Abstract
Changes in polysome-bound mRNA (translatome) are correlated closely with changes in the proteome in cells. Therefore, to better understand the processes mediating the response of glioblastoma (GBM) to ionizing radiation (IR), we used polysome profiling to define the IR-induced translatomes of a set of human glioblastoma stem-like cell (GSC) lines. Whereas cell line specificity accounted for the largest proportion of genes within each translatome, there were also genes that were common to the GSC lines. In particular, analyses of the IR-induced common translatome identified components of the DNA damage response, consistent with a role for the translational control of gene expression in cellular radioresponse. Moreover, translatome analyses suggested that IR enhanced cap-dependent translation processes, an effect corroborated by the finding of increased eIF4F-cap complex formation detected after irradiation in all GSC lines. Translatome analyses also predicted that Golgi function was affected by IR. Accordingly, Golgi dispersal was detected after irradiation of each of the GSC lines. In addition to the common responses seen, translatome analyses predicted cell line-specific changes in mitochondria, as substantiated by changes in mitochondrial mass and DNA content. Together, these results suggest that analysis of radiation-induced translatomes can provide new molecular insights concerning the radiation response of cancer cells. More specifically, they suggest that the translational control of gene expression may provide a source of molecular targets for GBM radiosensitization.
Keywords: radiation, gene expression, translational control, glioblastoma stem-like cells, polysomes
Introduction
Investigations into the molecular processes mediating the cellular response to ionizing radiation (IR) have, for the most part, focused on the post-translational modification of existing proteins. These studies have led to a detailed understanding of signaling pathways involved in such fundamental components of radioresponse as DNA repair, cell cycle checkpoint activation and apoptosis. As an additional regulatory process, modifications in gene expression have long been thought to contribute to cellular radioresponse. This premise was initially based on data from prokaryotes in which radiation-induced gene expression provides an adaptive or protective response against radiation-induced death. In eukaryotic cells IR has been shown to induce transcription of specific genes and at the whole genome level modify the cellular transcriptome (1, 2). Such results are reminiscent of the prokaryotic adaptive response and thus suggest that defining the inducible genes would not only provide information regarding the mechanisms determining radioresponse of mammalian cells but also identify molecular targets for modifying radiosensitivity.
Arguing against this scenario is the poor correlation between IR-induced changes in mRNAs and their corresponding proteins (3). It is this uncoupling of the transcriptome and proteome after irradiation that calls into question the functional significance of IR-induced gene expression. However, eukaryotic gene expression, in contrast to prokaryotes, is regulated not only through transcription but through a series of post-transcriptional events, a process referred to as translational control, a critical determinant of gene expression. Under a variety of biological and experimental conditions (4–8), translational control has been shown to account for the discrepancies between the transcriptome and proteome. An approach that bypasses post-transcriptional processes and identifies genes undergoing translation, i.e. the translatome, is the microarray analysis of polysome-bound RNA (9, 10). We initially applied this technique to long established glioma cell lines 6h after exposure to 7Gy and included a comparison to the traditional microarray analysis of total cellular RNA (11). The data generated showed that the number of genes whose translational activity was modified by IR was greater than those affected in the transcriptome analysis. Moreover, there were few, if any genes affected in both the transcriptome and translatome, indicating that the processes are not coordinated with each proceeding through different mechanisms. Of significance and in contrast to changes in the transcriptome, there was a correlation between the genes whose translational activity was affected by IR and the expression of the corresponding proteins. The implication of this study was that radiation does modify gene expression, but does so primarily via translational control.
The correlation between changes in polysome-bound mRNA and the corresponding protein suggests that defining the translatome in irradiated cells will generate unique insight into the processes comprising cellular radioresponse and, for tumor cells may suggest strategies for enhancing radiosensitivity. Towards this end, we have extended our initial investigation to glioblastoma stem-like cell (GSC) lines. Whereas the biology of long established glioma cell lines has little in common with glioblastomas in situ, GSCs are thought to be a clonogenic subpopulation critical to the development, maintenance and treatment response of glioblastomas (12–14). The data presented here defines the IR-induced translatomes for 3 GSC lines. Subsequent translatome analyses identified both common and line specific consequences of GSC irradiation, which were then validated at the functional level. These results further implicate translational control of gene expression as a fundamental component of cellular radioresponse.
Materials and Methods
Cell lines and treatments
Studies were performed using 3 neurosphere-forming cultures isolated from human GBM surgical specimens as described (15) and maintained as frozen stocks: NSC11 (kindly provided by Dr. Frederick Lang, MD Anderson Cancer Center in 2008), 0923 (16) was obtained from the Neuro-Oncology Branch, NCI in 2013, and GBMJ1 (17) was generated at Moffitt Cancer Center in 2008. Cell lines were revived every 2 months from frozen stocks made after receiving cell lines and were recently authenticated in May, 2015 by STR analysis (Idexx Laboratories, Columbia, MO). Neurospheres were maintained in stem cell medium consisting of DMEM/F12 (Invitrogen), B27 supplement (Invitrogen), and human recombinant bFGF and EGF (50 ng/ml each, R&D Systems) at 37°C, 5%CO2/5%O2. CD133+ cells (NSC11 and GBMJ1) (17, 18) or CD15+ cells (0923) (19, 20) were isolated from each neurosphere cultures by FACS and used as a source for the described experiments (17). The CD133+ and CD15+ cell cultures met the criteria for tumor stem-like cells including self-renewal, differentiation along glial and neuronal pathways, expression of stem cell related genes, and formation of brain tumors when implanted in immunodeficient mice. For use in an in vitro experiment, CD133+ or CD15+ neurosphere cultures were disaggregated into single cells as described (17) and seeded onto poly-L-ornithine (Invitrogen)/laminin (Sigma) coated tissue culture dishes in stem cell media. Under these conditions, single-cell GSCs attach and proliferate maintaining their CD133+ or CD15+ expression and stem-like characteristics (21). Radiation was delivered using a 320 kV X-ray machine (Precision XRay Inc.) at a dose rate of 2.3 Gy/min; control cultures were mock irradiated. Cell cultures were treated with 4μM INK128 (Chemietek) dissolved in dimethyl sulfoxide (DMSO) or vehicle control immediately after irradiation.
Polysome isolation and microarray analysis
Isolation of polysome-bound RNA was performed in triplicate as described by Galban et al (22) with slight modifications. Briefly, cells were grown to ~80% confluency in 150-mm2 culture dishes and irradiated or mock irradiated. At time of collection, cells were incubated with 100 μg/ml of cycloheximide for 15 minutes at 37°C, 5%CO2/5%O2. Cytoplasmic RNA was collected by lysing cells in polysome buffer [15 mmol/L Tris-HCl (pH 7.5), 300 mmol/L NaCl, 15 mmol/L MgCl2, 1% Triton X100, 100 μg/mL cycloheximide, 1 mg/mL heparin, and 500 units/mL RNasin (Promega)]. After 15 minutes on ice, lysates were centrifuged (12,000 × g for 15 minutes), and the resulting cytosolic supernatant was layered onto a 10% to 50% sucrose gradient. Gradients were then centrifuged at 35,000 × g for 3 hours at 4°C and polysome-bound fractions were collected using an ISCO Density Gradient Fractionation System (ISCO, Lincoln, NE) with continuous monitoring based on A254. The RNA in each fraction was extracted using TRIzol LS (Invitrogen). The integrity of the RNA was assured using a Bioanalyzer (Agilent). The isolated RNA was amplified with GeneChip 3′ IVT Express Kit (Affymetrix) and hybridized to GeneChip Human Genome U133A 2.0 Array chips (Affymetrix) per manufacturer’s protocol. Using Affymetrix Expression Console, MAS5 normalization was performed and the means of probeset intensities that were modified by at least ± 1.5 fold with a Student’s two tailed t-test value of p < 0.05 (vs. control) were identified and submitted to Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity) for core analysis and Gene Set Enrichment Analysis (GSEA) (23, 24) using the MSigDB gene set C5 (GO gene sets). Heatmaps were created using R version 3.0.1. Microarray data has been deposited in NCBI’s Gene Expression Omnibus (25) and are accessible through GEO Series accession number GSE74084 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74084).
Total RNA Isolation
Total RNA, also in biological triplicates, was extracted from cells using TRIzol (Invitrogen) followed by the RNeasy Mini Kit (Qiagen). The integrity of the RNA was assured using a Bioanalyzer (Agilent).
Cap-binding assay
eIF4F cap complex formation was measured with m7-GTP batch chromatography as described (26, 27). Briefly, cells were lysed in 20 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L β-glycerophosphate, 1 mmol/L sodium orthovanadate, 1% Triton X-100, 0.2 mmol/L PMSF, 1× phosphatase inhibitor cocktails II and III (Sigma-Aldrich), and 1× HALT protease inhibitor cocktail (Thermo Scientific) for 15 minutes on ice. Lysate (400 μg) was incubated with 200μL of Immobilized γ-Aminophenyl-m7-GTP Agarose (Jena Bioscience) overnight at 4°C. Beads were washed 3 times with lysis buffer; bound protein was eluted, denatured, and then separated with SDS-PAGE followed by immunoblotting for eIF4G (Santa Cruz), 4E-BP1, and eIF4E (Cell Signaling).
Immunofluorescent analysis of γH2AX foci
To visualize foci, cells grown in chamber slides were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 1% bovine serum albumin in PBS containing 5% goat serum. The slides were incubated with antibody to phospho-H2AX (Millipore) followed by incubation with goat-anti-mouse-Alexa488 (Invitrogen) and mounted with Prolong gold antifade reagent containing DAPI (Invitrogen) to visualize nuclei. Cells were analyzed on a Zeiss upright fluorescent microscope.
Measurement of Golgi area
Immunofluorescent images of cells stained with a conjugated antibody to the cis-Golgi marker GM130 (BD Biosciences) and ProLong Gold antifade reagent with DAPI (Life Technologies) and were measured by manual demarcation of the Golgi with a limiting polygon and calculation of its area using ImageJ.
Mitochondrial analyses
Mitochondrial mass was determined using MitoTracker Green FM (Invitrogen). Cells attached to poly-L-ornithine/laminin coated plates were stained with 80nM MitoTracker Green FM in stem cell medium for 15 minutes at 37°C, 5%CO2/5%O2. Cells were then trypsinized, rinsed with DPBS(−), resuspended in DPBS(−) and analyzed using a FACSCalibur flow cytometer. Mitochondrial DNA (mtDNA) was extracted using DNeasy Blood and Tissue kit (Qiagen) and quantified using qPCR using 7500 Real Time PCR System (Applied Biosystems) and RT2 SYBR Green ROX qPCR Mastermix (Qiagen). To evaluate mtDNA content, relative amounts of mtDNA-encoded cytochrome c oxidase subunit II (qHsaCED0048349, Bio-Rad) were determined by the ΔΔCt method using nuclear DNA-encoded β-globin (qHsaCED0048812, Bio-Rad) as an internal control.
Statistical analysis
Statistical significance was determined using Student’s t-test using Excel. Values of P<0.05 were considered significant. Data are expressed as means ± standard error of the mean.
Results
IR-induced translatomes
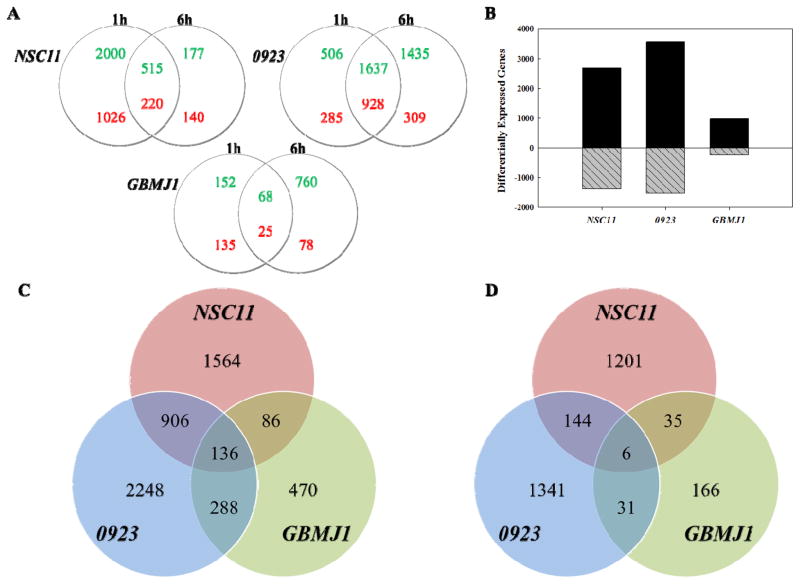
To investigate radiation-induced translational control in GSCs, polysome-bound mRNA was isolated by sucrose gradient fractionation and subjected to DNA microarray based gene expression analysis (11). Specifically, polysome-bound RNA was collected 1 and 6h after irradiation (2Gy) of 3 GSCs (NSC11, 0923 and GBMJ1) with gene expression levels compared to their respective, unirradiated controls. The number of genes whose polysome-association was increased or decreased (p-value ≤ 0.05, Fold Change ≥ ±1.5) at 1 or 6h after irradiation relative to controls is shown in Figure 1A. Previous studies of the IR-induced translatome involved evaluation of only the 6h time (11, 28). In NSC11 cells, more genes were affected at 1h after irradiation than at 6h. As indicated by the overlap in the NSC11 Venn diagram, most of the gene changes at 6h were also present at 1h after irradiation, suggesting that in these cells the processes mediating radiation-induced translational control are operative primarily within the first hour after exposure. However, in GBMJ1 cells more genes were affected at 6h than at 1h; in 0923 cells, whereas there was a substantial overlap in the gene changes detected at both time points, there were more genes unique to 6h than 1h post-irradiation.
Figure 1.
IR-induced translatomes. (A) Venn diagrams comparing genes identified by microarray analysis of polysome-bound RNA to be up- (green) or down- (red) regulated in each GSC line at 1 and 6h after exposure to 2Gy. (B) Combined number of genes whose polysome binding was modified at 1 and 6h after exposure to 2Gy (black bars: increased; grey hatched bars: decreased). Venn diagrams comparing the 3 GSC lines in terms of (C) up-regulated and (D) down-regulated genes in their respective IR-induced translatomes. p < 0.05, fold change of at least ±1.5
Given the disparate time courses among GSCs, we defined the radiation-induced translatome of each line by combining the gene lists obtained at 1 and 6h post-irradiation (Figure 1B). The number of genes comprising the radiation-induced translatome varied among the GSC lines with 0923 cells being the most susceptible to radiation-induced translation control and GBMJ1 the least. A direct comparison of GSC lines in terms of genes comprising the radiation-induced translatome is shown in Figures 1C and D. Whereas cell line specificity accounted for the largest proportion of genes within each translatome, there were also genes in common among the GSC lines. For genes whose polysome binding was increased after irradiation there were 136 in common between all 3 lines and 1416 increased in at least 2 lines (Figure 1C and Supplemental Table S1); for the genes whose polysome binding was decreased in response to radiation there were 6 in common between all 3 lines and 216 decreased in 2 of the 3 lines (Figure 1D and Supplemental Table S2).
Functional Analysis of the IR-induced translatome
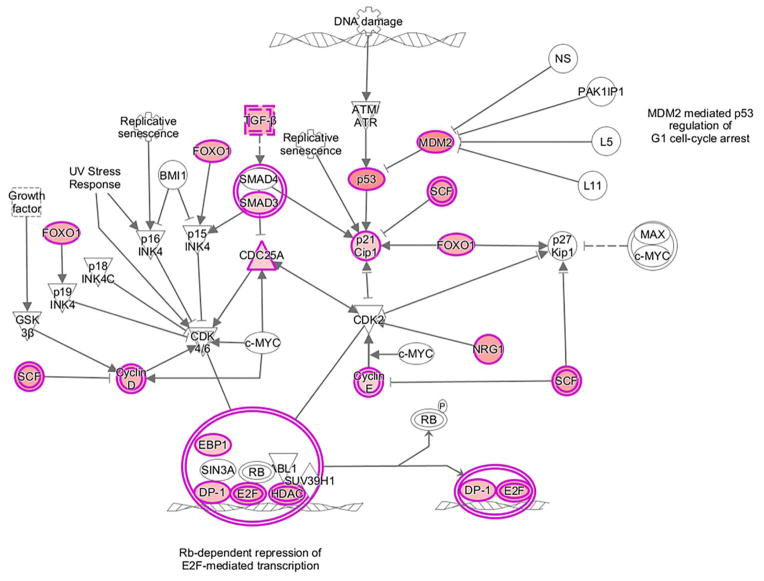
IR-induced gene expression has traditionally been evaluated using total cellular mRNA, which provides a measure of the cellular transcriptome. Consistent with our previous report using established cell lines (11), comparison of the IR-induced translatomes and transcriptomes from GSCs revealed few commonly affected genes (Supplemental Figure S1), suggesting that novel information pertaining to cellular radioresponse can be obtained from analysis of their IR-induced translatomes. Along these lines, the functional significance of IR-induced translational control was initially investigated using IPA, which assigns genes into networks and then associates the networks with functions and pathways. For this analysis, we used the common translatome defined as those transcripts whose polysome binding were enhanced after irradiation in at least 2 of the 3 GSC lines (1416 genes). Of the functions assigned to the top twenty networks enriched by IR (Table 1), 6 contained “DNA Replication, Recombination and Repair”, critical processes in cellular radioresponse. Among the canonical pathways upregulated by radiation was “Cell Cycle: G1/S Checkpoint Regulation” (p-value=1.96×10−03) (Figure 2), which is also an established component of cellular radioresponse. Of note, none of the 21 genes shown as induced in the common translatome in Figure 2 were also present in the common radiation-induced transcriptome of the GSCs. For the genes whose translation was down regulated after GSC irradiation, IPA revealed no obvious relationships to radioresponse (Supplemental Table S3). This initial analysis is thus consistent with translational control playing a role in the events mediating the cellular response to IR.
Table 1.
Top networks (IPA) enriched in the IR-induced common translatome, DNA Replication, Recombination and Repair is highlighted (bold).
| Score | Focus Molecules | Top Networks |
|---|---|---|
| 41 | 32 | Cell-To-Cell Signaling and Interaction, Cellular Assembly and Organization, Cellular Development |
| 41 | 32 | RNA Post-Transcriptional Modification, Dermatological Diseases and Conditions, Developmental Disorder |
| 41 | 32 | RNA Post-Transcriptional Modification, Cellular Assembly and Organization, Infectious Disease |
| 36 | 30 | Lipid Metabolism, Small Molecule Biochemistry, Carbohydrate Metabolism |
| 36 | 30 | Hereditary Disorder, Skeletal and Muscular Disorders, Dermatological Diseases and Conditions |
| 36 | 30 | Cellular Assembly and Organization, Carbohydrate Metabolism, Lipid Metabolism |
| 36 | 30 | DNA Replication, Recombination, and Repair, Cellular Compromise, Cell Cycle |
| 36 | 30 | Gene Expression, DNA Replication, Recombination, and Repair, Connective Tissue Development and Function |
| 34 | 29 | Post-Translational Modification, Developmental Disorder, Endocrine System Disorders |
| 34 | 29 | Cell Cycle, Cellular Assembly and Organization, Cellular Function and Maintenance |
| 32 | 28 | Post-Translational Modification, Protein Synthesis, Carbohydrate Metabolism |
| 32 | 28 | Cell Morphology, Cellular Assembly and Organization, RNA Damage and Repair |
| 32 | 28 | DNA Replication, Recombination, and Repair, Cell Signaling, Cell Cycle |
| 32 | 28 | Cell Morphology, Cell Cycle, Cellular Development |
| 30 | 27 | Gene Expression, Protein Synthesis, DNA Replication, Recombination, and Repair |
| 30 | 27 | Endocrine System Development and Function, Molecular Transport, Small Molecule Biochemistry |
| 30 | 27 | Organismal Development, Connective Tissue Disorders, Developmental Disorder |
| 29 | 27 | Cell-To-Cell Signaling and Interaction, Cellular Growth and Proliferation, Hematological System Development and Function |
| 29 | 27 | Cellular Assembly and Organization, DNA Replication, Recombination, and Repair, Connective Tissue Development and Function |
| 29 | 28 | Cell Cycle, DNA Replication, Recombination, and Repair, Cellular Assembly and Organization |
Figure 2.
IPA canonical pathway Cell Cycle: G1/S Checkpoint Regulation with up-regulated genes in the IR-induced common translatome highlighted in pink.
Cap-dependent translation
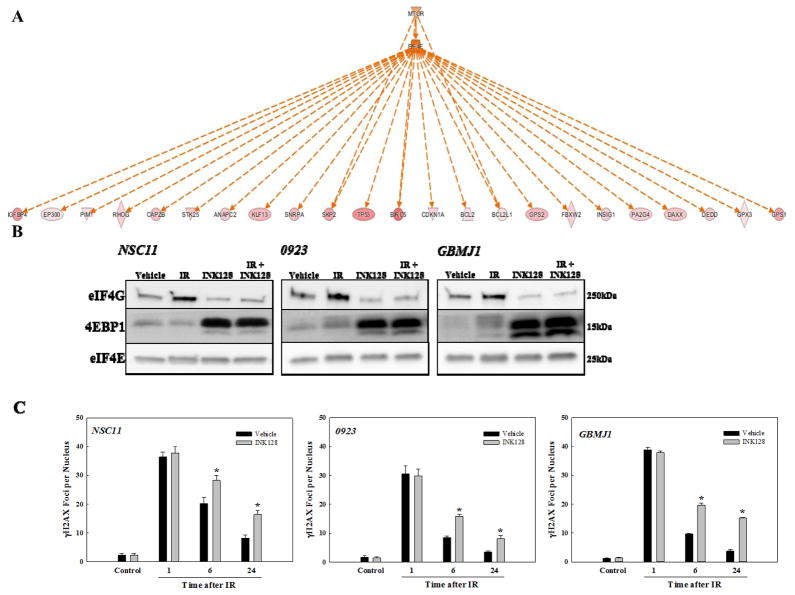
In addition to DNA repair and cell cycle checkpoint regulation, further interrogation of the radiation-induced GSC translatome suggested the activation of other cell processes not traditionally associated with radioresponse. For example, upstream signaling analysis (IPA) linked a set of 23 genes in the common IR-induced GSC translatome (Figure 3A, red symbols) to the activation of eIF4E and mTOR (Figure 3A, orange symbols), implying that cap-dependent translation is increased after GSC irradiation. Essential to cap-dependent translation is the interaction of eIF4E with eIF4G resulting in the formation of the eIF4F- 5′ mRNA cap complex (29). Thus, to test the accuracy of this prediction, we determined the effects of radiation on eIF4F-cap complex formation using m7-GTP batch chromatography to pull down the bound eIF4F complex, which was then subjected to immunoblotting for eIF4E, 4EBP1 and eIF4G. As shown in Figure 3B, treatment of GSCs with the ATP-competitive mTOR inhibitor INK128 reduced cap complex formation as indicated by the decrease in eIF4G and increase in 4EBP1 pulled down with eIF4E, consistent with previous studies (30, 31). In contrast, IR increased the amount of eIF4G pulled down, signifying an increase in cap complex formation in each of the GSC lines. The IR-induced increase was prevented, however, when INK128 was added immediately after irradiation suggesting that radiation-induced cap complex formation requires mTOR activity.
Figure 3.
(A) IPA upstream signaling analysis. Genes up-regulated in the common translatome are in pink (intensity of color is descriptive of the level of up-regulation). Orange indicates predicted activation. Solid and dotted lines indicate direct and indirect relationships, respectively. (B) eIF4F-cap complex formation. m7-GTP affinity chromatography was performed on each GSC at 1h after irradiation (5Gy). Cells were treated with INK128 (4μM) or vehicle control immediately after IR. m7-GTP bound proteins were resolved via SDS-PAGE followed by immunoblot analysis. Immunoblots are representative of 3 independent experiments. (C) Influence of INK128 on radiation-induced γH2AX foci. Cells were irradiated (2Gy), treated immediately after with INK128 (4μM) or vehicle, and collected at the specified times. Foci were counted for 25 nuclei per condition per experiment. Values are means ± SEM for 3 independent experiments. *p < 0.05 according to Student’s t-test (INK128 vs. vehicle).
To further investigate the potential link between mTOR-mediated cap-dependent translation and GSC radioresponse, the effect of INK128 on radiation-induced γH2AX foci induction and dispersal were determined (Figure 3C). The critical lesion responsible for radiation-induced cell death is the DNA double strand break (DSB); γH2AX foci induction corresponds to the induction DSBs and their dispersal correlates with their repair (32, 33). In this study, GSCs were exposed to 2Gy and either INK128 or vehicle (Control) was added immediately after irradiation with nuclear foci determined at time points out to 24 hours. For all three GSCs, no difference between INK128- and vehicle-treated cells was detected at 1h after irradiation, suggesting that INK128 had no effect on the initial level of IR-induced DSBs. However, at 6 and 24 hours after irradiation, the number of γH2AX foci remaining in the INK128-treated cells was significantly greater than in the control cells, consistent with an inhibition of DNA DSB repair. Consistent with the γH2AX foci results and a previous study using the mTOR inhibitor AZD2014 (34), addition of INK128 immediately after irradiation enhanced the radiosensitivity of each of the GSC lines (Supplemental Figure S2). Thus, although mTOR has other activities (35), given that the IR-induced GSC translatome contained genes associated with the DNA repair (Figure 2), the results in Figures 3B and C implicate cap-dependent translation as a mediator of GSC radioresponse.
Golgi Morphology
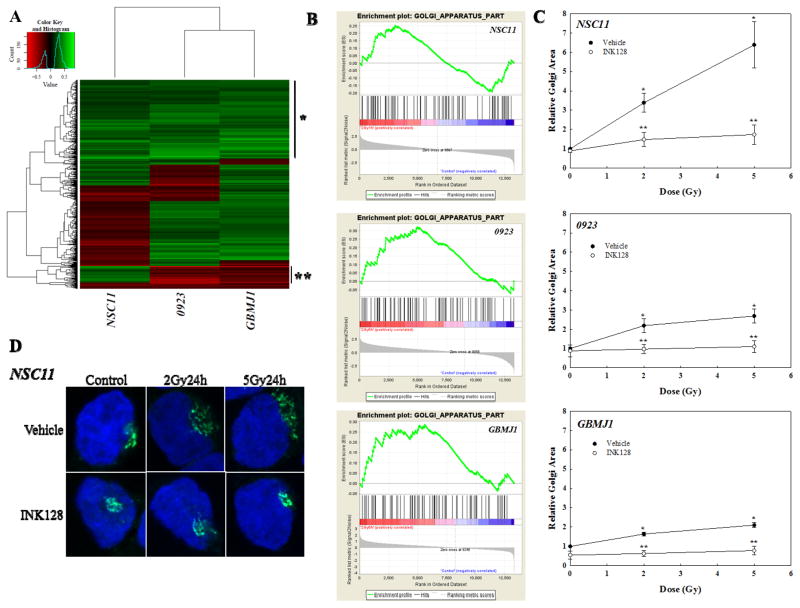
GSEA was used as an additional approach to evaluate the radiation-induced translatomes. Hierarchical clustering of normalized enrichment scores (NES) of GO terms illustrates the similarities and differences between GSC lines (Figure 4A). The top portion of the heatmap (demarcated by *) indicates the GO terms in which the radiation-induced translatomes of the 3 GSCs were most similar. Among the GO terms in this region were 6 of the 7 associated with the Golgi apparatus; enrichment plots for the specific GO term Golgi_Apparatus_Part show a similar pattern of enrichment across GSC lines (Figure 4B). Because this analysis predicts that radiation-induced translational control influences Golgi in the GSCs we determined the effects of radiation on Golgi morphology as previously described (36). For this assay GSCs were irradiated and 24h later subjected to immuno-fluorescent cytochemical analysis for GM130, a highly expressed cis-Golgi protein, allowing for visualization of Golgi morphology. Farber-Katz et al recently showed that after irradiation of a variety of established cell lines Golgi morphology shifts from a perinuclear to a more fragmented, cytoplasmic distribution, which was referred to as Golgi dispersal and quantified as Golgi area per cell (36). As shown in Figure 4C and D, irradiation of GSCs resulted in a dose-dependent increase in Golgi dispersal with NSC11 being the most susceptible. Treatment of GSCs with INK128 immediately after irradiation essentially eliminated the IR-induced change in Golgi morphology. These data indicate that the radiation-induced Golgi dispersal in GSCs required mTOR activity implying a role for radiation-induced translational control and consistent with GSEA.
Figure 4.
IR-induced translatomes and Golgi function. (A) Unsupervised hierarchical cluster analysis of normalized enrichment scores of GO terms. *, area containing Golgi-related GO terms; **, area differentially enriched in mitochondrial GO terms. (B) GSEA enrichment plots for GO term Golgi Apparatus Part in in each GSC line (C) Golgi area per cell relative to vehicle control. Cells were irradiated, immediately treated with INK128 (4μM) or vehicle, and collected after 24h. For each condition, 20 cells per condition per experiment were evaluated; values represent the mean ± SEM for 3 independent experiments. *p<0.05 according to Student’s t-test (IR vs. 0Gy control). **P<0.05 according to Student’s t-test (INK128 vs. vehicle control). (D) Representative micrographs (63×) of NSC11 cells treated with IR followed by either INK128 (4μM) or vehicle immediately after IR. Cells were fixed and stained with an antibody to GM130 (cis-Golgi) and DAPI (DNA).
Mitochondria
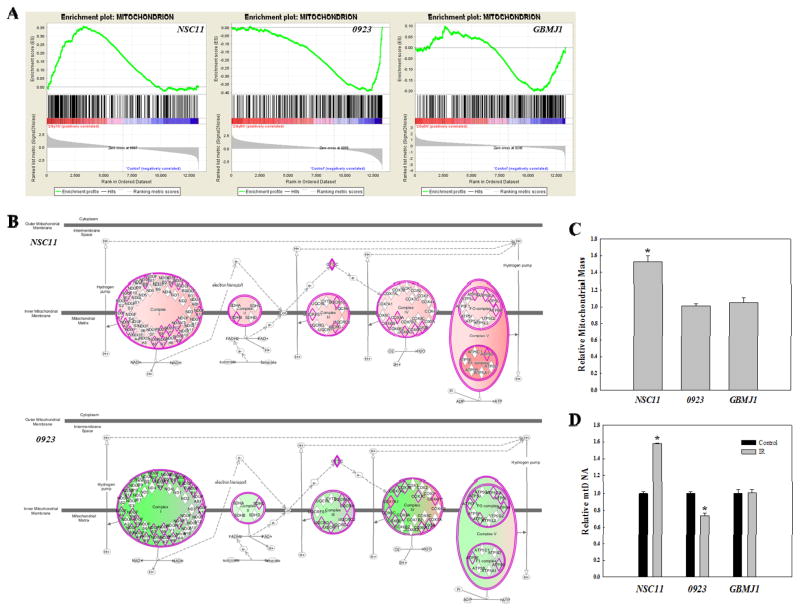
The hierarchical clustering of GO terms also suggested cell line specific consequences of radiation-induced translational control. Specifically, the region designated by ** on Figure 4A contains 10 of 13 GO terms associated with mitochondria, which were enriched in NSC11, diminished in 0923, and unchanged in GBMJ1. The normalized enrichment scores for all 13 mitochondrial GO terms (Supplemental Table S4) were positive for NSC11, negative for 0923 and a mixture of positive and negative for GBMJ1. Enrichment plots for Mitochondrion (Figure 5A), the mitochondria associated GO term encompassing the largest number of genes, illustrate the differences between the radiation-induced translatomes for each GSC line in terms of mitochondria. A cell line specific role for mitochondria was also suggested when radiation-induced translatomes from each of the GSCs were subjected to IPA. In contrast to Figures 2–3 evaluating commonly increased genes, this IPA included genes from each GSC that increased and decreased in response to radiation. The IPA canonical pathways “Mitochondrial Dysfunction” and “Oxidative Phosphorylation” were significantly enhanced in irradiated NSC11 cells with two of the three most significant p-values (4.34×10−07 and 3.66×10−06, respectively); a number of genes in these pathways were up-regulated as shown in Figure 5B (top). Both “Mitochondrial Dysfunction” and “Oxidative Phosphorylation” also appeared in IPA of irradiated 0923 cells (p-values of 8.44×10−10 and 1.84×10−6, respectively), however the associated genes were down-regulated (Figure 5B, bottom). Neither “Mitochondrial Dysfunction or “Oxidative Phosphorylation” appeared in IPA of irradiated GBMJ1 cells. Thus, both GSEA and IPA predicted that radiation enhances mitochondrial function in NSC11 cells, inhibits mitochondria in 0923 cells and has no effect on mitochondria in GBMJ1 cells.
Figure 5.
Cell line-dependent IR-induced changes in mitochondria. (A) GSEA enrichment plots for the GO term Mitochondrion in each GSC line. (B) IPA canonical pathway for “Oxidative Phosphorylation”. Proteins outlined in pink are those inputted to IPA from the IR-induced translatome of the either NSC11 (top) or 0923 (bottom). Pink shading indicates predicted activation and green shading indicates predicted repression, with the intensity describing the level of activation/repression. (C) Relative mitochondrial mass at 24h after IR (2Gy). (D) Relative mitochondrial DNA content 24h after IR (2Gy) was determined by qPCR using mitochondrial cytochrome c oxidase subunit II (MT-CO2) normalized to genomic DNA content. Data are expressed as means ± SEM for 3 independent experiments. *p < 0.05 according to Student’s t-test (IR vs. control).
To test this prediction, mitochondrial mass and mitochondrial DNA (mtDNA) content were quantified in the GSC lines (Figures 5C–D). IR induced an increase in both mitochondrial mass and mtDNA in NSC11, consistent with an increase in mitochondrial activity and thus the IR-induced translatome. No change in mitochondrial mass or mtDNA was detected after irradiation of GBMJ1, again consistent with translatome analysis. In 0923 cells, although mitochondrial mass was unchanged after irradiation, there was a significant decrease in mtDNA. In response to other types of injury, a decrease in mtDNA was associated with a shift to glycolytic energy production reflecting a reduction in mitochondrial function (37). Thus, as predicted by analysis of the radiation-induced translatomes, whether irradiation influences mitochondria and whether the influence was positive or negative was GSC line dependent. Supporting the validity of the microarray analysis of polysome-bound mRNA, the levels of proteins corresponding to representative genes within the IPA and GO networks described above (Figures 2–5) were increased after irradiation of NSC11 cells (Supplemental Figure S3).
Discussion
In the study reported here we used polysome profiling to define the radiation-induced translatome of 3 GSC lines. After exposure to a clinically relevant dose of 2Gy, each GSC line contained a substantially greater number of genes with increased as compared to decreased translational activity (i.e. polysome-binding). In the absence of apoptosis as the mode of radiation-induced GSC death (17), an increase in an energy consuming event such as gene translation suggests that it may contribute to GSC survival after irradiation, a situation consistent with the posttranscriptional operon model (38) in which functionally related genes are translated in a coordinated manner. To investigate the applicability of this model to GSC radioresponse, we used IPA and GSEA to predict potential cellular processes affected by the IR-induced changes in gene translation, which were then validated at the biochemical level. Thus, rather than determining the levels of corresponding proteins as an approach to validating microarray analysis of polysome bound RNA, given the multiple regulatory points within a network or process, this strategy was aimed at determining the functional consequences of the IR-induced GSC translatome.
If the changes in gene translation promote cell survival after irradiation, then networks and pathways associated with the DNA damage response would be expected to increase. As shown in figure 2, this was the case in analysis of the IR-induced GSC translatome. However, additional functions were detected in the translatome analyses that have not been typically associated with radioresponse. Along these lines, the IPA based identification of eIF4E activation suggested that IR enhances cap-dependent translation. Of note, knockdown of eIF4E was found to increase eIF4E binding to >1000 unique transcripts including many implicated in DNA replication, recombination and repair and to enhance the radiosensitivity of a pancreatic tumor cell line (26). Given that the majority of mRNAs in eukaryotic cells are translated in a cap-dependent manner, it would seem that this process would be required for modifications in the translatome. Previous studies showed that eIF4F-cap-complex formation after IR was reduced in a murine fibroblast cell line (39), increased in a normal human breast epithelial cell line (40) and unaffected in the human breast tumor cell line MDA-MB-231 (26). As shown here, IR increased cap-complex formation in each of the GSC lines indicative of increased cap-dependent translation, as predicted by translatome analysis. A critical regulator of cap-dependent translation is mTOR; its inhibition with INK128 reduced the repair of IR-induced DNA DSBs as expressed by the slower dispersal of H2AX foci and enhanced IR-induced cell killing, consistent with a determining role for cap-dependent translation in cellular radioresponse. However, as shown in figure 3B, INK128 not only prevented the IR-induced increase in cap-complex formation in GSCs but also reduced basal levels. Formation of the eIF4F-cap complex is a final and rate-limiting step in gene translation reflecting a culmination of the upstream regulatory events that comprise the post-transcriptional infrastructure. Thus, it is unclear at this point whether an increase in cap-binding or simply the maintenance of basal cap-binding activity is necessary for IR-induced translational control. Whereas the specific role of mTOR in the IR-induced translational control of gene expression in GSCs remains to be determined, the data presented suggest that it can serve as a target for GBM radiosensitization.
Analysis of the IR-induced translatome also suggested that the Golgi apparatus participates in the radioresponse of each of the 3 GSC lines. A recent study by Farber-Katz et al (36) reported that irradiation of a variety of cell lines results in Golgi dispersal, which they attributed to the DNA-PK mediated phosphorylation of GOLPH3, an oncogene that functions in the secretory pathway of the Golgi (41). It was also shown that knockdown of GOLPH3 enhanced and its overexpression reduced the sensitivity in HeLa cells to the DNA damaging agent doxorubicin, suggesting that Golgi dispersal contributes to cell survival. As shown here, IR induced Golgi dispersal in each of GSC lines, which was essentially eliminated by the mTOR inhibitor INK128. While the interaction between DNA-PK and GOLPH3 was not evaluated, these results suggest that cap-dependent translation is critical to the IR-induced Golgi dispersal in GSCs. The specific mechanism through which Golgi dispersal contributes to radioresponse remains to be determined. However, one possibility may involve modifications in protein processing and delivery necessitated by the IR-induced changes in gene translation.
In addition to fundamental components applicable to all GSCs, it is likely that their radioresponse also involves cell line specific aspects. Analysis of the IR-induced translatomes of the individual GSCs suggested that such cell line specificity applied to mitochondria. This would actually be consistent with the literature in that IR has been reported to increase (42, 43), and decrease (44, 45) mitochondrial function. The processes determining whether IR affects mitochondrial function have not been defined. However, the data presented here suggest that the type of mitochondrial response to IR is, at least in part, determined by translational control, as reflected by the IR translatomes of the 3 GSCs.
Because IR-induced polysome profiles cluster according to the tissue of origin (28), to investigate the significance of radiation-induced translational control of gene expression, the study described here focused on a set of 3 GSC lines. With respect to validating translational control of gene expression as defined by of a microarray analysis of polysome-bound mRNA, we previously showed a correlation with changes in corresponding proteins in established glioma cell lines (11), a correlation also shown in the present study (Supplemental Figure S3). However, whereas a necessary starting point, such measurements do not establish a functional role for translational control in cellular radioresponse. To determine whether the IR-induced modifications in the translatome are of biological consequence, we analyzed the affected genes in terms of cellular processes and pathways. While defining functional effects, this approach accounted for the ability of any number of proteins to influence a given process as well as the heterogeneity between GSC lines. The data presented indicate that IR-induced translational control plays a role in regulating a number of cellular processes that are likely to influence cell survival, which implicates the translational control of gene expression as a fundamental component of radioresponse and a potential source of targets for tumor radiosensitization.
Supplementary Material
Acknowledgments
Financial Support: Division of Basic Sciences, Intramural Program, National Cancer Institute (Z1ABC011372, Z1ABC011373)
Footnotes
Conflict of Interest: none
References
- 1.Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68(2):415–24. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 2.Short SC, Buffa FM, Bourne S, Koritzinsky M, Wouters BG, Bentzen SM. Dose- and time-dependent changes in gene expression in human glioma cells after low radiation doses. Radiat Res. 2007;168(2):199–208. doi: 10.1667/RR0940.1. [DOI] [PubMed] [Google Scholar]
- 3.Szkanderova S, Port M, Stulik J, Hernychova L, Kasalova I, Van Beuningen D, et al. Comparison of the abundance of 10 radiation-induced proteins with their differential gene expression in L929 cells. Int J Radiat Biol. 2003;79(8):623–33. doi: 10.1080/09553000310001606821. [DOI] [PubMed] [Google Scholar]
- 4.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6(5):1099–108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 5.Pradet-Balade B, Boulme F, Beug H, Mullner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci. 2001;26(4):225–9. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- 6.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 7.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28(4):182–8. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 9.Mikulits W, Pradet-Balade B, Habermann B, Beug H, Garcia-Sanz JA, Mullner EW. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 2000;14(11):1641–52. doi: 10.1096/fj.14.11.1641. [DOI] [PubMed] [Google Scholar]
- 10.Zong Q, Schummer M, Hood L, Morris DR. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc Natl Acad Sci U S A. 1999;96(19):10632–6. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu X, de la Pena L, Barker C, Camphausen K, Tofilon PJ. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006;66(2):1052–61. doi: 10.1158/0008-5472.CAN-05-3459. [DOI] [PubMed] [Google Scholar]
- 12.Bayin NS, Modrek AS, Placantonakis DG. Glioblastoma stem cells: Molecular characteristics and therapeutic implications. World J Stem Cells. 2014;6(2):230–8. doi: 10.4252/wjsc.v6.i2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 14.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6(6):425–36. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 15.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 16.Ene CI, Edwards L, Riddick G, Baysan M, Woolard K, Kotliarova S, et al. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS One. 2012;7(12):e51407. doi: 10.1371/journal.pone.0051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCord AM, Jamal M, Williams ES, Camphausen K, Tofilon PJ. CD133+ glioblastoma stem-like cells are radiosensitive with a defective DNA damage response compared with established cell lines. Clin Cancer Res. 2009;15(16):5145–53. doi: 10.1158/1078-0432.CCR-09-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamal M, Rath BH, Tsang PS, Camphausen K, Tofilon PJ. The brain microenvironment preferentially enhances the radioresistance of CD133(+) glioblastoma stem-like cells. Neoplasia. 2012;14(2):150–8. doi: 10.1593/neo.111794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440–52. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nevo I, Woolard K, Cam M, Li A, Webster JD, Kotliarov Y, et al. Identification of molecular pathways facilitating glioma cell invasion in situ. PLoS One. 2014;9(11):e111783. doi: 10.1371/journal.pone.0111783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–80. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Galban S, Fan J, Martindale JL, Cheadle C, Hoffman B, Woods MP, et al. von Hippel-Lindau protein-mediated repression of tumor necrosis factor alpha translation revealed through use of cDNA arrays. Mol Cell Biol. 2003;23(7):2316–28. doi: 10.1128/MCB.23.7.2316-2328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayman TJ, Williams ES, Jamal M, Shankavaram UT, Camphausen K, Tofilon PJ. Translation initiation factor eIF4E is a target for tumor cell radiosensitization. Cancer Res. 2012;72(9):2362–72. doi: 10.1158/0008-5472.CAN-12-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Proud CG. Methods for studying signal-dependent regulation of translation factor activity. Methods Enzymol. 2007;431:113–42. doi: 10.1016/S0076-6879(07)31007-0. [DOI] [PubMed] [Google Scholar]
- 28.Kumaraswamy S, Chinnaiyan P, Shankavaram UT, Lu X, Camphausen K, Tofilon PJ. Radiation-induced gene translation profiles reveal tumor type and cancer-specific components. Cancer Res. 2008;68(10):3819–26. doi: 10.1158/0008-5472.CAN-08-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh AC, Truitt ML, Ruggero D. Oncogenic AKTivation of translation as a therapeutic target. Br J Cancer. 2011;105(3):329–36. doi: 10.1038/bjc.2011.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayman TJ, Wahba A, Rath BH, Bae H, Kramp T, Shankavaram UT, et al. The ATP-competitive mTOR inhibitor INK128 enhances in vitro and in vivo radiosensitivity of pancreatic carcinoma cells. Clin Cancer Res. 2014;20(1):110–9. doi: 10.1158/1078-0432.CCR-13-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazan-Mamczarz K, Peroutka RJ, Steinhardt JJ, Gidoni M, Zhang Y, Lehrmann E, et al. Distinct inhibitory effects on mTOR signaling by ethanol and INK128 in diffuse large B-cell lymphoma. Cell Commun Signal. 2015;13:15. doi: 10.1186/s12964-015-0091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8(12):957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, et al. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9(4):662–9. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 34.Kahn J, Hayman TJ, Jamal M, Rath BH, Kramp T, Camphausen K, et al. The mTORC1/mTORC2 inhibitor AZD2014 enhances the radiosensitivity of glioblastoma stem-like cells. Neuro Oncol. 2014;16(1):29–37. doi: 10.1093/neuonc/not139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farber-Katz SE, Dippold HC, Buschman MD, Peterman MC, Xing M, Noakes CJ, et al. DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell. 2014;156(3):413–27. doi: 10.1016/j.cell.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akakura S, Ostrakhovitch E, Sanokawa-Akakura R, Tabibzadeh S. Cancer cells recovering from damage exhibit mitochondrial restructuring and increased aerobic glycolysis. Biochem Biophys Res Commun. 2014;448(4):461–6. doi: 10.1016/j.bbrc.2014.04.138. [DOI] [PubMed] [Google Scholar]
- 38.Keene JD, Lager PJ. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005;13(3):327–37. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- 39.Trivigno D, Bornes L, Huber SM, Rudner J. Regulation of protein translation initiation in response to ionizing radiation. Radiat Oncol. 2013;8:35. doi: 10.1186/1748-717X-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braunstein S, Badura ML, Xi Q, Formenti SC, Schneider RJ. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol. 2009;29(21):5645–56. doi: 10.1128/MCB.00711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott KL, Chin L. Signaling from the Golgi: mechanisms and models for Golgi phosphoprotein 3-mediated oncogenesis. Clin Cancer Res. 2010;16(8):2229–34. doi: 10.1158/1078-0432.CCR-09-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartoletti-Stella A, Mariani E, Kurelac I, Maresca A, Caratozzolo MF, Iommarini L, et al. Gamma rays induce a p53-independent mitochondrial biogenesis that is counter-regulated by HIF1alpha. Cell Death Dis. 2013;4:e663. doi: 10.1038/cddis.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamori T, Yasui H, Yamazumi M, Wada Y, Nakamura Y, Nakamura H, et al. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic Biol Med. 2012;53(2):260–70. doi: 10.1016/j.freeradbiomed.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 44.Barjaktarovic Z, Schmaltz D, Shyla A, Azimzadeh O, Schulz S, Haagen J, et al. Radiation-induced signaling results in mitochondrial impairment in mouse heart at 4 weeks after exposure to X-rays. PLoS One. 2011;6(12):e27811. doi: 10.1371/journal.pone.0027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida T, Goto S, Kawakatsu M, Urata Y, Li TS. Mitochondrial dysfunction, a probable cause of persistent oxidative stress after exposure to ionizing radiation. Free Radic Res. 2012;46(2):147–53. doi: 10.3109/10715762.2011.645207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.