop codon was inserted into appropriate locations using the QuikChange Site-Directed Mutagenesis Kit (Aligent Technologies, formerly Stratagene). To generate internal deletions, the targeted regions were looped out using primers complementary to their flanking sequences. Mutation of conserved 5 amino acids in the deacetylation domain of Tcf1 or Lef1 was achieved with the same mutagenesis approach using extra-long flanking primers. All the mutants were verified for the desired mutations without irrelevant changes. The pBlueScript constructs expressing WT or mutant p45 Tcf1, WT or mutant Lef1 were linearized, and corresponding proteins were generated by in vitro translation using the TNT T7 coupled Wheat Germ Extract System (Promega) following the manufacturer’s instructions.
Fluorometric deacetylation assay and HDAC inhibitors
The HDAC Activity Fluorometric Assay Kit (BioVision) was used to measure deacetylation activity of in vitro translated or recombinant proteins. The test samples were incubated with the provided fluorogenic substrate, Boc-Lys(Ac)-AMC [complete name: Nα-(t-butoxycarbonyl)-Nω-acetyl-L-lysine 7-amido-4-methylcoumarin] in 1×HDAC Assay Buffer at 37°C for 2.5 hrs, and then the reaction was stopped by adding the Lysine Developer and further incubated at 37°C for 30 minutes. The produced fluorescence was measured in 96-well black plate (Thermo Scientific) using the Synergy H1 Hybrid reader (BioTek Instruments) with excitation at 360 nm and emission at 460 nm. In some assays, the following HDAC inhibitors were used at concentrations that were 20–300 fold of their reported IC50, Vorinostat, MS-275 (also known as Entinostat), Tubacin, Sirtinol (all from Cayman Chemical) and Trichostatin A (Sigma-Aldrich).
HDAC assay using protein substrates
Recombinant H3K9Ac (Cat No. 31253) and H3K27Ac (Cat. No. 31290, Active Motif) were used as substrates for HDAC assay. The reaction contained 1×HDAC assay buffer [20 mM Tris/HCl (pH 8.0) and 150 mM NaCl], 0.2 mM ZnCl2, 0.2 µg substrate protein, and 2 µg each of GST, FPLC-purified WT Tcf1, or Tcf1-loop34 protein. Recombinant Hdac1 protein (Cat. No. 31342, Active motif) was used at 1 µg in each reaction as a positive control. The reaction was carried out at 25°C for 12 hrs and stopped by addition of SDS-PAGE loading buffer followed by treatment at 95°C for 5 minutes. The samples were then resolved on a 15% SDS-PAGE gel and immunoblotted with anti-H3K9Ac, H3K27Ac, or total H3 antibody as above.
HDAC assay using a peptide substrate
Histone H3 (1–21) peptide (ARTKQTARKSTGGKAPRKQLA, AS-61701) and its K9 acetylated form, [Lys9(Ac)]-Histone H3 (1–21) [H3K9Ac (1–21), AS-64191] were from AnaSpec. The H3K9Ac peptide was used in place of protein substrate in the HDAC assay described above, except that ZnCl2 was omitted. The reaction was carried out using varied amounts of FPLC-purified Tcf1 for test of dose dependency, or using varied reaction time and substrate concentrations for kinetic analysis. The reaction was stopped by snap-freeze on dry ice followed by mass spectrometry analysis.
Mass spectrometry using matrix-assisted laser desorption/ionization (MALDI)
Two µl of the HDAC reaction containing H3K9Ac peptide substrate was mixed thoroughly with 1 µl of supernatant from saturated α-cyano-4-hydroxycinnamic acid (CHCA, Sigma-Aldrich) in 50% acetonitrile and 0.2% trifluoroacetic acid (TFA, Sigma-Aldrich) and spotted on to a MTP 384 well Anchor Chip MALDI plate with hydrophilic centers of 0.8 mm (Bruker). Desalting in situ was performed by rinsing each dried spot with 3 µl of 0.1% TFA for 20 seconds.
MALDI-TOF spectra were acquired with a Bruker Ultraflex TOF/TOF mass spectrometer equipped with gridless delayed extraction ion source, frequency-tripled NdYAG laser (50 µJ, 3 nsec pulse) using a SmartBeam II controller to distribute power across the pulse, a low-mass ion suppressor, electronic precursor selection, collision cell, LIFT cell for fragment ion post-acceleration, a post-LIFT metastable suppressor and a gridless ion reflector48. Mass calibration solutions were prepared from Michrom bovine serum albumin digests (Bruker). Fourteen peptides ranging in m/z from 927.495 (Tyr-Leu-Tyr-Glu-Ile-Ala-Arg) to 2615.123 (Val-His-Lys-Glu-Cys-Cys-His-Gly-Asp-Leu-Leu-Glu-Cys-Ala-Asp-Asp-Arg-Ala-Asp-Leu-Ala-Lys), with carboxymethylcysteine as a global modification, established a mass accuracy of 4 ppm.
Peptide spectra
A gridless ion source, floats initially at a homogeneous 25,140 V positive bias. Following each laser pulse, a delay of 90 nsec transpired before the bias of the second lens was dropped 22,540 V to extract ions. Ions below 800 Da were electronically deflected out of the beam line. Ions traversed an Einsel lens held at 8.2 kV and a two-stage, grid-free reflector with mid mirror voltage of 13.44 kV and back mirror voltage of 26.66 kV. The SmartBeam II is usually run at 70% attenuation firing at 200 to 2000 Hz with a laser spot diameter of 60 µm. The linear and reflector detector biases were set at 2.4 kV and 2.1 kV, respectively. The digitizer is an Acqiris U1084A Ver. 1.16 with 524 Mb memory, a full-scale sensitivity set to 100 mV and analog offsets set to 50 mV. Mass spectral data were generated by summing 2000–4000 laser shots.
For relative quantification of H3 (1–21) in the K9Ac (substrate) and deacetylated (product) forms, a fresh preparation of saturated MALDI matrix supernatant was mixed with a fixed amount of Angiotensin I (m/z 1296.69 Da) such that 10 fmol was deposited per spot. Mass spectra data for both peptides were obtained in 8–11 replicates. Spectra were accumulated for each spot until the area of Angiotensin I isotype pattern reached 5,000 counts. The areas under 4 consecutive peaks for each peptide (e.g. encompassing the natural abundance of stable isotopes) were calculated in each replicate, and the relative quantity of the deacetylated H3 (1–21) product was determined for kinetics analysis.
Laser-Induced Fragmentation Time of Flight (LIFT)
The Ultraflex III also has a selected ion sequencing mode that is enabled when a second ion source is positioned into the beam line. This LIFT cell is situated beyond the collision cell and the Timed Ion Selection (TIS) gates. During LIFT acquisitions, laser power is increased about 40% to increase internal energy of intact peptides which accelerates Post Source Decay. The bias between the first source and LIFT cell is maintained at only 7.5 kV to improve the precision of TIS precursor isolation. Resolution of the TIS, R =m/Δm, is effectively 1000, meaning all ions are attenuated prior to entering the LIFT cell save a bolus of ±1.5 Da centered on the monoisotopic peak of the H3K9Ac peptide or its deacetylated product. In the four-stage LIFT cell the voltage potential is raised from ground to 19 kV and fragment ions are spatially focused via deceleration and full-bias pulse into the reflector (which itself compensates for energy dispersion). Following the LIFT cell another TIS acts as a metastable deflector for precursor ions. Hence a focused array of fragment ions arrive at the dual microchannel plate which is set off the beam line following the ion reflector. Ion mass is determined by previous calibration of arrival times.
Ladder sequence ions were assigned using BioTools 3.2 SR4 to pass centroid data (SNAP algorithm) to the MASCOT version 2.5. The August 8, 2015 version of SwissProt database restricted to human taxonomy was searched accepting variable lysine acetylation, excluding all global modifications and specifying no enzyme. MASCOT identified substrate H3K9Ac and deacetylated product peptides with a score of 86 and 84 respectively with a mass window of 0.1 Da.
Retroviral transduction and generation of bone marrow chimeras
WT p45 Tcf1 or Tcf1mut5aa was cloned into the MIG-R1 bicistronic retroviral vector with GFP as an expression indicator. Each retrovirus was packaged by transient transfection of 293T cells with the retroviral vector along with pCLeco using Lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions. The retrovirus-containing media were harvested for infection of BM stem/progenitor cells49.
BM cells were isolated from Tcf7−/−Lef1−/− mice and depleted of all lineage-positive cells by labeling with biotinylated lineage markers in combination with streptavidin-coupled M280 Dynabeads (Life Technologies). The Lin− BM cells were cultured overnight in IMDM medium with 15% fetal bovine serum (FBS), 50 µM 2-mercaptoethanol, 100 µg/ml streptomycin and penicillin, 50 ng/ml stem cell factor (SCF), and 20 ng/ml thrombopoietin (TPO) in a 24-well flat-bottom plate at the density of 0.5 × 106/ml. The cells were then resuspended in retrovirus-containing media with refreshed cytokines, loaded onto non-tissue culture 24-well plate coated with 10 µg/well retronectin (Takara), and spinofected at 1000 × g for 90 min at room temperature in the presence of 4 µg/ml Polybrene (hexadimethrine bromide). After spinofection, the cells were cultured with the retrovirus-containing media for an extended 2 hrs, and the media were then changed to the IMDM supplemented with cytokines as above. The spinofection was repeated on the following day, and 24 hrs later, the infected BM stem/progenitor cells were transplanted into lethally irradiated (1,050 rad) CD45.1+ B6.SJL mice. Six weeks after the transplantation, CD8+ T cell lineage integrity was analyzed using the thymuses and spleens from the recipients.
Purification of Tcf1 and other recombinant proteins
The Tcf1 cDNA was inserted in-frame with an N-terminal His tag in the pET-28 vector. The plasmid was transformed into Rosetta competent cells (EMD Millipore), and a single clone was cultured in the LB medium at 37°C. When the OD at 595 nm reached 0.6, isopropyl β-D-1-thiogalactopyronaside (IPTG, Thermo-Fisher Scientific) was added to the culture at a final concentration of 0.5 mM, and the culture was continued for additional 10 hours at room temperature. The bacterial pellet was washed with cold PBS and lysed in Lysis Buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.9, 0.2% Triton X-100, 10 mM imidazole, 1 mM PMSF and protease inhibitor cocktail from Sigma-Aldrich) by incubation on ice for 30 minutes. The lysates were then sonicated using a Q700 sonicator (QSonica) using a ¼ inch diameter probe at 30% input amplitude for 6 times. The translucent lysates were centrifuged and the supernatants were incubated with His60 Nickel Superflow Resin (Takara-Clontech) at 4°C for 4 hours. The resin was then collected and washed 3 times with the Lysis Buffer before loading onto a Microspin Chromatography Column (Bio-Rad Laboratories). The His-tagged Tcf1 protein was eluted with Elution Buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.9, 200 mM imidazole, 1 mM PMSF and protease inhibitor cocktail). The eluate fractions with high amounts of His-tagged Tcf1 proteins were collected and dialyzed against PBS pH 7.4 in Slide-A-Lyzer mini dialysis units (Thermo-Fisher Scientific) for 4°C for 4 hours. The protein purity was verified by SDS-PAGE followed by Coomassie staining, and proper expression of His-tagged Tcf1 was confirmed by immunoblotting with anti-Tcf1 (Cell Signaling Technologies, clone C46C7) and anti-His (Cell Signaling Technologies, clone 27E8) antibodies. The protein was quantified and used in the fluorometric deacetylation assay.
HDAC-like activity was previously reported in bacteria50, and we adopted the following approaches to further eliminate possible contamination by bacterial HDAC or other proteins. The pellet of Rosetta bacteria expressing Tcf1 was lysed in a Lysis Buffer containing 500 mM NaCl (all other components were identical as above). The same buffer was used for binding of His-tagged Tcf1 to the Nickel Superflow Resin. This modification allowed dissociation of bacterial DNA from the imidazole-eluted Tcf1 protein. The Tcf1 preparation was then run over a Superdex 75 10/300 GL column (GE Healthcare Life Sciences) using an AKTA-pure FPLC. The running buffer consisted of 50 mM Tris/HCl (pH 8.0) and 500 mM NaCl. The purity of select fractions was assessed by SDS-PAGE followed by Coomassie Blue Staining. The fraction(s) containing predominantly Tcf1 without apparent other contaminating proteins was dialyzed in 50 mM Tris/HCl (pH 8.0) and 150 mM NaCl, and used in HDAC assays using acetylated H3 proteins or peptide as substrates.
To provide proper negative controls for the HDAC assays, we used either His-tagged recombinant PHF8 protein (amino acids 539–689 segment) or GST protein isolated from the same host bacteria. To purify GST protein, pGEX4T-1 vector that express the GST tag was transformed into the Rosetta competent cells, cultured as above, and the resulting bacterial pellet was lyzed in Lysis buffer contain 150 mM NaCl and centrifuged as above. The lysates were incubated with Glutathione Magnetic Beads (Pierce) at 4°C for 2 hours, the beads were then collected and washed 3 times with the Lysis Buffer. The GST protein was eluted from the beads using 10 mM reduced glutathione (Research Products International), and dialyzed in 50 mM Tris/HCl (pH 8.0) and 150 mM NaCl.
Electrophoretic mobility shift assay (EMSA)
Recombinant WT p45 Tcf1 and its mutants (0.6 µg each) were incubated in an EMSA buffer containing 20 mM KCl, 0.5 mM dithiothreitol, 0.5 mM EDTA, 10% glycerol, and 1 µg of polyinosinic:polycytidylic acid (Sigma-Aldrich) on ice for 15 min followed by additional 15 min incubation with a Tcf/Lef binding TOP probe. The TOP oligonucleotides, 5’-gatctagggcaccctttgaagctct and 5’-agagcttcaaagggtgccctagatc, contain the consensus Tcf/Lef binding motif (underlined), and were synthesized in 5’-biotinylated form and annealed as the probe. In the competition assay, unmodified TOP oligonucleotides were synthesized separately and annealed as the competitor. The protein-DNA complex was resolved on native PAGE containing 6% acrylamide. The DNA was transferred to BrightStar-Plus positively charged nylon membrane (Ambion/Life Technologies) and then UV-crosslinked. The biotinylated probe, free or in the protein complex, was detected with streptavidin-conjugated horseradish peroxidase coupled with chemiluminescent substrate in the LightShift Chemiluminescent EMSA kit (ThermoFisher Scientific).
Homology modeling
SWISS-MODEL26 was employed to construct a 3D structure for Tcf1 using homology modeling. The sequence of Tcf1 (UniProt: Q00417) was obtained from the UniProt51. The known Hdac8 crystal structure (PDB: 3F0R) was used as the desired template. The structures were visualized using PyMOL (https://www.pymol.org/).
Multiple sequence alignment of Tcf1 and HDACs
The sequence alignment of Tcf1 and Hdac8 was based on comparison of the Hdac8 crystal structure and the modeled Tcf1 structure. Global alignment of conventional HDACs (excluding Hdac10) was performed using MUSCLE52. The 30 aa HDAC domain sequence in Tcf1 was also used to search homologous sequences in HDACs using the BLAST2 program. These approaches were combined to generate the multiple sequence alignments shown in Fig. 7b.
Coevolution analysis of Tcf1 and Lef1 orthologs
Homologous sequences of Tcf1 and Lef1 were obtained from the UniProt database. If multiple sequences for each protein were obtained from one species, only one was retained for downstream analysis. Sequences that contained more than 50% gaps in 1) the global alignment or 2) their corresponding regions that aligned to the Tcf1 HDAC domain were excluded from the analysis. The resulting 98 sequences from 56 species were used as input to direct-coupling analysis (DCA)30 for detection of co-evolution of residue pairs.
Supplementary Material
Acknowledgments
We thank Radiation Core facility (A. Kalen) for mouse irradiation, and I. Antoshechkin (Millard and Muriel Jacobs Genetics and Genomics Laboratory at the Caltech) for RNA-Seq. We thank Dr. H. Kawamoto (Kyoto University, Japan) for providing the Tcf1 antiserum, Dr. H. Habelhah for advice on purification of recombinant proteins, and Drs. J. T. Harty, J. D. Colgan, and A. Bhandoola for critical reading and insightful discussion of the manuscript. We thank the Flow Cytometry Core facility (J. Fishbaugh, H. Vignes and G. Rasmussen) for cell sorting, and the Facility is supported by Carver College of Medicine/Holden Comprehensive Cancer Center (the University of Iowa), Iowa City Veteran’s Administration Medical Center, and the National Center for Research Resources of the NIH (1 S10 OD016199). The Proteomics Facility is supported by an endowment from the Carver Family trust. This study is supported by grants from the American Cancer Society (RSG-11-161-01-MPC to H.-H.X.), the NIH (AI112579, AI105351, AI115149 and AI119160 to H.-H.X., AI113806 to W.P.), NSF (CAREER Award 1452411 to C.A.M.), and the Carver Trust Young Investigator Award (01–224) to H.H.Q.; J. Z. and C.L. are supported by the Intramural Research Program of the NHLBI.
Footnotes
Accession numbers. RNA-Seq and ChIP-Seq data are deposited at GEO, under accession number GSE73240.
Author contributions. S.X. and F. L. performed most of the experiments with the help of S.Y., Q.S., and F.C.P.; Z.Z. analyzed the high throughput data under the supervision of W.P.; Y.Z. performed homology modelling and coevolution analyses under the supervision of C.Z.; Y.L. and R.M.P. performed the proteomics analysis; P.K.M., H.H.Q., and C.A.M. helped purification of recombinant proteins; C.L. directed gene targeting; J.Z. directed high throughput sequencing; W.P. and H.H.X supervised the overall study, analyzed the data, and wrote the paper. All authors edited the manuscript.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Fisher AG. Cellular identity and lineage choice. Nature reviews Immunology. 2002;2(12):977–982. doi: 10.1038/nri958. [DOI] [PubMed] [Google Scholar]
- 2.Smale ST. The establishment and maintenance of lymphocyte identity through gene silencing. Nature immunology. 2003;4(7):607–615. doi: 10.1038/ni0703-607. [DOI] [PubMed] [Google Scholar]
- 3.Rothenberg EV, Zhang JA. T-cell identity and epigenetic memory. Current topics in microbiology and immunology. 2012;356:117–143. doi: 10.1007/82_2011_168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33(1):12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes & development. 1995;9(10):1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- 6.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes & development. 1998;12(15):2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449(7161):473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 8.Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329(5987):85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, et al. The transcriptional landscape of alphabeta T cell differentiation. Nature immunology. 2013;14(6):619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, et al. A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4+ T cells. Nature immunology. 2014;15(10):947–956. doi: 10.1038/ni.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boucheron N, Tschismarov R, Goeschl L, Moser MA, Lagger S, Sakaguchi S, et al. CD4(+) T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2. Nature immunology. 2014;15(5):439–448. doi: 10.1038/ni.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374(6517):70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 13.Steinke FC, Xue HH. From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunologic research. 2014;59(1–3):45–55. doi: 10.1007/s12026-014-8545-9. [DOI] [PubMed] [Google Scholar]
- 14.Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nature reviews Immunology. 2008;8(8):581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 15.Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8(1):11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 16.Germar K, Dose M, Konstantinou T, Zhang J, Wang H, Lobry C, et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A. 2011;108(50):20060–20065. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476(7358):63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37(5):813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiemessen MM, Baert MR, Schonewille T, Brugman MH, Famili F, Salvatori DC, et al. The nuclear effector of wnt-signaling, tcf1, functions as a T-cell-specific tumor suppressor for development of lymphomas. PLoS biology. 2012;10(11):e1001430. doi: 10.1371/journal.pbio.1001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nature reviews Immunology. 2008;8(10):788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinke FC, Yu S, Zhou X, He B, Yang W, Zhou B, et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nature immunology. 2014;15(7):646–656. doi: 10.1038/ni.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yankulov K. Dynamics and stability: epigenetic conversions in position effect variegation. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2013;91(1):6–13. doi: 10.1139/bcb-2012-0048. [DOI] [PubMed] [Google Scholar]
- 23.Taniuchi I, Ellmeier W. Transcriptional and epigenetic regulation of CD4/CD8 lineage choice. Adv Immunol. 2011;110:71–110. doi: 10.1016/B978-0-12-387663-8.00003-X. [DOI] [PubMed] [Google Scholar]
- 24.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nature reviews Molecular cell biology. 2008;9(3):206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wanczyk M, Roszczenko K, Marcinkiewicz K, Bojarczuk K, Kowara M, Winiarska M. HDACi--going through the mechanisms. Frontiers in bioscience. 2011;16:340–359. doi: 10.2741/3691. [DOI] [PubMed] [Google Scholar]
- 26.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors. Biochemistry. 2008;47(51):13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, et al. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO reports. 2007;8(9):879–884. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12(7):1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 30.Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci U S A. 2011;108(49):E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annual review of biochemistry. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 32.Haery L, Thompson RC, Gilmore TD. Histone acetyltransferases and histone deacetylases in B- and T-cell development, physiology and malignancy. Genes & cancer. 2015;6(5–6):184–213. doi: 10.18632/genesandcancer.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dovey OM, Foster CT, Conte N, Edwards SA, Edwards JM, Singh R, et al. Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood. 2013;121(8):1335–1344. doi: 10.1182/blood-2012-07-441949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabrero JR, Serrador JM, Barreiro O, Mittelbrunn M, Naranjo-Suarez S, Martin-Cofreces N, et al. Lymphocyte chemotaxis is regulated by histone deacetylase 6, independently of its deacetylase activity. Molecular biology of the cell. 2006;17(8):3435–3445. doi: 10.1091/mbc.E06-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasler HG, Young BD, Mottet D, Lim HW, Collins AM, Olson EN, et al. Histone deacetylase 7 regulates cell survival and TCR signaling in CD4/CD8 double-positive thymocytes. Journal of immunology. 2011;186(8):4782–4793. doi: 10.4049/jimmunol.1001179. [DOI] [PubMed] [Google Scholar]
- 36.Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103(6):885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 37.Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376(6543):791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 38.van Loosdregt J, Fleskens V, Tiemessen MM, Mokry M, van Boxtel R, Meerding J, et al. Canonical Wnt signaling negatively modulates regulatory T cell function. Immunity. 2013;39(2):298–310. doi: 10.1016/j.immuni.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 39.Hossain MB, Hosokawa H, Hasegawa A, Watarai H, Taniguchi M, Yamashita M, et al. Lymphoid enhancer factor interacts with GATA-3 and controls its function in T helper type 2 cells. Immunology. 2008;125(3):377–386. doi: 10.1111/j.1365-2567.2008.02854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hazzalin CA, Mahadevan LC. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS biology. 2005;3(12):e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
References associated with Methods
- 41.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med. 1996;184(3):1137–1147. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. Chapter 11: Unit 11 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25(15):1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Analytical and bioanalytical chemistry. 2003;376(7):952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- 49.Yu S, Jing X, Colgan JD, Zhao DM, Xue HH. Targeting tetramer-forming GABPbeta isoforms impairs self-renewal of hematopoietic and leukemic stem cells. Cell Stem Cell. 2012;11(2):207–219. doi: 10.1016/j.stem.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nature reviews Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 51.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.