Abstract
Background
In the vertebrate retina six neuronal and one glial cell class are produced from a common progenitor pool. During neurogenesis, adjacent retinal cells use Notch signaling to maintain a pool of progenitors by blocking particular cells from differentiating prematurely. In mice there are multiple Notch pathway ligands and receptors, but the role(s) of each paralogue during retinal histogenesis remains only partially defined.
Results
Here we analyzed the cell autonomous and nonautonomous requirements for the Deltalike1(Dll1) ligand during prenatal retinogenesis. We used the α-Cre driver to simultaneously delete a Dll1 conditional allele and activate the Z/EG reporter, then quantified Dll1 mutant phenotypes within and outside of this α-Cre GFP-marked lineage. We found that Dll1 activity is required for Hes1 expression, both autonomously and nonautonomously, but were surprised that retinal ganglion cell differentiation is only blocked cell autonomously. Moreover Dll1 does not act during cone photoreceptor neurogenesis. Finally, Dll1 mutant adult retinas contained small retinal rosettes, and RGC patterning defects but were otherwise normal.
Conclusions
Although Dll1 participates in bidirectional (cis + trans) Notch signaling to regulate Hes1 expression, it is only acts cell autonomously (in cis) to interpret inhibitory signals from other cells that block RGC neurogenesis.
Keywords: Notch signaling, retinal ganglion cell, cone photoreceptor, lateral inhibition, retinogenesis
Introduction
The Notch signaling pathway is employed by diverse cell types and tissues, and used repeatedly, even within a single cell type. During neurogenesis, this pathway transduces at minimum, a lateral inhibition signal that facilitates progenitor cell growth and blocks neuronal differentiation. This is the classic role of Notch, first characterized in the fly nervous system and nematode gonad (reviewed in Doroquez and Rebay, 2006; Artavanis-Tsakonas and Muskavitch, 2010; Treisman, 2013). Signaling initiates when a transmembrane ligand binds to a transmembrane Notch receptor. The ligands are of two main classes, Delta-Deltalike or Serrate-Jagged-Lag1. Ligand-receptor binding triggers a series of proteolytic events, the last of which releases the receptor intracellular domain (ICD), to form a protein complex with Rbpj and MAML that is active within the nucleus (reviewed in Kopan and Ilagan, 2009). This protein complex promotes gene transcription, of which Hes1 and Hes5 are two well-characterized transcriptional targets throughout vertebrate development (reviewed in Kageyama et al., 2008).
Loss- and gain-of-function studies emphasize the strong evolutionary conservation of Notch signaling during retinogenesis (reviewed in Gregory-Evans et al., 2013). In the chick and frog retina, knock-down of Delta function resulted in excess retinal ganglion cells (RGCs), while Delta overexpression induced prolonged mitotic activity by retinal progenitor cells (RPCs) (Austin et al., 1995; Henrique et al., 1995; Ahmad et al., 1997; Dorsky et al., 1997; Silva et al., 2003). These experiments were particularly insightful, demonstrating that vertebrate Delta laterally signals neighboring retinal cells (signals in trans), but also interprets a cell autonomous inhibitory signal (signals in cis) (Henrique et al., 1995). However, in the mouse eye, there are no analogous examinations of ligand activities reported, in part because there are three Delta-like genes, Dll1, Dll3 and Dll4 expressed in the retina (Nelson et al., 2009). Loss of function studies have defined the general requirements for Dll1 or Dll4 in the mouse retina (Rocha et al., 2009; Luo et al., 2012), and demonstrated that Dll3 has no role in this tissue (Nelson et al., 2009). Indeed, Rocha and colleagues showed that Dll1 and Dll4 initiate expression sequentially in the mouse optic cup, and that Dll1 is required by a subset of progenitor cells for their proliferation and to suppress RGC differentiation (Rocha et al., 2009). Interestingly, Dll4 can also promote RPC proliferation, but primarily blocks photoreceptor differentiation (Luo et al., 2012). However, deeper genetic dissection of the activities of each ligand has not been undertaken, particularly for subpopulations of retinal cells. Analogous to other tissues of the body, Notch signaling can occur 1) among RPCs; 2) from postmitotic or fully differentiated cells to RPCs or 3) among postmitotic and differentiating retinal neurons. In each context, it is unclear if Dll1 acts alone, Dll4 acts alone, or both ligands function in concert, and importantly when and where each ligand engages in cis and/or trans signaling.
Here we analyzed the cell autonomous consequences of removing Dll1 in vivo during early retinal development, and the resulting adult retinal phenotypes. We found that in the embryonic retina, Dll1 signaling can be bidirectional among RPCs, since Hes1 expression was reduced both autonomously and nonautonomously. Among RGCs, cones, amacrines and horizontal cells, we found that Dll1 is only required cell autonomously (in cis) to suppress RGC differentiation. We also demonstrate that Dll1 mutant retinas are thinner and contain rosettes, which phenocopies other Notch pathway mutants. However, just one early retinal class, RGCs, displayed a singular requirement for Dll1.
Results
Adult retinal phenotypes induced by deletion of Dll1
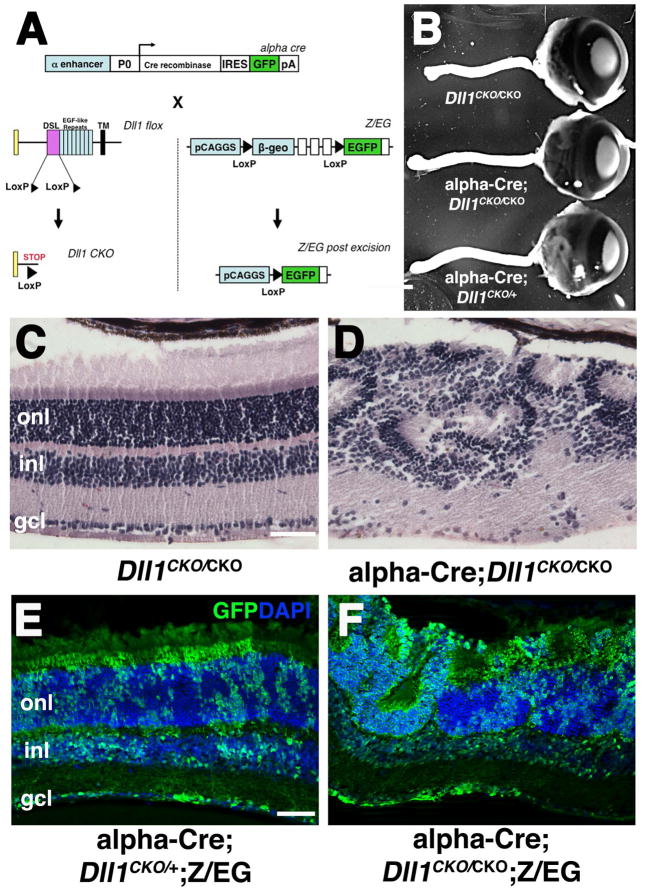
This study was undertaken with two objectives in mind first, to characterize the adult retinal phenotypes of Deltalike1 (Dll1) conditional mutants; and second, to learn which prenatal retinal cells require this ligand, both cell autonomously and nonautonomously. Particularly the second question was provoked by the striking salt-and-pepper expression pattern of Dll1 in prenatal retinal cells (Lindsell et al., 1996; Beckers et al., 1999; Rocha et al., 2009). To accomplish both goals we used a conditional mouse allele (Dll1CKO) (Hozumi et al., 2004; Brooker et al., 2006), the α-Cre retinal driver that initiates Cre expression in the distal optic cup at E10.5 (Marquardt et al., 2001) and a Z/EG lineage reporter (Novak et al., 2000)(Fig 1A). Throughout this paper, control animals were either Dll1CKO/CKO (lacking the Cre transgene), or αCre;Dll1CKO/+;Z/EG littermates. Neither genotype displayed histologic or molecular marker phenotypes at any age analyzed (n ≥ 5 animals per age). We compared α-Cre;Dll1CKO/CKO mutant and control adult eyes, and found them grossly indistinguishable (Fig 1B). This is in contrast to analogous deletion of Notch1 or Rbpj, which induced a pronounced microphthalmia (Jadhav et al., 2006; Yaron et al., 2006; Riesenberg et al., 2009; Zheng et al., 2009). However, histologic sections of α-Cre;Dll1CKO/CKO eyes revealed retinal rosettes in the outer nuclear layer (ONL), indicative of abnormal patterning during development (compare Figs 1C,1D). Although the Dll1 retinal rosette defect phenocopied Notch1 and Rbpj mutants, the rosettes were often smaller, and arose later in development, in agreement with a previous study using a different Cre driver (Rocha et al., 2009).
Figure 1. Adult retinal phenotypes of α-Cre;Dll1CKO/CKO conditionally mutants.
A) Deletion strategy for α-Cre-mediated removal of Dll1 and activation of GFP expression from the Z/EG transgene. B) α-Cre;Dll1CKO/+ and α-Cre;Dll1CKO/CKO adult eyes and optic nerves appear identical to Dll1CKO/CKO control littermates. No Dll1 heterozygous phenotypes were found by gross inspection or histology (n=6). C–D) Histologic sections of P21 eyes highlighted thinner retinal tissue with rosetting in the absence of Dll1 (n=9). E–F) Anti-GFP/DAPI double label of α-Cre;Z/EG;Dll1CKO/+ and α-Cre;Z/EG;Dll1CKO/CKO P21 retinal sections. In the distal retina, the α-Cre lineage (GFP-marked) in both control and Dll1CKO/CKO mutants is interspersed with GFP-negative cells (DAPI only), even within rosettes. This is vastly different than the segregation of wild type and mutant cells found in α-Cre;Z/EG;RbpjCKO/CKO eyes (Riesenberg et al., 2009). Anterior is right in B, scleral is up in C–F. Scale bars = 500μm in B, 20 μm in C,E. α= Pax6 intronic α enhancer; P0 = Pax6 promoter, IRES = internal ribosome binding site; pA = poly A sequence; DSL= Delta/Serrate/Lag domain; TM = transmembrane-spanning domain; ONL = outer nuclear layer, INL = inner nuclear layer, GCL = ganglion cell layer.
To score the cell autonomy of Dll1 retinal defects, and for meaningful comparison with Notch1 and Rbpj mutant analyses (Yaron et al., 2006; Riesenberg et al., 2009; Maurer et al., 2014), we incorporated the Z/EG transgene into our crossing scheme (Novak et al., 2000). Thus, retinal progenitor cells (RPCs) with α-Cre activity and their progeny were permanently marked with GFP expression (Fig 1A). This experimental strategy previously uncovered both abnormal morphology of adult Rbpj−/− cells, and inappropriate segregation from Rbpj wild type (GFP-) retinal cells (Riesenberg et al., 2009). However, GFP-marked, adult Dll1−/− cells were of normal size and shape, and randomly distributed throughout the retina, including within rosettes (Fig 1E,F).
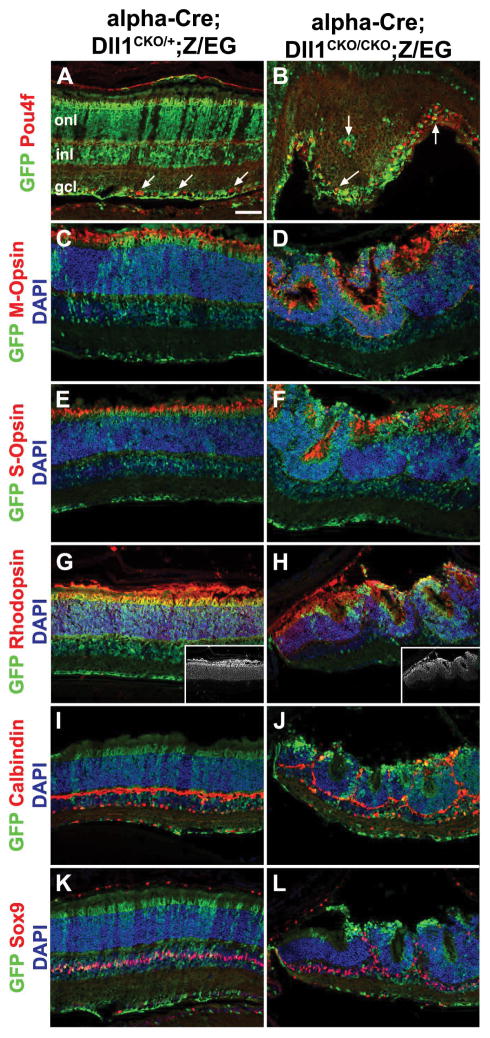
To identify which retinal cell classes require Dll1 activity, we compared terminal differentiation marker expression in P21 cryosections from control and Dll1 conditionally mutant eyes. For the major classes of neurons or glia, we confirmed that retinal ganglion cells (RGCs) exhibit the strongest requirement for Dll1 (Figs 2A–B; Fig 4). There were more GFP+Pou4f/Brn3+ RGCs in Dll1 mutants, both within the GCL and in ectopic locations (arrows in Fig 2B). We also observed that the distribution of cones (M- and S-Opsin+ or Arr3+) and rod (Rhodopsin+) photoreceptors (Figs 2D,2F,2H,5R) were indistinguishable between genotypes, although Dll1 mutants were mispatterned. The loss of Dll1 also had no effect on the laminar position or proportion of INL cell types, beyond a general displacement by the retinal rosettes. Representative markers illustrating this point include Calbindin, expressed by horizontals and amacrines (Figs 2I–2J), Calretinin, expressed by A2 amacrines (not shown) and Sox9, present in Müller glia (Figs 2K–2L). We conclude that the adult retinal phenotypes of Dll1 conditional mutants were less severe than those of Notch1 or Rbpj, with Dll1 specifically required to suppress RGC differentiation.
Figure 2. Retinal cell types in Dll1 conditionally mutant P21 eyes.
Retinal sections triple labeled with anti-GFP in green, retinal marker in red and DAPI counterstain in blue. A–B) After loss of Dll1, Pou4f+ RGCs are autonomously increased within the GFP-marked alpha-Cre lineage, with occasional RGCs misplaced outside of the GCL (arrows). C–F) Both red/green (M-Opsin+) and blue (S-Opsin+) cone photoreceptors are mispatterned within retinal rosettes of Dll1 mutants. G–H) Rhodopsin+ rods were also components of retinal rosettes that resulted from removal of Dll1. Insets show Rhodopsin pattern alone in white. I–L) Inner retinal cells, like Calbindin+ horizontals and amacrines, and Sox9+ Muller glia surrounded the rosettes, but otherwise were unaffected by loss of Dll1. Scale bar in A = 20 microns. onl = outer nuclear layer, inl = inner nuclear layer, gcl = ganglion cell layer. n=3 control and mutant animals examined for each marker.
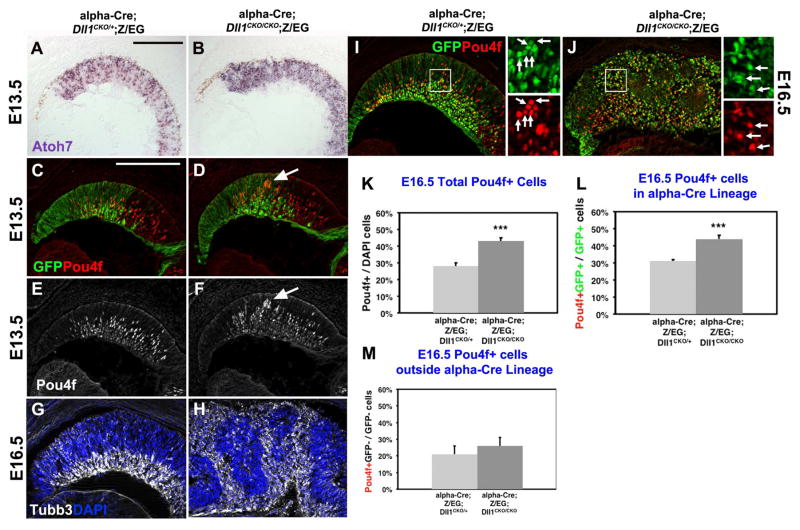
Figure 4. Dll1 regulation of RGC development.
A–B) E13.5 mRNA expression of Atoh7 at E13.5 shows abnormal pattern in the alpha-Cre domain, upon deletion of Dll1. C–F) At E13.5, Pou4f+ RGCs were largely unaffected by loss of Dll1, although sporadic clusters of nascent RGCs (arrows in D) occurred. G–H) Anti-Tubb3 (white) and DAPI double labeling at E16.5. In Dll1 retinal mutants, excess differentiated neurons surrounded forming retinal rosettes contained undifferentiated cells (see Fig 3). I–J) GFP and Pou4f colabeling was quantified for the α-Cre lineage (arrows). Boxed area shown at right is magnified 10×. K) The total number of Pou4f+ RGCs was significantly increased in Dll1 mutants. L) Dll1 normally suppresses RGC differentiation cell autonomously in Pou4f+GFP+/GFP+ cells. M) Whereas there was no nonautonomous affect for the Pou4f+GFP-/GFP- population. Bar in A,C = 20 microns, * = p <0.05; ** = p <0.01; *** = p< 0.001. For each marker n ≥ 3 embryos per age and genotype.
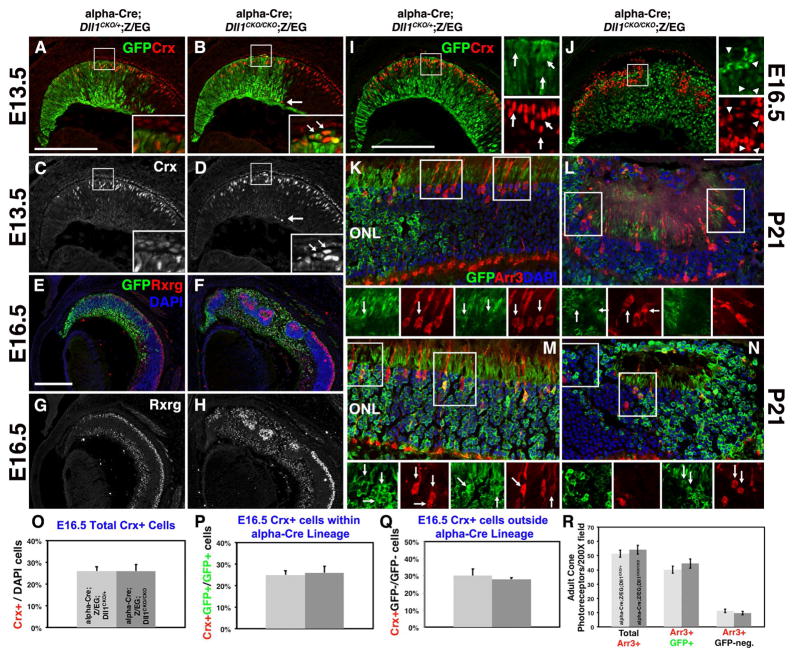
Figure 5. Embryonic Crx+ photoreceptor precursors do not require Dll1 function.
A–D) Mispatterning of E13.5 Crx+GFP+ retinal cells upon removal of Dll1. Local disruptions in the normal pattern of Crx+ bipotential photoreceptor precursors occurred at both the apical (boxed area) and basal (arrow) retina. E–H) Rxrg expression in nascent cone photoreceptors (outer, top) and RGCs (inner, lower retina) highlights the presence of cones in forming retinal rosettes and the expansion of RGCs upon conditional deletion of Dll1. I–J) GFP and Crx colabeling were quantified, by assessing Crx+ cells within (arrows) or outside (arrowheads) the α-Cre lineage. The boxed areas shown at right are magnified 10×. O,P,Q) The percentages of total Crx+, Crx+GFP+/GFP+ or Crx+GFP-/GFP- cells were unaffected by loss of Dll1. K-N,R) Quantification of P21 cone photoreceptors (Arr3+) showing no significant difference between genotypes (R). Two independent examples for genotype are provided, with boxed areas at higher mag below highlight the Arr3+GFP+ colabeled cones (arrows). Apical is up in all images. Bar in A,E, I = 20 microns, L = 50 microns; ONL = outer nuclear layer. Boxed areas are shown at 10× higher magnification at right (I–J), or below (K–N). For each marker n ≥ 3 embryos or adult eyes per age/genotype.
Retinal progenitor cells and Dll1 activity
Previous investigations of Notch pathway loss-of-function phenotypes in the mouse retina, showed that as early as E13.5 there is a strong loss of RPC proliferation, accompanied by premature neuron formation and retinal mispatterning (Jadhav et al., 2006; Yaron et al., 2006; Riesenberg et al., 2009; Zheng et al., 2009). Moreover, each phenotype is progressive as development proceeds. By contrast, in Chx10-Cre;Dll1CKO/CKO eyes there was no change in the ratio of proliferating RPCs to differentiating neurons prior to E14.5 (Rocha et al., 2009). We wondered whether mosaic expression of the Chx10-Cre driver (Rowan and Cepko, 2004) may have masked earlier requirements for Dll1 in the optic cup, since in theory, neighboring wild type cells could nonautonomously “rescue” Dll1−/− mutant cells. To test this idea, we evaluated α-Cre;Dll1CKO/CKO embryonic retinal phenotypes, since the mosaicism exhibited by this Cre is notably lower between E10.5 to E16.5 (Marquardt et al., 2001; Yaron et al., 2006; Riesenberg et al., 2009). We also quantified the consequences of removing Dll1 from α-Cre lineage cells (GFP+), versus wild type cells (GFP-neg.). First, we analyzed the expression pattern of the downstream gene Hes1 (Jarriault et al., 1995), which was not previously tested in Dll1 mutant eyes (Rocha et al., 2009). The removal of either Notch1 or Rbpj resulted in a dramatic, cell autonomous, downregulation of Hes1 mRNA and protein in E13.5 RPCs (Jadhav et al., 2006; Yaron et al., 2006; Riesenberg et al., 2009; Zheng et al., 2009). Yet at E13.5 Dll1 retinal mutants displayed subtle disruptions of Hes1 expression (arrows in Fig 3D). GFP/Ki67 or GFP/BrdU colabeling of nearby sections indicated that the loss of Dll1 had little or no effect on RPC proliferation at this age. This is in good agreement with the Chx10-Cre;Dll1CKO/CKO eyes which had no RPC proliferation defects until E14.5 (Rocha et al., 2009). Thus, we conclude that there is essentially no requirement for the Dll1 ligand activity by Hes1+ RPCs prior to E13.5.
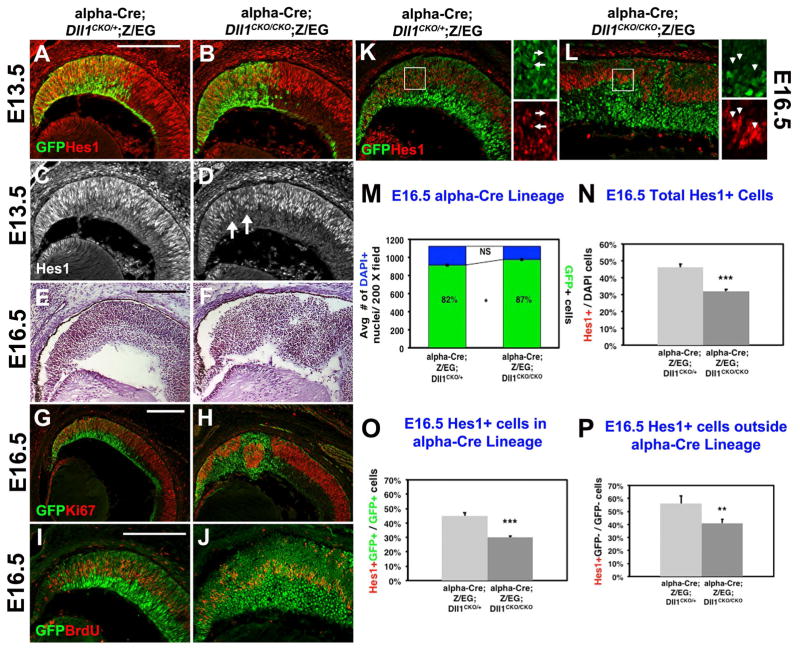
Figure 3. Retinal progenitor cell defects induced by loss of Dll1.
A–D) At E13.5 there is normally a high degree of GFP and Hes1 coexpression. Only a subtle loss of Hes1 expression occurred in GFP-marked Dll1−/− cells at this age (arrows in D). E–F) Hemotoxylin-stained paraffin sections highlighted a failure of retinal lamination in E16.5 Dll1 retinal mutants (F). G–H) Ki67 and GFP colabeling of nearby retinal sections further highlighted that cells within Dll1 mutant retinal rosettes are generally. I–J) BrdU and GFP are normally coexpressed in E16.5 RPCs in the α-Cre lineage, but in Dll1 retinal mutants, there was a loss of the BrdU+ GFP+ subpopulation. K–L) Control E16.5 α-Cre RPCs (GFP+) coexpress Hes1, with boxed area shown at 10× magnification at right. M) There was no difference in the average number of DAPI+ cells (blue bars), although the E16.5 alpha-Cre lineage (green subset within bars) increased when Dll1 was removed (81.6% ± 1.3% control; 86.8% ± 1.3% mutant). ( N) The total proportion of E16.5 Hes1+ RPCs was significantly reduced in Dll1 mutants. O,P) There was a significant loss of Hes1-expressing cell cell autonomously in Hes1+GFP+/GFP+ cells (O) and nonautomously in Hes1+GFP-/GFP-cells (P). Bar in A,G, I = 20 microns, * = p <0.05; ** = p <0.01; *** = p< 0.001. For each marker n ≥ 3 embryos per age and genotype.
To score the autonomy of early loss of Dll1 in the retina, we turned our attention to E16.5 conditional mutants, where retinal rosettes were fully penetrant in histologic sections (Fig 3E–F). This mispatterning included abnormal clusters of Hes1+ (RPC), Ki67+ (mitotic) and BrdU+ (S-phase) cells, within the rosettes of Dll1 mutant eyes (Figs 3G–3L). Although these defects appeared at a slightly older age, they are reminiscent of defects found in Rbpj and Notch1 retinal mutants, with one exception. There were statistically fewer Hes1+ RPCs within the α-Cre mutant lineage (Fig 3O), although this mutant lineage was 5% bigger (Fig 3M). By comparison, E16.5 Rbpj retinal mutants displayed a dramatic, 10-fold loss of Hes1+ RPCs that directly correlated with a smaller GFP-marked, mutant lineage (Riesenberg et al., 2009). We attribute the more severe phenotype of Rbpj mutants to the combined effects of fewer rounds of mitosis, plus an increase in apoptosis that contributes to a thinner optic nerve. So we compared the number of cPARP+GFP+ cells in E16.5 control and Dll1 conditional mutants, but found no increase in apoptosis in Dll1 mutants, either within the GFP+ (lineage-marked) or GFP-neg retinal populations (n=3 embryos per genotype, data not shown). This is consistent with the retinas from both genotypes containing the same average number of DAPI+ nuclei (Fig 3M, n= 3 eyes and 18 sections per genotype). We conclude that Dll1 does not participate in a Notch1-Rbpj-mediated block of apoptosis, since adult Dll1 mutant optic nerves were of normal size (Figs 1B,D). Interestingly, Six3-Cre; Dll4CKO/CKO adult retinas have thinner optic nerves (Luo et al., 2012), indicating that the Dll4 alone is sufficient to mediate this anti-apoptotic signal.
Next we scored the cell autonomy Hes1 expression in E16.5 control and Dll1 mutant RPCs. Classically, a lateral inhibition signal is sent from postmitotic or differentiated cells to mitotically active RPCs, to regulate the rate of subsequent neurogenesis. So, if Dll1 only transmits a lateral inhibition signal in trans to neighboring RPCs (Notch+ and Hes1+), we would expect to see cell nonautonomous consequences from its removal. This means there would be fewer Hes1+GFP-neg. mutant cells. Alternatively, if Dll1 acts cell autonomously (in cis) to interpret an incoming lateral inhibition signal, GFP+ mutant RPCs would have reduced Hes1 expression. At E16.5 there was an overall significant reduction in Hes1+ cells, in the absence of Dll1 (Fig 3N). Next, we quantified the cohorts of Hes1+ retinal cells within (GFP+) or outside (GFP-neg) the α-Cre lineage, in control and Dll1 mutants (Figs 3O,P). We found that both Hes1 subpopulations were significantly reduced (Figs 3O,P), indicating that the Dll1 ligand is active, both in cis and in trans, at this age. Although Dll1 mRNA and Dll1LacZ/+ expression is mostly found in nascent retinal neurons, Dll1 mRNA has also been reported in a subset of mitotic RPCs (Henrique et al., 1997; Nelson et al., 2006; Nelson et al., 2009; Rocha et al., 2009; Luo et al., 2012). Our interpretation is that both postmitotic/differentiated retinal cells and mitotic RPCs require Dll1, although to differing extents.
Dll1 acts cell autonomously during retinal ganglion cell genesis
Math5/Atoh7 is a bHLH factor that endows retinal cells with the competence to adopt an RGC fate (Brown et al., 2001; Wang et al., 2001; Brzezinski IV et al., 2012), and is required for Brn3b/Pou4f2 expression in differentiating RGCs (Liu et al., 2001; Wang et al., 2001; Prasov et al., 2012). Previously, Atoh7 mRNA and Pou4f2+ protein expression were surveyed in embryonic Chx10Cre;Dll1CKO/CKO retinas (Rocha et al., 2009). At E12.5 there was an upregulation of Atoh7 expression, and a 4-fold increase in the percentage of Brn3b/Pou4f2+ RGCs, and 4.5-fold increase in p27/Kip+ terminally exiting RPCs. But, the cell autonomy of these phenotypes was not determined. In our experiments we also saw mispatterning of Atoh7 mRNA in E13.5 α-Cre;Dll1CKO/CKO distal eyes (Figs 4A,B). However, there is no difference in the percentages of βgal+GFP+/GFP+ cells in E13.5 α-Cre;Z/EG; Atoh7LacZ/+;Dll1CKO/+and α-Cre;Z/EG; Atoh7LacZ/+;Dll1CKO/CKO eyes (Maurer et al., 2014). We conclude that changes in the Atoh7 lineage correlate with the onset of retinal mispatterning in Dll1 mutants, but that Dll1 normally suppresses RGC differentiation by acting parallel to, or downstream of, Atoh7 activity.
We then colabeled E13.5 control and Dll1 mutant retinal sections with a pan-Pou4f /Brn3 antibody, which recognizes Pou4f1, Pou4f2, Pou4f3 proteins in nascent RGCs (Figs 4C–4F). We consistently saw atypical, small clusters of Pou4f+ cells (arrows in Figs 4D,4F). But by E16.5, Dll1 mutants were more obviously mispatterned, with Tubb3+ neurons surrounding the forming rosettes (Figs 4G,H). Here too, Pou4f+ RGCs were significantly increased (Figs 4I,4J,4K), in line with the 34% increase previously found in E14.5 Chx10Cre;Dll1CKO/CKO eyes (Rocha et al., 2009). To score the cell autonomy of this phenotype, the proportions of Pou4f+ cells within (GFP+) and outside of (GFP-) the α-Cre lineage were quantified (Figs 4L,4M). Surprisingly, we found there is cell autonomous increase in Pou4f+GFP+ RGCs (Fig 4L), yet the Pou4f+GFP-negative cohort is unaffected. We interpret this to mean that Dll1 normally acts in cis to interpret a lateral inhibition signal emanating from other cells. This is in stark contrast to Dll1 regulation of Hes1 expression both autonomously and nonautonomously (Fig 3).
Dll1 is not required for prenatal photoreceptor neurogenesis
In the chick and frog retina Delta-Notch signaling is a major suppressor of photoreceptor cell fate (Austin et al., 1995; Henrique et al., 1995; Ahmad et al., 1997; Dorsky et al., 1997; Silva et al., 2003), and in the mouse retina Notch1 and Rbpj normally block photoreceptor development (Jadhav et al., 2006; Yaron et al., 2006; Riesenberg et al., 2009). Indeed, as early as E13.5 conditional Notch1 and Rbpj mutants display expanded Otx2+ and Crx+ expression domains, with an increase in differentiated cone photoreceptors apparent at E16.5 (Furukawa et al., 1997b; Chen et al., 2002; Nishida et al., 2003; Jadhav et al., 2006; Yaron et al., 2006; Riesenberg et al., 2009). However, only a general phototransduction marker, Recoverin, was examined in Dll1 conditional mutants (Rocha et al., 2009).
So we asked to what extent Dll1 regulates the earliest stages of photoreceptor development, by assaying Crx, expressed by tri-potential precursors that produce cones, rods or bipolars (Freund et al., 1997; Furukawa et al., 1997b), or Rxrg expressed by cones and RGCs, which are easily distinguished from one another (Roberts et al., 2005). At E13.5, αCre;Dll1CKO/CKO retinas had only minor mispatterning of Crx+ cells (insets in Figs 5A–4D). By E16.5, the Dll1 mutant retinal rosettes contained Crx+ and Rxrg+ cells (Figs 5E–5H). The clustering of Crx+ or Rxrg+ cells within retinal rosettes phenocopies Notch1 and Rbpj retinal mutants, as well as Dll4 retinal mutants (Yaron et al., 2006; Riesenberg et al., 2009)(Luo et al., 2012). However, there were no changes in the total population of Crx+ cells between control αCre;Dll1CKO/+;Z/EG and α-Cre;Dll1CKO/CKO;Z/EG littermates (Fig 5I,5J, 5O), or significant shifts in the Crx+ population, within (GFP+) and outside of (GFP-) the α-Cre lineage (Figs 5P,5Q). We also quantified cone Arrestin (Arr3+) photoreceptor cells in P21 α-Cre;Dll1CKO/+;Z/EG and α-Cre;Dll1CKO/CKO;Z/EG eyes (Figs 5K–5N, 5R), to verify that the removal of Dll1 did not induce a delay of photoreceptor genesis. We conclude that Dll1 has no role in regulating photoreceptor precursor cells, or during cone photoreceptor differentiation. This differs from Dll4 retinal mutants where cone and rod photoreceptor formation was significantly derepressed (Luo et al., 2012). Finally, it is plausible that Dll4 is affected by the loss of Dll1, so we compared the total Dll4+, Dll4+GFP+ and Dll4+GFP-neg cell populations between E13.5 α-Cre;Dll1CKO/+;Z/EG and α-Cre;Dll1CKO/CKO;Z/EG littermates. We observed no changes in the spatial patterning or statistically significant shifts in cell numbers (data not shown, n=6/genotype). Overall, we presume that Dll1 and Dll4 paralogues act distinctly to suppress RGC versus photoreceptor cell neurogenesis in the mammalian retina.
Discussion
Dll-Notch regulation of vertebrate retinogenesis has been under active investigation for more than 20 years in frog, chick, zebrafish and mouse model organisms. This signaling pathway maintains a balance between proliferating and differentiating cells, and releases waves of RPCs to undergo neurogenesis at the correct time, and in appropriate proportions. Yet, we still lack clear understanding of the underlying mechanisms as differentiating neurons signal to neighboring RPCs, or between postmitotic cells solidifying their final fate. It remains unclear which, and how many, ligand and receptor proteins are expressed by particular retinal cells, how their expression changes over time, and finally, how the bi-directional activities of ligands and receptors (cis versus trans signaling) are distilled into unidirectional outcomes. In the mammalian retina, parsing of particular ligand activities among different gene paralogues further complicates these mechanisms.
Cis and trans modes of ligand signaling
In the developing nervous system, there is prolonged regulation of the rate at which proliferating progenitor cells produce differentiated cells types, versus replenish their pool. The Notch pathway is a highly conserved signaling system in which differentiating cells laterally inhibit proliferating progenitors, thereby controlling the dynamics of each population. To fine tune and coordinate this signaling, there must also be feedback signaling, back to the cells that sent the lateral inhibition signal. Ligand mutations that abolish lateral inhibition render a sending cell mute (= block trans signaling). By contrast, ligand mutations that affect the feedback machinery make the same cell deaf (= block cis signaling) (Henrique et al., 1995; Henrique et al., 1997; Rocha et al., 2009). The cell autonomous requirements of Dl-Notch signaling were first identified in the Drosophila compound eye, using multiple cell-specific marking systems (reviewed in Baker, 2000). These studies mapped cis/cell autonomous versus trans/nonautonomous mutant phenotypes to individual photoreceptor neurons. To begin to approach this level of mechanistic understanding of Notch pathway ligands in the mouse eye, here we used α-Cre, a Dll1 conditional mutant and Z/EG lineage tracer, to score the cell autonomy of particular prenatal retinal phenotypes.
Maintaining the retinal progenitor cell pool
In the optic cup, Dll1 mRNA is first detectable at E10.5, just ahead of the first wave of neurons at E11.0 (Bao and Cepko, 1997; Rocha et al., 2009; Luo et al., 2012). By midgestation about 80% of Dll4+ retinal cells coexpress Dll1, suggesting that Dll1 activity starts earlier and acts more broadly (Rocha et al., 2009; Preusse et al., 2015). It further implies that Dll1 and Dll4 laterally inhibit neighboring RPCs, largely by signaling from postmitotic cells. Additional evidence supporting this idea, includes sparse expression of either ligand in mitotic RPCs (Rocha et al., 2009; Luo et al., 2012), and mild proliferation defects in Dll1 (Rocha et al., 2009) and Dll4 conditional mutants (Luo et al., 2012). By comparison, Notch1 receptor mutants have a profound reduction of proliferating RPCs (Jadhav et al., 2006; Yaron et al., 2006). A recent study also showed that Dll4 substitution into the Dll1 locus fully rescued the Dll1 RPC phenotype (Preusse et al., 2015). Thus, in the early retina it seems clear that Dll1 and Dll4 redundancy reinforces the lateral inhibition signal (in trans) to neighboring Notch+, Hes1+ RPCs. But more work is still needed, and future experiments could examine the consequences of simultaneously removing both ligands, plus score the cell autonomy of resulting phenotypes. It would also be highly informative to conduct a ligand protein co-localization time course during retinal development.
Data presented in this paper however, identify at least one other role for Dll1 in RPCs, since Hes1 expression was cell autonomously downregulated after Dll1 was removed (Fig 3). At the same time we also noted a small (5%) but significant expansion of the α-Cre lineage. These extra cells may represent RPCs that normally do not express Hes1, therefore respond to Dll1 signaling differently than do the Hes1+ RPCs. This may represent a limitation of our experimental approach, since the Dll1 mutant GFP-marked cells analyzed could either be those with Cre activity (IRES GFP+), or those erroneously differentiated into neurons (Z/EG lineage GFP+). This situation might be overcome by switching to a Cre-activatable tdTomato lineage reporter (Madisen et al., 2010) in the future. Nonetheless, we conclude that the mitotic RPCs that depend on Dll1 signaling do so in cis (autonomously), to prevent their own premature differentiation possibly by modulating Notch1 or Notch3 activities. Furthermore, these distinct roles for Dll1 appear to contribute to the diversification of RPCs. It would be extremely interesting to isolate, purify and molecularly profile the subset Dll1+ retinal cells that are mitotic. Moreover, it will finally be possible to observe Dll1 protein dynamics during retinal neurogenesis, using a brand new Dll1 mouse model (Shimojo et al., 2016).
Regulation of retinal neurogenesis
In the frog retina where there is just one Delta gene, its widespread overexpression caused 50% of Delta-expressing cells to differentiate as either RGCs or cones, which is significantly higher than normal (Dorsky et al., 1997). However, the later misexpression of Delta produced only excess rod photoreceptors. In the chick eye, retroviruses were used to deliver a Delta-1 cDNA or dominant-negative (dn) Delta-1 construct to proliferating RPCs (Henrique et al., 1997). Here, excess Delta-1 expression blocked differentiation, while dnDelta-1 interfered with RPC feedback inhibition, resulting in premature differentiation, especially as RGCs. Although excess RGC neurons were also found in Chx10-Cre;Dll1 mutant mouse retinas (Rocha et al., 2009), it has remained unknown which mode of Dll1 signaling suppresses RGC neurogenesis. A priori, Dll1 would be predicted to signal in trans (nonautonomously), as postmitotic/differentiated cells block neighboring (Notch+) RPCs from differentiating prematurely. Yet we found that Dll1, only acts to prevent cells from interpreting an inhibitory signal coming from their neighbors, not from sending a signal (in trans). Here, Dll1 may cell reinforce a selected fate, or influence (bias) the final fate choice. This is consistent with the idea that Dll1 and Dll4 trans signaling are redundant, yet each ligand exhibits a unique cis activity (Dll1 blocks RGC neurogenesis, Dll4 blocks photoreceptor neurogenesis). Over the longer term, elucidation of these simultaneous and complex modes of signaling will be advanced further by biochemical and molecular assays performed within the context of retinal development.
Experimental Procedures
Animals
Deltalike1CKO/CKO mice were maintained on a 129 background and genotyped using published protocols (Hozumi et al., 2004; Brooker et al., 2006). Throughout this paper the abbreviation CKO indicates a “Conditional Knockout Allele”. The α-Cre transgenic mice (Marquardt et al., 2001) and Z/EG lineage tracing mice (Novak et al., 2000) were each maintained on a CD-1 background and genotyped per published protocols. Images of whole adult eyes were captured on a Leica dissecting microscope with an Optronics digital camera and software. All mice were housed and cared for in accordance with the guidelines provided by the National Institutes of Health, Bethesda, MD and the Association for Research in Vision and Ophthalmology.
Retinal phenotype analyses
Embryonic and adult eye tissues were fixed in 4% paraformaldehyde/PBS for 40–60 minutes at 4°C, processed through a sucrose/PBS series, cryoembedded; or processed postfixation for paraffin embedding, sectioning and standard histology performed. Antibody labeling was performed on 10 micron cryosections used rabbit anti-Arr3+ (mCArr; Millipore AB15282, 1:10,000), rat anti-βgal (Tom Glaser, 1:1000)(Saul et al., 2008), rat anti-BrdU (Serotec clone BU1/75 1:100, unmasked 1 hour in 1N HCl); rabbit anti-Calbindin (Chemicon/Millipore, 1:1000), rabbit anti-cPARP (Cell Signaling 1:500), rabbit anti-Crx (Cheryl Craft, USC; 1:1000)(Zhu and Craft, 2000), goat anti-Dll4 (R&D Systems AF1389, 1:100 unmasked for 10 minutes in 0.5% Triton X-100/PBS), rabbit or chick anti-GFP (Invitrogen or Abcam 1:1000); rabbit anti-Hes1 (1:1000)(Lee et al., 2005); rabbit anti-Ki67 (Vector Labs, 1:1000); rabbit anti-S Opsin (Cheryl Craft, USC; 1:1000); rabbit anti-M/L Opsin (Cheryl Craft, USC; 1:1000)(Zhu and Craft, 2000; Zhu et al., 2003), goat anti-pan Pou4f/Brn3 (Santa Cruz sc6026, 1:50); mouse anti-Rhodopsin (Chemicon/Millipore, 1:1000); rabbit anti-Rxrg (Santa Cruz Biotech; 1:200); rabbit anti-Sox9 (Chemicon/Millipore; 1:200); rabbit anti-Tubb3 (Covance, 1:1000); sheep anti-Vsx2/Chx10 (N-terminus, Exalpha Biologicals,1:1000), 1 mg/ml DAPI stain (Sigma Chemical 1:1000). Secondary antibodies were directly conjugated to Alexa Fluor488, Alexa Fluor594 (Invitrogen) or biotinylated (Jackson Immunologicals) and sequentially labeled with Alexa 488- or 594-Streptavidin (Jackson Immunologicals), followed by DAPI nuclear labeling. For S-phase analyses, BrdU (Sigma) was injected intraperitoneally and animals sacrificed 1.5 hours later for tissue processing and anti-BrdU labeling, following the method of (Mastick and Andrews, 2001).
In situ hybridization on cryosections was performed as described (Brown et al., 1998) using a Math5 cDNA plasmid as a template for a digoxygenin-labeled antisense riboprobe. Microscopic imaging of retinal sections was performed on either a Zeiss microscope with color and black and white cameras and an Apotome deconvolution device or using a Leica DM5500 microscope, equipped with a SPEII solid state confocal. Images were processed using Axiovision (v7.0), Leica LASAF, Adobe Photoshop (CS4) software programs and electronically adjusted for brightness, contrast and pseudocoloring.
Cell Counting
Labeled cells in tissue sections were quantified with Axiovision (v6.0) or Photoshop CS4 software. Three or more animals were analyzed per genotype and age, with ≥2 sections per control or mutant littermate. Sections were judged to be of equivalent depth in the eye by anatomical landmarks in the head and other eye tissues, with only the nasal side of the retina imaged for consistency. The cell autonomy of Hes1+, Pou4f+, Crx+ or Atoh7LacZ (βgal+) cells was determined within 200× distal retinal fields. Quantification of Arr3+ or Dll4+ cells was determined in 400× distal retinal fields. Here the percentages of marker+GFP+/GFP+, marker-GFP+/DAPI, marker+GFP-/DAPI or GFP+/DAPI cells ± standard error of the mean (s.e.m.) were determined, with GFP expression representing the combination of IRES-GFP and Z/EG transgenes. A two-tailed Student’s T test and Welch posthoc test were used for p values (Excel, v14.3.4).
Key Findings.
Retinal progenitors require Dll1 both in cis and trans
RGC neurogenesis requires Dll1 in cis
Dll1 does not regulate prenatal photoreceptor development
Acknowledgments
Grant Sponsor: NIH/NEI; Grant Number: R01EY013618
The authors thank Julian Lewis and Noah Shroyer for Dll1CKO/CKO mice; Ruth Ashery-Padan for α-Cre transgenic mice; Tom Glaser, and Cheryl Craft for antibody reagents; Richard Lang for use of an Apotome deconvolution imaging system; Ashley Riesenberg, Tien Le, April Bird, Kelly McCulloh and Nadege Lum for technical assistance; Tom Glaser and Anna La Torre for valuable discussions.
References
- Ahmad I, Dooley CM, Polk DL. Delta-1 is a regulator of neurogenesis in the vertebrate retina. Dev Biol. 1997;185:92–103. doi: 10.1006/dbio.1997.8546. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol. 2010;92:1–29. doi: 10.1016/S0070-2153(10)92001-2. [DOI] [PubMed] [Google Scholar]
- Austin CP, Feldman DE, Ida JA, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Baker NE. Notch signaling in the nervous system. Pieces still missing from the puzzle. Bioessays. 2000;22:264–273. doi: 10.1002/(SICI)1521-1878(200003)22:3<264::AID-BIES8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. Journal of Neuroscience. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers J, Clark A, Wunsch K, Hrabe De Angelis M, Gossler A. Expression of the mouse Delta1 gene during organogenesis and fetal development. Mech Dev. 1999;84:165–168. doi: 10.1016/s0925-4773(99)00065-9. [DOI] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, IV, Prasov L, Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Developmental Biology. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang QL, Xu S, Liu I, Li LY, Wang Y, Zack DJ. Functional analysis of cone-rod homeobox (CRX) mutations associated with retinal dystrophy. Hum Mol Genet. 2002;11:873–884. doi: 10.1093/hmg/11.8.873. [DOI] [PubMed] [Google Scholar]
- Doroquez DB, Rebay I. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit Rev Biochem Mol Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Chang WS, Rapaport DH, Harris WA. Regulation of neuronal diversity in the Xenopus retina by Delta signalling. Nature. 1997;385:67–70. doi: 10.1038/385067a0. [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor specific expression and regulates photoreceptor differentiation. Cell. 1997b;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Gregory-Evans CY, Wallace VA, Gregory-Evans K. Gene networks: dissecting pathways in retinal development and disease. Prog Retin Eye Res. 2013;33:40–66. doi: 10.1016/j.preteyeres.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Henrique D, Hirsinger E, Adam J, Le Roux I, Pourquie O, Ish-Horowicz D, Lewis J. Maintenance of neuroepithelial progenitor cells by Delta-Notch signalling in the embryonic chick retina. Curr Biol. 1997;7:661–670. doi: 10.1016/s0960-9822(06)00293-4. [DOI] [PubMed] [Google Scholar]
- Hozumi K, Negishi N, Suzuki D, Abe N, Sotomaru Y, Tamaoki N, Mailhos C, Ish-Horowicz D, Habu S, Owen MJ. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5:638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006;133:913–923. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2008;50(Suppl 1):S97–103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Wroblewski E, Philips GT, Stair CN, Conley K, Reedy M, Mastick GS, Brown NL. Multiple requirements for Hes 1 during early eye formation. Dev Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci U S A. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Jin K, Xie Z, Qiu F, Li S, Zou M, Cai L, Hozumi K, Shima DT, Xiang M. Forkhead box N4 (Foxn4) activates Dll4-Notch signaling to suppress photoreceptor cell fates of early retinal progenitors. Proc Natl Acad Sci U S A. 2012;109:E553–562. doi: 10.1073/pnas.1115767109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Mastick GS, Andrews GL. Pax6 regulates the identity of embryonic diencephalic neurons. Mol Cell Neurosci. 2001;17:190–207. doi: 10.1006/mcne.2000.0924. [DOI] [PubMed] [Google Scholar]
- Maurer KA, Riesenberg AN, Brown NL. Notch signaling differentially regulates Atoh7 and Neurog2 in the distal mouse retina. Development. 2014;141:3243–3254. doi: 10.1242/dev.106245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Gumuscu B, Hartman BH, Reh TA. Notch activity is downregulated just prior to retinal ganglion cell differentiation. Dev Neurosci. 2006;28:128–141. doi: 10.1159/000090759. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Ray CA, Hayashi T, Bermingham-McDonogh O, Reh TA. Acheate-scute like 1 (Ascl1) is required for normal delta-like (Dll) gene expression and notch signaling during retinal development. Dev Dyn. 2009;238:2163–2178. doi: 10.1002/dvdy.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Prasov L, Masud T, Khaliq S, Mehdi SQ, Abid A, Oliver ER, Silva ED, Lewanda A, Brodsky MC, Borchert M, Kelberman D, Sowden JC, Dattani MT, Glaser T. ATOH7 mutations cause autosomal recessive persistent hyperplasia of the primary vitreous. Hum Mol Genet. 2012;21:3681–3694. doi: 10.1093/hmg/dds197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preusse K, Tveriakhina L, Schuster-Gossler K, Gaspar C, Rosa AI, Henrique D, Gossler A, Stauber M. Context-Dependent Functional Divergence of the Notch Ligands DLL1 and DLL4 In Vivo. PLoS Genet. 2015;11:e1005328. doi: 10.1371/journal.pgen.1005328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenberg AN, Liu Z, Kopan R, Brown NL. Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J Neurosci. 2009;29:12865–12877. doi: 10.1523/JNEUROSCI.3382-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci. 2005;46:2897–2904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- Rocha SF, Lopes SS, Gossler A, Henrique D. Dll1 and Dll4 function sequentially in the retina and pV2 domain of the spinal cord to regulate neurogenesis and create cell diversity. Dev Biol. 2009;328:54–65. doi: 10.1016/j.ydbio.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Saul SM, Brzezinski JAt, Altschuler RA, Shore SE, Rudolph DD, Kabara LL, Halsey KE, Hufnagel RB, Zhou J, Dolan DF, Glaser T. Math5 expression and function in the central auditory system. Mol Cell Neurosci. 2008;37:153–169. doi: 10.1016/j.mcn.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Isomura A, Ohtsuka T, Kori H, Miyachi H, Kageyama R. Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 2016;30:102–116. doi: 10.1101/gad.270785.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AO, Ercole CE, McLoon SC. Regulation of ganglion cell production by Notch signaling during retinal development. J Neurobiol. 2003;54:511–524. doi: 10.1002/neu.10156. [DOI] [PubMed] [Google Scholar]
- Treisman JE. Retinal differentiation in Drosophila. Wiley Interdiscip Rev Dev Biol. 2013;2:545–557. doi: 10.1002/wdev.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133:1367–1378. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- Zheng MH, Shi M, Pei Z, Gao F, Han H, Ding YQ. The transcription factor RBP-J is essential for retinal cell differentiation and lamination. Mol Brain. 2009;2:38. doi: 10.1186/1756-6606-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Brown B, Li A, Mears AJ, Swaroop A, Craft CM. GRK1-dependent phosphorylation of S and M opsins and their binding to cone arrestin during cone phototransduction in the mouse retina. J Neurosci. 2003;23:6152–6160. doi: 10.1523/JNEUROSCI.23-14-06152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Craft CM. Modulation of CRX transactivation activity by phosducin isoforms. Mol Cell Biol. 2000;20:5216–5226. doi: 10.1128/mcb.20.14.5216-5226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]