Abstract
The asymmetric partitioning of fate determining proteins has been shown to contribute to the generation of effector and memory CD8+ T cell precursors. Here, we demonstrate the asymmetric partitioning of mTORC1 activity upon activation of naïve CD8+ T cells. This results in the generation of one daughter T cell with increased mTORC1 activity, increased glycolytic activity and increased expression of effector molecules. The other daughter T cell inherits relatively low levels of mTORC1 activity, possesses increased lipid metabolism, expresses increased anti-apoptotic molecules and subsequently displays enhanced long-term survival. Mechanistically, we demonstrate a link between TCR-induced asymmetric expression of amino acid transporters and RagC-mediated translocation of mTOR to the lysosomes. Overall, our data provide important insight into how mTORC1-mediated metabolic reprogramming affects the fate decisions of T cells.
Asymmetric division is an evolutionarily conserved mechanism by which a single cell gives rise to two daughter cells of distinct fates 1. The asymmetric partitioning of fate determining proteins has been shown to contribute to the generation of effector and memory CD8+ T cell precursors based on differing levels of CD8 expression (CD8hi and CD8lo) 2,3. Previous work has identified that CD8hi T cells are derived from the daughter cell proximal to the antigen presenting cell (APC) and ultimately differentiate into effector CD8+ T cells; in contrast, CD8lo T cells are derived from the daughter cell distal to the APC and give rise to CD8+ memory T cells 2–5. Subsequent studies have further demonstrated asymmetry in critical transcription factors such as T-bet and TCF-1 in mediating the phenotypic differences between daughter cells 3,6.
The mTOR signaling pathway plays a critical role in regulating CD4+ T cell activation and differentiation 7–12 as well as regulating CD8+ T cell effector and memory generation 13–17. In part, the ability of mTOR to coordinate T cell differentiation and activation has been attributed to its ability to promote metabolic reprogramming 18–20. Robust mTORC1 activity promotes glycolytic activity and increased expression of effector molecules in CD8+ T effector cells 16. Indeed, T-Tsc2−/− CD8+ T cells, which display hyperactive mTORC1 activity, demonstrate enhanced cytotoxic T lymphocyte (CTL) activity but fail to develop into persistent memory T cells. Alternatively, antigen stimulation in the presence of the mTORC1 inhibitor rapamycin leads to enhanced generation of memory T cells, and CD8+ T cells with diminished mTORC1 activity derive energy utilizing fatty acid oxidation rather than glycolysis 13–16.
Here we tested the hypothesis that mTORC1 activity is asymmetrically inherited upon the first cell division of naïve CD8+ T cells. We demonstrate the asymmetric partitioning of mTORC1 activity upon activation of CD8+ T cells. This results in the generation of one daughter T cell with relatively increased mTORC1 activity, glycolytic activity, and expression of effector molecules. The other daughter T cell inherits relatively low levels of mTORC1 activity, possesses increased spare respiratory capacity (SRC) and lipid metabolism, expression of anti-apoptotic molecules and subsequently displays enhanced long-term survival. Mechanistically, we demonstrate a link between TCR-induced asymmetric expression of amino acid transporters and RagC-mediated translocation of mTOR to the lysosomes. Overall, our data provide important insight into how mTORC1-mediated metabolic reprogramming affects the fate decisions of CD8+ T cells.
Results
Divided CD8+ T cells possess asymmetric mTORC1 kinase activity
To determine if mTORC1 activity is asymmetrically inherited during cellular division, cells from mice with transgenic expression of major histocompatibility (MHC) class I-restricted (ovalbumin specific) TCR (OT-I) were labeled with cell proliferation dye-eFluor450 (eFluor450) and were intravenously (i.v.) transferred into C57BL/6 wild-type hosts. The next day, these mice were infected with Listeria monocytogenes expressing ovalbumin (LM-OVA) (i.v.) and splenocytes were harvested 48 h later. Consistent with previous studies 2–4,21, when examining CD8+ T cells from the first division (second brightest eFlour450 population), we observed two distinct populations based on CD8 surface expression, cells with high CD8 expression (hereafter CD8hi) and low CD8 expression (hereafter CD8lo) (Fig. 1a). Similarly, when comparing these two populations, we observed higher expression of CD25 and T-bet in the CD8hi T cells while the CD8lo T cells have higher expression of CD62L (Fig. 1a). Assessment of mTORC1 activity by flow cytometry based on phosphorylation of downstream target ribosomal S6 (p-S6) revealed that the CD8hi T cells had enhanced p-S6 expression compared to the CD8lo T cells, suggesting increased mTORC1 activity in the CD8hi T cells (Fig. 1a).
Figure 1. mTORC1 activity is asymmetrically inherited in dividing CD8+ T cells upon TCR stimulation.
(a) Flow cytometry analyzing adoptive transferred CD8hi and CD8lo eFluor450-labeled OT-I T cells gated on the first division from splenocytes of WT host mice at 48 h post LM-OVA infection. Histogram overlay of CD25, T-bet, CD62L, and p-S6 expression between CD8hi and CD8lo T cells. MFI, upper left corner. (b) Histogram overlay of mTOR pathway proteins between CFSE-labeled CD8hi and CD8lo T cells (gated as shown) that were stimulated in vitro for 36 h. MFI, upper left corner. (c) Immunoblot analysis of mTOR substrates of sorted in vitro activated CD8+ T cells. (d) Confocal images of dividing T cells that were activated in vitro. Quantification of the ratio of phosphorylated to total MFI of S6, n=39. (e) Quantification of MFI ratio of p-S6, β-tubulin, and CD90.2among dividing daughter pairs, n=132 (p-S6), 26 (β-tubulin and CD90.2). (f) Histogram overlay of CD8 and p-S6 expression from first-division adoptive transferred OT-I CD8+ T cells into either Rag2−/− or LM-OVA infected WT host. (g) Confocal images of in vitro stimulated WT, T-Rheb−/−, and T-Tsc2−/− CD8+ daughter T cells. Statistics analysis of p-S6 MFI ratio between CD8hi and CD8lo daughter T cells, n=26 (WT) and 37 (T-Rheb−/− and T-Tsc2−/−). *P < 0.05; **P < 0.0001; NS, not significant (Wilcoxon rank test (d) or One-way ANOVA (e, g)). Data are compiled from 3 independent experiments (d, e) or one experiment representative of at least 3 independent experiments (a–c, f, g) (mean in e, g). Scale bars, 10μm (d, g).
To confirm if differential inheritance of CD8 expression and mTORC1 activity is a consequence of cellular division or occurs prior to division, we compared expression levels in CD8+ T cells from the first division with undivided counterparts (brightest eFlour450 population). Undivided T cell expressed lower levels of CD8 than cells from the first division (Supplementary Fig. 1a), indicating that heterogeneous expression of CD8 is induced after cellular division. Likewise, upon the first division, but not in the undivided population, we observed distinct populations of T cells heterogeneous for p-S6, CD98 and T-bet expression (Supplementary Fig. 1b), suggesting two distinct populations of T cells emerging only after first division, and independent of CD8 expression within the undivided T cell population. Unlike T-bet, Eomesodermin (Eomes), a transcription factor essential for both CD8+ T effector and memory cells 22, was not asymmetrically distributed during the first division (Supplementary Fig. 1c), and both CD8hi and CD8lo T cell populations from the first division had higher expression of CD44 than naïve CD8+ T cells (Supplementary Fig. 1c), indicating that both populations were equally activated and not all proteins were asymmetrically inherited.
Next, we sought to determine whether the in vivo findings could be validated with an in vitro system using carboxyfluorescein succinimidyl ester (CFSE) labeled OT-I Rag2−/− splenocytes stimulated with ovalbumin MHC class I peptide (OVA-I). Consistent with our earlier finding (Fig. 1a), CD8hi T cells displayed higher p-S6 compared to CD8lo T cells (Fig. 1b). Similarly, phosphorylation of 4E-BP1, another downstream substrate of mTORC1, was also highly expressed in CD8hi T cells compared to CD8lo T cells (Fig. 1b). While both CD8hi and CD8lo T cells had similar expression of TSC2, a negative regulator of mTORC1 activity, expression of phosphorylated TSC2 was found higher in CD8hi T cells compared to CD8lo T cells (Fig. 1b), indicating a reduced inhibition of mTORC1 activity in CD8hi T cells.
To further confirm these results, we sorted in vitro generated CD8hi and CD8lo T cells to assess mTOR activity by immunoblotting. CD8hi T cells had increased p-S6, but no significant difference in phosphorylation of AKT, a downstream target of mTORC2, compared to CD8lo T cells (Fig. 1c). Compared to CD8lo T cells, CD8hi T cells also had higher phosphorylation of 4E-BP1, but expressed equivalent amounts of phosphorylated ERK (Supplementary Fig. 1d), suggesting the differences in signaling are selective for the mTORC1 pathway. Both the CD8hi and CD8lo daughter T cells showed increased expression of the activation-induced markers CD69 and CD44 (Supplementary Fig. 1e), suggesting that the differences in mTORC1 signaling are not due to lack of TCR activation.
We next assessed the partitioning of proteins in dividing T cells by confocal microscopy using an in vitro system with plate-bound anti-CD3, anti-CD28 and ICAM-1, which has previously been shown to induce CD8+ T cells asymmetric division 2,3. Both daughter CD8+ T cells displayed roughly equivalent amounts of S6 protein (Fig. 1d). However, the ratio between the mean fluorescence intensity (MFI) of p-S6 over total S6 was higher in CD8hi T cells compared to CD8lo T cells (Fig. 1d), indicating increased mTORC1 activity in CD8hi T cells. To control that these results are not skewed by a minor subpopulation displaying great asymmetry of mTORC1 activity, we quantified the frequency of dividing CD8+ T cells that asymmetrically partition p-S6 at first division to be 60% (see methods) (Fig. 1e), which is comparable with the frequency of T cell reported to undergo asymmetric distribution of T-bet (66%) 3. As a control, we observed no asymmetric partitioning of β-tubulin and CD90.2 between dividing CD8+ daughter T cells (Fig. 1e).
As previous studies have indicated that the interaction of LFA-1 on T cells with ICAM-1 on APCs (or plate-bound stimulation) is required for the asymmetric division of CD8+ T cells 2,3,5,23, we tested whether the asymmetric partitioning of mTORC1 activity was a consequence of polarity. Confocal imaging of a dividing T cell pair in contact with an APC indicated increased p-S6 in the proximal daughter T cell compared to the distal T cell (Supplementary Fig. 1f). To test whether non-polarized T cell division, such as during homeostatic T cell proliferation in the absence of antigen stimulation, would result in non-asymmetric inheritance of mTORC1 activity in daughter T cells, labeled OT-I T cells were adoptively transferred (i.v.) into Rag2−/− mice. Seventy-two hours later, we observed a relatively uniform distribution of CD8 expression on the transferred T cells that had undergone the first division, as opposed to the bimodal peaks of CD8 expression observed during antigen-induced (polarized) activation (Fig. 1f). Furthermore, none of the transferred CD8+ T cells recovered from the Rag2−/− recipients demonstrated differential expression of p-S6, CD25, CD62L, T-bet, MYC or CD98 within the first division, in contrast to T cells undergoing antigen-induced division during a LM-OVA infection (Fig. 1f and data not shown). Thus, asymmetric partitioning of mTORC1 activity in CD8+ T cells is not a generalized property of cell division but rather of APC-antigen-driven (polarized) activation.
To determine whether asymmetric cell division is dependent upon mTORC1 activity, we compared in vitro activated wild-type CD8+ T cells, T-Rheb−/− CD8+ T cells, which have markedly decreased mTORC1 activity, and T-Tsc2−/− CD8+ T cells, which have hyper-active mTORC1 activity. Using confocal microscopy, dividing daughter cells were identified via the tubulin bridge. Similar to wild-type CD8+ T cells, CD8hi and CD8lo daughter pairs were found in dividing T-Rheb−/− and T-Tsc2−/− CD8+ T cells (Fig. 1g), indicating that asymmetric division of CD8 expression is not controlled by mTORC1 activity. Furthermore, we observed no asymmetric partitioning of p-S6 in dividing T-Rheb−/− and T-Tsc2−/− CD8+ T cells compared to wild-type CD8+ T cells, thereby showing that differential mTORC1 activity in dividing T cells is required for asymmetric partitioning of mTORC1 activity to occur. Overall, these results so far indicate that mTORC1 activity is divided asymmetrically in the CD8hi and CD8lo daughter T cells during the first antigen-driven cell division.
Asymmetric mTORC1 activity promotes distinct metabolic programs
mTORC1 plays an important role in regulating the metabolic reprogramming of T cells, which in turn regulates T cell fate and function 19. To test whether the asymmetric partitioning of mTORC1 activity would lead to the generation of two daughter CD8+ T cells with distinct metabolic profiles, we compared the metabolic phenotype of sorted CD8hi and CD8lo T cells generated under in vitro activation. CD8hi T cells showed a higher basal and maximal extracellular acidification rate (ECAR) compared to CD8lo cells (Fig. 2a), indicative of increased glycolysis and consistent with the current view that CD8+ effector T cells are highly glycolytic 18. Alternatively, we observed that the CD8lo T cells have higher SRC (Fig. 2b), a metabolic feature of long-lived cells 18,24,25, compared to CD8hi T cells.
Figure 2. Asymmetric inheritance of mTORC1 results in CD8+ T daughter cells with distinct metabolic capabilities.
(a–b) CD8hi and CD8lo T cells from in vitro activation were assayed for ECAR (a) and OCR (b). Summary of basal ECAR, maximum ECAR, and SRC are shown on right, n=8 (a), 6 (b). (c) As in Fig. 1a, Histogram overlay showing MYC expression between CD8hi and CD8lo T cells. MFI, upper right corner. (d) Immunoblot analysis comparing expression of proteins involved in glycolytic pathway between in vitro stimulated CD8hi and CD8lo T cells. (e) Confocal images of in vitro stimulated daughter CD8+ T cells from GFPMYC mice stained with CD8 and p-S6. Statistical analysis of GFPMYC MFI between CD8hi and CD8lo T cells (right), n=18. (f) Quantification of mitochondrial and genomic DNA ratio between sorted in vitro activated CD8hi and CD8lo T cells, n=12. (g) Immunoblot analysis showing expression of mitochondrial associated proteins comparing between CD8hi and CD8lo T cells as in (d). (h) Confocal images of in vitro activated CD8hi and CD8lo T cells stained with MitoTracker dye and DAPI. CD8hi T cells (green) are further stained with a different CD8 antibody clone post-sort to distinguish between CD8lo T cells. Statistical analysis of MitoTracker MFI between CD8hi and CD8lo cells, n=60. *P < 0.05; **P < 0.005; ***P < 0.0005 (Mann-Whitney t test (a, b, f, h) or Wilcoxon rank test (e)). Data are from one experiment representative of at least 3 independent experiments (a–e, g) or compilation of 3 independent experiments (f) (mean ± s.d. in a, b left, mean ± s.e.m. in a, b, f). Scale bars, 10μm (e, h).
MYC plays an important role in the metabolic reprogramming of both CD4+ and CD8+ T cells upon TCR activation, especially controlling the expression of multiple glycolytic enzymes 26, and MYC expression is enhanced by mTORC1 activation 27,28. To test if MYC was preferentially upregulated in CD8hi T cells compared to CD8lo T cells, we adoptively transferred (i.v.) labeled OT-I T cells into wild-type hosts that were then infected with LM-OVA (i.v.). Upon flow cytometric assessment of CD8hi and CD8lo T cells from the first division, MYC protein was asymmetrically enriched in the CD8hi T cells compared to CD8lo T cells (Fig. 2c). Similarly, upon in vitro stimulation, asymmetric partitioning of MYC was observed by immunoblot analysis of sorted CD8hi and CD8lo T cells (Fig. 2d) and by confocal microscopy of CD8hi and CD8lo T cell dividing daughter pairs (Fig. 2e). In addition to MYC, CD8hi T cells also showed higher expression of hexokinase 2 (HK2) and GLUT-1, which are key proteins involved in the glycolytic pathway, compared to CD8lo T cells (Fig. 2d). Overall, our data demonstrate that upon CD8+ T cell division, the asymmetric partitioning of mTORC1 activity is associated with the generation of CD8hi mTORhi MYChi T cells that are highly glycolytic.
CD8lo daughter T cells generated by the asymmetric division contribute to the generation of memory CD8+ T cells 2,4, which have distinct metabolic and mitochondrial profiles compared to effector CD8+ T cells 14,24,25. To further assess the metabolic reprogramming of CD8lo T cells, we examined the relative mitochondrial content of these cells compared to the highly glycolytic CD8hi T cells from sorted in vitro generated T cells. CD8lo T cells had a higher mitochondrial to genomic DNA ratio compared to CD8hi T cells (Fig. 2f). In addition, by confocal microscopy, we observed increased mitochondrial mass in sorted CD8lo T cells compared to CD8hi T cells (Fig. 2g). Furthermore, immunoblot analysis of sorted in vitro generated CD8lo T cells also showed increased expression of the mitochondrial protein VDAC as well as proteins associated with oxidative phosphorylation (NDUFB8 and SDHA), and pro-survival proteins (BCL-2 and BCL-xL) compared to CD8hi T cells (Fig. 2h). In addition, CPT-1α, an enzyme that is critical in fatty acid oxidation and enriched in memory CD8+ T cells 14,24,25, was increased in CD8lo T cells compared to CD8hi T cells (Fig. 2h). Thus, while CD8lo T cells have decreased glycolysis, their increase in lipid metabolism and SRC is associated with an increase in mitochondrial mass and function that are critical in memory T cell formation 14,24,25.
CD8lo daughter T cells become long-lived memory T cells
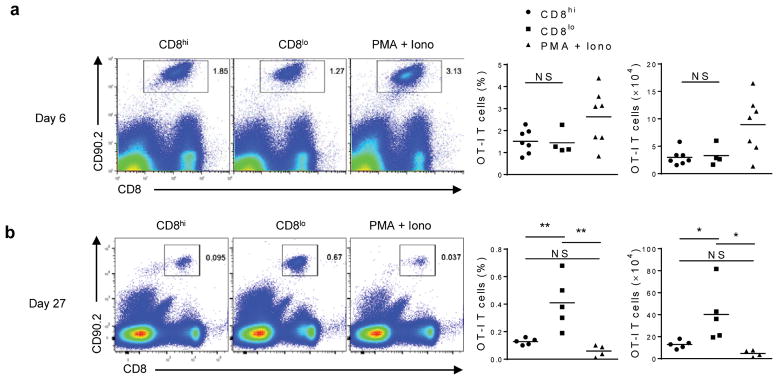
Next, we investigated if asymmetric partitioning of mTORC1 activity correlated with distinct immune function. Consistent with previous findings 2,4, sorted in vitro activated CD8hi T cells expressed increased levels of IFN-γ (Ifng) and perforin (Prf1) mRNA as assessed by qRT-PCR (Supplementary Fig. 1g). As measured by ELISA, sorted CD8hi T cells produced more IFN-γ than CD8lo T cells (Supplementary Fig. 1h). Next, to evaluate the impact of asymmetric mTORC1 activation on CD8+ T cells survival, we adoptively transferred (i.v.) congenically-marked sorted in vitro activated CD8hi or CD8lo T cells into wild-type hosts that were infected with vaccinia-OVA either the same day or 21 days after transfer. Upon assessment of splenocytes six days after the adoptive transfer, we detected similar numbers of antigen specific CD8+ T cells from the mice that received CD8hi or CD8lo T cell populations (Fig. 3a). However, CD8hi T cells were barely detectable after re-challenge in vivo at 21 days post-adoptive transfer (Fig. 3b), indicating that transferred CD8hi T cells failed to differentiate into proper memory T cells. In contrast, CD8lo T cells were able to persist and respond to secondary challenge (Fig. 3b). In addition, naïve CD8+ T cells stimulated non-specifically with PMA and ionomycin, which induce non-polarized cell division, resulted in symmetrical high mTORC1 activity between CD8hi and CD8lo T cells (Supplementary Fig. 2), and also showed decreased memory formation in vivo in response to secondary challenge (Fig. 3b). These observations suggest that CD8hi T cells possess robust effector function but fail to survive and thus are not detected upon re-challenge, whereas CD8lo cells initially possess reduced effector capacity, but survive and proliferate upon re-challenge.
Figure 3. Asymmetric inheritance of mTORC1 results in CD8+ T daughter cells with different in vivo survival.
PMA + Ionomycin-activated or sorted CD8hi and CD8lo T cells from in vitro activation were adoptively transferred into congenically distinct WT host, and then were infected with vaccinia-OVA the same day (a) or 21 days later (b). Six days after infection (day 6 or day 27), spleens were harvested and quantification of CD90.2+ CD8+ T cells was assessed by flow cytometry. Statistical quantification of percentage and absolute number of recovered CD90.2+ CD8+ T cells between CD8hi, CD8lo, and PMA + Iono activated T cells 6 days or 27 days after the adoptive transfer (six days after infection) are shown, n= 7,4,7 (a, CD8hi, CD8lo, PMA+Iono), 5,5,4 (b, CD8hi, CD8lo, PMA+Iono). *P < 0.05; **P < 0.005; NS, not significant (One-Way ANOVA). Data are from one experiment representative of at least 4 independent experiments (mean).
T cell activation leads to translocation of mTOR to the lysosome
We sought to define the cellular mechanism that accounts for the asymmetric partitioning of mTORC1 activity upon the first division in CD8+ T cells. Amino acid sufficiency has been shown to promote mTORC1 localization to the lysosomal surfaces, where mTORC1 can become activated 29,30. This occurs through the activation of the Rag GTPases, which upon amino acid sensing, disassociate from the lysosomal surface to capture mTOR in the cytoplasm and then shuttle back to the lysosomes 29,31. TCR-mediated activation in CD8+ T cells is known to elevate amino acid uptake 32, thus, we tested whether TCR activation regulates the ability of the Rag GTPases to promote the translocation of mTOR to the lysosomes. We stimulated naïve OT-I CD8+ T cells with OVA-I peptide in vitro for 0, 4 or 24 hours and assessed the co-localization of RagC (a member of the Rag GTPases) and the lysosome marker LAMP-2 by confocal microscopy. Upon four hours of T cell activation, there was an initial decrease in the association of RagC and LAMP-2 compared to unstimulated T cells (Fig. 4a), indicating that TCR stimulation leads to the translocation of RagC from the lysosomes to the cytoplasm. By 24 hours post-activation, RagC partially re-localized to the lysosomes (Fig. 4a).
Figure 4. The differential RagC mediated translocation of mTOR to the lysosome contributes to the asymmetric division of CD8+ T cells.
(a) OT-I Rag2−/− splenocytes were stimulated in vitro with OVA-I peptide for 0, 4, or 24 h. RagC and LAMP-2 localization was assessed by confocal microscopy. Statistical analysis of co-localization of RagC and LAMP-2 (r) was assessed, n=30, 22, 23 (0, 4, 24 h). (b) Same as (a), except co-localization analysis for mTOR and LAMP-2, n=22, 9, 18 (0, 4, 24 h). (c) Statistical analysis of co-localization of mTOR and LAMP-2 (r) in CD8+ T cells from T-Rheb−/− and T-Tsc2−/− at indicated time point after TCR activation, n=40 (T-Rheb−/−), 39 (T-Tsc2−/−). (d) Confocal images of in vitro activated CD8+ T cells showing localization of LAMP-2 and mTOR between CD8hi and CD8lo daughter T cells. Statistical analysis of co-localization of mTOR and LAMP-2 (r) between paired daughter cells are shown on the right, n=14. *P < 0.005; **P < 0.0005; ***P < 0.0001; NS, not significant (One-Way ANOVA (a, b), One-Way ANOVA (c) or Wilcoxon rank test (d)). Data are from one experiment representative of at least 2 independent experiments (mean in a–c). Scale bars, 10μm (a, b, d).
Next, we used confocal microscopy to determine the movement of mTOR to the lysosomes. Assessment of co-localization of mTOR and LAMP-2 in naïve unstimulated CD8+ T cells indicated minimal mTOR localization to the lysosomes; however, as early as four hours after antigen stimulation, mTOR co-localized with the lysosomes and this association further increased at 24 hours (Fig. 4b). We observed a similar increase of mTOR translocation to the lysosomes in T-Rheb−/− and T-Tsc2−/− CD8+ T cells as wild-type CD8+ T cells (Fig. 4c), indicating that translocation of mTOR to the lysosomes is independent of mTORC1 activity. Our data thus far establishes that TCR activation of CD8+ T cells initiates the translocation of mTOR to the lysosome.
To determine if mTOR localization to the lysosomes was observed in asymmetrically dividing CD8+ T cells, we performed confocal microscopy on in vitro activated T cells undergoing division. We observed increased co-localization of mTOR and LAMP-2 in CD8hi T cells compared to CD8lo T cells (Fig. 4d). These observations suggest that asymmetric inheritance of mTORC1 activity occurs as the result of the preferential translocation of mTOR to the lysosomes in the CD8hi T cell compared to the CD8lo T cell.
CD98 expression levels lead to asymmetric mTOR localization
We next investigated the mechanism accounting for the increased translocation of mTOR to the lysosomes in CD8hi T cells. RagC-mediated translocation of mTOR to the lysosomes is induced by amino acid influx (particularly the branch chain amino acid leucine) 29. To test whether differential mTORC1 activity among daughter T cells may be the result of the asymmetric partitioning of amino acid transporters upon cell division and differential leucine influx, we assessed the expression of the amino acid transporter SLC7A5, which controls leucine influx and subsequent mTORC1 activation, and as such has an essential role in CD8+ effector T cell generation 32. We observed increased expression of Slc7a5 mRNA in sorted in vitro generated CD8hi T cells compared to CD8lo T cells by qRT-PCR (Fig. 5a).
Figure 5. TCR-induced mTOR localization to lysosome is dependent on differential amino acid transporter expression.
(a) qRT-PCR analysis of Slc7a5 mRNA expression in sorted CD8hi and CD8lo T cells activated in vitro, n=8. (b) As in Fig. 1a, histogram overlay of CD98 expression in CD8hi and CD8lo T cells. MFI, upper left corner. (c) Confocal images of CD8 and CD98 expression between daughter cells activated in vitro. Statistical analysis of CD98 MFI between CD8hi and CD8lo daughter T cells, n=13. (d) Confocal images of mTOR and LAMP-2 localization between daughter CD8+ T cells from in vitro activation. Statistical analysis of co-localization of mTOR and LAMP-2 (r) between CD98hi and CD98lo CD8+ T cells, n=19. (e) Flow cytometry of p-S6 and CD98 expression in CD8+ T cells activated in vitro ± amino acid transporter inhibitor (BCH) and unstimulated (US). MFI, upper right corner. (f) Statistical analysis of mTOR and LAMP-2 co-localization in CD8+ T cells under US, no treatment, and BCH (top) or rapamycin (bottom) treatment by confocal microscopy, n=10 (BCH), 15 (rapamycin). (g) Confocal images of daughter CD8+ T cells showing localization of LAMP-2 and mTOR among no treatment, BCH, and rapamycin treatment. Statistical analysis of mTOR and LAMP-2 co-localization (r) is shown on the right, n=20. *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001; NS, not significant (Mann-Whitney t test (a, b), Wilcoxon rank test (c, d, g) or One-Way ANOVA (f)). Data are from compilation of 3 independent experiments (a), one experiment representative of at least 3 independent experiments (b–g) (mean ± s.e.m. in a, mean in f). Scale bars, 10μm (c, d, g).
To explore if the leucine amino acids transporter was preferentially upregulated in CD8hi T cells compared to CD8lo T cells, we adoptively transferred (i.v.) labeled OT-I T cells into WT hosts that were infected with LM-OVA (i.v.) 24 hours later. As reliable detection of SLC7A5 protein is currently unavailable, we instead assessed the expression of CD98, a glycoprotein that forms a heterodimer with SLC7A5 33. Flow cytometric observation of CD8hi and CD8lo T cells determined that CD98 expression was asymmetrically enriched in the CD8hi T cells compared to CD8lo T cells (Fig. 5b). Additionally, upon in vitro activation, we observed higher CD98 expression on dividing CD8hi than CD8lo T cells by confocal microscopy (Fig. 5c). This implies that CD98 is asymmetrically partitioned between dividing daughter CD8+ T cells.
Consistent with published findings 34, co-culture of OT-I CD8+ T cells with peptide-loaded bone marrow derived APCs induced the CD98 localization at the immunological synapse, as indicated by co-localization with talin (Supplementary Fig. 3). Because CD98 interacts with ICAM-1 to facilitate leucine uptake 34, we tested whether CD98, along with SLC7A5, was preferentially enriched in the CD8hi T cell compared to CD8lo T cell. We observed that in dividing T cell pairs, the daughter T cells with higher CD98 expression had greater co-localization of mTOR to the lysosomes than the daughter T cells with lower CD98 expression (Fig. 5d). Of note, T-Rheb−/− CD8+ T cells displayed asymmetric inheritance of CD98 (Supplementary Fig. 4), indicating that mTORC1 does not control the asymmetric distribution of CD98 during division (note though, they showed lower expression of CD98 than wild-type CD8+ T cells).
Next, we tested the functional consequences of inhibiting SLC7A5 on TCR-induced mTOR translocation. In vitro activation of naïve CD8+ T cells in the presence of the amino acid transport inhibitor 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid (BCH) led to decreased p-S6 in CD8+ T cells compared to untreated T cells but similar to expression in the unstimulated T cells (Fig. 5e). However, we observed no difference in CD98 expression in activated CD8+ T cells between the untreated and BCH-treated condition (Fig. 5e). Likewise, inhibition of SLC7A5 activity with BCH during CD8+ T cell activation led to decreased translocation of mTOR to the lysosome compared to the untreated condition (Fig. 5f). In contrast, rapamycin treatment, which inhibits mTORC1 activity independent of amino acid influx, had no effect on mTOR translocation to the lysosome in TCR-activated CD8+ T cells (Fig. 5f), indicating that amino acids play an essential role in initiating mTORC1 activity by translocation of mTOR to the lysosomes.
Finally, we used confocal microscopy to assess the consequences of BCH and rapamycin treatment on the asymmetric translocation of mTOR to the lysosome in dividing CD8+ T cell pairs. While untreated dividing CD8+ T cells showed punctate and asymmetric localization of mTOR to the lysosome (Fig. 5g), treatment with BCH resulted in a dispersed distribution of mTOR throughout the cytoplasm in both daughter cells, indicating abrogated mTOR translocation to the lysosome (Fig. 5g), whereas rapaymcin-treated daughter T cells showed a similar mTOR distribution as non-treated T cells (Fig. 5g). Overall, these data suggest that the asymmetric partitioning of CD98 regulates the leucine-mediated translocation of mTOR to the lysosomes preferentially in the CD8hi CD98hi daughter T cells compared to the CD8lo CD98lo T cells (Supplementary Fig. 5).
Discussion
mTOR has been identified to regulate CD4+ and CD8+ T cell differentiation 7–13,15–17,20. Upon T cell activation, mTORC1 activity can distinguish CD4+ T cells with distinct fates 12. Here we show that asymmetric partitioning of mTORC1 activity following initial division of CD8+ T cells leads to the generation of two daughter cells with distinct metabolic programming. The daughter T cell (CD8hi) proximal to the APC inherits increased mTORC1 activity and is highly glycolytic, two properties that are required for robust effector cell function 16,17,35,36. Alternatively, the daughter cell (CD8lo) distal to the APC inherits decreased mTORC1 activity and is less glycolytic, but possesses increased mitochondria and SRC, as well as increased expression of anti-apoptotic molecules, and as such, have a metabolic programming similar to that of long-lived memory cells 24,25.
Previous findings have identified two distinct daughter T cells based upon CD8 expression 2. Based on our observations that link mTORC1 activity and amino acid transporter CD98 to asymmetric division, we propose that tracking the asymmetric partitioning of mTORC1 activity or CD98 represent better, more functional marker of asymmetric division of CD8+ T cells. We also noticed the ability of mTORC1 to enhance MYC expression in CD8hi daughter T cells thereby promoting metabolic reprogramming downstream of mTORC1. Our observations that effector and memory T cell generation are intimately linked to metabolic reprogramming during early CD8+ T cells division underscore the integral role of metabolism in defining immune cell function 19. Consistent with such a model, a recent report linked PI3K signaling and nutrient sensing to T and B cell fate decisions 6.
Mechanistically, our study links asymmetric mTORC1 activation based on the differential RagC-mediated translocation of mTOR to the lysosomes between dividing daughter CD8+ T cells. Here we document that TCR-induced mTORC1 activation and mTOR lysosomal translocation is dependent on amino acid influx. We observed that the asymmetric partitioning of mTORC1 activity was facilitated by the asymmetric partitioning of TCR-induced amino acid transporters, which is consistent with the critical role of SLC7A5 in promoting T cell effector function 32
While our observations are consistent with previous reports demonstrating the generation of effector and memory T as a result of asymmetric division of CD8+ T cells 2,3,21,23, our work does not preclude a model whereby the generation of memory T cells is derived from the effector pool 37–39. Ultimately, we believe it is the level of mTORC1 activity a CD8+ T cell possesses and the subsequent downstream metabolic programming that influences its fate, thereby potentially integrating into all proposed models. For instance, in our current study, a dividing wild-type CD8+ T cell pair with different levels of mTORC1 activity will result in diverse T cell fate. Supporting this, our previous work proves that T cells with equal levels of high (T-Tsc2−/−) or low (T-Rheb−/−) mTORC1 activity will result in one fate at the expense of the other 16. Asymmetric partitioning of proteins and signaling molecules is thought to play a role in driving divergent cellular functions during differentiation. As such, examining the partitioning of mTORC1 activity in other cell types, tissues or organisms may help elucidate mechanisms of cellular differentiation.
Methods
Mice
Six- to eight-week-old male or female mice were used for performing all the experiments in this study. All mouse procedures were approved by the Johns Hopkins University Institutional Animal Care and Use Committee. C57BL/6, GFPMYC, Cd4-Cre and OT-I, Cd90.1, Rag2−/− mice were obtained from Jackson Laboratories. Mice with loxP-flanked Tsc2 alleles were kindly provided by the laboratory of Dr. Michael Gambello. Mice with loxP-flanked Rheb alleles were generously gifted from Dr. Paul Worley. No empirical test was performed for choosing sample size prior to experiments. No randomization of samples or animals was used nor were investigators blinded throughout the study.
Antibodies and Reagents
Antibodies against the following proteins were purchased from BD Biosciences: CD8α (53-6.7, 1:500), CD90.1 (OX-7, 1:1000), CD69 (H1.2F3, 1:300), CD90.2 (53-2.1, 1:500), CD25 (PC61, 1:500), CD62L (MEL-14, 1:400), Streptavidin-PerCP (554064, 1:300) and Fc Block (2.4G2, 1:100). Antibodies against CD8β (H35-17.2, 1:500), T-bet (eBio4B10, 1:500), CD44 (IM7, 1:400), CD98 (RL388, 1:500) and Eomes (Dan11mag, 1:400) were purchased from eBioscience. Fc-mICAM-1 Chimera from R&D systems. The following antibodies were purchased from Cell Signaling: TSC2 (D93F12, 1:300), p-TSC2 (S939, 1:300), p-S6 (S240/244, D68F8, 1:1000 for flow cytometry and 1:4000 for immunoblotting), AKT (11E7, 1:2000), p-AKT (S473, D9E, 1:2000), p-4E-BP1 (T37/46, 236B4, 1:300 for flow cytometry and 1:2000 for immunoblotting), mTOR (7C10, 1:300), RagC (D8H5, 1:300), p-ERK(T202/Y204, D13.14.4E, 1:2500), hexokinase 2 (C64G5, 1:1000), MYC (D84C12, 1:1000), GLUT-1 (D3J3A, 1:1000), VDAC (D73D12, 1:2500), SDHA (D6J9M, 1:2500), BCL-2 (D17C4, 1:2500), and BCL-xL (54H6, 1:2500). Anti-LAMP-2 (GL2A7, 1:300), CPT-1α (EPR12740(B), 1:1000), and NDUFB8 (20E9DH10C12, 1:1000) were purchased from Abcam. LaminB (C-20, 1:2000) was purchased from Santa Cruz Biotechnology. Anti-β-tubulin (AA2, 1:1000), talin (8d4, 1:500), and actin (A2066, 1:4000) was purchased from Sigma. OVA-I peptide (SIINFEKL) was purchased from AnaSpec. Stimulatory anti-CD3 (2C11) and anti-CD28 (37.51) were purified from hybridoma supernatants prepared “in-house.” CFSE and non-conjugated CD8α (5H10, 1:300) antibody used for microscopy were obtained from Invitrogen. Cell Proliferation Dye-eFluor450 and fixable viability dye eFluor780 were purchased from eBiosciences. Rapamycin was purchased from LC laboratories. 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid (BCH), PMA, and ionomycin were purchased from Sigma Aldrich. Secondary fluorophore conjugated antibodies were purchased from Invitrogen: anti-rabbit Alexa Fluor488, anti-rabbit Alexa Fluor647, anti-rabbit Alexa Fluor405, anti-rat Alexa Fluor555, and anti-mouse Alexa Fluor405 (all dilutions in 1:500 to 1:1000). MitoTracker dye was purchased from Invitrogen. All antibody validation profiles can be found via 1DegreeBio (http://1degreebio.org/) or CiteAb (http://www.citeab.com).
T cell in vitro Stimulation
In vitro stimulation was performed with plate-bound αCD3 (2μg/ml), mICAM-1/Fc chimera (2μg/ml) and αCD28 (2μg/ml) in indicated experiments. For PMA + Ionomycin activation, cells are stimulated with PMA (50ng/ml) and Ionomycin (500ng/ml) for 5 h and washed out. Otherwise, splenocytes were stimulated with 5 μg/ml OVA class I peptide (SIINFEKL) for indicated time intervals.
Flow Cytometry & Cell Sorting
All experiments were performed on a BD FACS Calibur, LSR II or Aria II and analyzed using FlowJo software. For all flow cytometry experiments gates were set appropriately with unstimulated T cells.
CD8+ T Cells Adoptive Transfer
For ex vivo phenotyping of CD8hi and CD8lo T cells in first division, splenocytes were harvested from OT-I mice and adoptively transferred (i.v.) into congenically mismatched WT hosts. Twenty-four hours post-transfer, mice were infected with WT non-attentuated LM-OVA. Forty-eight hours post-infection, spleens were harvested and processed for subsequent analysis.
For acute and memory experiment, OT-I Rag2−/− CD90.2+ splenocytes were stimulated with 5μg/ml OVA-I peptide for 36–40 h and sorted CD8hi and CD8lo T cells from first division. 5×104 CD8hi or CD8lo T cells were adoptively transferred (i.v.) into C57BL/6 WT CD90.1+ recipients. Recipient mice were either infected with 1×106 plaque-forming units vaccinia-OVA on day 0 or on day 21 post-transfer. Six days after infection (Day 6 or Day 27), spleens were harvested and quantification of CD90.2+ CD8+ T cells was assessed.
Immunoblot Analysis
CD8hi and CD8lo T cells are sorted from first division. Samples were flash frozen and lysed in RIPA lysis buffer with protease and phosphatase inhibitor cocktails. Proteins were detected by ECL Plus substrate (GE Healthcare). All images were obtained using UVP Biospectrum500 Imaging System.
Real Time PCR
T cells were stimulated and sorted as described under T cell in vitro stimulation. Total RNA was collected using TRIzol reagent (Life Technologies). cDNA was generated with ProtoScript II RT (New England BioLabs) and Real time PCR was performed using TaqMan Universal Master Mix II (Life Technologies). Real time PCR primers and probes obtained from Applied Biosystems: Ifng (Mm99999071_m1), Prf1 (perforin, Mm00812512_m1), and Slc7a5 (solute carrier family 7, member 5, Mm00441516_m1). Human18s (Life Technologies) is used as endogenous control.
ELISA
IFN-γ was analyzed by mouse ELISA Ready-SET-Go! kit as recommended by the manufacturer (eBioscience).
Mitochondrial and genomic DNA analysis
Total DNA was isolated from sorted cells using QIAamp DNA kit (QIAGEN) according to the manufacturer’s protocol. Isolated DNA were quantified by Nanodrop. Real time PCR was performed using SYBR Green (Life Technologies) according to the manufacturer’s protocol. Primers for amplifying genomic DNA and mitochondrial DNA are specific for H19 and Nd1 respectively. H19: Forward 5′-GTACCCACCTGTCGTCC-3′ and Reverse 5′-GTCCACGAGACCAATGACTG-3′. Nd1: Forward 5′-AATCGCCATAGCCTTCCTAACAT-3′ and Reverse 5′-GGCGTCTGCAAATGGTTGTAA-3′.
Metabolic Assays
Preparation of cells as described under T cell in vitro stimulation. 1.5×105 cells were plated per well on coated TC-treated XF96 cell culture microplate coated with poly-D lysine (50μg/ml). SRC was determined by dividing the maximal OCR (detected following FCCP administration) by the basal OCR. Glycolytic stress test was performed with media containing XF Assay Medium Modified DMEM. Plating media for mitochondrial stress test contained 25mM Glucose, 2mM L-Glutamine, 1mM Na pyruvate in XF Assay Medium Modified DMEM. Experiments were performed using XF 96 Extracellular Flux Analyzer (Seahorse Biosciences). The following were injected at the indicated time interval: D-glucose (5mM), 2-deoxyglucose (2DG, 0.1mM), oligomycin (2μM), FCCP (0.5mM), Rotenone (2μM) and Antimycin A (4μM).
Phospho-Flow Cytometry
CD8+ T cells were surface stained with anti-CD8 biotin (53-6.7, BD Bioscience), fixed with 2% formaldehyde for 15 min in 37°C and permeabilized with ice-cold 90% methanol at −20°C for 20 min. After washing, cells were blocked in 10% FBS solution with Fc Block. Staining was analyzed by flow cytometry.
Imaging APCs and T Cells Interaction
To image antigen presenting cells (APCs) and T cells interaction, APCs were derived from bone marrow cells from femur and tibia of C57BL/6 mice and cultured in the presence of GM-CSF (10ng/ml) and IL-4 (5ng/ml) for 6 days. On day 3 and 5, new medium supplemented with GM-CSF (10ng/ml) and IL-4 (5ng/ml) was added. On day 6, loosely adherent cells in the culture were harvested, and 7.5×104 cells were plated on slides overnight. After incubation with OVA-I peptide (1μg/ml) for 1 h at 37°C. After washing, 2.5×104 OT-I CD8+ T cells were overlaid. Cells are activated and fixed at indicated time points.
Confocal Microscopy
CD8+ T cells were activated in vitro with plate-bound stimulation for 28–36 h depending on genotypes and experimental treatment. Cells were treated with 10μM Cytochalasin B (Sigma) 2–3 h before fixation. Slides were coated with 50μg/ml Poly-D Lysine (Sigma) overnight and vigorously washed before addition of cells. Cells on slides were fixed with 4% paraformaldehyde (methanol free, Thermo Scientific) for 10 min before washing with PBS. Slides were then permeabilized with 90% methanol or 0.1% Triton X-100 (Sigma) followed by blocking in 5% goat serum (Gibco). Slides were then stained as described in phospho-flow cytometry methods above or in 0.01% Tween-20 (Sigma) in 5% goat serum. Slides were mounted with Slow Fade Gold anti-fade reagent with or without DAPI (Invitrogen). Cells were imaged with either an upright 710NLO-Meta confocal module (AxioExaminer; Zeiss) with a 63× /1.2W C-Apo objective or an inverted 780-Quasar confocal module (AxioObserver; Zeiss) with a 40× /1.2 PlanApo oil objective. Data were acquired with Zen imaging software (Zeiss) and analyzed with either ZEN (Zeiss) or Volocity (PerkinElmer) analysis software. Paired dividing T cells were identified based on the presence of tubulin bridge between conjoined cells or the shape of nucleus by DAPI staining. Molecules are considered to be asymmetrically divided if the ratio of dividing daughter cells exceeds two standard deviations above the ratio of the mean of β-tubulin, as described previously 6. For quantification of co-localization, Global pearson’s coefficient (r) was calculated from Volocity.
Statistical Analysis
All graphs were created using GraphPad Prism software, and statistical analysis was performed with GraphPad Prism. Paired cell analysis was assessed by Wilcoxon matched-pairs signed rank test; comparisons between 2 independent groups were assessed by Mann-Whitney t tests; comparisons between 3 or more independent groups were assessed by One-Way ANOVA with a Holm-Sidak’s multiple comparisons test. A p-value less than 0.05 was considered statistically significant. Error bars represent mean ± standard error of the mean.
Supplementary Material
Acknowledgments
We thank members of the Powell lab and C. Gamper for critical discussion of the manuscript; B. Smith and T. Stephens for their technical support on microscopy studies; M. Gambello (Emory University) for Tsc2loxP mice; and P. Worley (Johns Hopkins University) for RhebloxP mice. This work was supported by NIH grants AI072677 (to J.D.P.), AI77610 (to J.D.P.), AI091481 (to J.D.P.), S10 OD016374 (to the JHUSOM Microscope Facility) and S10 RR024550 (to the JHUSOM Microscope Facility).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Contributions
K.N.P., I.-H.S., C.H.P., and J.D.P planned the experiments, analyzed the data and wrote the manuscript, K.N.P., I.-H.S., C.H.P., Y.-C.L, M.-H.O., A.T.W and J.W. performed the experiments; G.M.D and A.T.W. contributed to the experimental design, execution and data interpretation; A.J.T. and R.L.B. provided support for flow cytometry experiments; G.M.D. and J.D.P conceived the idea for the study; K.N.P., I.-H.S., C.H.P., Y.-C.L, M.-H.O., G.M.D and J.D.P provided critical review of the manuscript.
References
- 1.Betschinger J, Knoblich JA. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol. 2004;14:R674–685. doi: 10.1016/j.cub.2004.08.017. S0960982204005986 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Chang JT, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. 1139393 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Chang JT, et al. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity. 2011;34:492–504. doi: 10.1016/j.immuni.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arsenio J, et al. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nature immunology. 2014;15:365–372. doi: 10.1038/ni.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliaro J, et al. Asymmetric cell division of T cells upon antigen presentation uses multiple conserved mechanisms. Journal of immunology. 2010;185:367–375. doi: 10.4049/jimmunol.0903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin WH, et al. Asymmetric PI3K Signaling Driving Developmental and Regenerative Cell Fate Bifurcation. Cell Rep. 2015;13:2203–2218. doi: 10.1016/j.celrep.2015.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. S1074-7613(09)00237-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nature immunology. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. S1074-7613(10)00205-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heikamp EB, et al. The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex. Nature immunology. 2014;15:457–464. doi: 10.1038/ni.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang K, et al. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity. 2013;39:1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollizzi KN, Waickman AT, Patel CH, Sun IH, Powell JD. Cellular Size as a Means of Tracking mTOR Activity and Cell Fate of CD4+ T Cells upon Antigen Recognition. PloS one. 2015;10:e0121710. doi: 10.1371/journal.pone.0121710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. nature08097 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollizzi KN, et al. mTORC1 and mTORC2 selectively regulate CD8(+) T cell differentiation. J Clin Invest. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finlay DK, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol. 2014;14:435–446. doi: 10.1038/nri3701. nri3701 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollizzi KN, Powell JD. Regulation of T cells by mTOR: the known knowns and the known unknowns. Trends Immunol. 2015;36:13–20. doi: 10.1016/j.it.2014.11.005. S1471-4906(14)00209-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metz PJ, et al. Regulation of Asymmetric Division by Atypical Protein Kinase C Influences Early Specification of CD8(+) T Lymphocyte Fates. Sci Rep. 2016;6:19182. doi: 10.1038/srep19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nature immunology. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 23.King CG, et al. T cell affinity regulates asymmetric division, effector cell differentiation, and tissue pathology. Immunity. 2012;37:709–720. doi: 10.1016/j.immuni.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Windt GJ, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci U S A. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R, et al. The Transcription Factor Myc Controls Metabolic Reprogramming upon T Lymphocyte Activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. S1074-7613(11)00515-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gera JF, et al. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J Biol Chem. 2004;279:2737–2746. doi: 10.1074/jbc.M309999200. [DOI] [PubMed] [Google Scholar]
- 28.West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- 29.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. 334/6056/678 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. S0092-8674(12)00998-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinclair LV, et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nature immunology. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torrents D, et al. Identification and characterization of a membrane protein (y+L amino acid transporter-1) that associates with 4F2hc to encode the amino acid transport activity y+L. A candidate gene for lysinuric protein intolerance. J Biol Chem. 1998;273:32437–32445. doi: 10.1074/jbc.273.49.32437. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Charrier L, Gewirtz A, Sitaraman S, Merlin D. CD98 and intracellular adhesion molecule I regulate the activity of amino acid transporter LAT-2 in polarized intestinal epithelia. J Biol Chem. 2003;278:23672–23677. doi: 10.1074/jbc.M302777200. [DOI] [PubMed] [Google Scholar]
- 35.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sukumar M, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science. 2009;323:505–509. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wherry EJ, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nature immunology. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 39.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.