Abstract
The most exciting recent advance for achieving durable management of advanced human cancers is immune therapy, especially the concept of immune checkpoint blockade. However, with the exception of melanoma, most patients do not respond to immune therapy alone. A growing body of work has shown that epigenetic drugs, specifically DNA methyltransferase inhibitors, can upregulate immune signaling in epithelial cancer cells through demethylation of endogenous retroviruses and cancer testis antigens. These demethylating agents may induce T-cell attraction and enhance immune checkpoint inhibitor efficacy in mouse models. Current clinical trials are testing this combination therapy as a potent new cancer management strategy.
Introduction
Arguably, the most exciting recent advance for achieving durable management of advanced human cancers is immune therapy, especially the concept of immune checkpoint blockade (1) (2-5) (6) (7). This immunotherapy explosion is a result of elegant fundamental discoveries of ligand receptor interactions that control the immune activity of T-cells against tumor cells (8-12). These basic advances and resulting translational applications constitute a key component of a paradigm that has been termed tumor “immune evasion” (13) (14). Interactions between the series of defined ligands and receptors on tumor cells and host immune cells render the latter immunologically inert or “tolerant”. This recognition and molecular dissection of the tolerant state completely resurrected the concept of targeting cancer immunologically and provided the tools to modulate immune signaling from both tumor and host immune cells, reversing a key element of immune evasion and promoting tumor elimination (14).
To this end, a growing body of clinical trials has shown exceptional promise. Antibodies blocking CTLA-4, an inhibitory molecule on T cells, produce durable responses for treatment of melanoma (6,7) and are currently in clinical trials for lung, prostate, and other cancers (15) (16,17). Antibodies targeting human PD-1 (receptor on T cells) and PD-L1, the inhibitory ligand for PD-1 that is expressed at varying levels by cancer cells, have produced exceptionally durable responses in patients with highly aggressive, treatment-resistant metastatic cancers. The effects may be most apparent in patients whose tumors express PD-L1 (1-5,18). While melanoma has been the most responsive solid tumor (5), exciting results have been achieved in the most lethal of cancers, advanced non-small cell lung cancer (NSCLC) (1,2,5). This is of special interest as this cancer was previously considered not to be immune responsive. FDA approval for melanoma and NSCLC has resulted from the above trials (5).
While these advances are very exciting, with the exception of melanoma, the majority of patients do not respond to immune checkpoint therapy alone (5,13). This raises the obvious question as to whether combinations of immune therapy with other agents could robustly extend clinical response and efficacy in a larger spectrum of cancer subtypes. Indeed, such concepts are evolving. First, combining immune checkpoint targeting agents in trials giving both anti-PD1 and anti-CTLA4 to patients, while mandating specialized care of toxicities, shows great promise for melanoma (7). Second, combination strategies with standard chemotherapy and targeted therapy approaches can be considered. In this regard, we consider the exciting possibility, gleaned from a signal seen by our group in the clinic and a growing body of pre-clinical data, that epigenetic therapy could robustly sensitize patients to immune checkpoint therapy.
Definition of epigenetic therapy
Although the term epigenetic therapy is now widely used, what defines and constitutes this term is a shifting concept. There has been an explosion over the last decade in our understanding of what constitutes the normal and cancer “epigenome” and how it is regulated (19-21). New insights are constantly emerging into functionally significant histone modifications, importance of DNA methylation patterns, and understanding of nucleosome occupancy dynamics (21). Epigenetic discoveries continually define not only promising new targets for cancer therapy but also ways to “re-use” older drugs already in use in the clinic (22). The above regulatory features, as they participate in abnormal epigenetic alterations in cancer, represent potentially reversible targets for existing drugs and an increasing repertoire of new drugs.
We concentrate in this review on the use of drugs already in the clinic that can induce epigenetic effects modulating immune parameters of tumor or host immune cells. These drugs are emblematic of the principal that epigenetic therapy generally targets three protein categories: Writers, enzymes that establish epigenetic marks; Readers, proteins that recognize histone modifications or DNA methylation, are recruited to these marks, and may bring in other protein complexes to change gene expression; and Erasers, enzymes that remove epigenetic marks (23). We will focus on drugs that inhibit writers of DNA methylation, DNA methyltransferases (DNMTs), and erasers (histone deacetylases or HDACs) that regulate histone lysine acetylation. The actions of DNMTs and HDACs are generally associated with transcriptional repression. Thus, the drugs targeting these proteins can augment expression of involved genes with many consequences for pathways downstream of this gene activation.
DNMT inhibitors (DNMTis) are cytidine analogues that, when incorporated into DNA, not only directly block the catalytic actions of DNMTs to trigger DNA demethylation but also cause their degradation (24). This latter loss of the protein, often not taken into account when considering use of DNMTis, can remove key scaffolding properties that may function for repression of transcription (25) (26,27). Cancers almost universally exhibit profound changes in DNA methylation of cytosines at CpG dinucleotides. These changes include global loss of methylation at regions such as repetitive elements that must be silenced for genome stability and gain of methylation at the promoter regions of tumor suppressor and other genes (19). DNMTis cause expression of genes that are silenced by promoter DNA methylation, reactivating tumor suppressor genes (28). Transient exposure of multiple types of tumor cells to low doses of DNMTis promotes induction of apoptosis, reduced cell cycle activity, and decreased stem cell functions in cancer cells (29). Clinical efficacy of DNA methyltransferase inhibitors (DNMTis), such as 5-azacytidine (Aza) and 5-aza-2’-deoxycytidine (Dac) (29,30) for treating hematologic neoplasms has led to FDA approval for the pre-leukemic disorder myelodysplasia (MDS) (31).
HDAC inhibitors (HDACis) are approved for treatment of cutaneous T cell lymphoma (CTCL) and peripheral T cell lymphoma (PTCL) (32) (33). It is, as yet, not clear why these tumors are so sensitive to HDACis (23). HDACis have pleiotropic effects, often very dose and compound dependent. Some of these affect histone acetylation and clearly induce epigenetic effects while others influence the acetylation status of non-histone and/or non-nuclear proteins, or cause off-target effects including DNA damage (23,34). Administered to tumor cells after low doses of DNMTis, HDACis can augment the re-expression of genes with promoter DNA hypermethylation (35). This combination is in clinical trials but it remains to be firmly established that it has clinical efficacy above the use of DNMTis and/or HDACis alone.
The intersection of epigenetic therapy with immunotherapy
Over the past several years, within the context of a Stand up to Cancer (SU2C) project to implement epigenetic therapy for cancer, our group has brought a low dose concept for use of DNMTis (Aza or Dac) with or without HDACis to clinical trials for multiple tumor types. Signals for potential efficacy have particularly appeared for advanced, multiply pre-treated non-small cell lung cancer (NSCLC) (36,37). One result in particular has driven much further emphasis in the clinic and the laboratory. A small number of patients with advanced NSCLC who progressed after receiving low-dose epigenetic therapy entered a trial for immune checkpoint therapy. Approximately 20% of the patients responded to the immune checkpoint therapy alone, passing 24 weeks without progression, with most achieving high-grade RECIST criteria responses (1,38). This is an astounding result for immunotherapy in NSCLC. All five patients who had received the prior epigenetic therapy passed the 24 week point without progression with subsequent immune checkpoint therapy and three of these developed high grade partial RECIST criteria responses that have all been durable over 2.5 years (36,37). These findings have prompted initiation of a larger clinical trial, which is now ongoing. Moreover, our laboratory group pursued studies to determine the mechanism(s) that might account for epigenetic sensitization to immune therapy. Our findings to date, and those of others, support the hypothesis that there may be extraordinary potential for combined epigenetic and immune therapy to increase the frequency of durable responses for immune checkpoint therapy in not only NSCLC but also other common tumor types.
Pre-clinical studies show that epigenetic therapy drugs boost immune attraction properties of epithelial cancer cells
DNMTis and HDACis have long been known to upregulate expression of individual components of immune signaling in epithelial cancer cells (39). Perhaps best recognized is induced expression of Cancer Testis Antigens (CTAs), including those on the X chromosome (CG-X antigens) and on autosomes (non-X CG antigens). CTAs are expressed in early embryonic and germ cells, but generally silenced in mature somatic cells by promoter CpG island DNA methylation (40). CTAs often remain DNA methylated and silenced in cancer cells although they can also lose methylation and be abnormally expressed (41). The promoter methylation of CTAs is controlled by interactions between DNMT1 and de novo DNMTs, principally DNMT3B. Inhibition of DNMTs can cause demethylation and re-expression of CTAs including the MAGE-A1 and NY-ESO-1 antigens (40) (42) in cancer cells but not normal fibroblasts (43). Hypomethylation of CTAs correlates with global hypomethylation in epithelial ovarian cancer (EOC), as well as BORIS upregulation (44). BORIS, a paralog of the CTCF insulator protein, is itself a CTA but is postulated to regulate other CTAs (41).
Since CTAs can be recognized by the host immune system, they represent good candidates for immune therapy, including vaccines. There is thus great potential for DNMT inhibitor treatment to upregulate CTAs on tumors, facilitating targeting by the host immune system (41). Guo et al. treated with the DNMTi Dac to demethylate and upregulate the murine CTA, P1A, in 4T.1 mammary carcinoma cells in syngeneic BALB/C mice. P1A was presented and recognized by H-2L d)-restricted P1A-specific T cells, and combined therapy with Dac and adoptive transfer of these T cells significantly reduced lung metastases in this mouse model (45). The novel DNMT inhibitor SGI110, which has longer in vivo stability than Aza or Dac and has shown clinical activity in patients with MDS and AML (46), also upregulates CTAs. In AML xenografts, SGI110 upregulates NY-ESO-1 and MAGE-A and induces cytotoxicity by CD8+ T cells specific for NY-ESO-1 (47). Similar results were observed in epithelial ovarian cancer (EOC) xenografts (48). These promising results led to a Phase I clinical trial in EOC in which Odunsi et al. added Dac to NY-ESO-1 vaccine combined with doxorubicin chemotherapy in patients with relapsed EOC. They observed DNA hypomethylation at the NY-ESO-1 promoter. NY-ESO-1 was upregulated and increased serum antibodies to NY-ESO-1 were detected, most importantly in two-thirds of the patients who previously were sero-negative for NY-ESO-1 antibodies. They observed specific T cell responses against NY-ESO-1 and stable disease or partial clinical response in 6/10 patients (49).
Our own data (37,50) validate the upregulation of CTAs by DNMT inhibitors. CTAs were significantly upregulated by Aza in the majority of 77 epithelial cancer cell lines. CTAs were most upregulated in colorectal (64% of cell lines) and ovarian (39%) cancer lines and less so for breast cancers (19%). We also noted an upregulation of genes involved in antigen processing and presentation by Aza treatment or in DNMT1−/− DNMT3B−/− DKO cells (51) compared to the parental HCT116 cell line (37,50). These include the MHC Class I proteins (B2M, HLA-A, HLA-B, HLA-C) that present antigens on the surface of epithelial cells for host immune cell recognition, as well as proteins involved in processing of antigens by the proteasome (PSMB8, PSMB9, TAP1) (50). This upregulation was previously noted by Karpf and colleagues after DNMTi treatment (42). Unlike CTAs, these genes are not initially methylated at their promoter regions, so a separate mechanism(s), likely downstream of epigenetic changes, is responsible for their upregulation.
The interferon response, upstream of antigen processing and presentation genes, is activated by DNMT inhibitors. This was first described by Karpf et al. (52); they observed an induction of STAT signaling and Type I interferon genes in colon cancer cells treated with Dac and showed that Dac could sensitize cells to treatment with interferon alpha. This activation was confirmed in a later study (42). Matei et al. observed upregulation of cytokines as well as JAK/STAT and interferon signaling pathways in tumor biopsies from ovarian cancer patients treated with a combination of Dac and carboplatin (30). High doses of DNMTi (10 μM Dac) induced an interferon response, apoptosis, and increased endogenous retroviral (ERV) transcripts and repetitive satellite RNAs in p53-null mouse fibroblasts (53). Leonova et al. attributed these latter responses to concordant regulation of satellite repeats by P53 and DNMTs and a buildup of repetitive RNAs that triggered the interferon response.
Against this above background, our group has observed a robust concordance for Aza and Dac induced increases in virtually all of the above immune parameters. We observed increased interferon signaling and concordant upregulation of surface antigens and their assembly proteins in 77 epithelial cancer cell lines treated with Aza (37,50). We defined a 300 gene expression signature that we termed Aza-Induced iMmune genes or AIM (50). In general, AIM genes were not induced by HDACi (TSA) treatment alone, but Aza plus TSA caused higher expression than Aza alone (50). We noted the highest AIM activation in EOC and NSCLC (50). Expression of AIM separated primary tumor samples from The Cancer Genome Atlas (EOC, NSCLC, and other cancers) into high and low expression groups (50). We hypothesize that the “low AIM” tumors represent an “immune evasion/ immune editing” pattern (54) (55) that Aza could reverse to sensitize patients to subsequent immune therapy (50).
Recent work from our group and the de Carvalho group (56,57) shows that one key way in which DNMTis upregulate immune signaling in cancer is through the viral defense pathway. In ovarian cancer cell lines, DNMTis activate a canonical interferon signaling pathway, inducing interferon beta and JAK/STAT signaling, through upregulation of dsRNA that activates the cytosolic dsRNA sensors TLR3 and MDA5. One type of RNA triggering this response is transcribed from hypermethylated endogenous retroviruses (ERVs) (56). Roulois et al. showed similar involvement of dsRNA and the MDA5 sensor in colon cancer cells and demonstrated that this interferon response was essential to the inhibition of colon cancer stem cells by DNMTis (57). Blocking the interferon response rescued about half of the DNMTi-induced apoptosis in ovarian cancer cells (56).
The ERVs that trigger the above DNMTi-induced immune response represent a significant fraction of repetitive elements in the human genome that are silenced in somatic cells by DNA methylation. In fact, up to 90% of methylated CpGs are located in the 45% of the human genome represented by repetitive sequences (58). ERVs are generally silenced in normal cells to promote genome stability, but are demethylated and re-expressed in some tumors. ERV demethylation and re-expression by DNMTis has been shown in human embryonic stem cells to cause upregulation of IFITM1, a protein involved in viral defense signaling (59). In melanoma, the ERV-K (HML-2) 5’LTR shows CpG hypomethylation and increased transcriptional activity (60). ERVs can be targeted as tumor-associated antigens on melanoma cells (61). Thus ERV activation promotes viral signaling and presents possible tumor-specific antigens to target.
While hypomethylation and upregulation of methylated regions encoding double-stranded RNA is a major contributor to the DNMTi-induced immune response, other target sites of demethylation can be important. IRF7, which encodes a master transcription factor activating the interferon response, is frequently silenced in association with promoter CpG island DNA hypermethylation in lung and other cancers (62) (37) (57) (50) (56). This loss of function can diminish interferon responses in tumor cells. Indeed, when this gene is methylated, its expression can be upregulated by Aza in squamous NSCLC (37) and EOC cells (50). Our group (56) and the De Carvalho group (57) found that when IRF7 is hypermethylated, knockdown of this gene significantly reduces the DNMTi-induced interferon response in ovarian (56) and colon (57) cancer cells, respectively.
How might epigenetic therapy then be combined with immune therapy to combat advanced cancers? As introduced earlier, we hypothesize that activation of the above viral defense gene signature by drugs like Aza might reverse elements of tumor immune evasion and enhance immune checkpoint therapy. In our recent study (56), basal expression levels of the Aza-induced viral defense gene signature in tumor samples correlate with long-term benefit in patients with advanced melanoma treated with the immune checkpoint inhibitor anti-CTLA-4 (63). Importantly, for virtually all of these melanoma patients, treatment benefit, high tumor mutational burden, and basal viral defense signature were all significantly associated (56). Moreover, low dose Aza plus anti-CTLA4 were significantly more effective at controlling tumor growth compared to each agent alone in the B16 mouse model of melanoma (56). These results point to the importance of immune/interferon signaling in the tumor cells, as B16 cells treated in vitro with Aza, then injected into mice who were then treated with anti-CTLA-4, showed the same effects (56). Melanoma has demonstrated the most impressive results for responses to immune checkpoint therapy (6) (5) (7). We would thus propose testing whether epigenetic therapy improves response to anti-CTLA-4 and/or anti-PD-1 therapies in clinical trials for melanoma.
Indeed, synergy of epigenetic and immune therapies was shown in ovarian cancer by Wang and colleagues (64). Treating a syngeneic mouse model of ovarian cancer with low-dose Dac treatment caused upregulation of chemokines that recruit host natural killer (NK) and effector CD8 + T cells to the tumor. In addition, Dac boosted the production of IFNγ and TNFα from effector T cells, while combining Dac with anti-CTLA-4 therapy promoted differentiation of naïve T cells into effector T cells. As a result, this combination reduced tumor burden in the mice and extended their survival.
Another way in which Aza may sensitize to immune checkpoint therapy is through upregulation of immune tolerance ligands on tumor cells. In EOC and NSCLC cell lines, transcript and surface protein levels of PD-L1 (37,50) were upregulated by Aza. Activation of this ligand is a downstream consequence of activating the viral/ interferon response pathway. Importantly, high versus low expression in tumor cells of this tumor ligand for the immune cell receptor PD-1 appears to correlate with good response to anti-PD-1 therapy (3) (1,2,4,5) (18). A thorough study of CD34+ blast cells from myelodysplastic syndrome (MDS), chronic myelomonocytic leukemia (CMML) and acute myeloid leukemia (AML) patients treated with Dac showed upregulation of PD-L1, PD-L2, PD-1, and CTLA-4. PD-1 upregulation was due to demethylation of the gene (65). Thus immune checkpoint blockade drugs targeting these pathways might benefit MDS/AML patients, especially those receiving Dac.
Epigenetic regulation of host immune cells
While all of the above research has been focused on the effects of epigenetic therapy agents on tumor cells, these drugs affect host immune cells as well. Epigenetic regulation during development and differentiation of host immune cells has been well described (66) (67) (68) (39,69). The gene encoding the Foxp3 transcription factor controls regulatory T cell (Treg) development and function (70) (71) (68). Tregs are necessary for control of autoimmunity but also dampen the host immune response against tumor cells. Foxp3 is methylated and not expressed in naïve CD4+CD25− T cells as well as activated CD4+ T cells but unmethylated and expressed in Tregs (72). The Foxp3 protein is stabilized by acetylation by HDAC9, promoting Treg development and preventing transcription of IL-2, the cytokine produced by CD8+ T effector cells (23). Thus DNMTs and HDACs have opposite effects on Treg development.
HDAC inhibitors boost anti-tumor immune responses. The HDACis panobinostat and vorinostat reduce tumor burden in immunocompetent mice, but not in immunocompromised RAG2γC−/− and IFNγR−/− mice (73). The authors found significant synergy of HDACi and interferon gamma (IFNy) treatment in mouse models of colon cancer and lymphoma. IFNy is secreted by cytotoxic T cells and natural killer (NK) cells and in these experiments it increased immunogenicity of tumor cells (73). Interestingly, B cells were a crucial component of the immune system in the response to HDACis (73). In addition, panobinostat significantly increased the effectiveness of adoptive cell transfer therapy (gp100 specific T cells) in the B16 mouse model of melanoma. Panobinostat enhanced gp100 specific T cell survival and decreased Tregs in the peripheral blood and the tumor microenvironment. This HDACi also induced significantly higher levels of the IL-2 receptor (CD25) and the co-stimulatory molecule OX-40 on T cells in the B16 mice. Taken together, these results suggest that HDACis boost the host immune response to tumors through B and T cells (74). In addition, inhibiting HDACs can also reduce myeloid-derived cells that can induce immune tolerance to help prevent the immune system from clearing tumors (75).
Epigenetic agents may also affect the development of natural killer (NK) cells, which recognize virus-infected cells or newly formed tumor cells and release cytokines to kill the infected cells. Specifically, Dac has been shown to sensitize AML blasts to lysis by NK cells. Kopp et al. showed that killer immunoglobulin-like receptors (KIR) and the activating receptor NKp44 were upregulated on NK cells expanded in vitro and treated with low doses of Dac. However, high doses of Dac decrease NK cell proliferation and viability (76). These data, along with use of drugs like DNMTis in MDS and recent pre-clinical studies, suggest that the beneficial immune effects of epigenetic therapy, like the beneficial effects on tumor cells, occur at low doses that avoid toxicities and immunosuppression (29) (56) (57) (22).
From these studies, it is apparent that epigenetic therapies will have effects on the host immune cells as well as the tumor cells. Thus, for full understanding of the potential for epigenetic therapy to sensitize to immune checkpoint therapy, it will be crucial in clinical trials to study biopsies from both the tumor and the peripheral or tumor infiltrating host immune cells before and after treatment.
Conclusions
We have put forth preclinical evidence to suggest how epigenetic therapy, via several signaling mechanisms involving both tumor cells and host immune cells, might enhance the efficacy of immune checkpoint therapy (Figure 1). Through coordinated upregulation of tumor antigens and MHC proteins, and interferon pathway induction by dsRNA transcripts including ERVs, DNA demethylating agents may induce T-cell attraction. Immune checkpoint efficacy in this setting may be enhanced when tolerance inducing ligand and receptor interactions are interrupted. Only clinical trials can prove the efficacy of this proposed paradigm. However, successes could establish epigenetic therapy as a relatively well tolerated addition to immune checkpoint therapy as a potent new cancer management strategy.
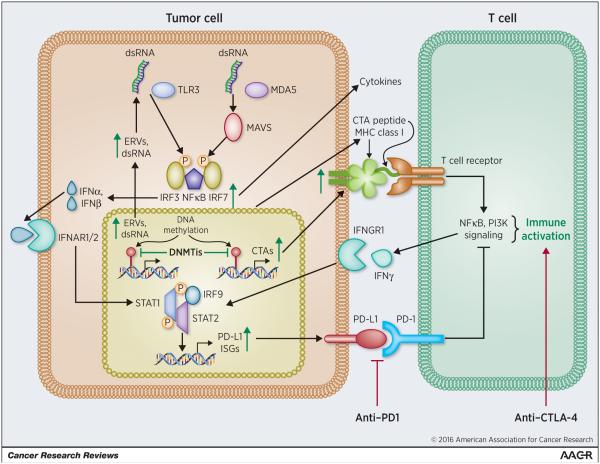
Figure 1. DNA methyltransferase inhibitors upregulate immune signaling in epithelial cells to synergize with immune checkpoint blockade therapy.
DNA methyltransferase inhibitors (DNMTis) remove methylation from promoter regions of silenced endogenous retroviruses (ERVs), causing double-stranded RNA (dsRNA) to activate sensors including TLR3 and MDA5, which signal through MAVS and IRF7 to cause transcription and secretion of interferon alpha and/or beta (IFNα, β). IFNα,β bind to the IFNAR1/2 receptor, activating JAK/STAT signaling and transcription of Interferon Stimulated Genes (ISGs) that include molecules involved in dsRNA destruction and apoptosis, cytokines that signal to host immune cells, as well as antigen processing and presentation (MHC Class I) genes. Separately, Cancer Testis Antigens (CTAs) are upregulated by DNMTi removal of methylation from their promoters and presented by MHC Class I on the cell surface and aid T cell recognition of cancer cells. Anti-CTLA-4 further aids activation of T cells and secretion of interferon gamma (IFNγ) that binds to its receptor IFNGR1 to activate STAT signaling and transcription of ISGs. The PD-L1 ligand is upregulated downstream of DNMTi treatment and the interferon response and binds to PD-1 on T cells to inhibit T cells; this interaction is disrupted by anti-PD-1 to promoter T cell activation.
References
- 1.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28(19):3167–75. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger R, Rotem-Yehudar R, Slama G, Landes S, Kneller A, Leiba M, et al. Phase I safety and pharmacokinetic study of CT-011, a humanized antibody interacting with PD-1, in patients with advanced hematologic malignancies. Clin Cancer Res. 2008;14(10):3044–51. doi: 10.1158/1078-0432.CCR-07-4079. [DOI] [PubMed] [Google Scholar]
- 4.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271(5256):1734–6. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 5.Topalian SL, Drake CG, Pardoll DM. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell. 2015;27(4):450–61. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber JS, D'Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16(4):375–84. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 8.Tivol EA, Borriello F, Schweitzer AN, Lynch WP, Bluestone JA, Sharpe AH. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 1995;3(5):541–7. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 9.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027–34. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11(2):141–51. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 11.Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270(5238):985–8. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 12.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8(8):793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 13.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161(2):205–14. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Squibb B-M. 2007 Phase II Study for Previously Untreated Subjects With Non Small Cell Lung Cancer (NSCLC) or Small Cell Lung Cancer (SCLC) ( NCT00527735) https://clinicaltrials.gov/show/NCT00527735.
- 16.Network HCR 2012 Phase II Trial of Gemcitabine, Cisplatin, Plus Ipilimumab as First-line Treatment for Patients With Metastatic Urothelial Carcinoma: Hoosier Cancer Research Network GU10-148 ( NCT01524991) https://clinicaltrials.gov/ct2/show/study/NCT01524991.
- 17.Squibb B-M. Study of MDX-010 in Patients With Metastatic Hormone-Refractory Prostate Cancer ( NCT00323882) 2006.
- 18.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–28. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 19.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153(1):38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allis C, Jenuwein T, Reinberg D, Caparros M. In: Epigenetics. Second Caparros M, editor. Cold Spring Harbor Laboratores Press; 2015. [Google Scholar]
- 22.Shen H, Laird PW. In epigenetic therapy, less is more. Cell stem cell. 2012;10(4):353–4. doi: 10.1016/j.stem.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13(9):673–91. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 24.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66(5):2794–800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 25.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25(3):269–77. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 26.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1(1):11–9. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 27.Clements EG, Mohammad HP, Leadem BR, Easwaran H, Cai Y, Van Neste L, et al. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baylin SB, Jones PA. Epigenetic Determinants of Cancer. In: Allis CD, Carparros M-L, Jenuwein T, Reinberg D, editors. Epigenetics. 2nd. Cold Spring Harbor Laboratories; Cold Spring Harbor, NY: 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, et al. Transient Low Doses of DNA-Demethylating Agents Exert Durable Antitumor Effects on Hematological and Epithelial Tumor Cells. Cancer Cell. 2012;21(3):430–46. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72(9):2197–205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11(10):3604–8. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- 32.Duvic M, Talpur R, Ni X, Zhang C, Hazarika P, Kelly C, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109(1):31–9. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.VanderMolen KM, McCulloch W, Pearce CJ, Oberlies NH. Romidepsin (Istodax, NSC 630176, FR901228, FK228, depsipeptide): a natural product recently approved for cutaneous T-cell lymphoma. J Antibiot (Tokyo) 2011;64(8):525–31. doi: 10.1038/ja.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert C, Rassool FV. HDAC Inhibitors: Roles of DNA Damage and Repair. Adv Cancer Res. 2012;116:87–129. doi: 10.1016/B978-0-12-394387-3.00003-3. [DOI] [PubMed] [Google Scholar]
- 35.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21(1):103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 36.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1(7):598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wrangle J, Wang W, Koch A, Easwaran H, Mohammad HP, Vendetti F, et al. Alterations of immune response of Non-Small Cell Lung Cancer with Azacytidine. Oncotarget. 2013;4(11):2067–79. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heninger E, Krueger TE, Lang JM. Augmenting antitumor immune responses with epigenetic modifying agents. Frontiers in immunology. 2015;6:29. doi: 10.3389/fimmu.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James SR, Link PA, Karpf AR. Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b. Oncogene. 2006;25(52):6975–85. doi: 10.1038/sj.onc.1209678. [DOI] [PubMed] [Google Scholar]
- 41.Karpf AR. A potential role for epigenetic modulatory drugs in the enhancement of cancer/germ-line antigen vaccine efficacy. Epigenetics. 2006;1(3):116–20. doi: 10.4161/epi.1.3.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karpf AR, Lasek AW, Ririe TO, Hanks AN, Grossman D, Jones DA. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2'-deoxycytidine. Mol Pharmacol. 2004;65(1):18–27. doi: 10.1124/mol.65.1.18. [DOI] [PubMed] [Google Scholar]
- 43.Cheng JC, Yoo CB, Weisenberger DJ, Chuang J, Wozniak C, Liang G, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6(2):151–8. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 44.Woloszynska-Read A, Zhang W, Yu J, Link PA, Mhawech-Fauceglia P, Collamat G, et al. Coordinated cancer germline antigen promoter and global DNA hypomethylation in ovarian cancer: association with the BORIS/CTCF expression ratio and advanced stage. Clin Cancer Res. 2011;17(8):2170–80. doi: 10.1158/1078-0432.CCR-10-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo ZS, Hong JA, Irvine KR, Chen GA, Spiess PJ, Liu Y, et al. De novo induction of a cancer/testis antigen by 5-aza-2'-deoxycytidine augments adoptive immunotherapy in a murine tumor model. Cancer Res. 2006;66(2):1105–13. doi: 10.1158/0008-5472.CAN-05-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taverna P, Lyons JF, Hao Y, Azab M, Kantarjian HM, Kropf P, et al. Determinants Of Demethylation and Clinical Response In AML Patients Treated With SGI-110, a Novel Subcutaneous (SQ) Hypomethylating Agent (HMA) In a Phase 1 Study. 2013. pp. 1442–42.
- 47.Srivastava P, Paluch BE, Matsuzaki J, James SR, Collamat-Lai G, Karbach J, et al. Immunomodulatory action of SGI-110, a hypomethylating agent, in acute myeloid leukemia cells and xenografts. Leuk Res. 2014;38(11):1332–41. doi: 10.1016/j.leukres.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srivastava P, Paluch BE, Matsuzaki J, James SR, Collamat-Lai G, Taverna P, et al. Immunomodulatory action of the DNA methyltransferase inhibitor SGI-110 in epithelial ovarian cancer cells and xenografts. Epigenetics. 2015;10(3):237–46. doi: 10.1080/15592294.2015.1017198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Odunsi K, Matsuzaki J, James SR, Mhawech-Fauceglia P, Tsuji T, Miller A, et al. Epigenetic potentiation of NY-ESO-1 vaccine therapy in human ovarian cancer. Cancer immunology research. 2014;2(1):37–49. doi: 10.1158/2326-6066.CIR-13-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RW, Vatapalli R, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5(3):587–98. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416(6880):552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 52.Karpf AR, Peterson PW, Rawlins JT, Dalley BK, Yang Q, Albertsen H, et al. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc Natl Acad Sci U S A. 1999;96(24):14007–12. doi: 10.1073/pnas.96.24.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110(1):E89–98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Advances in immunology. 2006;90:51–81. doi: 10.1016/S0065-2776(06)90002-9. [DOI] [PubMed] [Google Scholar]
- 55.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331(6024):1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 56.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162(5):974–86. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 2015;162(5):961–73. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20(2):116–7. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 59.Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015 doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stengel S, Fiebig U, Kurth R, Denner J. Regulation of human endogenous retrovirus-K expression in melanomas by CpG methylation. Genes Chromosomes Cancer. 2010;49(5):401–11. doi: 10.1002/gcc.20751. [DOI] [PubMed] [Google Scholar]
- 61.Cooper LJ, Krishnamurthy J, Rabinovich B, Mi T, Switzer K, Olivares S, et al. Genetic Engineering of T cells to Target HERV-K, an Ancient Retrovirus on Melanoma. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-14-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Q, Tainsky MA. Epigenetic silencing of IRF7 and/or IRF5 in lung cancer cells leads to increased sensitivity to oncolytic viruses. PLoS One. 2011;6(12):e28683. doi: 10.1371/journal.pone.0028683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L, Amoozgar Z, Huang J, Saleh MH, Xing D, Orsulic S, et al. Decitabine Enhances Lymphocyte Migration and Function and Synergizes with CTLA-4 Blockade in a Murine Ovarian Cancer Model. Cancer immunology research. 2015;3(9):1030–41. doi: 10.1158/2326-6066.CIR-15-0073. [DOI] [PubMed] [Google Scholar]
- 65.Oyan AM, Bo TH, Jonassen I, Ulvestad E, Gjertsen BT, Kalland KH, et al. CD34 expression in native human acute myelogenous leukemia blasts: differences in CD34 membrane molecule expression are associated with different gene expression profiles. Cytometry B Clin Cytom. 2005;64(1):18–27. doi: 10.1002/cyto.b.20044. [DOI] [PubMed] [Google Scholar]
- 66.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467(7313):338–42. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Youngblood B, Hale JS, Ahmed R. T-cell memory differentiation: insights from transcriptional signatures and epigenetics. Immunology. 2013;139(3):277–84. doi: 10.1111/imm.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morikawa H, Sakaguchi S. Genetic and epigenetic basis of Treg cell development and function: from a FoxP3-centered view to an epigenome-defined view of natural Treg cells. Immunol Rev. 2014;259(1):192–205. doi: 10.1111/imr.12174. [DOI] [PubMed] [Google Scholar]
- 69.Alvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol. 2015;15(1):7–17. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 70.Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108(5):1571–9. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5(2):e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114(18):3727–35. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.West AC, Mattarollo SR, Shortt J, Cluse LA, Christiansen AJ, Smyth MJ, et al. An intact immune system is required for the anticancer activities of histone deacetylase inhibitors. Cancer Res. 2013;73(24):7265–76. doi: 10.1158/0008-5472.CAN-13-0890. [DOI] [PubMed] [Google Scholar]
- 74.Lisiero DN, Soto H, Everson RG, Liau LM, Prins RM. The histone deacetylase inhibitor, LBH589, promotes the systemic cytokine and effector responses of adoptively transferred CD8+ T cells. J Immunother Cancer. 2014;2:8. doi: 10.1186/2051-1426-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waibel M, Christiansen AJ, Hibbs ML, Shortt J, Jones SA, Simpson I, et al. Manipulation of B-cell responses with histone deacetylase inhibitors. Nat Commun. 2015;6:6838. doi: 10.1038/ncomms7838. [DOI] [PubMed] [Google Scholar]
- 76.Kopp LM, Ray A, Denman CJ, Senyukov VS, Somanchi SS, Zhu S, et al. Decitabine has a biphasic effect on natural killer cell viability, phenotype, and function under proliferative conditions. Mol Immunol. 2013;54(3-4):296–301. doi: 10.1016/j.molimm.2012.12.012. [DOI] [PubMed] [Google Scholar]