Abstract
Mycobacterium tuberculosis (Mtb) survives within macrophages by evading delivery to the lysosome and promoting the accumulation of lipid bodies, which serve as a bacterial source of nutrients. Here we show that by inducing miR-33 and its passenger strand miR-33*, Mtb inhibits integrated pathways involved in autophagy, lysosomal function and fatty acid oxidation to support bacterial replication. Silencing of miR-33 and miR-33* by genetic or pharmacological means promotes autophagy flux through derepression of key autophagy effectors such as ATG5, ATG12, LC3B and LAMP1 and AMPK-dependent activation of the transcription factors FOXO3 and TFEB, enhancing lipid catabolism and Mtb xenophagy. These data define a mammalian miRNA circuit utilized by Mtb to coordinately inhibit autophagy and reprogram host lipid metabolism to enable intracellular survival and persistence in the host.
Mycobacterium tuberculosis (Mtb), the causative agent of the disease tuberculosis, remains the leading worldwide bacterial cause of death. Mtb survives within mononuclear cells, where it successfully combats macrophage microbicidal mechanisms by evading delivery to the lysosome, thereby limiting bacterial degradation1. Autophagy can contribute to the degradation of intracellular bacteria (xenophagy) through the formation of autophagosomes that sequester cytoplasmic bacilli and deliver them to lysosomes for degradation. Autophagy also plays important roles in innate and adaptive immune responses by promoting antigen presentation and modulating inflammatory responses. Its importance in host immunity is underscored by the observation that many intracellular pathogens have autophagy evasion strategies2. In the case of Mtb, when autophagy is stimulated by pharmacologic means or interferon-γ (IFN-γ) treatment, Mtb targeting to autophagosomes and bacterial killing is enhanced3, 4, 5. However, in the absence of such stimuli, the majority of Mtb do not associate with microtubule-associated protein 1 light chain 3 (LC3), a marker of autophagy. Moreover, in resting macrophages and in vivo in mice, autophagy only makes a modest contribution towards clearing Mtb3, 6, 7, 8. These observations raise the possibility that Mtb blocks autophagy, as has been shown for a number of human pathogens.
During Mtb infection, xenophagy is initiated when mycobacterial DNA is detected by the cytosolic DNA sensor cGAS6, 9, 10, 11. Once initiated, over 30 proteins, including the autophagy-related gene (ATG) proteins, orchestrate sequential membrane remodelling and trafficking events to complete autophagosomal membrane nucleation (Beclin-1, VPS34, ATG14L), autophagosomal elongation and maturation (ATG5, ATG12, ATG16L1, ATG4B, ATG3, ATG7, LC3), and lysosomal docking and fusion (Syntaxin17, UVRAG)12, 13. Transcriptional reinforcement is required to sustain autophagy by replenishing components that get degraded along with captured cargo, such as adapter proteins that bind cargo (eg. p62) and LC3, which in its phosphatidylethanolamine-conjugated form (LC3-II) is associated with the autophagosomal membrane. The transcription factors FOXO314 and TFEB15, which are activated by the adenosine 5’ monophosphate-activated protein kinase (AMPK), accomplish this by promoting the expression of genes involved in autophagosome and lysosomal biogenesis and function16, 17.
Mtb also alters macrophage cellular metabolism to promote the accumulation of lipid bodies, which serve as a source of nutrients in the form of cholesterol esters and fatty acids18, 19, 20. These “foamy” macrophages provide a secure niche for the bacterium by enabling survival and replication, and ultimately, persistence in the human host. The formation of mycobacterial lipid bodies has been shown to be dependent on Toll-like receptor signalling pathways, and isolated components of the mycobacterial cell wall such as lipoarabinomannan (LAM) can mimic the pathogen and induce lipid body formation21. However, the mechanisms regulating lipid body formation in Mtb-infected macrophages remain poorly understood.
MicroRNAs (miRNAs) have emerged as important post-transcriptional “fine-tuners” of gene expression in response to pathophysiological stimuli. These small noncoding RNAs bind to the 3’-untranslated region (3’UTR) of target mRNAs and reduce protein expression by blocking mRNA translation and/or by promoting mRNA degradation. A given miRNA can simultaneously regulate multiple target genes, often with related functions, resulting in potent cumulative effects on gene networks. Notable examples of this mechanism are miR-33a and miR-33b, intronic miRNAs embedded in the human sterol response element binding protein genes, SREBF2 and SREBF1, respectively (mice lack miR-33b)22, 23, 24, 25. miR-33a/b repress genes involved in cellular cholesterol export and fatty acid oxidation, including ABCA1, CROT, CPT1, HADHB and PRKAA1, thereby boosting cellular lipid levels and lowering plasma levels of high density lipoproteins22, 23, 24, 25. Furthermore, although generally one strand of a miRNA duplex preferentially associates with the RNA-induced silencing complex (RISC) and the “passenger” or miRNA* strand is degraded, both miR-33 and miR-33* accumulate and target an overlapping subset of lipid metabolism genes, suggesting that the two strands processed from the miR-33 precursor duplex may co-regulate biological pathways26.
Here we demonstrate that autophagy is a key pathway targeted by miR-33 and its passenger strand miR-33*. As autophagy promotes lipid catabolism (lipophagy) by delivering lipid droplet-stored triglycerides and cholesterol esters to lysosomes13, 27, autophagy suppression by miR-33 and miR-33* contributes to the coordinated actions of these miRNAs to promote cellular lipid accumulation. Moreover, we show that Mtb takes advantage of this host regulatory circuit by inducing the expression of miR-33 and miR-33* during macrophage infection to simultaneously impair xenophagy and promote fatty acid stores in lipid bodies to ensure its intracellular survival.
RESULTS
Mtb induces miR-33 and miR-33* expression
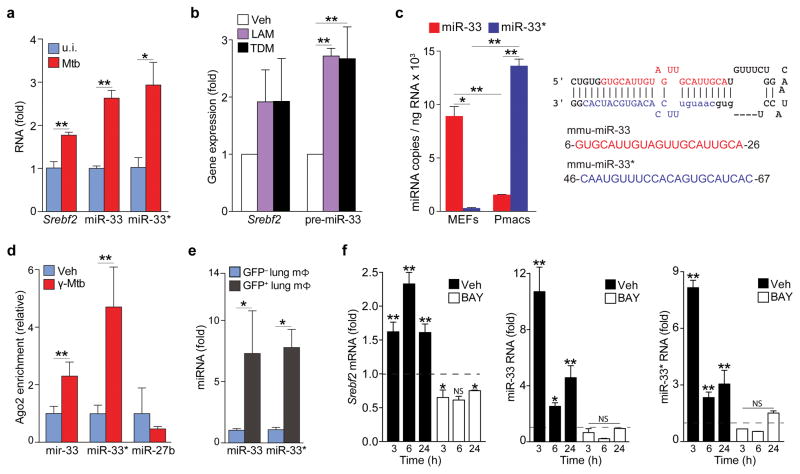
Because miR-33 boosts cellular sterol levels and is regulated by NF-κB28, we surmised that it might drive a metabolic response to Mtb infection. Infection of mouse peritoneal macrophages with the Mtb H37Rv strain increased expression of miR-33 and its host gene Srebf2 (Fig. 1a). Notably, Mtb H37Rv induced a parallel increase in the miR-33 passenger strand, miR-33*, in peritoneal macrophages (Fig. 1a), and similar results were observed in human THP-1 macrophages (Supplementary Fig. 1). Treatment of mouse bone marrow derived macrophages (BMDMs) with gamma-irradiated Mtb (γ-Mtb) or purified components of the mycobacterial cell wall LAM and trehalose dimycolate (TDM) increased expression of Srebf2 and the miR-33 precursor transcript from which mature miR-33 and miR-33* arise (Fig. 1b). Whereas copy numbers of miR-33* were low compared to miR-33 in mouse embryonic fibroblasts (MEFs), miR-33* was relatively abundant in mouse peritoneal macrophages (Fig. 1c), suggesting that it may have important roles in this cell type. Ago2 immunoprecipitated from peritoneal macrophages treated with γ-Mtb showed increased association with miR-33 and miR-33* compared to uninfected macrophages (Fig. 1d) demonstrating that both strands of the miR-33 duplex were loaded onto the RNA-induced silencing complex (RISC) upon Mtb infection.
Figure 1. M. tuberculosis infection upregulates the miR-33 locus in macrophages.
(a) qPCR quantification of miR-33, miR-33*, and Srebf2 levels in peritoneal macrophages infected with H37Rv Mtb for 48 h. (b) qPCR quantification of Srebf2 and pre-miR-33 in BMDMs treated with lipoarabinomannan (LAM; 5 μg/mL) or trehalose dimycolate (TDM; 2.5 μg/mL) 3 h (Srebf2) or 24 h (pre-miR-33) after treatment. (c) qPCR quantification of miR-33 and miR-33* expression in mouse embryonic fibroblasts (MEF) and peritoneal macrophages (Pmac). The mmu-miR-33 stem loop structure with the guide (miR-33; blue) and passenger (miR-33*; red) strand are shown at right. (d) Co-precipitation of endogenous miR-33, miR-33* or miR-27b (control) with Argonaute2 in peritoneal macrophages treated with vehicle (control) or gamma-irradiated Mtb (γ-Mtb; 10 μg/mL). (e) Quantification of miR-33 and miR-33* expression in GFP+ (infected) or GFP− (uninfected) alveolar macrophages isolated from mice infected with a GFP-expressing H37Rv Mtb strain for 2 weeks. (f) Quantification of Srebf2, miR-33 and miR-33* levels in BMDMs treated with γ-Mtb in the absence (Unstim) or presence of the NFκB inhibitor BAY11-7082 (BAY; 1 μM). Data are expressed as fold-change for the γ-Mtb treatment relative to untreated for each time point and dotted lines dileneate the threshold of control. *P≤0.1, **P≤0.05 (Students’ t-test (a, d, e), one-way ANOVA (b), two-way ANOVA (c, f)). Data are from one experiment representative of 3 independent experiments with similar results (a, f; mean ± s.e.m). Data are from 2 experiments (b, c, d; mean ± s.e.m). Data are from 3 mice (e; mean ± s.e.m).
To investigate whether miR-33 and miR-33* are regulated by Mtb in vivo, we isolated alveolar macrophages from the lungs of mice infected with Mtb expressing green fluorescent protein (GFP). We observed a five-fold increase in miR-33 and miR-33* expression in infected (GFP+) compared to uninfected (GFP−) macrophages from the same mice (Fig. 1e). To assess whether NF-κB was required for the induction of miR-33 and miR-33*, we treated BMDMs with the NF-κB inhibitor, BAY11-7082, and quantified miRNA expression in response to γ-Mtb infection. BAY11-7082 prevented induction of Srebf2, miR-33 and miR-33* expression by γ-Mtb (Fig. 1f). Together, these data indicate that miR-33 and miR-33* expression is upregulated in macrophages by Mtb and its cell wall constituents via an NF-κB dependent mechanism.
miR-33 and miR-33* regulate lipid body catabolism
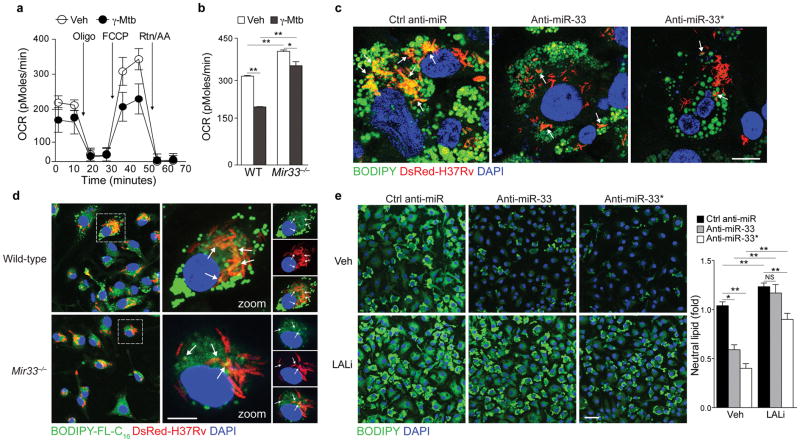
Next, we investigated whether miR-33 and miR-33* expression in Mtb-infected macrophages contributes to the formation of fatty acid-rich lipid bodies. Using Seahorse extracellular flux analysis, we quantified cellular oxygen consumption rates (OCR) as a measure of fatty acid β-oxidation in unstimulated or γ-Mtb-treated macrophages. Notably, γ-Mtb reduced OCR in wild-type BMDMs, whereas OCR was largely restored in Mir33−/− BMDMs that lack expression of both miR-33 and miR-33* (Fig. 2a, b). We quantified neutral lipids in Mtb-infected macrophages treated with anti-sense oligonucleotide (ASO) inhibitors of miR-33 or miR-33* or a non-targeting control ASO. In control ASO treated macrophages we observed numerous BODIPY-labeled lipid bodies that co-localized with intracellular bacteria in the cytoplasm (Fig. 2c). Upon inhibition of miR-33 or miR-33*, the size and number of lipid bodies in Mtb-infected macrophages were reduced (Fig. 2c). To test whether miR-33 and miR-33* regulated the utilization of host cell triglycerides by Mtb, we labeled macrophages with a BODIPY-C16 fatty acid analog prior to infection. Wild type macrophages showed incorporation of lipid droplet derived BODIPY-labeled fatty acid into the intracellular bacteria, and this was greatly reduced in Mir33−/− macrophages (Fig. 2d). Combined, these data suggest that Mtb-induced expression of miR-33 and miR-33* in macrophages impairs mitochondrial fatty acid oxidation to augment cellular lipid stores that are imported into the bacilli to serve as a nutrient source during chronic infection.
Figure 2. miR-33 and miR-33* reduce fatty acid oxidation and promote lipid body formation in M. tuberculosis-infected macrophages.
(a) Oxygen consumption rate (OCR) of BMDMs cultured with γ-Mtb for 6h and sequentially treated with Oligo, FCCP and rotenone plus antimycin (Rtn/AA) at the indicated times (arrows), measured in real time and presented as change in pMoles per unit time. (b) Maximal OCR of WT or miR-33−/− BMDMs cultured with γ-Mtb for 6h measured in real time and presented as change in pMoles per unit time (OCR). (c) Immunofluorescence imaging of BODIPY-stained lipid droplets in control anti-miR, anti-miR-33 or anti-miR-33* treated THP-1 macrophages infected with H37Rv for 24h. Scale bar = 25μm. (d) Immunofluorescence (IF) imaging of WT or miR-33−/− macrophages incubated with BODIPY-FL-C16 and BSA-conjugated oleic acid for 48 h to label nascent lipid droplets and then infected with DsRed-H37Rv Mtb for 6h. Scale bar = 25μm. (e) IF imaging of BODIPY-stained lipid droplets in ctrl anti-miR, anti-miR33 or anti-miR33* treated macrophages, incubated with or without an inhibitor of lysosomal acid lipase (LALi, 10μM) for 8h. Scale bar = 10μm. NS, not significant, *P≤0.05, **P≤0.005 (Student’s t-test (b), one-way ANOVA (e)). Data are from one experiment representative of 3 (a, b; mean ± s.e.m, and d) or 2 (c and e; mean ± s.e.m) independent experiments with similar results.
miR-33 regulates genes in the autophagy pathway
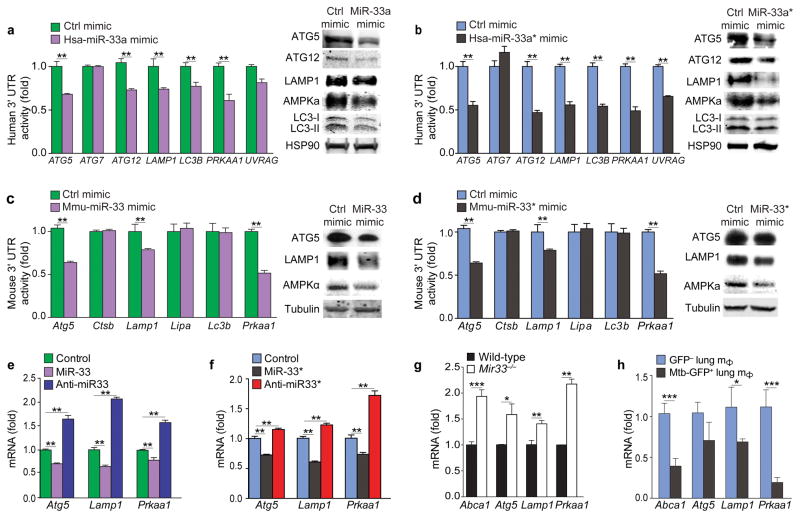
Autophagy participates in lipid metabolism by promoting the catabolism of lipid droplets in lysosomes27, 29. To test whether miR-33 and miR-33* regulate lipid droplet turnover via autophagy, we treated BMDMs with oleic acid to form lipid droplets and tested the ability of miR-33 and miR-33* inhibitors to promote their clearance in the presence or absence of an inhibitor of lysosomal acid lipase (LAL). Compared to BMDMs treated with control ASO, BMDMs treated with anti-miR-33 or anti-miR-33* had reduced lipid droplet content (Fig. 2e); however this was prevented upon inhibition of LAL (Fig. 2e), the lysosomal enzyme required for autophagy-mediated lypolysis. Using the prediction algorithms TargetScan and miRanda, we identified genes in the autophagy pathway as potential targets of human and mouse miR-33 and miR-33*, including genes involved in autophagosome formation, such as ATG5, ATG7, ATG12, MAP1LC3B, lysosomal function, such as lysosomal-associated membrane protein 1 (LAMP1), lysosomal acid lipase (LAL), and regulation of autophagy, such as UVRAG, PRKAA1, PRKAA2 (Supplementary Fig. 2). To investigate whether miR-33 and miR-33* regulate autophagy, we selected a subset of putative target genes involved in different stages of autophagy for further validation. Human embryonic kidney 293 (HEK293) cells were transfected with 3’UTR-luciferase reporter genes for human ATG5, ATG7, ATG12, LAMP1, UVRAG and MAP1LC3B (encoding LC3B) in the presence of hsa-miR-33a or control mimic. The 3’UTR-luciferase activity of ATG5, ATG12, LAMP1, UVRAG and MAP1LC3B, but not ATG7, was lower in cells treated with hsa-miR-33a mimic as compared to control mimic (Fig. 3a). Furthermore, THP-1 macrophages treated with hsa-miR-33a mimic had lower expression of ATG5, ATG12, LAMP1 and MAP1LC3B protein compared to control mimic-treated macrophages (Fig. 3a and Supplementary Fig 3), confirming these autophagy effectors as targets of miR-33. Similarly, the hsa-miR-33a* mimic reduced the 3’UTR-luciferase activity of ATG5, ATG12, LAMP1, UVRAG and MAP1LC3B and the protein amounts of these gene targets (Fig. 3b and Supplementary Fig 3), pointing to a common role for hsa-miR-33a and hsa-miR-33a* in regulation of the autophagy pathway.
Figure 3. Regulation of human and mouse autophagy gene targets by miR-33 and miR-33*.
(a–b) 3’UTR-luciferase reporter activity of human autophagy gene targets in HEK293 cells transfected with control (ctrl) mimics or (a) hsa-miR-33a or (b) hsa-miR-33a* (left panels). Western blotting of lysates from human THP-1 macrophages treated with control (ctrl) mimics or (a) hsa-miR-33a or (b) hsa-miR-33a* (right panels)., HSP90 is shown as an internal control (right). (c–d) 3’UTR-luciferase reporter activity of mouse autophagy gene targets in HEK293 cells transfected with control mimics or (c) mmu-miR-33 or (d) mmu-miR-33* (left panels). Western blotting of lysates from mouse peritoneal macrophages treated with control mimics or (c) mmu-miR-33 or (d) mmu-miR-33* (right panels). (e, f) mRNA levels of Atg5, Lamp1 and Prkaa1 in peritoneal macrophages transfected with (e) mmu-miR-33 or anti-miR33 or (f) mmu-miR-33* or anti-miR-33*. (g) qPCR analysis of miR-33 and miR-33* target gene mRNA expression in wild type (WT) and Mir33−/− BMDMs. (h) Quantification of miR-33 and miR-33* gene targets in GFP+ (infected) or GFP− (uninfected) alveolar macrophages isolated from mice infected with a GFP-expressing H37Rv Mtb strain for 2 weeks. *P≤0.1, **P≤0.05, ***P≤0.005 (Student’s t-test (a–d, g, h), one-way ANOVA (e, f)). Data are from one experiment representative of 3 (a–f; mean ± s.e.m) or 2 (g; mean ± s.e.m) independent experiments. Data are from 3 mice (h; mean ± s.e.m).
Of the human autophagy targets of miR-33 and miR-33*, Atg5 and Lamp1 contain conserved miR-33 and miR-33* binding sites in mouse. Mmu-miR-33 and mmu-miR-33* mimic reduced the activity of the Atg5 and Lamp1 3’UTR-luciferase reporter in HEK293 cells, as compared to control mimic treated HEK293 cells (Fig. 3c, d). Furthermore, peritoneal macrophages transfected with miR-33 and miR-33* mimics had lower expresion of ATG5 and LAMP1 protein compared to control mimic-treated macrophages (Fig. 3c, d; Supplementary Fig 3), thus validating mouse Atg5 and Lamp1 as miR-33 and miR-33* targets. Despite predicted miR-33 and miR-33* binding sites in the genes encoding cathepsin B (Ctsb) and LAL (Lipa), no effect of miR-33 or miR-33* was observed on Ctsb or Lipa 3’UTR activity (Fig. 3c, d). mRNA profiling of peritoneal macrophages transfected with miR-33 and miR-33* mimics or inhibitors showed a reciprocal regulation of Atg5, Lamp1 and Prkaa1 mRNA (Fig. 3e, f). Furthermore, compared to wild type macrophages, Mir33−/− macrophages lacking both miR-33 and miR-33* displayed elevated expression of Abca1, Atg5, Lamp1 and Prkaa1 mRNA (Fig. 3g). Finally, consistent with the observed induction of miR-33 and miR-33* by Mtb, expression of miR-33 and miR-33* targets in lipid metabolic and autophagy pathways were reduced in vivo in Mtb-infected (GFP+) macrophages compared to the uninfected (GFP−) macrophages from the same mice (Fig. 3h). Together these data indicate that in Mtb-infected macrophages elevated expression of miR-33 and miR-33* represses key genes in the autophagy pathway.
miR-33 and miR-33* regulate activation of FOXO3 and TFEB
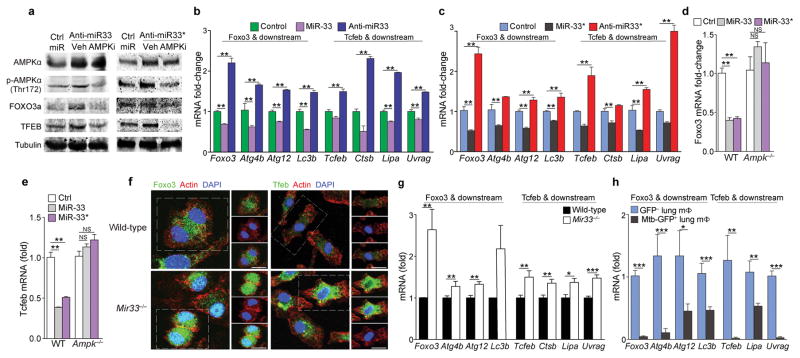
PCR array profiling revealed that miR-33 and miR-33* overexpression and inhibition reciprocally regulated several genes in the autophagy pathway that did not contain miR-33 or miR-33* binding sites and this was validated by quantitative PCR (Supplementary Fig. 4). AMPK is a target of miR-3322 and has dual functions in promoting autophagy: it represses mTORC1 and promotes the activity of TFEB30 and FOXO314. As such, we focused on the AMPK-dependent activation of TFEB and FOXO3, transcriptional regulators of autophagy and lysosomal biogenesis gene programs. Compared to control ASO treatment, miR-33 mimic repressed human and mouse Prkaa1 3’UTR activity in HEK293 cells and AMPKα protein expression in macrophages (Fig. 3a, c). Furthermore, anti-miR-33 treated macrophages showed increased Prkaa1 mRNA and AMPKα protein and phosphorylation, compared to control ASO treated macrophages (Fig. 3e, Fig. 4a). miR-33* is also predicted to target AMPKα and miR-33* overexpression and inhibition regulated human and mouse Prkaa1 3’UTR activity, mRNA, and protein expression and phosphorylation (Fig. 3, Fig. 4a). Although mouse Foxo3 and Tcfeb lack miR-33 or miR-33* binding sites, transfection of peritoneal macrophages with miR-33 or miR-33* reduced expression of Foxo3 and Tcfeb and their downstream target genes (FOXO3: Atg4b, Atg12, Map1lc3b; TFEB: Ctsb, Lipa and Uvrag), while treatment with anti-miR-33 or anti-miR-33* increased mRNA expression of these genes (Fig. 4b, c). Furthermore, macrophages treated with anti-miR-33 or anti-miR-33* show increased FOXO3 and TFEB protein and nuclear localization compared to control ASO treated macrophages (Supplementary Fig. 5), and these effects were abrogated in the presence of an AMPK inhibitor (Supplementary Fig. 5). Likewise, the effects of miR-33 and miR-33* overexpression or silencing on Foxo3 and Tcfeb mRNAs were abrogated in AMPK-deficient macrophages (Fig. 4d, e). Similar to our findings using miR-33 and miR-33* inhibitors, Mir33−/− macrophages displayed elevated expression and activation of FOXO3 and TFEB (Fig. 4f), as well as increased expression of their downstream target genes (Fig. 4g), compared to wild type macrophages. Finally, Foxo3 and Tcfeb mRNA and genes that they regulate were reduced in vivo in Mtb-infected (GFP+) macrophages compared to the uninfected (GFP−) macrophages from the same mice (Fig. 4h), consistent with elevated expression of miR-33 and miR-33* in Mtb-infected macrophages. These observations indicate that miR-33 and miR-33* regulate autophagy through both direct targeting of autophagy effectors and by repressing AMPK-dependent activation of autophagy and lysosomal gene transcription, and these regulatory mechanisms are engaged in Mtb infection.
Figure 4. miR-33 and miR-33* repress AMPKα and downstream transcription factors controlling autophagy and lysosomal gene programs.
(a) Western blotting of lysates from peritoneal macrophages treated with anti-miR-33 or anti-miR-33* or control (ctrl) miR in the presence or absence of the AMPK inhibitor compound C (Ampki, 5 μM). (b, c) mRNA levels of Foxo3, Tcfeb and their downstream transcriptional targets in peritoneal macrophages transfected with (b) miR-33 and (c) miR-33* mimics or inhibitors as indicated. (d) Foxo3 and (e) Tcfeb mRNA in wild-type (WT) or Ampk−/− BMDMs treated with mmu-miR-33, -33*, or Ctrl mimic. (f) Immunofluorescence (IF) imaging of FOXO3a and TFEB in green, F-actin (red) and DAPI (blue) in wild-type (WT) and Mir33−/− BMDMs. (g) Quantification of Foxo3, Tcfeb and their downstream transcriptional targets (right) in WT and Mir33−/− BMDMs. (h) Quantification of Foxo3, Tcfeb and their downstream transcriptional targets in GFP+ (infected) or GFP− (uninfected) alveolar macrophages isolated from mice infected with a GFP-expressing H37Rv Mtb strain, 2 weeks post-infection. *P≤0.1, **P≤0.05, ***P≤0.005 (Student’s t-test (b–c, g, h), one-way ANOVA (d, e)). Data are from one experiment representative of 3 ((b, c, g; mean ± s.e.m)) or 2 ((a, f) independent experiments. Data are from 2 experiments (d, e; mean ± s.e.m). Data are from 3 mice (h; mean ± s.e.m).
The miR-33 locus regulates cellular autophagy
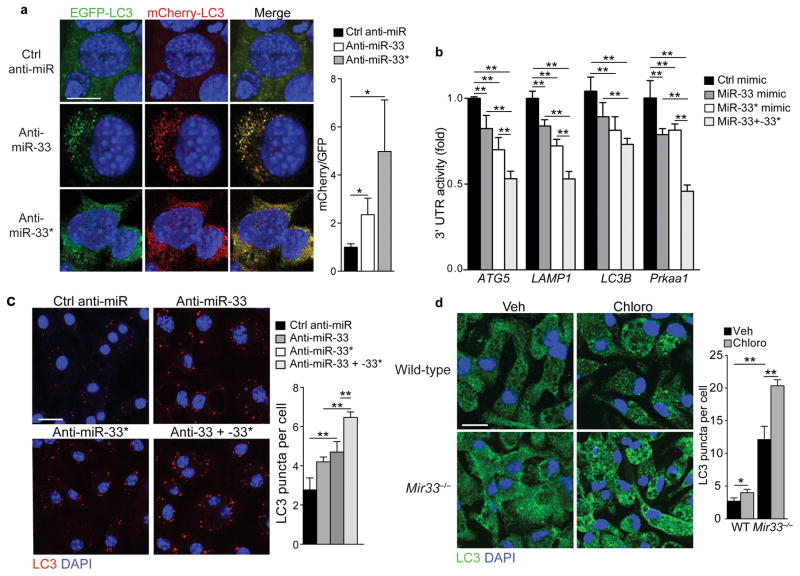
To test the functional effects of miR-33 and miR-33* targeting of autophagy, we inhibited miR-33 or miR-33* in MEFs stably expressing mCherry-GFP-LC3, which can be used to track LC3 expression in autophagosomes and autophagolysosomes; the GFP and mCherry tags together emit a yellow signal in pH-neutral autophagosomes, however upon autophagosome fusion with acidic lysosomes the pH-sensitive GFP signal diminishes, resulting in stronger red fluorescence31, 32. In control ASO treated MEFs, LC3-GFP and LC3-mCherry were diffusely expressed in the cytoplasm, whereas they were localized to punctate structures in anti-miR-33 and anti-miR-33* treated MEFs (Fig. 5a) consistent with enhanced autophagy initiation. Compared to control ASO treated MEFs, the anti-miR-33 and anti-miR-33* treated MEFs showed a greater number of yellow autophagosomes and red autophagosome-lysosome resulting in an increased mCherry:GFP ratio (Fig. 5a) indicative of enhanced autophagy flux. Because co-transfection of HEK293 cells with miR-33 and miR-33* repressed the 3’UTR of autophagy target genes more than either miR-33 and miR-33* alone (Fig. 5b), we examined whether the two strands of pre-miR-33 have additive effects on autophagy. Treating peritoneal macrophages with inhibitors of both miR33 and miR-33* resulted in an additive increase in LC3 puncta compared to inhibition of either miRNA alone (Fig. 5c), indicating that miR-33 and miR-33* cooperatively regulate autophagy. Mir33−/− macrophages had more LC3 puncta than wild type macrophages (Fig. 5d). Furthermore, the number of LC3 puncta in both wild type and Mir33−/− macrophages was increased by blocking lysosomal degradation using chloroquine indicating functional autophagy flux (Fig. 5d).
Figure 5. miR-33 and miR-33* cooperatively inhibit macrophage autophagy.
(a) Immunofluorescence (IF) imaging of mCherry-EGFP-LC3 mouse embryonic fibroblasts treated with anti-miR33, anti-miR33* or Ctrl anti-miR to assess autophagy flux. GFP and mCherry co-expression in autophagosomes is visible as yellow puncta in the merged imaged. Fusion of autophagosomes with lysosomes diminishes the pH-sensitive GFP signal increasing red fluorescence in autophagolysosomes. (b) 3’UTR-luciferase reporter activity in HEK293 cells transfected with control (ctrl) mimic, miR-33 or miR-33*, or both. (c) IF imaging of LC3 in peritoneal macrophages treated with control mimic, miR-33, miR-33* or both together. Quantification of cellular LC3 puncta is shown at right, (d) IF imaging of LC3 in WT and miR-33−/− macrophages. Quantification is shown at right. *P≤0.1, ** P≤0.05 (one-way ANOVA (a, b, c), two-way ANOVA (d)). Data are from one experiment representative of 2 (a, c, d; mean ± s.e.m)) or 3 (b; mean ± s.e.m) independent experiments. Scale bar = 10 μm (a, b, d).
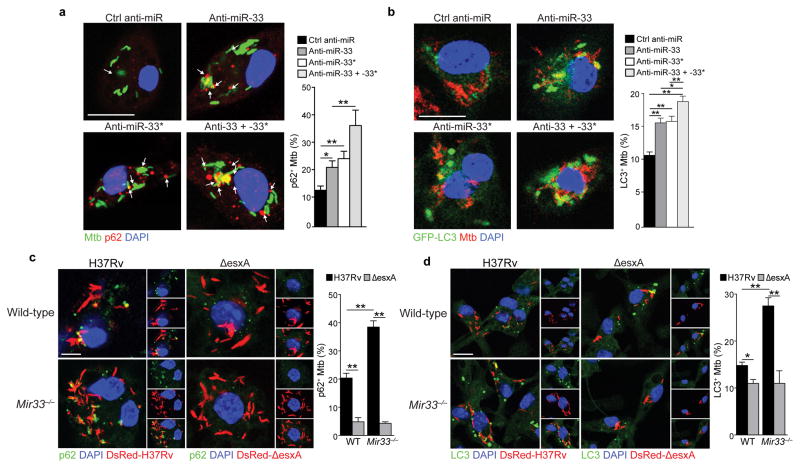
To investigate whether Mtb limits autophagy-dependent antimicrobial responses by inducing miR-33 and miR-33*, we measured the colocalization of Mtb H37Rv with p62, which tags bacteria for sequestration within LC3-positive autophagosomes. Peritoneal macrophages treated with anti-miR-33 or anti-miR-33* contained more p62-positive Mtb compared to control ASO treated macrophages, and p62-Mtb colocalization was further augmented by treating peritoneal macrophages with anti-miR-33 and anti-miR-33* together (Fig. 6a). Likewise, peritoneal macrophages treated with anti-miR-33 or anti-miR-33* contained more LC3-positive bacteria compared to macrophages treated with control ASO, an effect that was further enhanced by treating with both anti-miR-33 and anti-miR-33* (Fig. 6b). Finally, compared to wild type macrophages, Mir33−/− macrophages exhibited increased p62- and LC3-association with Mtb when cells were infected with the virulent H37Rv but not the mutant ΔesxA strain, which fails to permeabilize the phagosome and trigger xenophagy6 (Fig. 6c, d). Together, these data suggest that Mtb limits xenophagy by inducing miR-33 and miR-33* expression in macrophages.
Figure 6. Silencing of miR-33 and miR-33* enhances Mtb targeting by the autophagy machinery.
(a) Immunofluorescence (IF) imaging of peritoneal macrophages infected with GFP-tagged H37Rv Mtb (green), p62 (red) and DAPI (blue) for 24 h and treated with control (ctrl) miR, anti-miR33 or anti-miR33*, or both. Quantification of p62 co-localization with Mtb shown at right. (b) IF imaging of peritoneal macrophages infected with GFP-tagged LC3 (green), H37Rv Mtb (red) and DAPI (blue) for 24h and treated with control (ctrl) miR, anti-miR33 or anti-miR33*, or both. Quantification of LC3 co-localization with Mtb shown at right. (c–d) IF imaging of (c) p62 (green) or (d) LC3 in wild type (WT) and Mir33−/−macrophages infected with DsRed-H37Rv or DsRed-ΔesxA Mtb strains. Quantification is shown at right. *P≤0.1, **P≤0.05 (one-way ANOVA (a–b), two-way ANOVA (c–d)). Data are from one experiment representative of 2 (a, b, d) or 3 (c) independent experiments with similar findings. Scale bar = 10 μm (a–d).
Inhibition of miR-33 and miR-33* promotes Mtb clearance
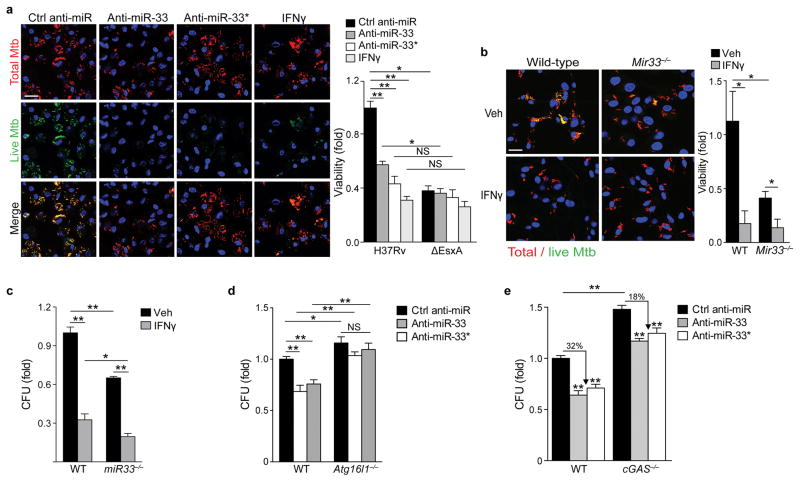
To investigate whether miR-33- and miR-33*-dependent repression of autophagy and lipid catabolism enhance Mtb survival, we assessed Mtb viability and growth in vitro and in vivo. We infected BMDMs with an Mtb strain expressing mCherry constitutively and GFP under control of a tetracycline-inducible promoter33. After treatment with anhydrotetracycline (AnTc), metabolically active bacteria express both GFP and mCherry, whereas dead or metabolically inactive bacteria are only mCherry-positive. Compared to control ASO treatment, anti-miR-33 and anti-miR-33* treatment reduced the proportion of metabolically active Mtb when BMDMs were infected with wild type Mtb, but not the ΔesxA mutant (Fig. 7a). The magnitude of this decrease was similar to that seen in BMDMs treated with IFN-γ prior to infection (Fig. 7a), which activates autophagy3. Furthermore, Mir33−/− BMDMs contained fewer metabolically active Mtb compared to wild type BMDMs (Fig. 7b) and similar results were observed when Mtb viability was quantified by bacterial colony formation (Fig. 7c). Notably, anti-miR-33 and anti-miR-33* failed to reduce bacterial viability in Atg16l1−/− BMDMs (Fig. 7d), which lack an essential component of the autophagy elongation complex, confirming that the antimicrobial effects of miR-33 and miR-33* silencing are mediated by autophagy. Because the autophagy pathway is critical to both lipid catabolism (lipophagy), as well as bacterial degradation (xenophagy), the Atg16l1−/− BMDMs would be impaired in both. To determine the role of miR-33 and miR-33* specifically in xenophagy, we examined Mtb survival in mouse Cgas−/− BMDMs, which lack the cGAS cytosolic sensor required to initiate xenophagy of Mtb9, 10, 11. As recently reported9, 10, 11, Mtb grew better in Cgas−/− BMDMs as compared to wild-type (Fig. 7e, Supplementary Fig. 6). Compared to control ASO treatment, anti-miR-33 and anti-miR-33* treatment restricted Mtb viability by 32% in wild-type BMDMs, but only 18% in the xenophagy-deficient Cgas−/− BMDMs (Fig. 7e). Collectively, these experiments demonstrate that miR-33 and miR-33* regulate intracellular Mtb survival, in part through xenophagy.
Figure 7. Inhibition of miR-33 and miR-33* enhances targeting of M. tuberculosis for autophagy and bacterial killing.
(a) Immunofluorescence imaging of peritoneal macrophages treated with control anti-miR, anti-miR-33, anti-miR-33*, or IFN-γ (200 units/mL) and infected with an Mtb H37Rv strain co-expressing mCherry and anhydrotetracycline-inducible GFP for 48 h. Metabolically active Mtb are GFP and mCherry positive, whereas inactive or non-viable bacteria are mCherry positive only. Scale bar = 25 μm. Quantification of bacterial viability in peritoneal macrophages infected with Mtb H37Rv or ΔesxA mutant strain shown on the right. (b–c) Quantification of Mtb survival in wild type (WT) or miR-33−/− BMDMs treated with or without IFN-γ (200 units/mL) using (b) the dual fluorescence viability assay as in (a), or (c) colony forming units (CFU) 5 days post-infection. (d) Mtb survival in wild type (WT) or Atg16l1−/− BMDMs treated with control anti-miR, anti-miR-33 or anti-miR-33*, measured by CFU 5 days post-infection. (e) Mtb survival in wild type (WT) or cGAS−/− BMDMs treated with control anti-miR, anti-miR-33 or anti-miR-33*, measured by CFU 5 days post-infection. NS, not significant; *P≤0.05, **P≤0.005 (Two-way ANOVA (a–e)). Data are from one experiment and are representative of 2 independent experiments (a-e; mean ± s.e.m) with similar findings.
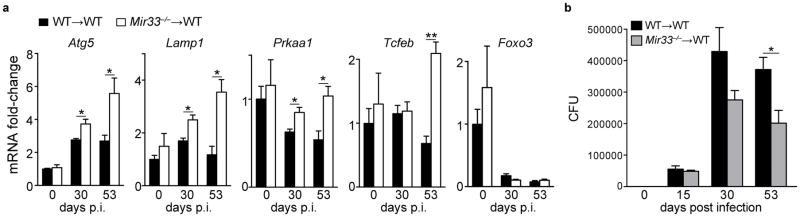
To assess the effects of Mtb induction of miR-33 and miR-33* expression in vivo, we generated a model of macrophage miR-33 deficiency using bone marrow transplantation, which has been shown to mediate donor engraftment of alveolar macrophages by more than 90%34. Following reconstitution of C57BL/6 mice with wild-type (WT→WT) or Mir33−/− (Mir33−/−→WT) hematopoietic cells, mice were infected with Mtb H37Rv. Analysis of total lung mRNA expression after 30 and 53 days of Mtb infection showed that Mir33−/−→WT mice had higher expression of miR-33- and miR-33*-regulated genes, including Prkaa1, Lamp1, Atg5 and Tcfeb, compared to lungs of WT→WT mice (Fig. 8a). Furthermore, Mir33−/−→WT mice had lower bacterial burden in the lungs compared to WT→WT mice at 15, 30 and 53 days post infection, indicating that mice with hematopoietic miR-33 deficiency had an improved ability to clear Mtb in vivo (Fig. 8b). Collectively, these data support a role for miR-33 in limiting macrophage autophagy, lipid catabolism, and consequently Mtb elimination (Supplementary Fig. 7).
Figure 8. Hematopoietic miR-33 deficiency enhances autophagy gene expression in the lung and reduces Mtb burden in mice.
Analysis of (a) gene expression and (b) bacterial counts in the lungs of wild type (WT→WT) and Mir33−/− (Mir33−/−→WT) bone marrow chimeric mice infected with H37Rv Mtb for the indicated time post-infection (p.i). *P≤0.05, **P≤0.005 (Student’s t-test (a, b)). Data are from 5 mice (a, b; mean ± s.e.m).
DISCUSSION
M. tuberculosis is a persistent intracellular pathogen that survives within the hostile microenvironment of the macrophage by avoiding lysosomal degradation and establishing a lipid riche niche. Although the autophagy machinery can deliver Mtb to autophagosomes for degradation, for unclear reasons, only a fraction of infecting tubercle bacilli are cleared by this mechanism. Our data demonstrate that by inducing expression of the miR-33 locus in macrophages, Mtb provides brakes on the autophagy pathway. We demonstrate that miR-33 and its passenger strand, miR-33*, repress a complex molecular cascade of key effectors (ATG5, ATG12, LAMP1, LC3B), activators (AMPK), and transcriptional regulators (FOXO3, TFEB) of autophagy and lysosome pathways. Repression of autophagy by miR-33 and miR-33* benefits Mtb in two ways: it inhibits xenophagy-mediated bacterial degradation and blocks lipophagy-regulated catabolism of host-derived lipid stores that are in turn used by the bacterium as a nutrient source. Silencing of miR-33 and miR-33* promotes p62- and LC3-targeting of Mtb to the autophagy machinery for degradation and enhances the anti-mycobacterial capacity of macrophages. Our studies demonstrate that miR-33 and miR-33* cooperate to regulate integrated pathways involved in autophagy, lysosomal function and lipid homeostasis, and highlight how manipulation of these host cell miRNAs during infection can promote Mtb survival.
Our identification of miR-33’s role in regulating autophagy is a prime example of how miRNAs can fine-tune biological pathways by repressing multiple genes with related functions. An added dimension to our findings is that this inhibitory activity arises not only from the actions of the miR-33 guide strand but also from its passenger strand miR-33*. Not all miRNAs conform to the conventional model in which the “guide strand” is preferentially loaded into RISC, and recent studies have identified a number of active miRNA* strands, including miR-126-3p and miR-126-5p26, 35, 36, 37. Unlike miR-33 and miR-33*, the two strands of the miR-126 duplex regulate distinct sets of target genes to protect from endothelial damage and atherosclerosis37. Our findings that miR-33 and miR-33* repress an overlapping set of target genes in the autophagy pathway suggest a “double hit” model in which both strands of the miR-33 locus cooperate to regulate a common biological pathway.
We show that γ-irradiated Mtb and the cell wall constituents LAM and TDM are sufficient to induce the miR-33 locus, a response that depends upon NF-κB and leads to impaired fatty acid β-oxidation and enhanced lipid droplet accumulation. This is in accordance with previous studies that found that lipid body formation in response to mycobacteria does not require live bacilli, can be driven by LAM, and is partially dependent upon TLR221, 38, as well as the observation that miR-33 expression is regulated by NF-κB39. Lipids play an important and incompletely understood role in Mtb physiology, metabolism and virulence. Lipid bodies serve as a secure sanctuary that shelters the bacteria from innate immune defenses and provide a nutrient-rich environment for replication40, 41, 42, 43. Our results show that by inducing the miR-33 locus Mtb promotes macrophage lipid stores through two mechanisms: reduction of autophagy-mediated delivery of lipid droplets to lysosomes for LAL-mediated lipolysis, and inhibition of cellular fatty acid oxidation. miR-33-driven lipid body accumulation may work in concert with other anti-lipolytic mechanisms engaged by Mtb, such as activation of the G protein-coupled receptor GPR109A that blocks the action of hormone sensitive lipase20. In miR-33 and miR-33*-deficient macrophages, the importation of cellular fatty acids by Mtb is greatly reduced, highlighting the importance of these host miRNAs in Mtb metabolism. Access to host lipids may play a particularly important role during latent infection, as lipid-laden bacteria acquire characteristics of dormant bacilli, including resistance to antimycobacterial drugs19, 44. These findings provide insight into the molecular pathways leading to the foamy phenotype of Mtb-infected macrophages.
Trafficking through the autophagy pathway depends upon the Mtb esxA gene6 and a prevailing idea is that EsxA damages the phagosomal membrane45, 46 leading to activation of cytosolic sensors that promote xenophagy6, 9, 10, 11, 47, 48. Notably, the microbicidal effects of miR-33 and miR-33* inhibitors were markedly reduced in macrophages infected with the ΔesxA mutant, demonstrating that autophagy induction by miR-33 inhibition does not override the EsxA requirement. The importance of autophagy in the microbicidal effects of miR-33 and miR-33* inhibitors is indicated by their diminished activity in the absence of Atg16L1 and cGAS. The retention of antimicrobial activity by miR-33 and miR-33* inhibitors in cGAS−/− macrophages, which are deficient in xenophagy but not lipophagy, likely reflects their additional effects on lipid metabolism.
Our data show that Mtb employ a novel mechanism of autophagy evasion by inducing the miR-33 locus to post-transcriptionally limit the autophagy machinery. Although our study does not discriminate the individual roles of AMPK and other autophagy effectors targeted by miR-33 and miR-33*, it highlights the importance of miRNAs as orchestrators of pathway control which can have profound effects on biological processes by causing modest decreases in multiples genes within a given pathway. It is likely that the influence of miR-33 in innate immune responses extends beyond Mtb given that autophagy has well-described roles in host defense and inflammatory disorders and that lipid bodies are increasingly appreciated to play important roles in innate immunity.
ONLINE METHODS
Reagents
For a list of antibodies and primers used, see Supplementary Table 1. The lysosomal acid lipase inhibitor Lalistat1 was provided by Drs. F. Maxfield and A. Rosenbaum (Weill Cornell Medical College) and the Mtb live-dead plasmid was provided by I. Keren (Northeastern University)33.
Mice
The Institutional Animal Care Use Committee of New York University Medical Center approved all animal experiments. Six-week old male C57BL/6 mice were obtained from Jackson Laboratory. Prkab1−/− mice, which lack AMPK activity as previously described49 were provided by G.Steinberg (McMaster University). Mir33−/− mice, which lack the miR-33 locus as previously described50 were kindly provided by P. Zamore (University of Massachusetts). Atg16l1flox/flox-Lyz-Cre mice, kindly provided by K. Caldwell (New York University), were described previously51. Cre-negative Atg16L1flox/flox littermates were used as controls. Bone marrow cells isolated from 8-week-old wild-type or Mir33−/− mice were injected (retro-orbital) in lethally irradiated 6–8 week old C57BL/6 males, and transplanted mice were allowed to recover for 7 weeks prior to infection. LC3-GFP mice were generously provided by A. Yamamoto (Columbia University). cGAS−/− mice, which lack the cytosolic DNA sensor cyclic GMP-AMP synthase (cGAS) as described previously52, were generously provided by H. Virgin (Washington University School of Medicine).
Cell culture
THP-1 and HEK293 cells were obtained from American Type Tissue Collection and were tested monthly for mycoplasma contamination. HEK293 cells were maintained in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S). THP-1 cells were maintained in RPMI 1640 media (Sigma) supplemented with 10% FBS and 1% P/S. THP-1 differentiation into macrophages was induced using 100 nM phorbol-12-myristate acetate (PMA) for 72 h. Immortalized mouse embryonic fibroblasts (MEFs) were cultured in high-glucose DMEM supplemented with 10% FBS, 1% P/S, and 1% Glutamax. Stably expressing dual-tagged LC3 (mCherry-EGFP-LC3) mouse embryonic fibroblast were a gift from the Taylor laboratory and maintained as previously described32. Peritoneal macrophages from adult mice were harvested by peritoneal lavage four days after intraperitoneal injection of thioglycollate, as previously described53. The cells were maintained in culture as adherent monolayer in medium containing DMEM, 10% FBS, and 20% L929-conditioned medium. To obtain bone marrow derived-macrophages (BMDMs), marrow was flushed from the tibia and femurs of 6–8 week old mice and differentiated into macrophages in DMEM media supplemented with 10% FBS, 1% P/S, and 15% L929-conditioned media for 7 days. For treatment of macrophages with Mtb ligands, BMDMs were treated for 24h with 5 μg/mL of purified Lipoarabinomannan (LAM), 2.5 μg/mL of purified Trehalose Dimycolate (TDM) or 10 μg/mL of gamma-irradiated M. tuberculosis (γ-Mtb), strain H37Rv (BEI Resources).
Bacterial strains and growth conditions
M. tuberculosis H37Rv was grown at 37°C to log phase in Middlebrook 7H9 broth supplemented with 0.05% Tween 80, BBL Middlebrook OADC Enrichment, and 0.2% (v/v) glycerol. The medium contained 25 μg/ml kanamycin for GFP- or dsRed-overexpressing bacteria or 50 μg/ml hygromycin when containing the live-dead plasmid.
Bacterial infection
For in vivo M. tuberculosis H37Rv infections, mice were infected via the aerosol route using an inhalation exposure system from Glas-Col, as previously described54. A log-phase Mtb culture was pelleted and subsequently resuspended in PBS containing 0.5% Tween80. After a centrifugation step for 8min at 120g, the supernatant (single cell suspension) was used to prepare the inoculum. Bacteria were diluted in 10 mL of sterile water and 5 mL of this inoculum were loaded into the inhalation exposure nebulizer unit. The aerosol unit was programmed to deliver ~100 colony forming units (CFU) per animal. The dose of infection was confirmed on day 1 post-infection by plating whole lung homogenates from 3 mice on Middlebrook 7H11 agar. Mice were euthanized and the left lung was excised, incubated for 48 h at 4°C in RNAlater (Ambion) and homogenized in Trizol. The right lung was homogenized in PBS + 0.5% Tween80 through a 70 μm cell strainer (BD Falcon). Bacterial colonies were counted after incubation at 37°C for 15–21 days. The bacterial load throughout the infection was monitored by harvesting lungs at the indicated times and plating serial dilutions on Middlebrook agar.
Isolation of alveolar macrophages
On day 17 post-infection, lungs from mice infected with GFP-expressing H37Rv were harvested54. The lungs were placed in digestion buffer (RPMI 1640, 5% FBS, 10mM HEPES), cut into 2 mm3 pieces, and incubated with Collagenase D (1 mg/ml, Roche) and DNaseI (30 μg/ml, Roche) at 37°C for 45 min. The tissues were forced through a 70 μm cell strainer (BD Falcon) to obtain a single cell suspension. Red blood cells were removed using ACK lysis buffer (155 mM NH3Cl, 10 mM KHCO3, 88 μM EDTA). For surface staining, cells were washed and suspended in FACS buffer (PBS, 1% FBS, 1 mM EDTA) containing antibodies and incubated at 4°C for 20 min. Cells were washed twice in FACS buffer prior to cell sorting. Alveolar macrophages (CD11chighCD11blow) were live-sorted from lung homogenates stained with CD11b-PE (1:500), SiglecF-BV421 (1:200), CD11c-APC (1:500), ZombieNIR-APCCy7 (1:1500) using an iCyt Synergy sorter in BSL-3 containment. Dead cells (ZombieNIR+) were gated out while leaving CD11c+ in. Subsequently CD11b+ cells were gated out. SiglecF+ cells were collected as FITC+ (H37Rv-GFP infected) or FITC− (uninfected) fractions. Cells were resuspended in Quiazol for subsequent sequential miRNA and total RNA isolation using the miRNeasy Micro Kit (Qiagen).
miR-33/-33* and anti-miR-33/-33* transfections
Mouse peritoneal macrophages, differentiated THP-1 macrophages, or MEFs were transfected with 80–120 nM miRIDIAN miRNA mimics (miR-33) or with 80–120 nM miRIDIAN miRNA inhibitors (anti-miR-33) (Dharmacon) utilizing Lipofectamine RNAiMax Transfection Reagent (Invitrogen). Control samples were treated with an equal concentration of a non-targeting control mimics sequence (ctrl miR) or inhibitor negative control sequence (ctrl inh), to control for non-specific effects in miRNA experiments.
RNA isolation and qRT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) and Direct-zol RNA MiniPrep columns (Zymo Research). For mRNA quantification, cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative real-time PCR was performed in triplicate using KAPA SYBR FAST Universal 2X qPCR Master Mix (Kapa Biosystems) on the iCycler Real-Time Detection System (BioRad). The mRNA level was normalized to GAPDH as a house keeping gene. The following primer sequences were used:
miRNA Quantification
For mature miRNA quantification, the miScript II RT Kit (Qiagen) was used for reverse transcription reactions, using HiSpec Buffer, according to the manufacturer’s protocol. miR-33 and miR-33* miScript miRNA Mimics were used to generate a standard curve of CT values (y axis) against log copy number (x axis), according to instructions in Qiagen’s miScript PCR System Handbook, to ensure equivalent miR-33 and miR-33* primer efficiencies and for absolute quantification. miScript Primer Assays and miScript SYBR Green PCR were used on the iCycler Real-Time Detection System (BioRad). For relative quantification, miRNA levels were normalized to RNU6-2 miScript PCR control (Qiagen).
Argonaute pull-down
RNA Immunoprecipitation (RIP) was performed using an antibody targeting murine Argonaute-2 (Ago2, Abcam) or control antibodies (IgG) according to the manufacturers’ instructions (Millipore) on lysates from mouse peritoneal macrophages treated with 10 μg/mL of gamma-irradiated Mycobacterium tuberculosis, γ-Mtb (strain H37Rv, BEI Resources) for 24h. Briefly, Ago2 antibodies were bound to magnetic beads and incubated with peritoneal macrophage lysates at 4°C for 24 h. Coprecipitated RNA was purified, cleaved from Ago2 by proteinase K. Total RNA was used for quantification by PCR using Taqman assays (Life Technologies) for miR-33, miR-33*, miR-27b and RNU6-2. The effect of γ-Mtb on the loading of miR-33, miR-33* and miR-27b into RNA Binding Protein complex (RBP) was determined by correcting for both RNU6-2 levels and the fraction that coprecipitated with IgG antibodies.
Fluorescence microscopy
Cells were fixed in 4% PFA and neutral lipids were stained using BODIPY 493/503 (10 μg/mL) or Nile Red (50 ng/mL), as previously described55. For immunofluorescence, cells were fixed in 4% paraformaldehyde (PFA), blocked/permeabilized in 2.5% BSA/0.1% Triton X-100, and stained with the indicated primary antibodies for 1 h at 37°C or at 4°C overnight. Fluorophore-conjugated secondary antibodies were incubated in the presence of a neutral lipid stain where applicable. Confocal images of lipid droplets were obtained using a Leica SP5 confocal microscope with appropriate lasers. Fluorescence microscopy of FOXO3 and TFEB was observed using a Nikon Eclipse microscope. mCherry-EGFP-LC3-expressing cells (provided by Dr. J. Paul Taylor, St. Jude Children’s Research Hospital) and phagocytosis of fluorescently-labeled apoptotic cells were imaged using a Zeiss Axiovert 200M fluorescent microscope, with Axio Vision 4.8.
3’UTR luciferase reporter assays
Mouse Atg5 (MmiT036882), Ctsb (MmiT026884), Lamp1 (MmiT092570), Lipa, Map1lc3b (MmiT034146), Prkaa1 (MmiT024101), and human ABCA1 (HmiT004727), ATG5 (HmiT022804), ATG7 (HmiT054679), ATG12 (HmiT055328), LAMP1 (HmiT010534), MAP1LC3B (HmiT019948), and UVRAG (HmiT018457) pEZX-MT01 miRNA 3’UTR target clones were purchased from GeneCopoeia. HEK 293T cells were plated in 96-well plates and co-transfected with 0.2 μg of the indicated 3’UTR luciferase reporter vectors and the miR-33 mimic or negative control mimic (Dharmacon) utilizing Lipofectamine 2000 (Invitrogen). Luciferase activity was measured using the Luc-Pair miR Luciferase Assay Kit (GeneCopoeia). Firefly luciferase activity was normalized to the corresponding Renilla luciferase activity and plotted as a fold-change relative to control-treated cells. Experiments were performed in quadruplicate wells of a 96-well plate and repeated at least three times.
Western blotting
Cells were washed twice with ice-cold PBS, scrapped in RIPA buffer with protease and phosphatase inhibitors, or directly in 2X Laemmli sample buffer (Bio-Rad) for LC3 western blots. Total protein samples (25–30 μg/well; 60 μg/well for phosphorylated proteins) were electrophoresed on 8%, 10%, or 18% SDS-polyacrylamide gels and transferred to nitrocellulose or PVDF membranes at 125V for 2h. For LC3 detection, 18% gels were used and transferred onto 0.2 μm PVDF membranes. All membranes were incubated overnight with the appropriate antibodies, and proteins were visualized using appropriate secondary antibodies conjugated to IR-dyes (Rockland) and scanned using the Odyssey Imaging System (Licor) as described previously53, or alternatively proteins were detected using an enhanced HRP-based chemiluminescence detection system (HRP-conjugated secondary antibodies from Amersham Biosciences and SuperSignal West Femto Maximum Sensitivity Substrate from Pierce) and analyzed using a FluorChem Imager (Alpha Innotech).
Seahorse Extracellular Flux Analysis
BMDMs were seeded into XF24 cell culture microplates (Seahorse Bioscience) and transfected with anti-miR-33 or control anti-miR as described above, and 48 h later oxygen consumption and extracellular acidification rates (OCR and ECAR respectively) were quantified using the XFe24 instrument (Seahorse Bioscience, Boston, MA) according to the manufacturer’s protocol. For glycolysis assessments, the XF Glycolysis Stress Test Kit was used according to the manufacturer’s protocol. For cellular fatty acid oxidation (FAO) quantification, the OCR was measured following exposure to BSA-conjugated palmitate (200 μM) in Krebs-Henseleit Buffer (KHB; 111 mM NaCl, 4.7 mM KCl, 2mM MgSO4, 1.2 mM Na2HPO4, 2.5 mM Glucose, 0.5 mM Carnitine) using the XFe24 instrument and the XF Cell Mito Stress Test kit.
Intracellular bacterial survival assays
To assess bacterial survival in vitro, BMDMs were infected with a single cell suspension of mycobacteria at a multiplicity of infection (MOI) of ~3 as previously described56. 4h post-infection, macrophages were extensively washed with PBS and were lysed with 0.2% Triton X-100 at the indicated time points. Serial dilutions of the lysates were plated on 7H11 agar plates and CFU were counted 15–21 days later. Alternatively, Mtb viability was assessed by performing a ‘live-dead assay’ using wild type H37Rv or H37Rv ΔesxA constitutively expresssing mCherry and inducibly express GFP under control of a tertracycline-inducible promoter17. The ΔesxA mutant was generously provided by William Jacobs Jr. (Albert Einstein College of Medicine) and the “live-dead” plasmid was generously provided by Iris Keren (Northeastern University). For these survival assays, primary mouse peritoneal macrophages or BMDMs were seeded in 8-well Lab-Tek chamber slides (Thermo Scientific) and treated with miRNA inhibitors (Dharmacon) 2 days prior to infection with a single cell suspension of wild type or ΔesxA Mtb at a MOI of 5. For the IFN-γ-treated sample, macrophages were treated with 200 units/mL of IFN-γ 24 h prior to infection. Four hours after infection, the media was removed, and macrophages were washed, as previously described56. 24 h prior to fixation, anhydrotetracycline (200nM) was added to the cells to induce bacterial GFP expression. Metabolically active Mtb will express both mCherry and GFP, whereas bacteria that are dead or metabolically inactive only fluoresce red. At 24 h and 48 h post-infection, cells were fixed in 4% PFA for 24 h, and counterstained with 4’,6-Diamidino-2-Phenylindole (DAPI - Sigma) at 0.5 μg/mL prior to mounting with Fluorescent Mounting Medium (Dako). Cells were visualized using Confocal Microscopy (Zeiss 600 System) and bacterial viability was scored in 6 different fields as the percentage of red bacteria that also exhibited green fluorescence. For the p62 staining, wild type H37Rv Mtb expressing GFP was used.
PCR array gene expression profiling
Total RNA was extracted from peritoneal macrophages transfected with miR-33, miR-33* or control mimics, or alternatively with anti-miR33, anti-miR33* or control anti-miR, as described above. Reverse transcription was performed on 1μg total RNA using the RT2 First Strand kit and quantitative real-time PCR (qRT-PCR) analysis of 84 key genes involved in autophagy was performed using Mouse Autophagy RT2 Profiler PCR Arrays (PAMM-084, SABiosciences) as per the manufacturer’s protocol. Data analysis was performed using the manufacturer’s integrated web-based software package for the PCR Array System using ΔΔCt based fold-change calculations. The array data have been deposited in the Gene Expression Omnibus Database under accession number GSE79055.
Statistics
Data are presented as mean ± the standard error of the mean (s.e.m), and the statistical significance of differences was evaluated with the Student’s t test. Significance was accepted at the level of P 0.05. For multiple group comparisons, analysis of variance (ANOVA) tests were performed. The Tukey and Dunnett tests were used as follow-up tests to the ANOVAs, where the Tukey test was used to compare every mean with every other mean, and the Dunnett test was used when comparing every mean to a control mean.
Supplementary Material
Acknowledgments
Support for this work came from the National Institutes of Health (R01 HL108182 and HL119047 to K.J.M; R01 AI087682 and R21 AI105298 to J.A.P), the American Heart Association (13POST14490016 to B.R., 14POST20180018 to CvS), the NYU Physician-Scientist Training Program (C.P.C.), The Potts Memorial Foundation (S.K.), Edward J. Mallinckrodt, Jr. Foundation (J.A.P.), Science Foundation Ireland grant 13/SIRG/2136 (F.J.S.) and Canadian Institutes of Health Research (postdoctoral fellowship to M.O.; MOP130365 and MSH130157 to K.J.R.). We thank the New York University Langone Medical Center Immune Monitoring Core for the use of the XFe24 Extracellular Flux Analyzer (supported by the NYU-HHC CTSI Grant UL1 TR000038 and the NYU Cancer Institute's Cancer Center Support Grant P30CA016087). We thank H. Virgin and D. MacDuff for generously providing cGAS−/− mice, K. Caldwell for Atg16l1flox/flox mice, B. Norris for help with flow cytometry, and C. O’Shaughnessy for help with mouse harvests.
Footnotes
AUTHOR CONTRIBUTIONS
M.O., S.K., J.A.P. and K.J.M. designed and analyzed the experiments. T.D.R., S.O., D.K. and K.J.R. assisted with gene expression analyses; B.R. performed immunostaining; C.v.S. performed immunoprecipitation assays; S.K., E.S., C.P.C, and F.J.S. performed M. tuberculosis studies; K.C. and P.D.Z. provided Mir33−− bone marrow cells. Y.L.M., K.J.M. and J.A.P. supervised the experiments; M.O., J.A.P. and K.J.M. wrote the manuscript.
References
- 1.Stanley SA, Cox JS. Host-pathogen interactions during Mycobacterium tuberculosis infections. Curr Top Microbiol Immunol. 2013;374:211–241. doi: 10.1007/82_2013_332. [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol. 2014;12:101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 4.Sakowski ET, et al. Ubiquilin 1 Promotes IFN-gamma-Induced Xenophagy of Mycobacterium tuberculosis. PLoS pathogens. 2015;11:e1005076. doi: 10.1371/journal.ppat.1005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanley SA, et al. Identification of host-targeted small molecules that restrict intracellular Mycobacterium tuberculosis growth. PLoS pathogens. 2014;10:e1003946. doi: 10.1371/journal.ppat.1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo EF, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A. 2012;109:E3168–3176. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimmey JM, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015 doi: 10.1038/nature16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins AC, et al. Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell host & microbe. 2015;17:820–828. doi: 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majlessi L, Brosch R. Mycobacterium tuberculosis Meets the Cytosol: The Role of cGAS in Anti-mycobacterial Immunity. Cell host & microbe. 2015;17:733–735. doi: 10.1016/j.chom.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Watson RO, et al. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell host & microbe. 2015;17:811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science (New York, NY) 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 16.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature reviews. Drug discovery. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez AM, et al. The role of AMP-activated protein kinase in the coordination of skeletal muscle turnover and energy homeostasis. Am J Physiol Cell Physiol. 2012;303:C475–485. doi: 10.1152/ajpcell.00125.2012. [DOI] [PubMed] [Google Scholar]
- 18.Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nature immunology. 2009;10:943–948. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS pathogens. 2011;7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh V, et al. Mycobacterium tuberculosis-driven targeted recalibration of macrophage lipid homeostasis promotes the foamy phenotype. Cell host & microbe. 2012;12:669–681. doi: 10.1016/j.chom.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 21.D'Avila H, et al. Mycobacterium bovis bacillus Calmette-Guerin induces TLR2-mediated formation of lipid bodies: intracellular domains for eicosanoid synthesis in vivo. Journal of immunology. 2006;176:3087–3097. doi: 10.4049/jimmunol.176.5.3087. [DOI] [PubMed] [Google Scholar]
- 22.Davalos A, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science (New York, NY) 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science (New York, NY) 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rottiers V, et al. MicroRNAs in metabolism and metabolic diseases. Cold Spring Harbor symposia on quantitative biology. 2011;76:225–233. doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goedeke L, et al. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Molecular and cellular biology. 2013;33:2339–2352. doi: 10.1128/MCB.01714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouimet M, et al. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao GJ, TS, Lv YC, Ouyang XP, He PP, Yao F, Tang YY, Zhang M, Tang YL2, Tang DP, Cayabyab FS, Tian GP, Tang CK. NF-kappaB suppresses the expression of ATP-binding cassette transporter A1/G1 by regulating SREBP-2 and miR-33a in mice. Int J Cardiol. 2014;171:e93–95. doi: 10.1016/j.ijcard.2013.1011.1093. Epub 2013 Dec 1017. [DOI] [PubMed] [Google Scholar]
- 29.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science (New York, NY) 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tresse E, et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin CJ, et al. Efferocytosis is an innate antibacterial mechanism. Cell host & microbe. 2012;12:289–300. doi: 10.1016/j.chom.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy J, Summer R, Wilson AA, Kotton DN, Fine A. The prolonged life-span of alveolar macrophages. Am J Respir Cell Mol Biol. 2008;38:380–385. doi: 10.1165/rcmb.2007-0224RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamura K, et al. The regulatory activity of microRNA* species has substantial influence on microRNA and 3' UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voellenkle C, et al. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA. 2012;18:472–484. doi: 10.1261/rna.027615.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schober A, et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med. 2014;20:368–376. doi: 10.1038/nm.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattos KA, et al. Lipid droplet formation in leprosy: Toll-like receptor-regulated organelles involved in eicosanoid formation and Mycobacterium leprae pathogenesis. Journal of leukocyte biology. 2010;87:371–384. doi: 10.1189/jlb.0609433. [DOI] [PubMed] [Google Scholar]
- 39.Zhao GJ, et al. NF-kappaB suppresses the expression of ATP-binding cassette transporter A1/G1 by regulating SREBP-2 and miR-33a in mice. Int J Cardiol. 2014;171:e93–95. doi: 10.1016/j.ijcard.2013.11.093. [DOI] [PubMed] [Google Scholar]
- 40.Brzostek A, Pawelczyk J, Rumijowska-Galewicz A, Dziadek B, Dziadek J. Mycobacterium tuberculosis is able to accumulate and utilize cholesterol. Journal of bacteriology. 2009;191:6584–6591. doi: 10.1128/JB.00488-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci U S A. 2010;107:9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez JG, et al. Global adaptation to a lipid environment triggers the dormancy-related phenotype of Mycobacterium tuberculosis. mBio. 2014;5:e01125–01114. doi: 10.1128/mBio.01125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Wel N, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 46.Simeone R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS pathogens. 2012;8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong KW, Jacobs WR., Jr Critical role for NLRP3 in necrotic death triggered by Mycobacterium tuberculosis. Cellular microbiology. 2011;13:1371–1384. doi: 10.1111/j.1462-5822.2011.01625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandey AK, et al. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS pathogens. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galic S, et al. Hematopoietic AMPK beta1 reduces mouse adipose tissue macrophage inflammation and insulin resistance in obesity. The Journal of clinical investigation. 2011;121:4903–4915. doi: 10.1172/JCI58577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horie T, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchiando AM, et al. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell host & microbe. 2013;14:216–224. doi: 10.1016/j.chom.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoggins JW, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metab. 2010;21:699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf A, et al. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. Journal of immunology. 2007;179:2509–2519. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 55.Listenberger LL, BD Fluorescent detection of lipid droplets and associated proteins<LD staining with Bodipy and Nile Red.pdf>. Curr Protoc Cell Biol. 2007 doi: 10.1002/0471143030.cb2402s35. [DOI] [PubMed] [Google Scholar]
- 56.Mehra A, et al. Mycobacterium tuberculosis type VII secreted effector EsxH targets host ESCRT to impair trafficking. PLoS pathogens. 2013;9:e1003734. doi: 10.1371/journal.ppat.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.