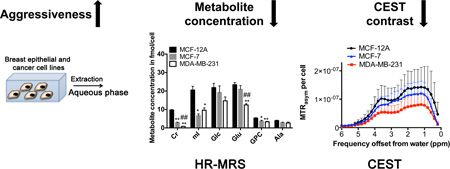
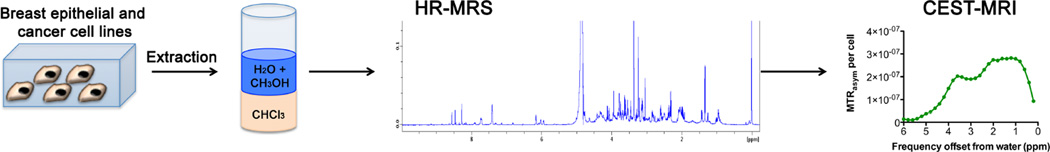
Abstract
Chemical exchange saturation transfer (CEST) is a magnetic resonance imaging (MRI) contrast mechanism that detects the exchange of protons from distinct hydroxyl, amine, and amide groups to tissue water through transfer of signal loss, with repeated exchange enhancing their effective signal. We applied CEST to systematically detect 15 common cellular metabolites in a panel of differentially aggressive human breast cancer cell lines. The highest CEST contrast was generated by creatine, myo-inositol, glucose, glutamate, and glycerophosphocholine, whose cellular concentrations decreased with increasing breast cancer aggressiveness. These decreased metabolite concentrations resulted in turn in a decreased CEST profile with increasing breast cancer aggressiveness in water-soluble extracts of breast cell lines. Treatment of both breast cancer cell lines with the chemotherapy drug doxorubicin resulted in increased metabolic CEST profiles, which correlated with significant increases in creatine, phosphocreatine, and glycerophosphocholine. CEST is able to detect breast cancer aggressiveness and response to chemotherapy in water-soluble extracts of breast cell lines. The presented results help shed light on possible contributions from CEST-active metabolites to the CEST contrast produced by breast cancers. The metabolic CEST profile may improve detection sensitivity over conventional magnetic resonance spectroscopy, and may have the potential to assess breast cancer aggressiveness and response to chemotherapy noninvasively using MRI if specialized metabolic CEST profile detection can be realized in vivo.
Keywords: CEST, metabolites, metabolism, breast cancer, cells, chemotherapy
Graphical abstract
Introduction
Metabolic alterations in glucose (Glc) and lactate (Lac) (1, 2), glutamine (Gln) (3), and choline phospholipid (4) metabolism are a hallmark of cancer (5). Cancer cells utilize less oxygen and radically alter their energy production, thereby shifting their mitochondrial function from energy production to the creation of biosynthetic intermediates that support cancer cell growth and proliferation (6). As a consequence, cancer cells typically display an increased consumption of Glc (1, 2), Gln (3, 7), and choline (Cho) (4), and an increased production of Lac (2). In addition, phosphocholine (PC) (4) and glycerophosphocholine (GPC) (8) are altered due to upregulation of choline transporters, choline kinase α, and phosphatidylcholine-specific phospholipases D and C (4). Proton magnetic resonance spectroscopy (MRS) and magnetic resonance spectroscopic imaging (MRSI), which can detect these metabolites in vivo, are starting to be used in the clinic in addition to conventional magnetic resonance imaging (MRI) for cancer diagnosis and treatment monitoring (9, 10). In vivo 1H MRS detects sums of overlapping metabolites, such as tCho (PC plus free choline (Cho) plus GPC), total creatine (creatine (Cr) plus phosphocreatine (PCr)), separate CH2 and CH3 peaks from mixed fatty acid signals in lipids (Lip), as well as individual metabolites, such as N-acetylaspartate (NAA) in the brain and Lac (11). However, MRS and MRSI have always been limited by their relatively low sensitivity, which results in low spatial resolution in vivo. With an increasing interest in monitoring metabolites for cancer diagnosis, treatment planning (10, 12), and treatment response (13), there is a need for developing alternative approaches to detect endogenous metabolites in the absence of contrast agents or tracers with improved spatial resolution, especially in vivo.
Chemical Exchange Saturation Transfer (CEST) is an emerging imaging approach with high sensitivity and specificity, which detects exchangeable protons in hydroxyl (-OH), amine (-NH2), and amide (-C(O)NH-) groups in distinct molecules (14) through transfer of signal loss between these protons and water. Large CEST contrast is generated if these protons at their respective frequency offsets (Δω) exchange rapidly and satisfy the condition Δω >> exchange rate (ksw) (14–17). CEST imaging could be a potentially more sensitive method to monitor endogenous metabolites in cancers because it can result in an up to several fold increase of metabolite signal, due to exchange with water (14, 18). This signal amplification through proton exchange with water enables the detection of metabolites at millimolar concentrations with high-resolution MRI. Chemical exchange with water produces line broadening, with the amount of broadening related to the rate of proton exchange. As such, the resonances detectable through CEST imaging are generally broader than those detected through 1H MRS with the width of the lines also affected by the saturation field strength. CEST contrast is also sensitive to physiological parameters such as pH and ionic strength, which affect the proton exchange rate with water, and to the water relaxation rates within tissue, which affect the signal loss (14). There has been an increasing interest in using CEST to monitor changes in the concentration and pH of metabolites (19, 20). Recent studies have shown that CEST can detect Cr (20), glutamate (Glu) in the brain (20), infused Glc in breast (19) and other tumor models (21), among other applications. CEST is also able to detect amide protons in proteins in the brain in vivo (22), which can be applied for distinguishing between radiation necrosis and recurrence of gliomas (23).
CEST imaging has been applied to grade brain tumors using Amide Proton Transfer (APT) contrast to detect the presence of soluble proteins with amide protons exchanging at slow to intermediate exchange rates (22, 23). We were interested in evaluating how CEST imaging might be applied to breast cancer, and have collected in vivo CEST images of orthotopic human MBA-MD-231 tumors in mice. Based on our observation that the maximum CEST signal in these breast tumor models did not correspond to amide proton frequencies, but rather amines and possibly hydroxyl protons, we were interested in determining the identity of the compounds responsible for this contrast.
To this end, we have investigated if CEST-MRI can detect metabolites that are elevated or down-modulated in water-soluble extracts of differentially aggressive human breast cancer cells as compared to nonmalignant human breast epithelial cells. We have compared highly aggressive, triple-negative human MDA-MB-231 breast cancer cells with weakly aggressive, estrogen-receptor positive (ER+) human MCF-7 breast cancer cells with nonmalignant human MCF-12A breast epithelial cells to cover different breast cancer subtypes of differential aggressiveness. For comparison and to delineate the contributions from individual metabolites found in these breast cells, we have performed high-resolution 1H MR Spectroscopy (HR-MRS) of metabolites, amino acids, as well as water-soluble dual-phase extracts from this panel of breast epithelial and differentially aggressive breast cancer cells, which was followed by measurement of CEST-MRI of the exact same samples. This approach, which is depicted in Figure 1, has given us a better understanding of the nature of endogenous metabolite CEST contrast, and signatures of particular pools of exchangeable protons in metabolites. By studying HR-MRS and CEST-MRI data from the water-soluble extracts of three different breast epithelial and cancer cell lines, we were able to identify several CEST-MRI features. These CEST features may enable the use of CEST-MRI in detecting metabolites for breast cancer diagnosis and treatment monitoring.
Figure 1.
Schematic depicting the work flow starting with cell culture and dual phase extraction of metabolites from breast epithelial and breast cancer cell lines. The water and methanol (CH3OH) phases containing the metabolites were measured consecutively, first by HR-MRS and then by CEST-MRI.
Materials and Methods
Phantom preparations
All compounds were purchased from Sigma-Aldrich (Sigma-Aldrich Corp., St. Louis, MO, USA) unless otherwise specified. Compounds were dissolved in standard, 1x-diluted phosphate buffered saline (PBS) at 20 mM, and pH was titrated to physiological pH of 7.3 using 1 M hydrochloric acid and 1 M sodium hydroxide. We prepared an MCF-12A model mixture based on the concentrations of the 15 metabolites shown in Table 1, which were identified by HR-MRS of MCF-12A cell extracts. All samples were placed in 3 mm capillary tubes for CEST imaging (24).
Table 1.
Concentrations of individual metabolites in the MCF-12A model mixture as used for the optimization of saturation parameters in Figure 3.
| Metabolites | Concentration [mM] |
|---|---|
| Cr | 0.66 |
| mI | 1.79 |
| Glc | 3.54 |
| Glu | 2.51 |
| GPC | 0.70 |
| Ala | 0.60 |
| Gly | 0.09 |
| Thr | 0.44 |
| PCr | 0.22 |
| Gln | 1.82 |
| Cho | 0.09 |
| PC | 0.16 |
| Lac | 1.89 |
| Tau | 0.25 |
| reference TSP | 0.55 |
Cell culture
MCF-12A is a non-tumorigenic, spontaneously immortalized human mammary epithelial cell line. MCF-7 is an estrogen receptor positive, weakly aggressive, non-metastatic human breast cancer cell line. MDA-MB-231 is a highly aggressive, triple-negative, metastatic human breast adenocarcinoma cell line. All cell lines were purchased from the American Type Culture Collection (ATCC). They were tested and authenticated by ATCC using two independent methods: the ATCC cytochrome C oxidase I PCR assay, and short tandem-repeat profiling using multiplex PCR. Cell lines were cultured as previously described (25).
Doxorubicin treatment
MCF-7 and MDA-MB-231 cells were treated with 5 µM doxorubicin hydrochloride in standard cell culture medium for 24 h or 48 h, as in the clinic, doxorubicin is often supplied as a bolus that achieves plasma concentrations of 5 µM to 10 µM (26). Doxorubicin was dissolved in dimethyl sulfoxide (DMSO) to give a 10 mM stock solution, which is 2000× of the final treatment concentration. For each experiment, 20 ml of fresh cell culture media was added to each T175 flask together with 10 µl of DMSO for vehicle controls, or 10 µl of 10 mM doxorubicin DMSO stock solution for doxorubicin treatment, which were then incubated for 24 h or 48 h before cells were harvested and dual phase extraction was performed. All untreated vehicle controls were prepared at the same time from the same respective cell batch. Only viable cells were collected by means of cell centrifugation while damaged cells were floating in the supernatant. Upon re-suspension of the pelleted cells prior to extraction, cell viability was verified by trypan blue exclusion to ensure that all extracted cells were viable.
For IC50 value measurements, 8,000 MCF-7 cells or 5,000 MDA-MB-231 cells were seeded in each well of a 96-well plate. After 3 days of incubation, cells reached a cell density of 80–90%. A series of doxorubicin concentrations in DMSO was applied to the cells, i.e. 0.0 µM, 0.5 µM, 1.0 µM, 2.0 µM, 4.0 µM, 6.0 µM, 8.0 µM, or 12.0 µM. After 24 h or 48 h of drug treatment, doxorubicin-containing medium was removed and fresh medium with 10 µl of WST-1 reagent (Sigma-Aldrich) was added to each well. The cell density was obtained by reading out the absorption at 450 nm on a spectrophotometer. Experiments were performed in quadruplicate. IC50 values for doxorubicin were calculated from regression lines of a plot of cell density versus doxorubicin concentration using the free pharmacological analysis software tool ED50plus v1.0 (http://www.free-downloads-center.com/download/ed50plus-v1-0-2434.html) for use within Microsoft Excel worksheets. IC50 values are reported as mean ± standard deviation.
Dual-phase extraction
All cell lines were cultured to 70% confluence. Approximately 108 viable, treated or untreated, cells were harvested, and both lipid and water-soluble cell extract fractions were obtained using a dual-phase extraction method as previously described (25). Briefly, cells were harvested by trypsinization, washed twice with 10 ml of saline at room temperature, and pooled into a glass centrifuge tube. Cells were counted directly after trypsinization. Four ml of ice-cold methanol were added to the cells, vigorously vortexed, and kept on ice for 10 min. Four ml of chloroform were added and vigorously vortexed. Finally, 4 ml of water were added, the sample was vortexed and left overnight at 4°C for phase separation. On the next day, the samples were centrifuged for 30 min at 35,000 g at 4°C, and the phases were carefully separated using Pasteur pipettes. The water-methanol phase containing the water-soluble cellular metabolites was treated with 10 mg of chelex for 10 min on ice to remove divalent cations. The chelex beads were then removed. Methanol was removed by rotary evaporation. The remaining water phases were lyophilized and stored at −20°C. This extraction effectively removed denatured proteins, DNA, RNA, and lipids.
High-resolution MRS
The lyophilized water-soluble cell extracts were dissolved in D2O containing the concentration and chemical shift reference 3-(trimethylsilyl) propionic-2, 2, 3, 3-d4 acid (TSP). Fully relaxed 1H MR spectra of the water-soluble extracts were acquired on a Bruker Avance 500 nuclear magnetic resonance spectrometer operating at 11.7 T using a 5-mm HX inverse probe as previously described (25) with the following acquisition parameters: TR = 9.5 s, TD = 64K, Number of Signal Averages (NSA) = 64. The following metabolites in the 1H MR spectra of the water-soluble extracts were fitted and integrated by the software MestRec 4.9 (Mestrelab Research, Escondido, CA) and normalized to the cell number, volume, and the TSP concentration standard as previously described (25): Thr doublet at 1.26 ppm, Lac doublet at 1.33 ppm, Ala doublet at 1.45 ppm, Glu multiplet at 2.34 ppm, Gln multiplet at 2.45 ppm, GSH multiplet at 2.56 ppm, Cr singlet at 3.03 ppm, PCr singlet at 3.04 ppm, Cho singlet at 3.21 ppm, PC singlet at 3.23 ppm, GPC singlet at 3.24 ppm, Gly singlet at 3.56 ppm, Tau triplet at 3.40 ppm, mI triplet at 4.07 ppm, and the TSP singlet at 0.00 ppm. After completing the HR-MRS measurements, each sample was lyophilized and stored at −20°C until it was prepared for CEST-MRI.
CEST-MRI acquisition of phantoms and extracts
Samples after HR-MRS were hydrated with 100 µl of PBS, adjusted to pH 7.3, and placed in a multi-sample holder as previously reported (24). All MRI images were acquired at 37°C (310 K) using an 11.7 T Bruker Avance system. The B0 field was shimmed using the shimming toolbox in Paravision Version 5.1 (Bruker BioSpin MRI GmbH). A modified rapid acquisition with relaxation enhancement (RARE) sequence including a saturation pulse was used to acquire saturation images at different irradiation frequencies, which were used to generate the Z-spectrum in each voxel. A slice thickness of 1 mm was used, and the typical imaging parameters were: TE = 4.3 ms, RARE factor = 16, matrix size of 128×64, number of averages (NA) was 2, and the typical field of view was 13×13×1 points. Two sets of saturation images were acquired. First, we acquired the frequency map images to map the spatial distribution of B0. Secondly, we acquired the CEST images to characterize the CEST properties of the samples. The acquisition time per frequency point was 12 s for frequency maps (TR = 1.5s) and 48 s for CEST images (TR = 6 s).
To produce the frequency maps, WAter Saturation Shift Referencing (WASSR) mapping was employed (18), for which we used a saturation pulse length of 500 ms, saturation field strength (B1) of 0.5 µT (21.3 Hz), and saturation frequency increment of 50 Hz (spectral resolution=0.1 ppm). We kept the image readout between frequency map images and CEST images identical. For CEST images, we used a saturation pulse length of 4 s, B1 of 1.6 (68 Hz), 2.4 (102 Hz), 3.6 (150 Hz), 4.7 (200 Hz), and 5.9 µT (250 Hz), respectively, and a frequency increment of 0.2 ppm.
Data analysis for CEST-MRI
All CEST-MRI data processing was performed using custom-written scripts in Matlab (Mathworks, Natick, MA, USA). WASSR images were fit to Lorentzians to find the frequency offset (Δω) for each voxel and to generate a saturation-based B0 shift map as described previously (24). Then, CEST images were corrected voxel-wise using the WASSR frequency maps.
For analysis, region of interest (ROI) masks were manually drawn covering each sample in its entirety, and the mean intensity was used for plotting the Z-spectra as a function of saturation frequency offset with respect to water. The CEST contrast was quantified by determining the asymmetry in the magnetization transfer ratio (MTRasym) as defined by the following expression:
where Ssat(+Δ ω) and Ssat(−Δ ω) are the signal intensities obtained by saturating at the frequency of Δω downfield and upfield from the water proton resonance frequency, respectively.
Orthotopic MDA-MB-231 breast tumor xenografts
All animal experiments were approved by the Johns Hopkins Institutional Animal Care and Use Committee. Approximately, 2×106 MDA-MB-231 breast cancer cells were orthotopically implanted into the fourth right mammary fat pad of 6 weeks old female athymic nu/nu mice (NCI) as previously described by us (27, 28). Tumor growth was monitored with standard calipers. Tumors reached their experimental size of approximately 600 mm3 within 8 weeks of inoculation. Three mice with one MDA-MB-231 tumor each were assessed with in vivo CEST-MRI as described below.
In vivo CEST-MRI
In vivo images were acquired on a Bruker Biospec 11.7 T horizontal MR scanner. The mice were anesthetized by using 0.5–2% isoflurane and imaged using a 15-mm planar surface coil. Breath rate was monitored throughout in vivo MRI experiments using a respiratory probe, and animals were kept warm with a heating bed. Initially, anatomical reference images were acquired using rapid acquisition with relaxation enhancement (RARE), TR = 2.5 s, TE = 4.7 ms, RARE factor = 16, and matrix size = 128×64. Then, a modified RARE sequence including a saturation pulse was used to acquire saturation images at different irradiation frequencies, to generate the Z-spectrum in each voxel. Two sets of saturation images were acquired as described previously (19, 31, 32). First, we acquired the frequency map images to map the spatial distribution of B0. In the frequency maps, WASSR mapping was employed (33), with a saturation pulse length of 500 ms, saturation field strength (B1) of 0.5 µT, and saturation frequency increment of 0.1 ppm. Secondly, we acquired the CEST images to characterize the CEST contrast. For CEST images, a saturation pulse length of 3 s, B1 of 3.6 µT, and Z-spectra acquired by incrementing saturation frequency every 0.2 ppm from −5 to +5 ppm; TR = 5 s, effective TE = 5 ms, matrix size = 96×48, slice thickness of 1.0 mm. CEST images were overlaid with the anatomical reference images.
Statistical analysis
For comparison of each metabolite among the three different cell lines MCF-12A, MCF-7, and MDA-MB-231 in our HR-MRS data, statistical analysis was performed using a two-tailed t-test with unequal variance (α = 0.05), and was considered significant for P-values less than 0.05. As discussed above, CEST MTRasym profiles were obtained for all the samples from each of the three cell lines. Within the CEST data, a one-way nonparametric ANOVA (Kruskal-Wallis test) was performed to determine whether differences in contrast were statistically significant among the three different cell lines (α = 0.05).
Results
CEST-MRI of triple-negative breast tumor models in vivo
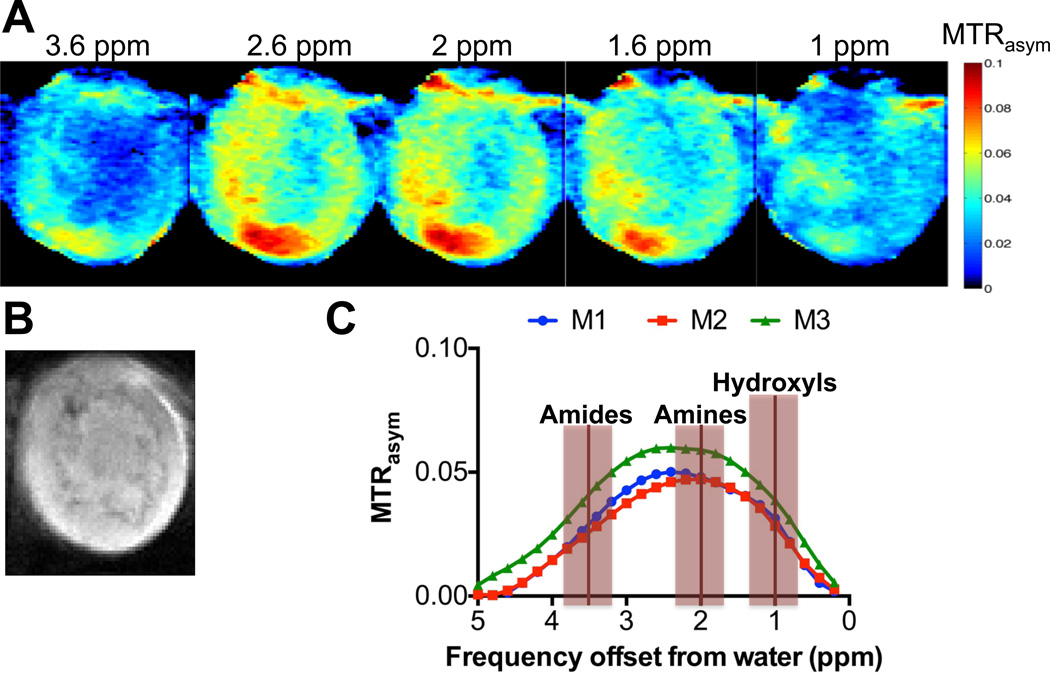
We have collected in vivo CEST images of orthotopic human MBA-MD-231 tumors in mice (Fig. 2). As is shown, the maximum signal in this broad signal spanning 1.0 – 4.0 ppm is observed at 2.25 ppm from water, which does not correspond to amide proton frequencies, which peak at 3.6 ppm, but is typical of amine protons. Based on this observation, we were interested in determining the identity of the compounds responsible for this contrast and to evaluate if CEST-MRI can detect metabolites that are elevated or down-modulated in human breast cancer cells as compared to nonmalignant human breast epithelial cells. In order to accomplish this, we prepared dual phase extracts of breast cancer cells adjusted to pH 7.3, and measured the CEST spectra of the water-soluble metabolites, small peptides, and amino acids.
Figure 2.
In vivo CEST-MRI of MDA-MB-231 breast tumor xenografts. (A) Representative CEST MRI maps, (B) T1-weighted RARE MRI, and (C) MTRasym for three individual mice with orthotopic human MDA-MB-231 breast tumor xenografts, which were labeled M1 for mouse 1, M2 for mouse 2, and M3 for mouse 3. CEST shifts of amide, amine, and hydroxyl resonances are highlighted in C.
Data acquisition parameters for CEST-MRI
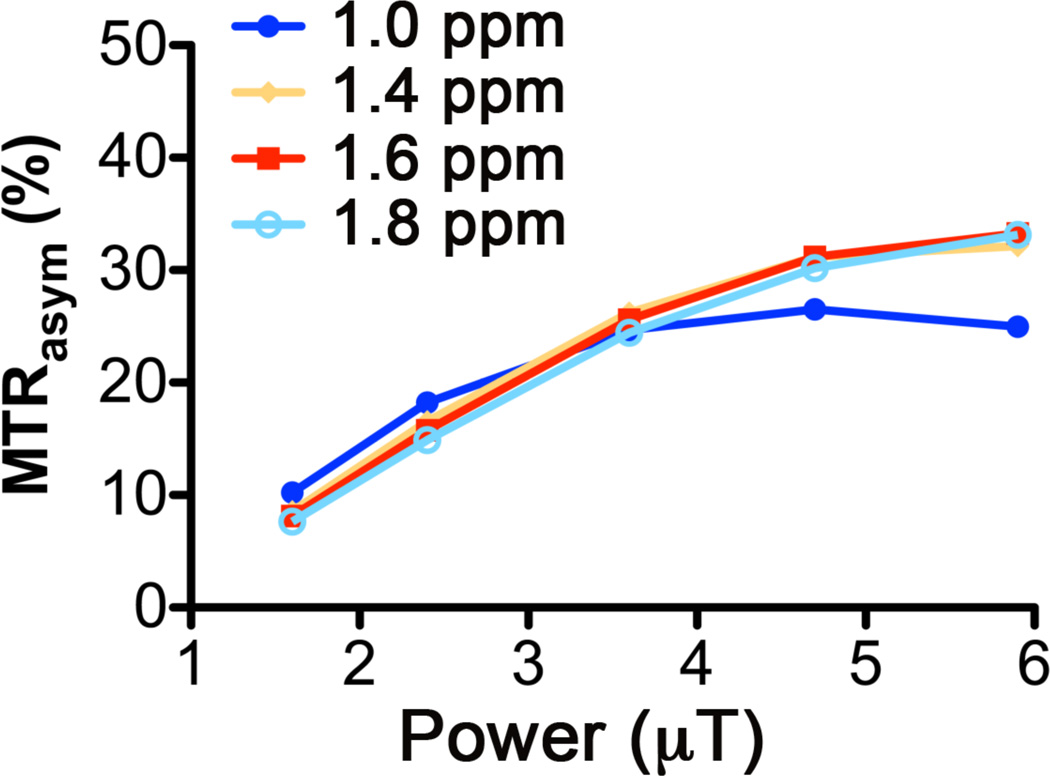
The magnitude of the CEST contrast of exchangeable protons in -OH, -NH2, and -NH- groups depends on the field strength and time of the saturation pulse applied (34). We therefore used a long saturation pulse of 4s and measured CEST-MRI of a model mixture of MCF-12A cells (Table 1) using several saturation field strengths of (ω1) = 1.6, 2.4, 3.6, 4.7, and 5.9 µT as shown in Figure 3. We chose ω1 = 3.6 µT for all further experiments because it was the highest power that showed a linear CEST response at all offsets (Fig. 3). At this power, the contrast for many metabolites was near maximum, entering the non-linear range above this power at all offsets. Note that CEST-MRI offsets are given with respect to the water resonance, not relative to TSP as is common with MRS.
Figure 3.
Optimization of saturation parameters using an MCF-12A model mixture showing the power dependence of MTRasym (%) at 1.0, 1.4, 1.6, and 1.8 ppm and its linearity at B1 < 3.6 µT.
CEST-MRI of phantom solutions of amino acids and metabolites
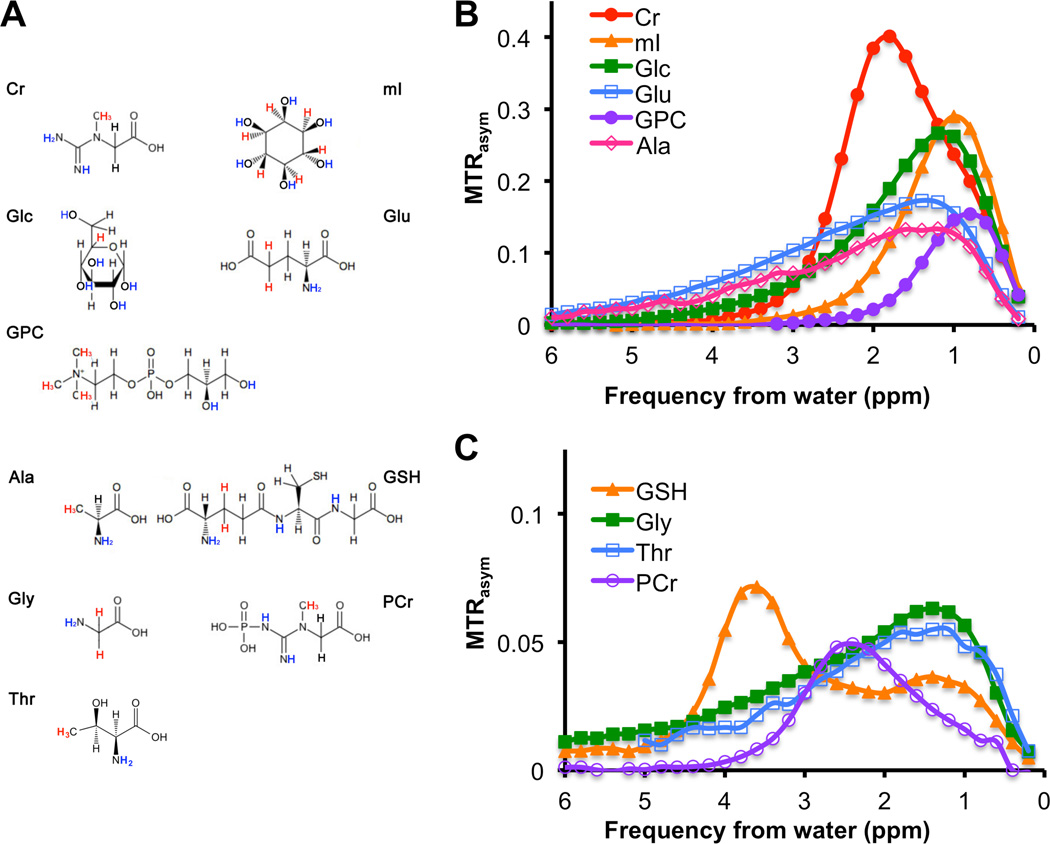
Solutions (20 mM in PBS) of amino acids and metabolites that are typically detected by HR-MRS in human breast epithelial and breast cancer cells were measured individually by CEST-MRI and are shown as MTRasym plots in Figure 4. Figure 4A shows the chemical structures of all measured compounds that produced detectable CEST contrast. Chemical structures are displayed with the CEST-visible exchangeable protons labeled in blue, and the protons used for 1H HR-MRS quantification labeled in red (Fig. 4A). Figure 4B shows the large (MTRasym > 0.2) to intermediate (MTRasym ≥ 0.1) CEST contrast from 20 mM solutions of separately measured creatine (Cr), myo-inositol (mI), Glc, glutamate (Glu), GPC, and alanine (Ala). CEST contrast was generated from the -OHs of mI, Glc, and GPC at Δω = 1.0 ppm, -NH=C(NH2)2s of Cr at Δω = 2.0 ppm, and -NH2s of Glu and Ala at Δω = 1.0 – 3.0 ppm (broad signals) (Fig. 4B). Small but detectable CEST contrast with an MTRasym < 0.1 was detected for the -NH=C(NH2)2s of PCr at Δω = 2.4 ppm, the -NH2s of Gly and Thr at Δω = 1.0 – 3.0 ppm (broad signal), and -C(O)NH-s of GSH at Δω = 3.6 ppm (Fig. 4). Gln, Cho, PC, Lac, and Tau resulted in negligible CEST MTRasym contrast for the saturation parameters used. For different sample conditions, the MTRasym profiles of Glu, Ala, and Tau were found to be similar previously (21).
Figure 4.
CEST contrast as MTRasym,, and chemical structures of metabolites as detected in breast cell lines, which were measured at 20 mM, pH 7.3, and 37°C. (A) CEST-visible exchangeable protons of -OH, -NH-, and -NH2 groups are shown in blue and the protons used for the HR-MRS quantification are shown in red. (B) MTRasym of cellular metabolites with strong CEST contrast of MTRasym > 0.2 and intermediate CEST contrast of 0.1 < MTRasym < 0.2. (C) MTRasym of cellular metabolites with weak CEST contrast of MTRasym ≤ 0.1.
CEST-MRI of water-soluble extracts from breast epithelial and breast cancer cells
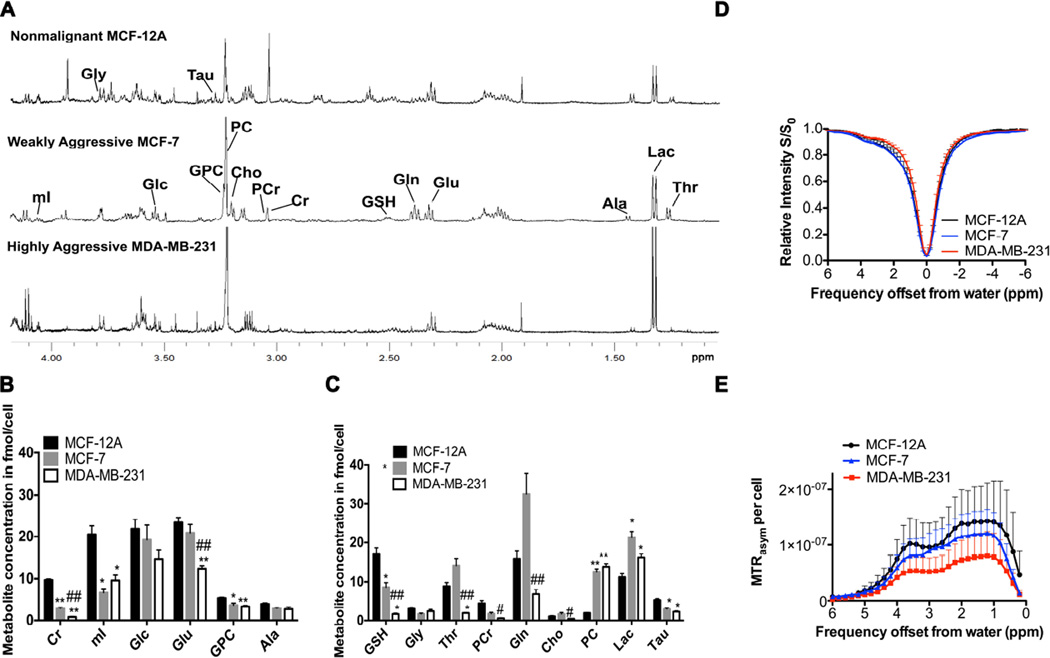
Figure 5A displays representative 1H MR spectra of the extracted water-soluble metabolites from nonmalignant MCF-12A, weakly aggressive MCF-7, and highly aggressive MDA-MB-231 cells. Metabolites were extracted using dual phase extraction, and the aqueous phases were first measured by HR-MRS, lyophilized after HR-MRS, and then the same samples were measured by CEST-MRI as depicted in the workflow diagram in Figure 1. Considering only CEST-active metabolites, the quantification (n=6 each) of 1H MR spectra revealed that the highly aggressive MDA-MB-231 cells and weakly aggressive MCF-7 cells contained significantly lower amounts of Cr, mI, GPC, GSH, and PCr levels than the nonmalignant MCF-12A cells as shown in Figures 5B, C. Glu was significantly decreased in MDA-MB-231 versus MCF-12A cells (Figs. 5B). CEST-MRI contrast of the same water-soluble extracts from these breast epithelial and breast cancer cells was measured at physiological pH at 37°C. The average CEST-MRI contrast quantified as MTRasym per cell (n=6 each) at 1.0 ppm, 1.8 ppm, and 3.6 ppm was the highest in MCF-12A, intermediate in MCF-7, and the lowest in MDA-MB-231 cell extracts, indicating that metabolite CEST-MRI can detect metabolite concentration differences resulting from differences in the aggressiveness of the tested water-soluble extracts of breast cancer cell lines. This finding was most likely driven by the -OHs of mI and GPC at 1.0 ppm; -NH2 of Glu and -NH=C(NH2)2s of Cr at 2.0 ppm; and -C(O)NH-s of GSH at 3.6 ppm (Fig. 5D, E) as these metabolites with high concentration and high to intermediate CEST contrast showed the same trend of decrease in MDA-MB-231 versus MCF-12A in our HR-MRS data (Fig. 5B, C). Statistical analysis of the overall CEST contrast for the three cell lines revealed a significant difference (p=0.02) between MCF-12A versus MDA-MB-231 cell extracts, indicating that the metabolite CEST contrast correlates inversely with breast cancer cell aggressiveness, while the differences between MCF-12A versus MCF-7, and between MCF-7 versus MDA-MB-231 did not reach statistical significance. Additional metabolic changes such as decreased levels in Tau, and increased Lac and PC levels were detected in MDA-MB-231 cells as compared to MCF-12A cells (Fig. 5C), but did not contribute to the changes in the CEST profile as these metabolites did not produce any CEST contrast (data not shown).
Figure 5.
HR-MRS and CEST-MRI of human breast epithelial and breast cancer cell extracts. (A) HR-MR spectra of extracts from nonmalignant MCF-12A breast epithelial cells, weakly aggressive MCF-7 breast cancer cells, and highly aggressive MDA-MB-231 breast cancer cells. (B) HR-MRS quantification of metabolites with strong CEST contrast and (C) weak CEST contrast in water-soluble extracts from nonmalignant MCF-12A breast epithelial cells, and MCF-7 and MDA-MB-231 breast cancer cells (n=6 each). (D) CEST-MRI Z-spectra for the same cell extracts as in A–C. (E) CEST contrast in MTRasym per cell (n=6 each), with MCF-12A showing significantly (p=0.02) higher CEST contrast than MDA-MB-231. Values are mean ± standard error. * p < 0.05, ** p < 0.01 versus MCF-12A. # p < 0.05, ## p < 0.01 versus MCF-7.
CEST-MRI of water-soluble extracts from chemotherapy-treated breast cancer cells
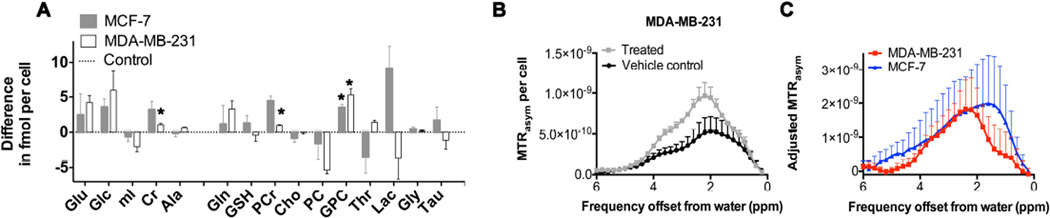
To test if metabolite CEST has potential for monitoring chemotherapy response in breast cancer, we treated the two breast cancer cell lines MCF-7 and MDA-MB-231 with 5 µM of doxorubicin, a drug commonly used for chemotherapy of breast cancer patients. Patient plasma concentrations following a doxorubicin bolus typically are in the range of 5 µM to 10 µM (26), which informed our media concentration of 5 µM doxorubicin for our cell culture treatment experiments. We measured the IC50 values for 24 and 48 hours of doxorubicin treatment in cell culture media to be 4.0 ± 0.3 µM and 2.7 ± 0.5 µM for MCF-7 cells, respectively, and 4.0 ± 0.1 µM and 1.4 ± 0.2 µM for MDA-MB-231 cells, respectively, which is quite similar to each other, and lies within the overall range of previously reported values (35). Figure 6 shows the HR-MRS detected metabolic MRS and CEST-MRI changes (MTRasym per cell) in water-soluble cell extracts obtained following 24 h of doxorubicin treatment of breast cancer cells. Only viable cells were extracted by dual phase extraction following treatment with doxorubicin. Cells treated with doxorubicin for 24 h contained significantly (p<0.05) increased levels of Cr, PCr and GPC (Fig. 6A). The CEST-MRI contrast quantified as MTRasym profile per cell (Fig. 6B) and adjusted MTRasym profile per cell (Fig. 6C) significantly (p=0.03) increased following doxorubicin treatment in both breast cancer cell lines (Fig. 6C). This increase in the MTRasym profile upon doxorubicin treatment was likely driven by the -OHs of GPC at 1.0 ppm, -NH=C(NH2)2s of Cr at 2.0 ppm, and -NH=C(NH2)2s of PCr at 2.6 ppm. Comparable results were obtained for 48 h of doxorubicin treatment in both cell lines as shown in Supplementary Figure 1.
Figure 6.
HR-MRS and CEST-MRI of cell extracts from doxorubicin treated MCF-7 and MDA-MB-231 breast cancer cells compared to the respective vehicle controls. (A) HR-MRS quantification (n=3 each) shown as difference between doxorubicin treated cell extracts minus untreated vehicle controls (dotted horizontal line at 0) in fmol per cell following treatment with 5 µM doxorubicin for 24 hours in MCF-7 (gray bars) and MDA-MB-231 (white bars) cells. (B) CEST-MRI of MDA-MB-231 cell extracts showing doxorubicin treated cells (gray squares, n=3) versus vehicle controls (black circles, n=3). (C) CEST-MRI quantified as adjusted MTRasym, which is defined as MTRasym per cell of treated cells minus vehicle controls (n=3 each), in MCF-7 (blue triangles) and MDA-MB-231 (red squares) cells. Values are mean ± standard error. * p< 0.05, treated versus control.
Discussion
Consecutive HR-MRS and CEST-MRI of water-soluble cell extracts revealed metabolic differences in breast epithelial and breast cancer cell lines that can be detected by metabolite CEST-MRI. The CEST MTRasym profiles of these cell extracts correlated inversely with breast cancer aggressiveness and were driven by concentration differences in the highly to intermediately CEST-active metabolites Cr, mI, Glu, GPC, and GSH. Doxorubicin treatment resulted in consistently increased MTRasym profiles in water-soluble breast cancer cell extracts, which was driven predominantly by increases in Cr, PCr, and GPC following treatment.
While all of the metabolites that we systematically tested possess exchangeable protons, not all of them generate large CEST contrast because the exchange rate (ksw) of these protons needs to be in the appropriate range to be detected by CEST as determined by the requirement Δω >> ksw for rapidly exchanging protons (14–17). In general, the CEST MTRasym profiles for individual metabolites depend on the type of exchangeable proton, i.e. -OH, -NH2, -C(O)NH-s, or -NH=C(NH2)2, groups, its exchange rate with water, its concentration, and the saturation pulse parameters, e.g. saturation ω1 and duration. The exchangeable protons in hydroxyl (-OH), amine (-NH2, -NH-), amide (-C(O)NH-), and guanidyl (-NH=C(NH2)2) groups of the measured metabolites covered offset frequencies between 1.0 – 4.0 ppm from water depending on their chemical environment. Taken together, our results from metabolites, amino acids, and differentially aggressive breast cell extracts help shed light on contributions from CEST-active metabolites that may be an important part of the overall CEST-MRI signal of breast cancers obtained with CEST imaging in vivo.
Our in vivo CEST-MRI data of orthotopic MDA-MB-231 breast tumor xenografts show that the maximum CEST signal from these tumors is consistently at 2.25 ppm, within the overall signal spanning the range of 1.0 – 4.0 ppm from the water signal. This peak is typical of amines, although hydroxyls can also resonate at this position. CEST-MRI of cell extracts from MDA-MB-231 breast cancer cells, in which all proteins, DNA, RNA, lipids, and pH effects have been removed, showed a similar MTRasym profile, covering 1.0 – 4.0 ppm as well, with peaks at 1.0 ppm, 2.0 ppm, and 3.5 ppm. Our comparative phantom data revealed that these signals in cell extracts originated from Cr, mI, Glc, Glu, GPC, Ala, and GSH, which was also confirmed with comparative MTRasym profiles and metabolite concentrations determined by 1H HR-MRS in two additional breast cell line extracts. While it is well known that water-soluble proteins generate CEST contrast in vivo, we are the first to demonstrate that the above metabolites and amino acids most likely contribute to the CEST-MRI signal of breast tumors in vivo. A recent study suggested that CEST-MRI in breast tumors may detect NADH, which may involve redox states coupled to NADH-dependent cellular redox reactions (36). This study relied on a limited number of phantoms and in vivo CEST-MRI of breast tumor xenografts (36). Based on our quantitative, combined HR-MRS and CEST results from cell extracts of the same breast cancer cell lines MCF-7 and MDA-MB-231, we conclude that detecting [NADH] or the NADH redox ratio in these breast tumor models through CEST-MRI might be challenging because metabolites such as Cr, mI, Glc, and GPC, which are not directly coupled to NADH redox reactions, also generate significant contrast. Tumor cell density and other factors as discussed below are also expected to contribute to the observed contrast changes.
While HR-MRS uses spectrally isolated, non-exchangeable protons for detection and quantification (marked in red in Fig. 4), several CEST-active metabolites that dominate the MTRasym profile of the tested breast cell extracts give signals at overlapping offset frequencies. For example, the -OHs of Glc, mI, and GPC overlap at 1.0 ppm, and the -NH2 of Glu and -NH=C(NH2)2 of Cr overlap at 2.0 ppm. A somewhat more isolated effect detected in our study was caused by -C(O)NH-s of GSH at 3.6 ppm, however, the broad signal from -NH2 of Glu may also have overlapped with GSH at 3.6 ppm. These overlaps in exchangeable proton frequencies reduce the specificity of metabolite CEST-MRI. Nevertheless, the evaluation of the MTRasym profile in its entirety may be of value in cases where several CEST-active metabolites show the same trend of change, as was the case in our study.
In vivo 1H MRS and MRSI currently suffer from some lack of specificity as it is not possible to resolve certain overlapping metabolites such as tCho and total creatine (11). Consecutive or simultaneous in vivo CEST-MRI and 1H MRSI may provide complementary information about metabolite concentrations in tumors. It is also possible to make CEST-MRI measurements exchange rate specific, and thereby increase the CEST-MRI detection specificity to only one or a few metabolites with a particular exchange rate, using for example the recently suggested Frequency Labelled Exchange (FLEX) (37), Chemical Exchange Rotation Transfer (CERT) (38), or Variable Delay Multi-Pulse (VDMP) (39) schemes. In addition, while MTRasym is used in this study due to the absence of conventional magnetization transfer contrast (cMTC) in our samples, for in vivo measurements where cMTC effects play a role it is possible to use lineshape analysis instead to extract CEST contrast (40, 41).
Importantly, the exchange of protons between metabolites, which are present in cells in millimolar concentrations, and the water pool, which has a 110 molar proton concentration, can enhance the sensitivity of detecting these metabolites using CEST by factors up to several hundredfold (16). This enhancement in CEST-MRI provides the potential to assess MTRasym profiles in tumors in vivo with increased sensitivity to aid cancer diagnosis and treatment monitoring. It is well known that for example brain tumors in vivo display CEST contrast from amide proton transfer (APT) at 3.5 ppm when using relatively weak saturation pulses (ω1 ~ 2 µT), which is produced from intracellular proteins and peptides (22, 23), and that this contrast can change in response to treatment (42, 43). In breast tumors, APT contrast was recently demonstrated as well (44). In the in vivo setting, additional CEST contributions such as from APT may further reduce the specificity of metabolite CEST, unless specialized MR sequences as discussed above are applied. We optimized the saturation pulse parameters for detecting metabolites and also removed larger proteins, DNA, RNA, and lipids by dual phase extraction, and therefore APT should not contribute significantly to the MTRasym profiles observed in our study. When moving CEST-MRI to the in vivo setting, additional complexity may be added to the MTRasym profile if the same metabolites occupy two or more different compartments with a different pH or different ionic strength, which would affect their exchange rates. Such compartments could be different cellular organelles, different cell types within the tumor, or different tumor microenvironments such as acidic, hypoxic, or necrotic tumor regions.
We observed that a reduction in the metabolite concentrations of Cr, mI, Glc, Glu, GPC, Ala, and GSH resulted in a decrease in the MTRasym profile. Highly aggressive MDA-MB-231 breast cancer cells contained significantly lower concentrations of these metabolites than nonmalignant MCF-12A breast epithelial cells, because they were possibly not imported into the cell, not synthesized, broken down at a greater rate, or excreted at a higher rate due to changes in the expression and activity of the respective transporters, catabolic or anabolic enzymes, or membrane pumps. Some of our findings are consistent with several previous reports about the aberrant Glc (1, 2, 6), Gln (3, 6, 7), and phospholipid (4) metabolism in cancer.
Cr uptake and levels (45) were increased in tumor cells in some studies. Other studies demonstrated decreased tumoral Cr and PCr levels in rectal cancers (46) and glioblastomas (47). In breast cancer, Cr concentrations did not correlate with prognosis, but with progesterone receptor status (48). We observed in our study that Cr and PCr were significantly decreased in MCF-7 and MDA-MB-231 breast cancer cells compared to nonmalignant MCF-12A cells, which is not consistent with progesterone receptor status dependence as both MCF-7 and MCF-12A cells are progesterone receptor positive, and MDA-MB-231 cells are triple-negative. Cr CEST has been previously developed in a feasibility study (20).
In breast cancer, tumor mI levels were previously shown to be significantly decreased in HER2-negative versus HER2-positive breast cancer (48). This is in partial agreement with our findings, as MDA-MB-231 and MCF-7 breast cancer cells are HER2-negative, and displayed significantly decreased mI compared to nonmalignant MCF-12A breast epithelial cells, which, however, are also HER2-negative (49). CEST measurement of endogenous mI has previously been performed in human brain, where mI is an abundant metabolite, glial marker, and osmolyte that is altered in brain disorders including Alzheimer's disease and brain tumors (50).
The presence of Glc in our cell culture studies was much higher than that in tumors in vivo because there was a constant supply of 25 mM Glc in the cell culture medium available to the cells. However, in densely packed tumors in vivo, Glc concentrations will typically vary between 0–1 mM, and negatively correlate with proliferation rate, as highly proliferating cancer cells use more Glc as shown by high-resolution magic angle spinning (HR MAS) MRS of breast cancer biopsies (51). Decreased Glc levels in the more aggressive breast cancer cell line MDA-MB-231 were consistent with a higher glycolytic rate and increased Lac production through lactate dehydrogenase in aggressive breast cancer cells, which is in agreement with previous HR MAS MRS findings (52). Different types of glucose infusion protocols have already been reported to obtain the Glc-based CEST (glucoCEST) signal from breast and colorectal tumor models (19, 21).
We observed in our study that highly aggressive MDA-MB-231 cells contained significantly decreased Gln and Glu concentrations compared to nonmalignant MCF-12A cells, whereas less aggressive MCF-7 cells exhibited increased Gln and only slightly decreased Glu concentrations compared to MCF-12A cells. Previous studies have shown that several cancer cell lines heavily rely on glutamine uptake (7). In many cancer cells, glutamine is a necessary carbon source and supports NADPH production for redox control (7). Moreover, previous reports have shown that breast cancer cells exhibit elevated glutaminase activity, which can be targeted to inhibit oncogenic transformation (53). Our findings are consistent with a recent publication, demonstrating that patients (n=136) with low tumor Gln levels exhibited poor outcome compared to patients with high tumor Gln levels (54). CEST of Glu following systemic Glu injection has previously been used for detecting brain tumor models as Glu only crosses the compromised blood brain barrier in the tumor, but not in healthy brain (20). Our findings indicate that endogenous Glu significantly contributes to the CEST profile in breast cancers.
The increases in GPC, Cr, and PCr following doxorubicin chemotherapy treatment of MCF-7 and MDA-MB-231 breast cancer cells resulted in a significant increase in the MTRasym profile following treatment in water-soluble extracts of the breast cancer cells. A previous report showed that an increase in the relative peak area of PCr was seen in breast cancer patients who responded to treatment (55), which is in good agreement with our study. Similar to our observation of a GPC increase following doxorubicin treatment, docetaxel (56), the anti-inflammatory agent indomethacin (57), and some targeted chemotherapeutic agents (58) have also been shown to increase cellular GPC levels in breast cancer cells. A recent feasibility study of detecting APT during the chemotherapy response of breast cancers in vivo indicated that APT increased in one patient with progressive disease and decreased in two patients with a partial or complete response (44). The significant increase in the MTRasym profile following doxorubicin treatment of breast cancer cells may be further developed for CEST-MRI monitoring of chemotherapy response in breast cancer patients.
In summary, we have characterized the CEST-MRI profiles of metabolites and amino acids, which have been shown with HR-MRS to be able to discriminate between nonmalignant human breast epithelial cells, weakly aggressive, and highly aggressive human breast cancer cells. By comparing the water-soluble extracts of this panel of human breast cell lines using consecutive HR-MRS and CEST-MRI, we were able to semi-quantitatively assess the levels of metabolites by CEST-MRI as a profile, which was able to serve as readout for malignancy. We detected significant increases in Cr, PCr, and GPC along with an increased metabolite CEST profile following doxorubicin chemotherapy treatment of breast cancer cells in water-soluble extracts. Hence, CEST-MRI could potentially be a good approach for detecting breast cancer aggressiveness and response to therapy based on altered metabolite and amino acid profiles if it will be possible to develop specialized metabolic CEST profile detection in vivo. The presented results help shed light on possible contributions from CEST-active metabolites in the overall CEST signal of breast cancers.
Supplementary Material
HR-MRS and CEST-MRI of cell extracts from 48 hours doxorubicin treated MCF-7 and MDA-MB-231 breast cancer cells compared to the respective vehicle controls. (A) HR-MRS quantification (n=3 each) shown as difference between doxorubicin treated cell extracts minus untreated vehicle controls (dotted horizontal line at 0) in fmol per cell following treatment with 5 µM doxorubicin for 48 hours in MCF-7 (gray bars) and MDA-MB-231 (white bars) cells. (B) CEST-MRI of MDA-MB-231 cell extracts showing doxorubicin treated cells (gray squares, n=3) versus vehicle controls (black circles, n=3). (C) CEST-MRI quantified as adjusted MTRasym, which is defined as MTRasym per cell of treated cells minus vehicle controls (n=3 each), in MCF-7 (blue triangles) and MDA-MB-231 (red squares) cells. Values are mean ± standard error. * p< 0.05, treated versus control.
Acknowledgments
This study was supported by NIH R01 CA134695, R01 CA154725, R01 EB015032, R01 EB015031, and P50 CA103175.
Abbreviations
- HR-MRS
high resolution magnetic resonance spectroscopy
- CEST
chemical exchange saturation transfer
- Cho
choline
- Gln
glutamine
- Glu
glutamate
- mI
myo-inositol
- Glc
glucose
- Cr
creatine
- Ala
alanine
- Thr
threonine
- Gly
glycine
- Tau
taurine
- PCr
phosphocreatine
Footnotes
Conflict of interest disclosure statement: None of the authors have any conflict of interest with regard to the manuscript submitted for review.
References
- 1.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 3.Kaelin WG, Jr, Thompson CB. Q&A: Cancer: clues from cell metabolism. Nature. 2010;465:562–564. doi: 10.1038/465562a. [DOI] [PubMed] [Google Scholar]
- 4.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11:835–848. doi: 10.1038/nrc3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moestue SA, Giskeodegard GF, Cao MD, Bathen TF, Gribbestad IS. Glycerophosphocholine (GPC) is a poorly understood biomarker in breast cancer. Proc Natl Acad Sci U S A. 2012;109:E2506. doi: 10.1073/pnas.1208226109. author reply E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danishad KK, Sharma U, Sah RG, Seenu V, Parshad R, Jagannathan NR. Assessment of therapeutic response of locally advanced breast cancer (LABC) patients undergoing neoadjuvant chemotherapy (NACT) monitored using sequential magnetic resonance spectroscopic imaging (MRSI) NMR Biomed. 2010;23:233–241. doi: 10.1002/nbm.1436. [DOI] [PubMed] [Google Scholar]
- 10.Haddadin IS, McIntosh A, Meisamy S, Corum C, Styczynski Snyder AL, Powell NJ, et al. Metabolite quantification and high-field MRS in breast cancer. NMR Biomed. 2009;22:65–76. doi: 10.1002/nbm.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howe FA, Barton SJ, Cudlip SA, Stubbs M, Saunders DE, Murphy M, et al. Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 2003;49:223–232. doi: 10.1002/mrm.10367. [DOI] [PubMed] [Google Scholar]
- 12.Serkova NJ, Brown MS. Quantitative analysis in magnetic resonance spectroscopy: from metabolic profiling to in vivo biomarkers. Bioanalysis. 2012;4:321–341. doi: 10.4155/bio.11.320. [DOI] [PubMed] [Google Scholar]
- 13.Nelson MT, Everson LI, Garwood M, Emory T, Bolan PJ. MR Spectroscopy in the diagnosis and treatment of breast cancer. Semin Breast Dis. 2008;11:100–105. doi: 10.1053/j.sembd.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): what is in a name and what isn't? Magn Reson Med. 2011;65:927–948. doi: 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherry AD, Woods M. Chemical exchange saturation transfer contrast agents for magnetic resonance imaging. Annu Rev Biomed Eng. 2008;10:391–411. doi: 10.1146/annurev.bioeng.9.060906.151929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143:79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 17.Terreno E, Castelli DD, Aime S. Encoding the frequency dependence in MRI contrast media: the emerging class of CEST agents. Contrast Media Mol Imaging. 2010;5:78–98. doi: 10.1002/cmmi.369. [DOI] [PubMed] [Google Scholar]
- 18.Liu G, Song X, Chan KW, McMahon MT. Nuts and bolts of chemical exchange saturation transfer MRI. NMR Biomed. 2013;26:810–828. doi: 10.1002/nbm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan KW, McMahon MT, Kato Y, Liu G, Bulte JW, Bhujwalla ZM, et al. Natural D-glucose as a biodegradable MRI contrast agent for detecting cancer. Magn Reson Med. 2012;68:1764–1773. doi: 10.1002/mrm.24520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haris M, Nanga RP, Singh A, Cai K, Kogan F, Hariharan H, et al. Exchange rates of creatine kinase metabolites: feasibility of imaging creatine by chemical exchange saturation transfer MRI. NMR Biomed. 2012;25:1305–1309. doi: 10.1002/nbm.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker-Samuel S, Ramasawmy R, Torrealdea F, Rega M, Rajkumar V, Johnson SP, et al. In vivo imaging of glucose uptake and metabolism in tumors. Nat Med. 2013;19:1067–1072. doi: 10.1038/nm.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou JY, Payen JF, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003;9:1085–1090. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, Tryggestad E, Wen Z, Lal B, Zhou T, Grossman R, et al. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nature Medicine. 2011;17:130–134. doi: 10.1038/nm.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu G, Gilad AA, Bulte JW, van Zijl PC, McMahon MT. High-throughput screening of chemical exchange saturation transfer MR contrast agents. Contrast Media Mol Imaging. 2010;5:162–170. doi: 10.1002/cmmi.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer research. 2004;64:4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 26.Robert J, Gianni L. Pharmacokinetics and metabolism of anthracyclines. Cancer surveys. 1993;17:219–252. [PubMed] [Google Scholar]
- 27.Rizwan A, Cheng M, Bhujwalla ZM, Krishnamachary B, Jiang L, Glunde K. Breast cancer cell adhesome and degradome interact to drive metastasis. Npj Breast Cancer. 2015;1:15017. doi: 10.1038/npjbcancer.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizwan A, Bulte C, Kalaichelvan A, Cheng M, Krishnamachary B, Bhujwalla ZM, et al. Metastatic breast cancer cells in lymph nodes increase nodal collagen density. Scientific reports. 2015;5:10002. doi: 10.1038/srep10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yarden RI, Lauber AH, El-Ashry D, Chrysogelos SA. Bimodal regulation of epidermal growth factor receptor by estrogen in breast cancer cells. Endocrinology. 1996;137:2739–2747. doi: 10.1210/endo.137.7.8770893. [DOI] [PubMed] [Google Scholar]
- 30.Rizwan A, Serganova I, Khanin R, Karabeber H, Ni X, Thakur SB, et al. Relationships between LDH-A, Lactate and Metastases in 4T1 Breast Tumors. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, Moake M, Har-el YE, Long CM, Chan KW, Cardona A, et al. In vivo multicolor molecular MR imaging using diamagnetic chemical exchange saturation transfer liposomes. Magn Reson Med. 2012;67:1106–1113. doi: 10.1002/mrm.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan KW, Yu T, Qiao Y, Liu Q, Yang M, Patel H, et al. A diaCEST MRI approach for monitoring liposomal accumulation in tumors. Journal of controlled release : official journal of the Controlled Release Society. 2014;180:51–59. doi: 10.1016/j.jconrel.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim M, Gillen J, Landman BA, Zhou J, van Zijl PC. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med. 2009;61:1441–1450. doi: 10.1002/mrm.21873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahon MT, Gilad AA, Zhou J, Sun PZ, Bulte JW, van Zijl PC. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): Ph calibration for poly-L-lysine and a starburst dendrimer. Magn Reson Med. 2006;55:836–847. doi: 10.1002/mrm.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tassone P, Tagliaferri P, Perricelli A, Blotta S, Quaresima B, Martelli ML, et al. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. British journal of cancer. 2003;88:1285–1291. doi: 10.1038/sj.bjc.6600859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai K, Xu HN, Singh A, Moon L, Haris M, Reddy R, et al. Breast cancer redox heterogeneity detectable with chemical exchange saturation transfer (CEST) MRI. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2014;16:670–679. doi: 10.1007/s11307-014-0739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yadav NN, Bar-Shir A, Jones CK, Lin CY, Hua J, Gilad AA, et al. Exchange rate filtering of CEST agents using frequency-labeled exchange (FLEX) transfer. Melbourne, Australia: International Society of Magnetic Resonance in Medicine; 2012. p. 4189. [Google Scholar]
- 38.Zu Z, Janve VA, Li K, Does MD, Gore JC, Gochberg DF. Multi-angle ratiometric approach to measure chemical exchange in amide proton transfer imaging. Magnetic Resonance in Medicine. 2012;68:711–719. doi: 10.1002/mrm.23276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Yadav NN, Bar-Shir A, Jones CK, Chan KWY, Zhang J, et al. Variable delay multi-pulse train for fast chemical exchange saturation transfer and relayed-nuclear overhauser enhancement MRI. Magnetic Resonance in Medicine. 2014;71:1798–1812. doi: 10.1002/mrm.24850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaiss M, Schmitt B, Bachert P. Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of z-spectra. J Magn Reson. 2011;211:149–155. doi: 10.1016/j.jmr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Sheth VR, Li Y, Chen LQ, Howison CM, Flask CA, Pagel MD. Measuring in vivo tumor pHe with CEST-FISP MRI. Magnetic Resonance in Medicine. 2012;67:760–768. doi: 10.1002/mrm.23038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagiyama K, Mashimo T, Togao O, Vemireddy V, Hatanpaa KJ, Maher EA, et al. In vivo chemical exchange saturation transfer imaging allows early detection of a therapeutic response in glioblastoma. Proceedings of the National Academy of Sciences. 2014;111:4542–4547. doi: 10.1073/pnas.1323855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hectors SJCG, Jacobs I, Strijkers GJ, Nicolay K. Amide proton transfer imaging of high intensity focused ultrasound-treated tumor tissue. Magnetic Resonance in Medicine. 2013 doi: 10.1002/mrm.25000. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 44.Dula AN, Arlinghaus LR, Dortch RD, Dewey BE, Whisenant JG, Ayers GD, et al. Amide proton transfer imaging of the breast at 3 T: establishing reproducibility and possible feasibility assessing chemotherapy response. Magn Reson Med. 2013;70:216–224. doi: 10.1002/mrm.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanokura M, Tsukada K. Decreased activities of glycine and guanidinoacetate methyltransferases and increased levels of creatine in tumor cells. Biochem Biophys Res Commun. 1982;104:1464–1469. doi: 10.1016/0006-291x(82)91415-2. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Wang L, Zhang H, Deng P, Chen J, Zhou B, et al. (1)H NMR-based metabolic profiling of human rectal cancer tissue. Mol Cancer. 2013;12:121. doi: 10.1186/1476-4598-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei L, Hong S, Yoon Y, Hwang SN, Park JC, Zhang Z, et al. Early prediction of response to Vorinostat in an orthotopic rat glioma model. NMR Biomed. 2012;25:1104–1111. doi: 10.1002/nbm.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi JS, Baek HM, Kim S, Kim MJ, Youk JH, Moon HJ, et al. HR-MAS MR spectroscopy of breast cancer tissue obtained with core needle biopsy: correlation with prognostic factors. PLoS One. 2012;7:e51712. doi: 10.1371/journal.pone.0051712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Crowley P, et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl) 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- 50.Haris M, Cai K, Singh A, Hariharan H, Reddy R. In vivo mapping of brain myo-inositol. Neuroimage. 2011;54:2079–2085. doi: 10.1016/j.neuroimage.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sitter B, Bathen TF, Singstad TE, Fjosne HE, Lundgren S, Halgunset J, et al. Quantification of metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR Biomed. 2010;23:424–431. doi: 10.1002/nbm.1478. [DOI] [PubMed] [Google Scholar]
- 52.Serganova I, Rizwan A, Ni X, Thakur SB, Vider J, Russell J, et al. Metabolic imaging: a link between lactate dehydrogenase A, lactate, and tumor phenotype. Clin Cancer Res. 2011;17:6250–6261. doi: 10.1158/1078-0432.CCR-11-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terunuma A, Putluri N, Mishra P, Mathe EA, Dorsey TH, Yi M, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124:398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Twelves CJ, Porter DA, Lowry M, Dobbs NA, Graves PE, Smith MA, et al. Phosphorus-31 metabolism of post-menopausal breast cancer studied in vivo by magnetic resonance spectroscopy. Br J Cancer. 1994;69:1151–1156. doi: 10.1038/bjc.1994.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morse DL, Raghunand N, Sadarangani P, Murthi S, Job C, Day S, et al. Response of choline metabolites to docetaxel therapy is quantified in vivo by localized (31)P MRS of human breast cancer xenografts and in vitro by high-resolution (31)P NMR spectroscopy of cell extracts. Magn Reson Med. 2007;58:270–280. doi: 10.1002/mrm.21333. [DOI] [PubMed] [Google Scholar]
- 57.Glunde K, Ackerstaff E, Natarajan K, Artemov D, Bhujwalla ZM. Real-time changes in 1H and 31P NMR spectra of malignant human mammary epithelial cells during treatment with the anti-inflammatory agent indomethacin. Magn Reson Med. 2002;48:819–825. doi: 10.1002/mrm.10295. [DOI] [PubMed] [Google Scholar]
- 58.Brandes AH, Ward CS, Ronen SM. 17-allyamino-17-demethoxygeldanamycin treatment results in a magnetic resonance spectroscopy-detectable elevation in choline-containing metabolites associated with increased expression of choline transporter SLC44A1 and phospholipase A2. Breast Cancer Res. 2010;12:R84. doi: 10.1186/bcr2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HR-MRS and CEST-MRI of cell extracts from 48 hours doxorubicin treated MCF-7 and MDA-MB-231 breast cancer cells compared to the respective vehicle controls. (A) HR-MRS quantification (n=3 each) shown as difference between doxorubicin treated cell extracts minus untreated vehicle controls (dotted horizontal line at 0) in fmol per cell following treatment with 5 µM doxorubicin for 48 hours in MCF-7 (gray bars) and MDA-MB-231 (white bars) cells. (B) CEST-MRI of MDA-MB-231 cell extracts showing doxorubicin treated cells (gray squares, n=3) versus vehicle controls (black circles, n=3). (C) CEST-MRI quantified as adjusted MTRasym, which is defined as MTRasym per cell of treated cells minus vehicle controls (n=3 each), in MCF-7 (blue triangles) and MDA-MB-231 (red squares) cells. Values are mean ± standard error. * p< 0.05, treated versus control.