Abstract
Triple-negative breast cancer (TNBC) has the worst prognosis of all breast cancers, and women diagnosed with TNBC currently lack targeted treatment options. To identify novel targets for TNBC, we evaluated phosphatase expression in breast tumors and characterized their contributions to in vitro and in vivo growth of TNBC. Using Affymetrix microarray analysis of 102 breast cancers, we identified 146 phosphatases that were significantly differentially expressed in TNBC compared to estrogen receptor (ER)-positive tumors. Of these, 19 phosphatases were upregulated (0.66-fold; FDR=0.05) in TNBC compared to ER-positive breast cancers. We knocked down 17 overexpressed phosphatases in four triple-negative and four ER-positive breast cancer lines using specific small interfering RNAs and found that depletion of six of these phosphatases significantly reduced growth and anchorage-independent growth of TNBC cells to a greater extent than ER-positive cell lines. Further analysis of the phosphatase PTP4A3 (also known as PRL-3) demonstrated its requirement for G1/S cell cycle progression in all breast cancer cells, but PTP4A3 regulated apoptosis selectively in TNBC cells. In addition, PTP4A3 inhibition reduced the growth of TNBC tumors in vivo. Moreover, in silico analysis revealed the PTP4A3 gene to be amplified in 29% of basal-like breast cancers, and high expression of PTP4A3 could serve as an independent prognostic indicator for worse overall survival. Collectively, these studies define the importance of phosphatase overexpression in TNBC, and lay the foundation for the development of new targeted therapies directed against phosphatases or their respective signaling pathways for TNBC patients.
Keywords: phosphatase, PTP4A3, triple-negative breast cancer, cell growth, apoptosis
INTRODUCTION
Breast cancer is the second leading cause of cancer-related death in women (1). Targeted therapies can significantly reduce recurrence for ER-positive (e.g., selective estrogen receptor modulators, aromatase inhibitors) and Human epidermal growth factor receptor 2 (HER2)-positive (e.g., HER2-specific inhibitors) breast cancers (2–4). However, 15-to-20 percent of breast cancers do not express ER, progesterone receptor (PR) or HER2 (5,6). These triple-negative breast cancers (TNBCs) are highly aggressive, have poor prognoses, lack targeted therapies (7–9), and are currently treated with toxic, non-specific chemotherapy drugs. Consequently, there is an urgent need to identify more effective, less toxic therapeutic strategies.
In addition to ER and HER2 signaling pathways, other growth regulatory pathways have been shown to regulate growth and development of cancers. One of our major goals is to identify signaling pathways and molecules critical for growth of TNBCs. Previously, we identified a set of kinases overexpressed in ER-negative compared to ER-positive breast cancers (10). Further analysis demonstrated that many of these overexpressed kinases are essential for growth of ER-negative breast cancer cell lines (10). Aberrant phosphorylation has been implicated in many diseases, including cancer (11,12), and regulate a number of signaling pathways, including those associated with growth, survival, metabolism, and the cell cycle (13–15).
In this study, we hypothesized that phosphatases are differentially expressed in triple-negative as compared to ER-positive tumors. Using transcriptional profiling data to evaluate phosphatase expression in a breast cancer data set, we identified two sets of phosphatases (those highly expressed or under-expressed) in TNBC as compared to ER-positive tumors. Further analysis of the phosphatases upregulated in triple-negative compared to ER-positive breast cancers, identified several phosphatases critical for anchorage-independent growth in TNBC cells. We selected those phosphatases (PTP4A3, PPAP2B, CDC25B, and TIMM50) that were found critical for the tumorigenicity of TNBC cells for further study. Of these, PTP4A3 (protein tyrosine phosphatase type IVA, a.k.a., PRL-3) was required for in vitro and in vivo TNBC cell growth. PTP4A3 has previously been implicated in the regulation of migration, invasion, and metastasis (16), but its role in regulating cancer cell growth is less understood. PTP4A3 inhibition induces G1 arrest in breast cancer cells, reduces growth and Ki-67 expression in xenograft tumors, and loss of PTP4A3 in breast cancer cells induces apoptosis specifically in triple-negative breast cancer cells. Our results demonstrate that phosphatases that represent promising targets for the treatment and prevention of TNBC, and provide the rationale for further investigations supporting the development of inhibitors of phosphatases or their signaling pathways as viable strategies for women with or at high-risk of TNBC.
MATERIALS AND METHODS
Study population and design
Tissue samples used for this study were obtained from the Lester and Sue Smith Breast Cancer Tumor bank at Baylor College of Medicine. We studied 102 breast tumors from this bank (collected from sites in the US and Europe according to institutionally-approved guidelines) for the analysis of gene expression, DNA copy number, and protein change (GEO accession number GSE76275). We used a subset of these tissue samples, which are listed in Supplementary File – Tissue Sample IDs. All studies associated with this research were conducted with the approval of the Baylor College of Medicine and University of Texas MD Anderson Cancer Center Institutional Review Boards. Study demographics define the majority of the population to be postmenopausal women, 97% Caucasian, and a median subject age of 53 years (Supplementary Table 1). After collecting and freezing tumors in liquid nitrogen, DNA, RNA and protein content was individually isolated. In this cohort, 96% of tumors were invasive ductal carcinomas, 35% presented no lymph node involvement, none were metastasic, and 75% of study participants had tumors ≥2cm3 at the time of diagnosis. The ER and PR status of each tissue sample was determined through immunohistochemistry analysis, and HER2 status was determined by fluorescent in-situ hybridization, identifying 49 ER-positive tumors and 53 triple-negative tumors. Of the ER-positive breast cancers, 4 tumors were also HER2-positive (5%), and 42 PR positive (41%) (Supplementary Table 1).
Selection of genes for further study
49 ER-positive and 53 triple-negative breast tumors were profiled using the Affymetrix U133 Plus 2.0 gene expression array. Data analysis was limited to 332 genes identified as phosphatase or phosphatase-interacting proteins. If a gene was represented by more than one probeset on the U133 Plus2 chip, we used the most variable probeset. The list of genes included in the analysis and gene-identifiers are presented in Supplementary Table 3. Analysis (expression estimation and group comparison) was performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team (http://linus.nci.nih.gov/BRB-ArrayTools.html). dChip software was used for preparing the clustering picture (http://www.hsph.harvard.edu/cli/complab/dchip). Expression was estimated using Robust Multi-array Average (RMA) procedure. Criterion for group comparison was False Discovery Rate (FDR)=0.05. Genes with ≥1.5-fold increased expression in triple-negative compared to ER-positive breast tumors and an FDR<0.05 were selected for further study (Table 1).
Table 1.
Phosphatases differentially expressed ≥1.5-fold in triple-negative compared to ER-positive breast tumors (all FDR≤0.05)
| Type of Expression | Gene | Fold change | FDR | |
|---|---|---|---|---|
| Name | Symbol | |||
| Over-expressed | inositol(myo)-1(or 4)-monophosphatase 2 | IMPA2 | 2.43 | <0.001 |
| protein tyrosine phosphatase-like (proline instead of catalytic arginine), mem A | PTPLA | 2.32 | <0.001 | |
| protein phosphatase 1, regulatory (inhibitor) subunit 14B | PPP1R14B | 2.05 | <0.001 | |
| protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | PTPRZ1 | 2.01 | 0.007 | |
| protein tyrosine phosphatase type IVA, member 3 | PTP4A3 | 1.95 | <0.001 | |
| phosphatidic acid phosphatase type 2B | PPAP2B | 1.93 | <0.001 | |
| protein tyrosine phosphatase, receptor type, F | PTPRF | 1.85 | <0.001 | |
| cyclin-dependent kinase inhibitor 3 | CDKN3 | 1.83 | <0.001 | |
| CDC14 cell division cycle 14 homolog B (S. cerevisiae) | CDC14B | 1.80 | <0.001 | |
| discs, large (Drosophila) homolog-associated protein 5 | DLGAP5 | 1.74 | 0.007 | |
| nudix (nucleoside diphosphate linked moiety X)-type motif 11 | NUDT11 | 1.68 | <0.001 | |
| cell division cycle 25 homolog B (S. pombe) | CDC25B | 1.66 | <0.001 | |
| phosphoserine phosphatase | PSPH | 1.63 | 0.034 | |
| phospholipase D family, member 3 | PLD3 | 1.61 | <0.001 | |
| cell division cycle 25 homolog A (S. pombe) | CDC25A | 1.59 | <0.001 | |
| protein phosphatase 1, regulatory (inhibitor) subunit 11 | PPP1R11 | 1.56 | <0.001 | |
| nudix (nucleoside diphosphate linked moiety X)-type motif 5 | NUDT5 | 1.55 | <0.001 | |
| protein tyrosine phosphatase, non-receptor type 14 | PTPN14 | 1.53 | <0.001 | |
| translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) | TIMM50 | 1.52 | <0.001 | |
| Under-expressed | inositol polyphosphate-4-phosphatase, type II, 105kDa | INPP4B | 0.27 | <0.001 |
| protein phosphatase 1, regulatory (inhibitor) subunit 3C | PPP1R3C | 0.37 | <0.001 | |
| dual specificity phosphatase 4 | DUSP4 | 0.37 | <0.001 | |
| nudix (nucleoside diphosphate linked moiety X)-type motif 12 | NUDT12 | 0.38 | <0.001 | |
| fructose-1,6-bisphosphatase 1 | FBP1 | 0.38 | <0.001 | |
| ectonucleotide pyrophosphatase/phosphodiesterase 1 | ENPP1 | 0.39 | <0.001 | |
| protein phosphatase 2 (formerly 2A), regulatory subunit B, gamma isoform | PPP2R2C | 0.41 | <0.001 | |
| ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative function) | ENPP4 | 0.43 | <0.001 | |
| protein tyrosine phosphatase, receptor type, B | PTPRB | 0.48 | <0.001 | |
| cartilage intermediate layer protein, nucleotide pyrophosphohydrolase | CILP | 0.49 | <0.001 | |
| protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform | PPM1A | 0.51 | <0.001 | |
| phosphatase, orphan 2 | PHOSPHO2 | 0.51 | <0.001 | |
| dual specificity phosphatase 5 | DUSP5 | 0.55 | <0.001 | |
| protein tyrosine phosphatase, receptor type, T | PTPRT | 0.55 | <0.001 | |
| chromosome 12 open reading frame 51 | C12orf51 | 0.58 | <0.001 | |
| protein phosphatase 1H (PP2C domain containing) | PPM1H | 0.58 | 0.001 | |
| dual specificity phosphatase 28 | DUSP28 | 0.59 | <0.001 | |
| protein phosphatase 2, catalytic subunit, alpha isozyme | PPP2CA | 0.59 | <0.001 | |
| protein phosphatase 1D magnesium-dependent, delta isoform | PPM1D | 0.59 | <0.001 | |
| protein tyrosine phosphatase, receptor type, N polypeptide 2 | PTPRN2 | 0.59 | 0.002 | |
| protein tyrosine phosphatase, non-receptor type 13 | PTPN13 | 0.60 | <0.001 | |
| myotubularin related protein 9 | MTMR9 | 0.61 | <0.001 | |
| SAC1 suppressor of actin mutations 1-like (yeast) | SACM1L | 0.60 | <0.001 | |
| protein phospahatase 2, regulatory subunit B, alpha | PPP2R2A | 0.62 | <0.001 | |
| multiple inositol polyphosphate histidine phosphatase, 1 | MINPP1 | 0.62 | <0.001 | |
| FANCD2/FANCI-associated nuclease 1 | FAN1 | 0.63 | <0.001 | |
| protein phosphatase 6, catalytic subunit | PPP6C | 0.63 | <0.001 | |
Cell lines, reagents, and plasmids
All cell lines were purchased from American Type Culture Collection (ATCC; Manassas, VA), and were DNA fingerprinted by the Characterized Cell Line Core Facility at MDA Anderson Cancer Center in 2012 to insure correct identity. MCF-7, MDA-MB-231, MDA-MB-468, and HS578T cell lines were cultured in DMEM (Cellgro by Mediatech, Inc., Manassas, VA), and ZR-75-1, HCC1143, T47D, and BT474 cell lines were cultured in RPMI-1640 medium (Cellgro by Mediatech, Inc., Manassas, VA). Growth media for all cell lines was supplemented with 10% FBS, penicillin (100mg/ml), and streptomycin (100mg/ml). The siRNA oligos were purchased from Sigma-Aldrich (St. Louis, MO). A pool of three siRNA oligos was used at a final concentration of 30nM using DharmaFECT1 Transfection Reagent (Dharmacon, Inc., Lafayette, CO), following manufacturer’s instructions. pGIPZ lentiviral shRNAs for PTP4A3 (Open Biosystems, Inc., Huntsville, AL) were sub-cloned into a pTRIPZ lentiviral expression system (Open Biosystems, Inc., Huntsville, AL). The PTP4A3 overexpression vector and control vector (pCMV6) were commercially obtained and sequence-verified.
RNA preparation and quantitative RT-PCR (qRT-PCR)
RNA was isolated using the RNeasy Kit (QIAGEN, Valencia, CA). cDNA was generated using random primers, and Superscript II Reverse Transcriptase (Invitrogen, Life Technologies, Grand Island, NY). TaqMan assays were designed and qRT-PCR was performed. Cyclophilin was used as an endogenous control, and results were normalized to cyclophilin ± standard deviation (SD).
Generation of stable cell lines
Lentiviral particles were generated by transfecting, 293T cells with lentiviral constructs and packaging plasmids encoding VSV-G, Gag, Pol and Tat using FuGENE 6 Transfection Reagent (Roche Applied Science, Indianapolis, IN). Media overlaying the cells was harvested at 24, 48 and 72hrs post-transfection, and filtered through a 0.45µm MEC filter. Polyethylene glycol (PEG) precipitation was used to concentrate the virus. Stable cell lines expressing inducible shRNAs were generated by lentiviral infection using the pTRIPZ lentiviral expression system in the presence of 4ug/ml polybrene, followed by puromycin selection. All pTRIPZ stable cell lines were maintained in media with Tet-safe Serum (Clontech Laboratories, Inc., Mountain View, CA).
MDA MB231 and MCF-7 cell lines stably transfected with control vector or PTP4A3 were generated using FuGENE 6 Transfection Reagent. After 48hrs, transfected cell were selected by the addition of G418 (1mg/ml). Colonies visible by eye were expanded and tested for expression of PTP4A3.
Growth assays
Cells were transfected with 30nM final concentration of siRNA. After 24hrs cells were plated in triplicate in 48-well plates. Cell proliferation was measured using the Countess Automated Cell Counter (Invitrogen, Life Technologies, Grand Island, NY) and trypan blue staining. Cell count was assessed at days 4 and 6. Each data point was performed in triplicate, with results reported as average percentage ± SD.
For the growth assays with PTP4A3 overexpression, cells were plated in triplicate in 48-well plates. Two clones stably transfected with control vector and 4 clones stably transfected with CMV-PTP4A3 were used for the growth assay. Cells count was assessed at days 1, 2 and 4. Each data point was performed in triplicate, with results reported as average percentage ± SD.
Anchorage-independent growth assays
Cells were transfected with 30nM final concentration of siRNA. Anchorage-independent growth assays were performed by plating cells 24hrs after siRNA transfection in 0.35% SeaPlaque GTG Agar (FMC Corp., Philadelphia, PA) in the appropriate medium. Depending on the cell line, either 104 or 2×104 cells were plated per well. Suspended cells were plated on a 0.7% agar base in the same medium. Colonies were counted using the GelCount Colony Counter (Oxford Optronix, Oxford, UK) 2 and 3 weeks post-plating. All experiments were performed in triplicate, and results reported as average colony number ± SD.
Apoptosis assays
Apoptosis assays were performed using Annexin V staining (eBioscience, San Diego, CA) following manufacturer’s instructions, and replacing propidium iodide (PI) with DAPI staining. Cells were plated on 6-well plates, transfected with siRNA, and harvested after 3 days, then stained and analyzed in triplicate using a Gallios Flow Cytometer (Beckman Coulter, Inc., Brea, CA). Results were reported as average percentage of each population ± SD.
Cell cycle analysis
Cells were plated on 6-well plates, transfected with siRNA, and harvested after 4 days, at which time they were fixed, stained with PI, and analyzed in triplicate using a Gallios Flow Cytometer (Beckman Coulter, Inc., Brea, CA). Results are reported as the average percentage of each population ± SD.
Immunofluorescence staining and microscopy
Cells were plated on coverslips in 6-well plates. The next day cells were transfected with 30nM final concentration of siRNA. After 48hrs cells were washed in PBS, fixed for 20 minutes in 4% PFA at room temperature, and blocked for 1 hour at room temperature in 5% goat serum and 1% Triton X-100 in 1×PBS. Coverslips were incubated with primary antibody (phospho-Histone H3 (1:500) from Cell Signaling, and Ki-67 (Clone SP6) from Thermo Scientific) overnight at 4°C in 1% goat serum and 1% Triton X-100 in 1×PBS, incubated with secondary antibody (Alexa-488 goat anti-rabbit; 1:200) for 1 hour at room temperature, and then mounted with DAPI-containing mounting media (Vectashield). Fluorescent images were captured with a Nikon microscope. Experiments were performed in triplicate.
Immunohistochemistry and staining analysis
Tumor sections of 5um were stained for either Ki-67 (Clone SP6 from Thermo Scientific) or with Cleaved Caspase 3 (Cell signaling). Antigen retrieval was performed in citrate buffer, and endogenous peroxidase was blocked with 3% hydrogen peroxide for 10 min. Primary antibodies were incubated overnight at 4C, followed by incubation with biotinylated antirabbit antibody (1:100) for 30 minutes. Peroxidase activity was visualized using the Vector NovaRED Substrate Kit (PK-6101, Vector Laboratories, Inc.).
Western blotting
Cells were transfected with 30nM final concentration of siRNA. After 48hrs cells were washed in PBS, lysed in RIPA buffer, run on a 4–20% gradient gel, and transferred onto nitrocellulose. Primary antibodies were incubated with the membrane overnight at 4°C and included: p-ERK1/2, ERK, Cleaved caspase 7, Caspase 7, p-p38, and p38 (Cell Signaling; 1:1000), and Vinculin and Actin (Sigma; 1:5000). Blots were washed with PBS-T and incubated with secondary antibodies for 1 hour at room temperature. Bends were detected using ECL method.
Xenograft experiments
Nude mice were obtained from Charles River Laboratories (Wilmington, MA) and experiments were performed in accordance with MD Anderson Institutional Animal Care and Use Committee (IACUC)-approved protocols. For MDA MB231 xenograft studies, 1.5×106 pTRIPZ control or pTRIPZ shPTP4A3 cells were injected into mouse mammary fat pads. For MDA MB468 xenograft studies, 5×106 pTRIPZ control or pTRIPZ shPTP4A3 cells were mixed with 50% Matrigel and injected into mouse mammary fat pads. For MCF-7 nude mouse xenograft studies estrogen pellets were given 2 days before injecting 5×106 pTRIPZ control or pTRIPZ shPTP4A3 cells mixed with 50% matrigel into mouse mammary fat pads. When tumors reached >30mm3 in size, mice were randomized to doxycycline-treatment or control groups and monitored for tumor growth. Tumors were measured with digital calipers every other day for MDA MB231, and every third day for MDA MB468 and MCF-7. Volumes (mm3) were calculated as volume=(width2×length)/2. Statistical differences were calculated by comparison of growth slopes from log transformed tumor volumes plotted versus time.
Assessment of PTP4A3 amplification
PTP4A3 and Myc gene amplification status was obtained from the cBioPortal database (17,18). Breast cancer samples were assessed for PTP4A3 gene amplification, and represented in percentage of total tumor samples. Co-amplification of PTP4A3 and Myc in TNBC samples was determined using three independent data sets: TCGA, Curtis, and Nikolsky. The amplification status of PTP4A3, Myc, and four genes in between the PTP4A3 and Myc loci (ADCY8, KHDRBS3, FAM135B, and PTK2) was used to determine the independent amplification of PTP4A3.
Survival Analysis
The prognostic importance of PTP4A3 expression was evaluated using gene expression profiles and survival data generated by Curtis et al. and Kao et al. (5,19). R statistical software was used to analyze data obtained from the Oncomine database to generate Kaplan-Meier survival curves, to determine statistical significance using the log rank (Mantel-Cox) method, and to perform Cox proportional hazards models analyses. The patients were dichotomized at the mean expression level.
RESULTS
Affymetrics gene expression profiling identifies phosphatases under- and over-expressed in human triple-negative breast tumors
We analyzed the expression levels of phosphatases in 102 breast tumors, 53 triple-negative and 49 ER-positive, to identify genes differentially expressed in TNBCs. Data analysis was limited to 332 genes identified as phosphatase or phosphatase-interacting proteins. We identified 146 phosphatase genes differently expressed (82 overexpressed, 64 underexpressed) in triple-negative compared to ER-positive breast tumors (FDR=5%; Supplementary Figure 1). Further analysis showed 19 of the 82 overexpressed phosphatases had ≥1.5-fold increases in expression, and 27 of the 64 underexpressed phosphatases had ≥1.5-fold reductions in expression, and (FDR<0.05) (Table 1). We selected the 19 phosphatases differentially overexpressed by ≥1.5-fold for further study.
Depletion of phosphatases inhibits TNBC growth
We transfected 4 triple-negative (MDA MB468, MDA MB231, HS578T, HCC1143) and 4 ER-positive (MCF-7, T47D, ZR75-1, BT474) breast cancer cell lines with a pool of 3 siRNA oligos targeting the 17 phosphatases expressed ≥1.5-fold higher in triple-negative than in ER-positive breast tumors to determine if they are critical for breast cancer cell growth. Growth rate comparisons of the siRNA oligos- and control-transfected cells identified 12 phosphatases that effected ≥2 breast cancer cell lines. Knockdown of 6 phosphatases (PTP4A3, PPAP2B, DLGAP5, CDC25B, PSPH, TIMM50) showed a growth inhibitory effect in ≥2 of the 4 TNBC cell lines tested (Table 2A), and PTP4A3 showed a growth inhibitory effect in ≥3 of the 4 TNBC cell lines tested. In addition, growth inhibitory effects following phosphatase knockdown were more profound in the triple-negative compared to ER-positive cell lines.
Table 2.
| a. Growth of triple-negative and ER-positive cell lines treated with individual phosphatases or control (siLuc) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Triple-negative cell lines (% of control) |
ER-positive cell lines (% of control) |
||||||
| MDA MB468 | HS578T | HCC1143 | MDA MB231 | MCF-7 | T47D | BT474 | ZR-75-1 | |
| IMPA2 | 75 * | 74* | 78* | 84* | 160* | 42* | 71* | 92 |
| PTPLA | 92 | 139* | 141 | 88 | 101 | 43* | 107 | 89 |
| PPP1R14B | 70* | 73* | 95 | 62* | 80 | 78* | 126* | 169* |
| PTP4A3 | 43* | 55* | 48* | 47* | 107 | 73* | 83 | 121* |
| PTPRZ1 | 81* | 108 | 156* | 118* | 130* | 46* | 76 | 76* |
| PPAP2B | 70* | 22* | 144* | 27* | 49* | 29* | 124 | 174* |
| CDKN3 | 76* | 88 | 90 | 67* | 84* | 72* | 90 | 95 |
| DLGAP5 | 52* | 36* | 170* | 14* | 89 | 61* | 86 | 40* |
| PTPRF | 72* | 78 | 95 | 100 | 107 | 77 | 40* | 61* |
| CDC14B | 90 | 112 | 161* | 198* | 130* | 70 | 63* | 160* |
| CDC25B | 48* | 34* | 34* | 58* | 47* | 34* | 50* | 121 |
| PLD3 | 67* | 88 | 141 | 58* | 82* | 51* | 56* | 66* |
| NUDT11 | 55* | 115 | 80 | 68* | 117* | 69* | 92 | 73* |
| CDC25A | 88 | 149* | 258* | 117* | 142* | 25* | 70 | 94 |
| PPP1R11 | 85* | 90 | 58* | 111 | 51* | 51* | 94 | 94 |
| PSPH | 36* | 7* | 37* | 5* | 13* | 59* | 61* | 33* |
| TIMM50 | 45* | 34* | 86* | 52* | 32* | 80 | 70* | 62* |
| b. Anchorage-independent growth of triple-negative and ER-positive cell lines treated with individual phosphatases or control (siLuc) | |||||||
|---|---|---|---|---|---|---|---|
| Gene | Triple-negative cell lines (% of control) |
ER-positive cell lines (% of control) |
|||||
| MDA MB468 | SUM159 | HCC1143 | MDA MB231 | MCF-7 | T47D | BT474 | |
| PTP4A3 | 25* | 67* | 39* | 40* | 110 | 55* | 68* |
| PPAP2B | 55* | 86 | 123* | 15* | 106 | 39* | 94 |
| DLGAP5 | 51* | 136* | 70* | 238* | 118* | 63* | 94 |
| CDC25B | 35* | 98 | 55* | 70 | 136* | 61* | 121 |
| PSPH | 58* | 62* | 36* | 11* | 64* | 53* | 91 |
| TIMM50 | 44* | 82 | 82* | 48* | 156* | 71 | 95 |
= p<0.05
Depletion of phosphatases reduces TNBC anchorage-independent growth
To determine whether the 6 phosphatases whose inhibition reduced cell growth were also important for the anchorage-independent growth of TNBC cells, we treated 4 triple-negative (MDA MB468, SUM159, HCC1143, MDA MB231), and 3 ER-positive (MCF-7, T47D, BT474) breast cancer cell lines with siRNA targeting these phosphatases. 5 phosphatases (PTP4A3, PPAP2B, CDC25B, PSPH, TIMM50) showed an inhibitory effect on anchorage-independent growth in ≥2 of the TNBC cell lines (Table 2B). PTP4A3 depletion had a larger inhibitory effect on triple-negative as compared to ER-positive breast cancer cell lines, and PTP4A3 depletion inhibited the growth of TNBC cell lines by ≥one third as compared to control. PPAP2B depletion showed a growth inhibitory effect in MDA MB468 and MDA MB231 TNBC cells, and in T47D ER-positive cells. Depletion of DLGAP5 had minimal effect on the growth of triple-negative and ER-positive breast cancer cell lines and thus, was not studied further. Depletion of CDC25B and TIMM50 inhibited anchorage-independent growth in two TNBC cell lines, with no or minimal effect on ER-positive breast cancer cell lines. PSPH knockdown affected both triple-negative and ER-positive cell lines equally, and showed no selectivity for TNBC cell lines (Table 2B).
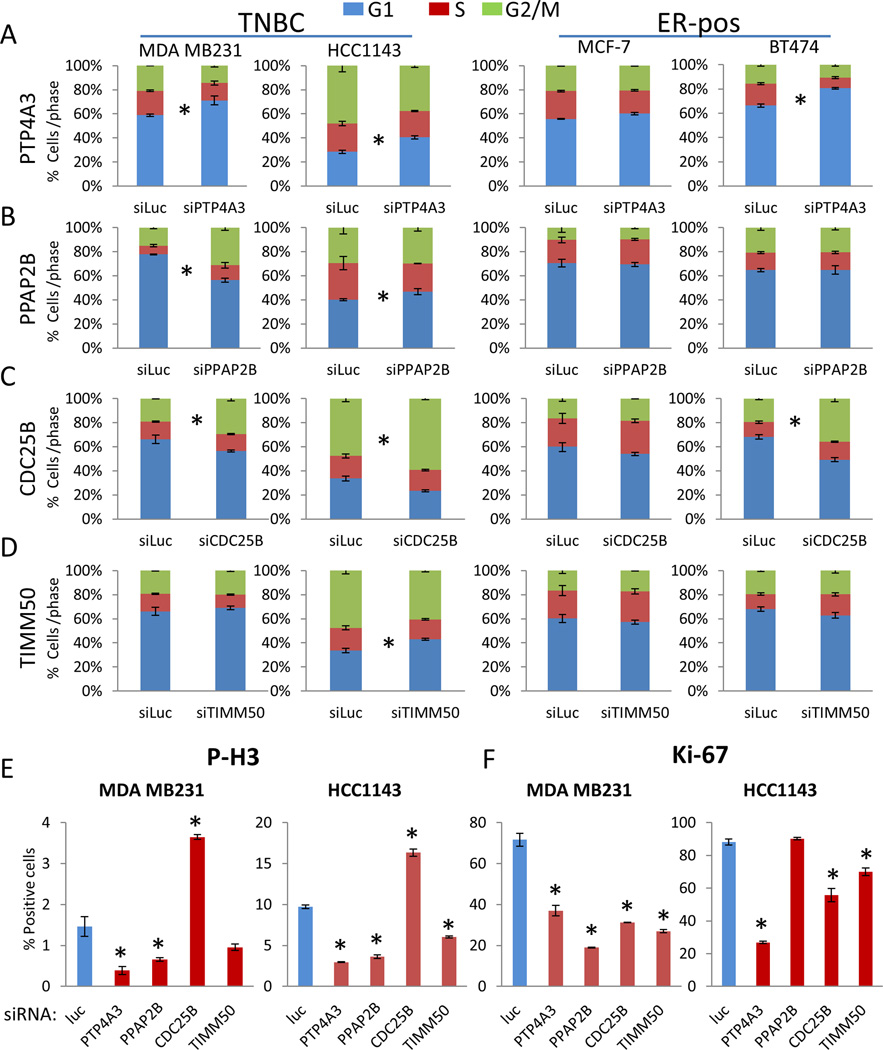
PTP4A3 depletion induces G1 arrest
To investigate the effect of depletion of the phosphatases (PTP4A3, PPAP2B, CDC25B, TIMM50) that showed an effect on breast cancer anchorage-independent cell growth selectively in TNBC, two triple-negative (MDA MB231, HCC1143) and two ER-positive breast cancer cell lines (MCF-7, BT474) were treated with siRNA, and assayed for cell cycle distribution. Both triple-negative and ER-positive BT474 breast cancer cells (and to a lesser extent ER-positive MCF-7 cells) transfected with PTP4A3 siRNA showed an increase of cells in G1 phase accompanied by a significant reduction of cells in S and G2/M (Figure 1A). PPAP2B depletion caused an increase in the proportion of cells in the G2/M phase in MDA MB231 cells (Figure 1B). Not surprisingly, siRNA knockdown of CDC25B increased the proportion of cells in the G2/M phase; CDC25B is a critical phosphatase for the progression of cells through mitosis (Figure 1C). Knockdown of TIMM50 showed little to no effect on the cell cycle (Figure 1D). We then performed immunofluorescent staining after siRNA transfection of PTP4A3, PPAP2B, CDC25B, and TIMM50. Knockdown of each phosphatase except CDC25B suppressed p-Histone H3 in MDA MB231 and HCC1143 cells (Figure 1E). Knockdown of each of the phosphatases reduced Ki67 in MDA MB231 and knockdown of all but PPAP2B reduced Ki67 in HCC2243 cells (Figure 1F). This indicates that depletion of PTP4A3 has a much greater impact on TNBC cell proliferation than depletion of the other phosphatases tested.
Figure 1. PTP4A3 inhibition induces G1 arrest in triple-negative breast cancer cell lines.
Breast cancer cell lines, MDA MB231, HCC1143, MCF-7 and BT474, were transfected with siRNA targeting 5 phosphatases or control siRNA targeting Luciferase. A–E) After 72hrs cells were fixed in ethanol, and stained with PI (* indicates p<0.05). The 5 phosphatases were: A) PTP4A3 B) PPAP2B C) CDC25B D) TIMM50 E) MDA MB231 and HCC1143 cells transfected with siRNA targeting phosphatases, were fixed 72hrs after siRNA transfection and stained for phospho-Histone H3 using immunofluorescence techniques. Cells were counted in 10 fields, and the experiment was performed in triplicate. F) Cells were fixed 72hrs after siRNA transfection and stained for Ki-67 using immunofluorescence techniques. Cells were counted in 10 fields, and the experiment was performed in triplicate. Error bars show standard deviation of the mean.
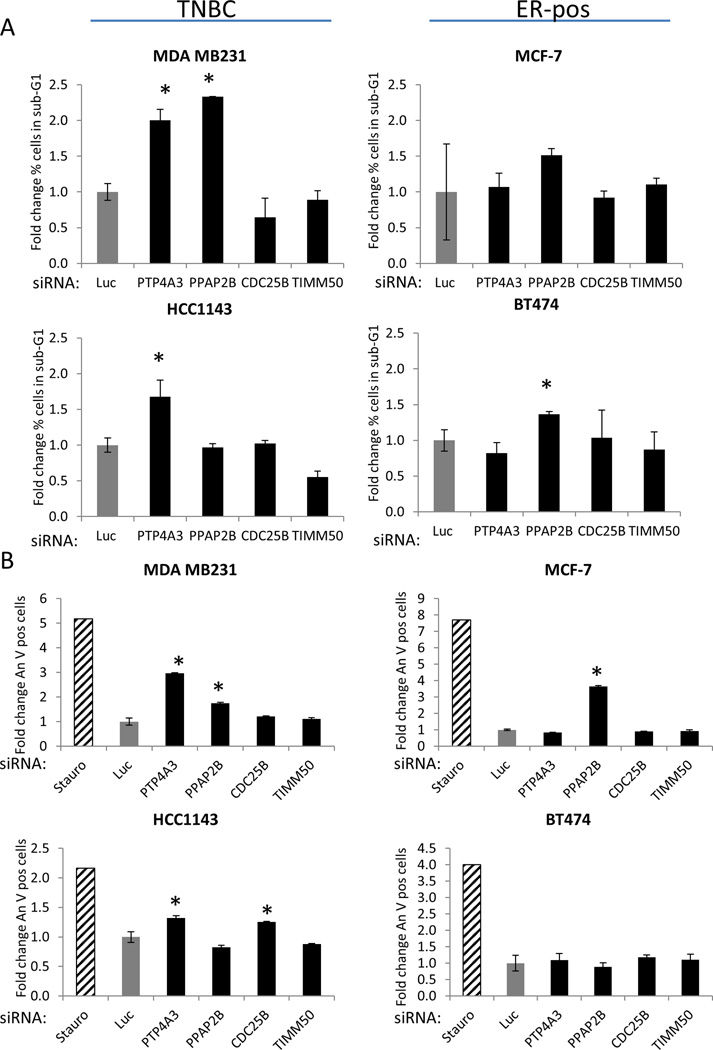
PTP4A3 depletion induces apoptosis in TNBC cell lines
We then studied cell death using the flow cytometry data and found that TNBC cell lines treated with PTP4A3 siRNA had an increase in the proportion of cells in the sub-G1 fraction, suggesting that PTP4A3 and PPAP2B knockdown induces apoptosis in MDA MB231 cells (Figure 2A). Knockdown of PTP4A3 also increased the sub-G1 proportion in HCC1143 cells. To further investigate whether the depletion of PTP4A3, PPAP2B, CDC25B, and TIMM50 induce apoptosis in TNBCs, we performed Annexin V assays of breast cancer cells transfected with phosphatase siRNAs. Knockdown of PTP4A3 increased Annexin V staining in TNBC cell lines, but not in the ER-positive cell lines (Figure 2B). Conversely, PPAP2 depletion increased Annexin V staining in MDA MB231 cells and in the ER-positive MCF-7 cells. These data suggest loss of PTP4A3 induces apoptosis selectively in TNBC cell lines. Based on the effect of PTP4A3 knockdown on cell growth (Table 2), cell cycle, and apoptosis, we chose to further investigate the role of PTP4A3 in TNBC.
Figure 2. PTP4A3 inhibition induces apoptosis selectively in triple-negative breast cancer cell lines.
Breast cancer cell lines, MDA MB231, HCC1143, MCF-7 and BT474, were transfected with siRNA targeting 5 phosphatases or control siRNA targeting Luciferase. (* indicates p<0.05). A) Cells were fixed 72hrs after siRNA transfected in ethanol, and stained with PI. The sub-G1 population is graphed here. Experiment was performed in triplicate, and average with SDEV was graphed. B) Cells were stained for Annexin V 72hrs after transfection. Experiment was performed in triplicate, and average with SDEV was graphed.
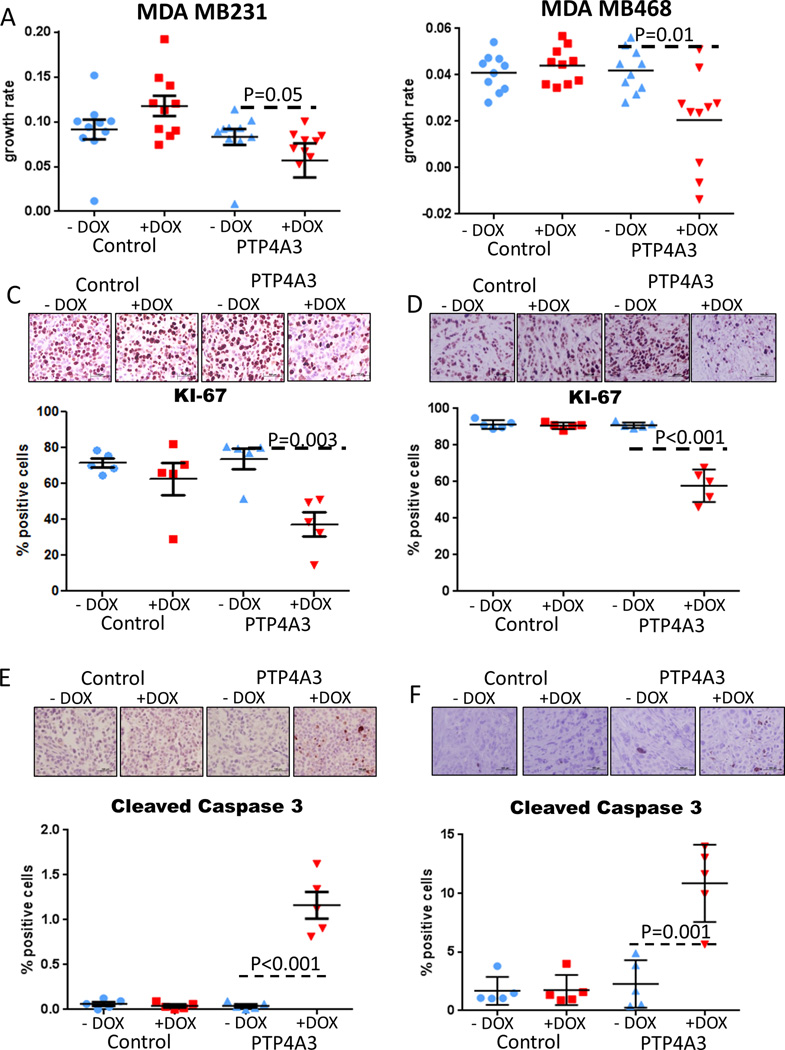
Depletion of PTP4A3 reduces TNBC tumor growth in vivo
We next investigated the effect of PTP4A3 knockdown on in vivo tumor growth. MDA MB231 cells stably expressing doxycycline-inducible PTP4A3 shRNA constructs or empty vector control constructs were injected into the mammary fat pads of nude mice. Once tumors were established, mice were randomized into two groups and treated with doxycycline-containing or control sucrose water. Tumor growth was significantly inhibited in tumors with PTP4A3 knockdown compared to control (Figure 3A and Supplementary Figure 3A–B). qRT-PCR confirmed that reduced PTP4A3 expression occurs in tumors treated with doxycycline (Supplementary Figure 3C). To conclusively determine that PTP4A3 depletion inhibits xenograft growth of TNBC cell lines, we conducted a similar experiment with MDA MB468 cells, and found that tumor growth was indeed significantly inhibited in tumors with PTP4A3 knockdown compared to controls (Figure 3B and Supplementary Figure 3D). As PTP4A3 knockdown affected the cell cycle and apoptosis in vitro and reduced tumor growth in vivo, we studied the effects of PTP4A3 inhibition in vivo by IHC analysis of Ki-67 and cleaved caspase 3 levels of xenograft tumors. PTP4A3 knockdown significantly reduced the number of Ki67-positive and increased the percentage of cleaved caspase 3-positive TNBC cells (Figure 3C–F). These results confirm our in vitro findings, and show that PTP4A3 inhibition reduces proliferation and induces apoptosis. We also performed an additional xenograft experiment with MCF-7, and found no significant inhibition of tumor growth following depletion of PTP4A3 (Supplementary Figure 3E–F). These results demonstrate that PTP4A3 is necessary for in vivo TNBC tumor growth.
Figure 3. Inhibition of PTP4A3 reduces tumor growth in triple-negative breast cancer mouse models.
A–B)) PTP4A3 tumors treated with doxycycline affects tumor growth due to the knockdown of PTP4A3. MDA MB231 cells (A) and MDA MB468 (B) stably transfected with shRNA-PTP4A3 were injected in the mammary fat pad of nude mice. Mice were randomized when tumor reached 30mm3. Tumors volumes were measured every other day. Tumor growth rate is significantly reduced after knockdown of PTP4A3 in MDA MB231. Tumor growth rates were calculated from the slopes of the growth curves for each tumor. Error bar shows standard deviation. C–D) KI-67 staining by IHC of xenograft tumors in control and PTP4A3 knockdown mice treated with doxycycline or vehicle. E–F) Cleaved caspase 3 staining by IHC of xenograft tumors in control and PTP4A3 knockdown mice treated with doxycycline or vehicle.
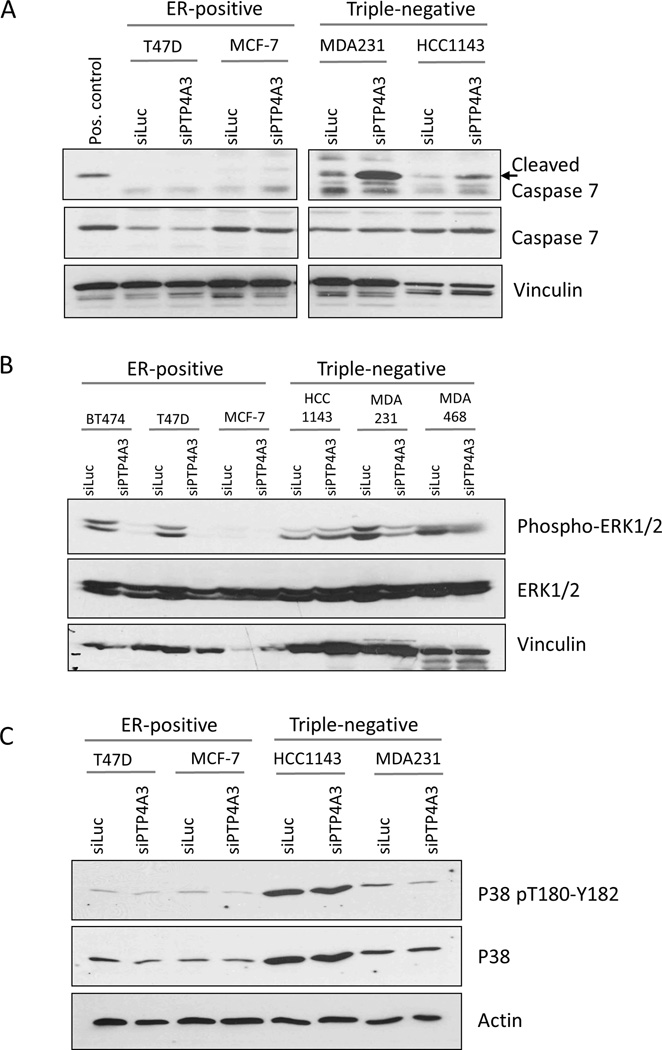
PTP4A3 depletion alters critical proliferation and apoptosis signaling pathways
To further explore the mechanism of growth suppression by PTP4A3 knockdown, we analyzed the Reverse Phase Protein Array (RPPA) data from TCGA breast cancer samples. Breast tumors with high (upper tertile) and low (lower tertile) PTP4A3 mRNA expression were selected for further analysis. We compared protein phosphorylation and total protein levels between the two groups, and identified those that significantly differ (p<0.1) between the high and low PTP4A3-expressing groups. As expected given our in vitro and in vivo findings, cleaved caspase-7 was elevated in tumors with low PTP4A3 expression selectively in basal-like (primarily triple-negative) but not luminal (primarily ER-positive) breast tumors (Supplementary Figure 2A and B). Western blot analysis confirmed increased cleaved caspase-7 expression following PTP4A3 knockdown in triple-negative but not ER-positive breast cancer cell lines (Figure 4A). These results show that loss of PTP4A3 suppresses TNBC cell growth by inducing apoptosis.
Figure 4. Signaling proteins involved in proliferation and programmed cell death are altered with PTP4A3 knockdown.
Two TNBC cell lines (MDA MB231 and HCC1143), and two ER-positive cell lines (MCF-7 and T47D) were transfected with PTP4A3 or control siRNA. After 72hrs cell were harvested and lysed for Western Blot analysis. A) Cleaved caspase 7 is increased after knockdown of PTP4A3 in TNBC cell lines only. B) ERK1/2 phosphorylation and protein levels are reduced after knockdown of PTP4A3. C) Phosphorylation of p38 is reduced after knockdown of PTP4A3.
We next investigated the effect of PTP4A3 loss on MAPK signaling. We preformed Western blot analyses to test whether PTP4A3 depletion affects the phosphorylation status of ERK and p38. ERK phosphorylation and total protein levels of ERK were reduced after PTP4A3 depletion in both ER-positive and ER-negative breast cancer cell lines (Figure 4B). Conversely, p38 total protein was not affected by loss of PTP4A3; however, p38 phosphorylation was suppressed in MDA MB231, the TNBC cell line with the largest phenotypic effect (Figure 4C). These changes in ERK and p38 expression and activity may contribute to the selective induction of apoptosis in triple-negative compared to ER-positive breast cancer cell lines.
PTP4A3 is amplified in triple-negative breast cancer and associated with poor overall survival
As loss of PTP4A3 suppresses growth and induces apoptosis in TNBC, we analyzed PTP4A3 expression levels in publically available human breast cancer data sets (5,19). PTP4A3 expression is significantly elevated in breast cancer as compared to normal cells, and is increased in triple-negative as compared to ER-positive breast cancers (Supplementary Figure 4A–D).
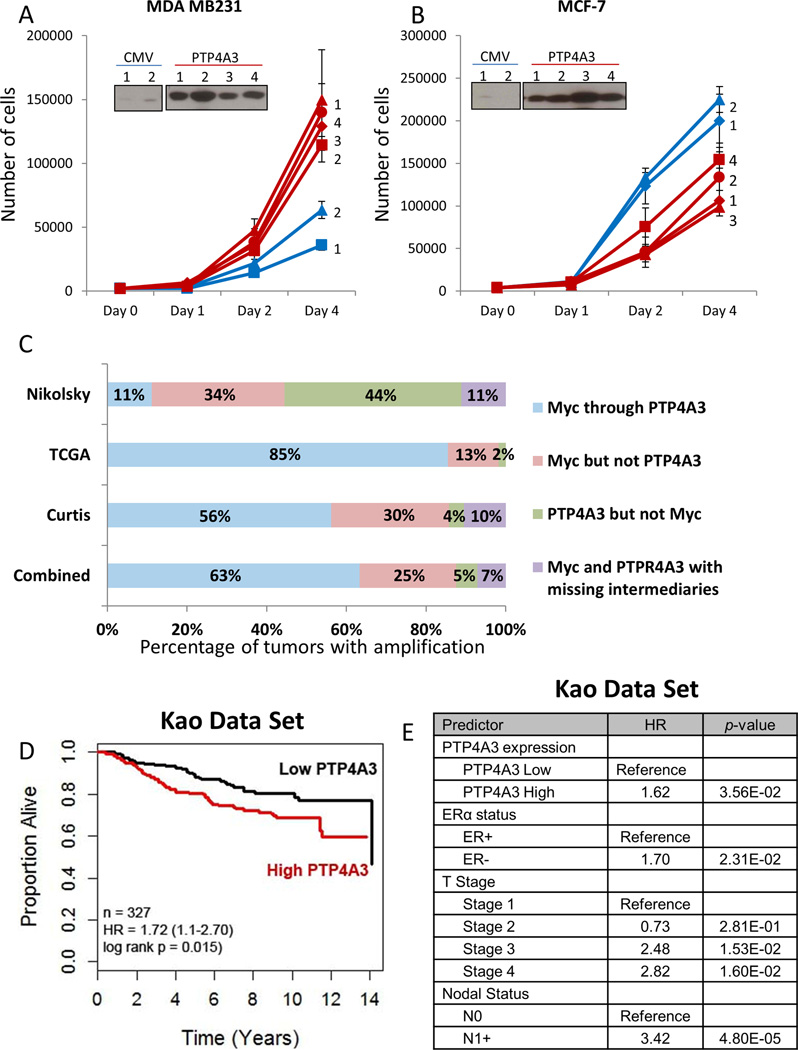
To determine if increased PTP4A3 expression increases cancer cell growth, we created stably transfected cell lines (MDA MB231, MCF-7) overexpressing PTP4A3. MDA MB231 overexpressing PTP4A3 showed increased growth rates, while MCF-7 clones overexpressing PTP4A3 exhibited reduced growth compared to vector transfected clones (Figure 5A–B). Given the high expression of PTP4A3 in TNBC, we investigated the amplification status of PTP4A3 in the TCGA dataset using cBioPortal (17,18). PTP4A3 is located on 8q.24.3, a region frequently amplified in breast cancer which also includes Myc (20). We discovered that 8q.24.3 is amplified in 21% of all breast cancers and 45% of basal-like breast cancers. In addition, PTP4A3 is amplified in 17% and Myc in 22% of all breast cancers, while PTP4A3 is amplified in 29% and Myc in 44% (often concomitantly with PTP4A3) of basal-like breast cancers. The analysis of the TCGA data set also revealed PTP4A3 amplification without the presence of Myc amplification in 2% of basal-like tumors. This is in contrast to Luminal A/B (primarily ER-positive) breast tumors, in which PTP4A3 is amplified in only 12% (Supplementary Figure 5A). Similar analysis of the TCGA ovarian cancer data set gave similar results: PTP4A3 amplification in 34%, Myc amplification in 41%, and PTP4A3 amplification without Myc amplification in 7% of serous ovarian cancers (Supplementary Figure 5B).
Figure 5. High levels of PTP4A3 promote TNBC cell growth, and associated with poor overall survival in breast cancer patients.
A) Overexpression of PTP4A3 caused an increased growth in the TNBC cell line MDA MB231. Equal amount of cells of MDA MB231 cells stably transfected with control or PTP4A3 were plated and assayed for growth for 4 days using trypan blue staining. B) Overexpression of PTP4A3 reduced growth of the ER-positive breast cancer cell lines MCF-7. Equal amount of cells of MCF-7 cells stably transfected with control or PTP4A3 were plated and assayed for growth for 4 days by trypan blue staining. C) Comparative analysis of gene amplification of PTP4A3, Myc and intermediate genes. The implication status of PTP4A3, Myc and genes located in between the PTP4A3 and Myc loci was assessed, and graphed in percentage of concurrent of independent amplification D). Kaplan-Meier curves of overall survival of all breast cancer patients in the Kao data set stratified by PTP4A3 expression E) Proportional hazard ratio analysis demonstrates that high PTP4A3 levels are an independent predictor for worse over-all survival in breast cancer patients in the Kao data set.
Due to the large size of the 8q24 genomic region, we hypothesized that PTP4A3 and Myc genes might be represented by two different amplicons. To test this hypothesis, we investigated the amplification status of PTP4A3, Myc and 4 different genes located between PTP4A3 and Myc in the 8q24 region in the Curtis, TCGA and Nikolsky data sets (Supplementary Figure 5C). This analysis revealed that amplification of PTP4A3 but not Myc occurs in 3% and 44% of the Curtis and Nikolsky data set tumors, respectively (Figure 5C). We also found that PTP4A3 and Myc co-amplification occurs without amplification of the region between PTP4A3 and Myc in 10% and 11% in the Curtis and Nikolsky data sets, respectively (Figure 5C, purple bars). These data suggest that PTP4A3 amplification is not a passenger of Myc amplification, and can be independently involved in breast cancer pathogenesis. Correlation of PTP4A3 copy number and expression levels demonstrated that mRNA expression increases with elevated copy numbers (Supplementary Figure 5D).
Next, we investigated the importance of PTP4A3 overexpression on survival of breast cancer patients (Kao data set) (19). Kaplan-Meier analysis of patients dichotomized on median PTP4A3 expression demonstrates poorer overall survival for patients with high versus low PTP4A3-expressing tumors (Figure 5D). Cox Proportional Hazard analysis of PTP4A3 above and below median, age, grade, lymph node status, and ER status, indicate PTP4A3 is an independent predictor of decreased overall survival in breast cancer (Kao data set, Figure 5E; Curtis data set, Supplementary Table 2). These results demonstrate that PTP4A3 is an independent predictor of poor overall survival, and suggests that PTP4A3, a critical growth regulatory phosphatase, is a novel potential target for the treatment of breast cancer, particularly TNBC.
DISCUSSION
In this study we identified phosphatases differentially over- or under-expressed in triple-negative breast cancer as compared to ER-positive breast cancer, and showed that several highly expressed phosphatases are essential for growth of TNBCs. This is the first report using an unbiased global approach demonstrating the growth-promoting effect of phosphatases overexpressed in TNBC. We identified 4 phosphatases (PTP4A3, PPAP2B, CDC25B, and TIMM50) that are critical for TNBC anchorage-independent growth. We further demonstrated that PTP4A3 is required for proliferation of TNBC cell lines, and that PTP4A3 knockdown induces a G1 cell cycle block in both TNBC and ER-positive breast cancer cell lines, and apoptosis specifically in TNBC cell lines.
We also demonstrated that PTP4A3 knockdown reduces phosphorylation of p38 and ERK1/2 in all breast cancer cell lines, consistent with the observed reduced proliferation and growth arrest. p38 inhibition has been shown to induce sensitization to apoptosis and increase sensitivity to cisplatin (21). These results suggest that loss of PTP4A3 causes apoptosis through p38 inactivation. This also suggests that high levels of PTP4A3 might cause resistance to cisplatin or other chemotherapy drugs in TNBCs. Other phosphatases have been shown to induce resistance to cancer treatments, such as mitogen-activated protein kinase phosphatase 3 (MKP3). Increased levels of MKP3 have been associated with tamoxifen resistance in ER-positive breast cancers (22). These findings demonstrate that phosphatases are critical signal transduction regulators in cancer.
PTP4A3 overexpression in TNBC has been confirmed in multiple data sets, and is associated with reduced survival. Using the TCGA data set and cBioPortal for cancer Genomics, we identified that PTP4A3 gene amplification is enriched in basal-like versus luminal breast cancers, and in some cases is independent of Myc amplification. These results indicate selective importance of PTP4A3 in TNBC. Previous studies have reported PTP4A3 overexpression in several cancers, including breast, gastric and colon cancer (23,24), particularly with metastatic colon and breast cancer. Furthermore, PTP4A3 expression is higher in invasive cancer than normal breast tissue, and protein expression is associated with lymph node positivity (25), as well as reduced disease-free survival (23).
PTP4A3 has a well-established role in cell motility and metastasis. Ectopic PTP4A3 overexpression influences migration and invasion of colon cancer cells, while PTP4A3 elimination decreases cell motility and metastatic capability in mice (26). PTP4A3 overexpression is primarily observed in metastases (distant or lymph node) versus primary tumors (27). The overexpression of PTP4A3 in triple-negative compared to ER-positive breast cancer supports the concept that high PTP4A3-expressing tumors are more aggressive. We have demonstrated that loss of PTP4A3 reduces cell proliferation and induces apoptosis, and are currently studying PTP4A3 overexpression in cancerous and precancerous cells to test whether PTP4A3 transforms premalignant cells into fully-invasive cancer cells.
While PTP4A3 regulation of molecular pathways is not well-studied and has focused primarily on migration and invasion, Ezrin and Integrin-beta 1 are direct targets of PTP4A3 in adhesion and motility (28,29). Recently, PTP4A3 has been shown to participate in src-mediated Rho signaling, thereby inducing cell motility and invasion (30). Similarly, PTP4A3 regulation of cancer cell growth is poorly understood. Our present studies show that loss of PTP4A3 alters ERK and p38 signaling. In addition, PTP4A3 has been shown to regulate p53 and p21 reporter activity (31). These results provide a potential mechanism for PTP4A3 regulation of proliferation and apoptosis in cancer cells. Other studies have shown that PTP4A3 activates the NF-kB and PI3K pathways, which are important for survival and proliferation of cancer cells (32,33). PTP4A3 overexpression is associated with increased p65 phosphorylation (32), and reduced PTEN expression (33). Alteration of these multiple pathways may collectively contribute to the reduced proliferation and apoptosis following PTP4A3 knockdown in breast cancer cells. We are currently performing experiments to elucidate direct targets of PTP4A3 in breast cancer cells that regulate cell cycle and cell death.
Here, we have identified phosphatases overexpressed in triple-negative as compared to ER-positive breast cancer, many of which are critical for TNBC cell growth. One of these phosphatases, PTP4A3, is a critical regulator of proliferation and apoptosis, and inhibition of PTP4A3 induces cell cycle arrest, apoptosis, and inhibition of TNBC tumor growth in vivo. These data strongly suggest that targeting PTP4A3 or its signaling pathway in TNBC is a promising strategy for women with these aggressive breast cancers.
Supplementary Material
Acknowledgments
Financial Support:
This work was funded by an NCI Cancer Center Support Grant (P30CA016672, P.H. Brown, G.B. Mills), a Susan G. Komen Promise Grant (KG081694, P.H. Brown, G.B. Mills), a Komen SAB Grant (P.H. Brown), and the Norman Brinker Award for Research Excellence (P.H. Brown). In addition, this research utilized the shared resources of the Cancer Center Support Grant (CCSG)-funded Characterized Cell Line Core (P30CA016672 G.B. Mills).
PH Brown is on the Scientific Advisory Board of Susan G. Komen for the Cure.
We would like to thank Svasti Haricharan for the data analysis of the 8q24 region, Michelle Savage for editing the manuscript, and Sam Short for assisting in the submission.
Footnotes
Conflict of interest:
All remaining authors declare no actual, potential, or perceived conflict of interest that would prejudice the impartiality of this article.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman B, Trudeau M, Awada A, Blackwell K, Bachelot T, Salazar V, et al. Lapatinib monotherapy in patients with HER2-overexpressing relapsed or refractory inflammatory breast cancer: final results and survival of the expanded HER2+ cohort in EGF103009, a phase II study. Lancet Oncol. 2009;10(6):581–588. doi: 10.1016/S1470-2045(09)70087-7. [DOI] [PubMed] [Google Scholar]
- 4.Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer. 2009;9(Suppl 2):S73–S81. doi: 10.3816/CBC.2009.s.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speers C, Tsimelzon A, Sexton K, Herrick AM, Gutierrez C, Culhane A, et al. Identification of novel kinase targets for the treatment of estrogen receptor-negative breast cancer. Clin Cancer Res. 2009;15(20):6327–6340. doi: 10.1158/1078-0432.CCR-09-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 14(4):197–210. [PubMed] [Google Scholar]
- 12.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 10(3):143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 13.Yasutis KM, Kozminski KG. Cell cycle checkpoint regulators reach a zillion. Cell Cycle. 12(10):1501–1509. doi: 10.4161/cc.24637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertucci MC, Mitchell CA. Phosphoinositide 3-kinase and INPP4B in human breast cancer. Ann N Y Acad Sci. 1280:1–5. doi: 10.1111/nyas.12036. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes S, Iyer S, Kerr WG. Role of SHIP1 in cancer and mucosal inflammation. Ann N Y Acad Sci. 1280:6–10. doi: 10.1111/nyas.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Q, Hong W, Tan YH. Mouse PRL-2 and PRL-3, two potentially prenylated protein tyrosine phosphatases homologous to PRL-1. Biochem Biophys Res Commun. 1998;244(2):421–427. doi: 10.1006/bbrc.1998.8291. [DOI] [PubMed] [Google Scholar]
- 17.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10(6):529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Pereira L, Igea A, Canovas B, Dolado I, Nebreda AR. Inhibition of p38 MAPK sensitizes tumour cells to cisplatin-induced apoptosis mediated by reactive oxygen species and JNK. EMBO Mol Med. 2013;5(11):1759–1774. doi: 10.1002/emmm.201302732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui Y, Parra I, Zhang M, Hilsenbeck SG, Tsimelzon A, Furukawa T, et al. Elevated expression of mitogen-activated protein kinase phosphatase 3 in breast tumors: a mechanism of tamoxifen resistance. Cancer Res. 2006;66(11):5950–5959. doi: 10.1158/0008-5472.CAN-05-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radke I, Gotte M, Kersting C, Mattsson B, Kiesel L, Wulfing P. Expression and prognostic impact of the protein tyrosine phosphatases PRL-1, PRL-2, and PRL-3 in breast cancer. Br J Cancer. 2006;95(3):347–354. doi: 10.1038/sj.bjc.6603261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu L, Luo H, Wang W, Li H, He T. Poor prognosis of phosphatase of regenerating liver 3 expression in gastric cancer: a meta-analysis. PLoS One. 8(10):e76927. doi: 10.1371/journal.pone.0076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao RT, Zhang XH, Pan YF, Liu HG, Xiang YQ, Wan L, et al. Prognostic and metastatic value of phosphatase of regenerating liver-3 in invasive breast cancer. J Cancer Res Clin Oncol. 136(9):1349–1357. doi: 10.1007/s00432-010-0786-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmerman MW, Homanics GE, Lazo JS. Targeted deletion of the metastasis-associated phosphatase Ptp4a3 (PRL-3) suppresses murine colon cancer. PLoS One. 8(3):e58300. doi: 10.1371/journal.pone.0058300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzinska-Ustymowicz K, Pryczynicz A. PRL-3, an emerging marker of carcinogenesis, is strongly associated with poor prognosis. Anticancer Agents Med Chem. 11(1):99–108. doi: 10.2174/187152011794941145. [DOI] [PubMed] [Google Scholar]
- 28.Forte E, Orsatti L, Talamo F, Barbato G, De Francesco R, Tomei L. Ezrin is a specific and direct target of protein tyrosine phosphatase PRL-3. Biochim Biophys Acta. 2008;1783(2):334–344. doi: 10.1016/j.bbamcr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Tian W, Qu L, Meng L, Liu C, Wu J, Shou C. Phosphatase of regenerating liver-3 directly interacts with integrin beta1 and regulates its phosphorylation at tyrosine 783. BMC Biochem. 13:22. doi: 10.1186/1471-2091-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiordalisi JJ, Dewar BJ, Graves LM, Madigan JP, Cox AD. Src-mediated phosphorylation of the tyrosine phosphatase PRL-3 is required for PRL-3 promotion of Rho activation, motility and invasion. PLoS One. 8(5):e64309. doi: 10.1371/journal.pone.0064309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min SH, Kim DM, Heo YS, Kim HM, Kim IC, Yoo OJ. Downregulation of p53 by phosphatase of regenerating liver 3 is mediated by MDM2 and PIRH2. Life Sci. 86(1–2):66–72. doi: 10.1016/j.lfs.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Lian S, Meng L, Liu C, Xing X, Song Q, Dong B, et al. PRL-3 activates NF-kappaB signaling pathway by interacting with RAP1. Biochem Biophys Res Commun. 430(1):196–201. doi: 10.1016/j.bbrc.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res. 2007;67(7):2922–2926. doi: 10.1158/0008-5472.CAN-06-3598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.