Abstract
Cancer stem cells exert enormous influence on neoplastic behavior, in part by governing asymmetric cell division and the balance between self-renewal and multipotent differentiation. Growth is favored by deregulated stem cell division, which enhances the self-renewing population and diminishes the differentiation program. Mutation of a single gene in Drosophila, Brain Tumor (Brat), leads to disrupted asymmetric cell division resulting in dramatic neoplastic proliferation of neuroblasts and massive larval brain overgrowth. To uncover mechanisms relevant to deregulated cell division in human glioma stem cells, we first developed a novel adult Drosophila brain tumor model using brat-RNAi driven by the neuroblast specific promoter inscuteable. Suppressing Brat in this population led to accumulation of actively proliferating neuroblasts and a lethal brain tumor phenotype. brat-RNAi caused upregulation of Notch signaling, a node critical for self-renewal, by increasing protein expression and enhancing nuclear transport of NICD. In human glioblastoma, we demonstrated that the human ortholog of Drosophila Brat, TRIM3, similarly suppressed NOTCH1 signaling and markedly attenuated the stem cell component. We also found that TRIM3 suppressed nuclear transport of active NOTCH1 (NICD) in glioblastoma, and demonstrated that these effects are mediated by direct binding of TRIM3 to the Importin complex. Together, our results support a novel role for Brat/TRIM3 in maintaining stem cell equilibrium and suppressing tumor growth by regulating NICD nuclear transport.
Keywords: Stem cell, Glioblastoma, Brat, TRIM3, Notch, Drosophila, Importin α and β
INTRODUCTION
Glioma stem cells (GSCs) are a driving force underlying the clinically devastating growth properties of glioblastoma (GBM) (1). While GSCs account for only a small fraction of total GBM cells, they confer a disproportionate influence on malignant behavior and therapy resistance (2-4). As opposed to normal stem cells, GSCs show disrupted asymmetric cell division properties that favor the creation of stem cells over those that are programmed for differentiation (5). Current understanding of asymmetric cell division as it relates to tumorigenesis has been derived largely from studies of Drosophila neuroblasts. In this model, mutation of a single gene, Brain Tumor (brat), causes altered stem cell division in a manner that generates a massively enlarged brain formed entirely of neoplastic neuroblasts during larval development (6,7).
Mammalian Tripartite Motif Containing Protein 3 (TRIM3) is a human ortholog of Drosophila Brat (48% homology) expressed solely in brain and is deleted in 25% of GBM samples; other mechanisms are responsible for its reduced gene and protein expression in nearly all GBMs (5). We previously demonstrated that restored expression of TRIM3 reduced neurosphere formation, attenuated the GSCs population, promoted normal asymmetric cell division and diminished in vitro and in vivo growth properties of human GBMs. Initial studies implicated NOTCH1 as a potential mediator of these effects (5).
Notch signaling is a central node that directs self-renewing proliferation of neural stem cells (8,9). Notch was first described as oncogenic in T- cell acute lymphoblastic leukemia, in which a specific translocation t(7;9)(q34;q34.3) generates a fusion protein with a truncated, active Notch Intra-Cellular domain (NICD) (10), Notch is now appreciated as a key pro-tumorigenic signaling protein in numerous cancers, including GBM (11-13), (14).
Activation of Notch signaling is complex and requires receptor activation, endocytosis followed by gamma-secretase-mediated cleavage to generate active NICD and finally, transport of NICD into the nucleus by Importins to initiate transcription (15-19).
In the current study, we investigated mechanisms by which Brat/TRIM3 regulates Notch signaling in brain tumors. We generated a novel Drosophila model using brat-RNAi expressed in neuroblasts, which results in a fatal adult brain tumor phenotype, and demonstrated that active Notch is a primary driver. We extended investigations to human GBM neurospheres and show a similar relationship between TRIM3 and NOTCH1 signaling. Finally, we present data supporting a mechanism in which Brat/TRIM3 suppresses Notch signaling by attenuating its nuclear transport through Importin α and β.
MATERIALS AND METHODS
Drosophila strains and genetics
UAS-bratRNAi and insc-GAL4; UAS-tub-cherry flies were obtained from Bloomington Drosophila stock center at Indiana University. UAS-MamH fly stock was a generous gift from Dr. Barry Yedvobnick (20). All Drosophila stocks were reared at 25°C with standard cornmeal/yeast/agar medium. UAS-bratRNAi and insc-GAL4; UAS-tub-cherry were crossed to generate adult flies with brain tumor phenotype. UAS-MamH flies were brought into same genetic background to suppress Notch signaling pathway in brat-RNAi tumor flies. Repo–GFP flies from Bloomington Drosophila Stock Center was crossed into brat-RNAi tumor model to check Repo positive cells. To suppress nuclear transport, we used UAS-ket-RNAi stock of Importin β ortholog Drosophila Ketel in brat-RNAi background (Bloomington Stock Center of Indiana University).
Drosophila dissection and immunocytochemistry
Adult Drosophila brains were dissected and fixed in 4% paraformaldehyde in PBS for 60-90 mins (21). These were then treated with 0.5% TritonX-100 in PBS for 30 mins and placed in primary antibodies in 1X PBS with 0.5% TritonX-100 and 10% BSA over night at 4°C. Brains were washed for at least 30 mins, added to secondary antibody solution for over night at 4°C, washed for the final time and kept in vectashield (Vector Laboratory) for 2 days at 4°C, and mounted for microscopy.
The following primary antibodies were used for the immunohistochemistry: NICD (1:100) [Cell signaling Technology], Miranda (1:100) and Deadpan (1:100) [a gift from Dr. Renee Read], Phospho-Histone H3 [Abcam], Musashi (1:100) [a gift from Dr. Hideyuki Okano] and Brat (1:100) [a gift from Dr. Jorgen Knoblich]. Alexa flour 488, 555 and 647 were used as secondary antibodies.
Drosophila western blot
30 normal brains and 30 brat-RNAi tumor brains were collected and nuclear extracts were prepared using Thermo scientific NE-PER Nuclear Extraction kit (Cat# 78833). Proteins were run on a 4-15% gradient gel and visualized using ECL. Primary antibodies used: NICD [Cell Signaling Technology], Cut (1:100) [a gift from Dr. Kenneth Moberg], Histone H3 (1:1000) [Abcam] and Actin (1:2000).
Neurosphere sources and culture
Neurosphere cultures were established from Normal Human Neuro-progenitor cells (NHNP) obtained from Lonza (Cat# PT-2599) and patient GBM samples N08-74 and 13113, as previously described, and experiments were performed using passage numbers 10-40 (5). The diagnosis of GBM was established by the senior author, who is a diagnostic neuropathologist (DJB) in accordance to criteria by the World Health Organization. The cell lines are routinely tested for GFAP, Olig2, Sox2 and CD133expression by western blot and immunocytochemistry. The cells were last tested within last 6 months. The cells were tested and cleared for mycoplasma using Lonza MycoAlert mycoplasma detection kit (Cat# LT07-703). GBM neurospheres and NHNPs were cultured using Neurobasal-A media (Invitrogen) with added Epidermal Growth Factor (Stem Cell Technologies), Fibroblast Growth Factor (Stem Cell Technologies) and Heparin (Stem Cell Technologies), as previously described (5).
Lentiviral constructs
FUW-TRIM3 construct was used in all experiments (5). The TRIM3 cDNA was obtained from pCMV6–AC–TRIM3–GFP TrueORF clone (RG211739; Origene). To clone the TRIM3 cDNA, the gene was amplified from the TrueORF clone and re-cloned in FUW vector using BamH1 and Hpa1 sites. The following primers were used: BamHI–TRIM3 50-GGATCCGCCATGGCAAAGAGGGAGGACAGC-30 and HpaI–TRIM3 50-GTTAACCTACTGGAGGTAGCGATAGGCTTT-30.
Lentiviral particle generation and infection
For GBM neurosphere, viral particles were collected and concentrated by ultracentrifugation at 25,000 rpm for 4 hours at 4°C. Viral pellets were resuspended in neurobasal-A media to prevent serum effects. Infection of cells and lentiviral-mediated gene expression were confirmed by demonstrating GFP expression by fluorescence microscopy and TRIM3 expression by Western blot analysis.
Flow cytometry and FACS sorting
GBM stem cells were sorted using BD FACSCanto II (BD Biosciences) based on stem cell marker CD133. Analysis of stem cell population in neurospheres was performed using antibodies against CD133 (Miltenyi Biotech) in BD FACSCanto II (BD Biosciences) flow cytometry. Antibody used: CD133 - PE conjugated [Miltenyi Biotech].
Co-Immunoprecipitation
Antibodies against endogenous Importin β1 [Sigma] and endogenous TRIM3 [Abcam] were used for co-immunoprecipitation and reverse co-immunoprecipitation respectively. Pierce Co-immunoprecipitation was used for the pull down and Pierce mild elution buffer was used to release the pritein complex from the beads. Protein was boiled at 95°C for 10 minutes after adding the non-reducing buffer with 20μM DTT. Proteins were run in 4-15% SDS gel along with appropriate controls.
Western blot analysis
Western blots were performed using 4-15% gradient gels from BioRad. Total proteins was isolated using RIPA buffer (Sigma) and nuclear and cytosolic extracts were obtained using the NE-PER buffer (Thermo scientific). The following antibodies were used to detect proteins: TRIM3 (Genetex), HES1 (5), NOTCH1, NICD, KPNA4 (Importin α3) [Everest Scientific] (18), KPNB1 (Importin β1) [Sigma](16), Tubulin (Genetex), GAPDH (Genetex), Histone H3. (Unless mentioned, antibodies were obtained from Cell signaling Technology).
Immunofluorescence analysis
Cells were isolated from media and a single cell suspension was created using Accutase. Cells were then fixed with 4% paraformaldehyde, cytospun on slides, permeabilized using 1X PBS with 0.3% TX100, and subjected to blocking at 0.1% BSA in 1X PBS for an hour. Primary antibody incubation was over night at 4°C followed by washing. After secondary antibody incubation for an hour at room temperature, cells were washed and mounted with vectashield with Dapi. Primary antibodies included: NICD [Cell Signaling Technology], KPNB1 (Importin β1) [Sigma](16), CD133 [Miltenyi Biotech]. Alexa flour 488, 555 and 647 were used as secondary antibodies.
Statistics
GraphPad prism and Flo-Jo software were used for statistical analysis. For densitometric analysis, Image J was used to quantify the western blot bands and to quantify the net fluorescent intensity in the cellular immunofluorescence. Imaris 3D imaging software was used to quantify the brain volume and total tumor cell population in flies. Obtained results were then analyzed for significance using non-parametric t-test at GraphPad Prism.
Microscopy
Drosophila adult brain tumor imaging and human patient-derived neurosphere cell imaging were achieved with the assistance of the Integrated Cellular Imaging core of Emory University. An FV1000 Olympus Confocal microscope was used to collect pictures.
RESULTS
Neuroblast-specific brat-RNAi induces an adult brain tumor phenotype
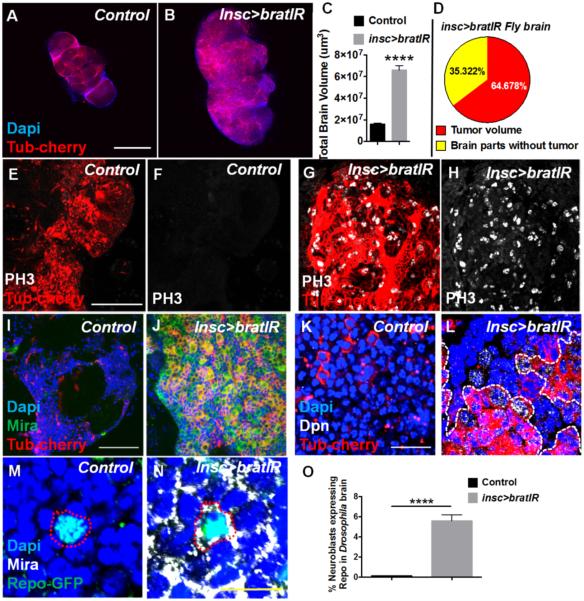
Brat protein localizes to the basal side of asymmetrically dividing neuroblasts in Drosophila, initiating a differentiation signal, whereas the apical side lacking Brat generates a neuroblast through a self-renewal program (1). brat null mutants lose the capacity to divide asymmetrically and accumulate proliferative neuroblasts that leads to a brain tumor phenotype that is lethal at the larval and pupal stage (7). To establish an adult brain tumor model in Drosophila that could be used to study the temporal evolution of signaling events and to translate to mammalian systems, we used brat-RNAi specifically targeted to Drosophila neuronal stem cells using the inscuteable-Gal4 driver that is expressed only during neuroblast division (1,22). We found that suppressing Brat in neuroblasts resulted in an adult Drosophila brain that was 3 times the size of a normal (Fig:1A-C). Tumoral neuroblasts resulting from brat-RNAi were noted in over 60% of the brain and involved nearly all regions equally (Fig: 1D), expanding it globally (Fig: 1B). To ensure that brat-RNAi resulted in reduced Brat protein, we used a Brat antibody kindly provided by Dr. Jorgen Knoblich. In the 3rd inster larval brain and in the adult brain (5 day), we found that brat-RNAi expression in neuroblasts consistently and significantly reduced Brat protein expression (Supplementary Fig: 1 A-X). Control flies, which expressed Insc-GAL4 driven m-cherry, were completely normal.
Figure 1. brat-RNAi mutation in Drosophila neuroblasts generate adult brain tumor with glial markers.
inscuteable-GAL4 driven UAS-TubCherry expression (A,B,E,F,G,H,I,J,K,L,M,N) as well as UAS-brat-RNAi expression in neuroblasts (B,G,H,J,L,N). (A,B,C, D) brat-RNAi expression in neuroblasts generates brain tumor in adult Drosophila with substantial accumulation of neuroblasts. N = number of adult flies > 100. (C) brat-RNAi tumor brains in adults become 3 times larger than normal brains. (D) insc>brat-RNAi Drosophila adults harbor tumors that represent nearly 65% of the total brain volume. (G,H) Neoplastic cells in brat-RNAi (labeled as brat IR) adult tumor show increased expression of phospho-histone 3 indicating active proliferation, as compared to control brain (E,F). N = 20. Tumor cells also express neuroblast markers Miranda (I,J) N = 20 and Deadpan (K,L) N = 20. (M,N,O) GFP reporter driven by Repo promoter shows significant glial cell characteristic in tumor neuroblasts, also marked with Miranda. N =20; n (number of cells counter per N) > 50 . insc = inscuteable; Scale bar: 150 um for (A,B), 35 um for (E,F,G,H), 30 um for (I,J), 10 um for (K,L), 100 um for (J,K), 5 um for (M,N). Mira = Miranda; Dpn = Deadpan. * P<0.05, **** P<0.0001. Statistics: non-parametric t-test. bratIR is brat-RNAi.
Previous studies and our phospho-histone 3 (PH3) staining indicated that adult Drosophila brain does not contain dividing cells, whereas neuroblasts induced by brat-RNAi showed a marked increase in proliferation (Fig: 1E,F vs G,H; Supplementary Fig: 2C-F). As opposed to normal adult Drosophila brain, which do not contain neuroblasts and lack expression of the stem cell markers markers Miranda (Mira) and Deadpan (Dpn), brat-RNAi tumor cells showed consistent Mira (Fig: 1I,J and Supplementary Fig: 2I,J) and Dpn upregulation (Fig: 1K,L), indicating a high degree of stemness. One unexpected observation was that some brain cells expressing Miranda did not show a corresponding expression of Tub-cherry (Fig 1I,J). Only cells that express inscuteable would be expected to also express UAS-tub-cherry transgene, since it is driven by inscuteable-GAL4. However, daughter cells that do not have inscuteable expression may still harbor enough brat-RNAi to suppress Brat and allow the expression of the stem cell marker Miranda. To determine whether proliferating stem cells demonstrate a glial lineage, we used the marker Repo-GFP together with the neuroblast marker Miranda and found that approximately 6% of proliferating neuroblasts also showed Repo expression, as compared to none in control brains (Fig:1M-O and Supplementary Fig: 2G,H).
We also examined the effects of brat-RNAi-driven brain tumors on function and survival in adult flies and found that this phenotype was associated with poor motor function, as determined by negative geotaxis (climbing) assay, and that the median post-eclosion survival was dramatically reduced (14 days in brat-RNAi flies) as compared to controls (45 days; p< 0.0001), indicating that this phenotype is functionally severe and eventually lethal (supplementary Fig: 2A,B) (23-25). These data confirm that brat-RNAi specifically directed to neuroblasts in Drosophila results in a brain tumor phenotype with disrupted asymmetric cell division that could be used to further explored mechanisms related to disrupted stem cell dynamics in brain tumors.
brat-RNAi upregulates Notch signaling and nuclear transport in neuroblasts
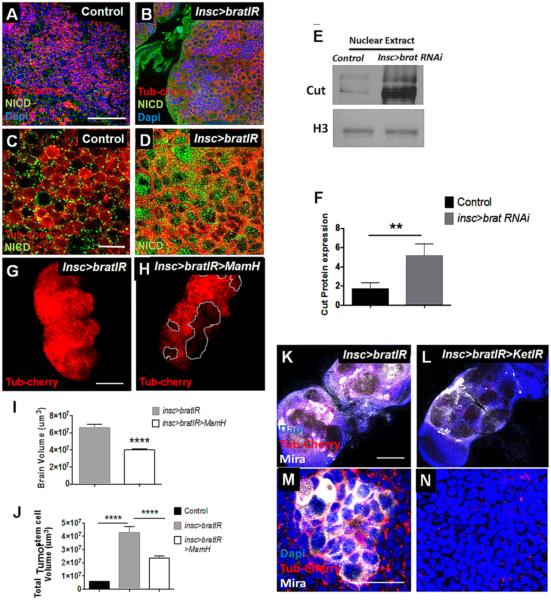
Brain tumor phenotypes developed by brat and Notch pathway mutants are similar, suggesting shared common pathways (26). For example, Numb is a Notch suppressor and numb mutants develop a phenotype similar to brat, although these fail to complement each other (27). We hypothesized that Brat suppresses Notch, given that Numb-independent Notch pathway activation has been recognized (28). To date, no direct link has been established between Notch and Brat in Drosophila (26). In human glioblastoma, the Brat ortholog TRIM3 suppresses NOTCH1 target HES1, suggesting an effect on Notch signaling (5). In the Drosophila brat-RNAi adult brain tumor model, we found that suppression of Brat in proliferating neuroblasts resulted in increased levels of active Notch (NICD) within these cells (Fig: 2A-D), and that much more NICD localized to the nuclei when Brat was suppressed (Supplementary Fig: 3A-C). We next asked whether the increased active NICD in nuclei following Brat suppressing was responsible for initiating a transcriptional program. Nuclear extract from brain tumor neuroblasts showed that Brat suppression led to increased expression of the transcription factor Cut (Fig: 2E,F), a Notch target, indicating active Notch signaling. These results suggest that Brat negatively regulates Notch signaling. To further investigate, we used a mutant form of Mastermind, the transcriptional partner of NICD that is required for a functional Notch transcriptional complex. The truncated form of Mastermind (MamH) (20) lacks the NICD binding domain and prevents Notch signaling. When we expressed MamH in neuroblasts along with brat-RNAi, the brat RNAi phenotype was diminished, with a significant reduction in both tumor cell numbers and tumor volume (Fig: 2G-J). Combined, our results indicate that Brat mediates its effects on neuroblast proliferation and brain tumor formation at least partially through suppression of Notch signaling.
Figure 2. Suppression of Notch signal and NICD nuclear transport reduces tumor burden in brat-RNAi adult brains.
Inscuteable-GAL4 driven UAS-TubCherry expression (A,B,C,D,G,H,K,L,M,N) as well as UAS-brat-RNAi expression (B,D,G,H,K,L,M,N), UAS-MamH in neuroblasts (H) and Ket-RNAi in neuroblasts (L,N). (A,B,C,D) Suppression of Brat causes accumulation of neuroblasts with more active Notch (NICD) in nuclei. Western blot was performed using nuclear extract only from control and tumor brain to detect Cut protein expression (E, F). R (replicates) = 3; N (number of flies per replicate) = 30. (G,H) A truncated form of Mastermind protein called MamH successfully reduces the brain volume (I) as well as the tumor cell volume marked with Tub-Cherry (J) in the tumor. N = 20. Nuclear transporter Ketel was suppressed in brat-RNAi tumor background using Ket-RNAi that reduced tumor volume (K,L). N =15. (M,N) Magnified picture shows tumor cells in brat-RNAi adult brain and absence of tumor cells after suppressing Ketel in the tumor. N = 15; n>30 cells per animal. Ket = Ketel. Scale bar: 25um for (A,B); 10um for (C,D), 150um for (G,H), 100um for (K,L), 10um for (M,N). VC = Vector Control. bratIR is brat-RNAi. * P<0.05, ** P<0.01, **** P<0.0001. Statistics: non-parametric t-test.
The increased accumulation of NICD in brat-RNAi tumor cell nuclei, beyond the level of increases in total protein expression, suggested that nuclear transport of NICD might be enhanced. Nuclear transport of NICD in Drosophila and mammalian systems is controlled by the Importin complex, consisting of the partners Importin α and β (29). To determine the significance of nuclear transport in Brat regulation of NICD, we genetically manipulated the Drosophila homolog of Importin β gene (Ketel) in the background of brat-RNAi. We found that RNAi of Ketel reduced the expression of Ketel protein and partially reversed the phenotype of brat-RNAi (Fig: 2K-N). Together, our results indicate that brat-RNAi increases the expression of NICD and also enhances its transport to the nucleus.
TRIM3, the human ortholog of Brat, suppresses NOTCH1 signaling and stemness in GBM-derived neurospheres
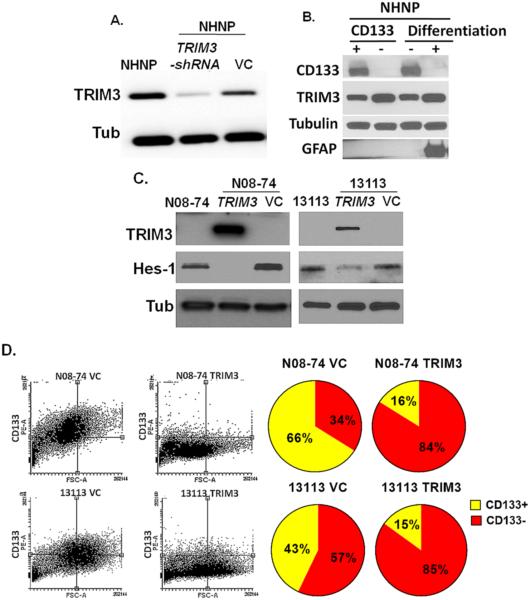
In order to translate our findings from Drosophila, we used neurosphere cultures derived from human GBM explants to study TRIM3, the human ortholog of Brat. TRIM3 is deleted in 25% of GBMs and is expressed at low levels in nearly all (5,30). Previous work has shown that restoration of TRIM3 attenuates in vitro and in vivo growth properties of GBM (5). First, we investigated TRIM3 expression in normal human neural progenitor cells (NHNP) and found strong TRIM3 protein expression (Fig. 3A). In Drosophila neuroblasts, Brat knockdown increased both total active Notch NICD expression and its nuclear accumulation. To determine whether this also occurred human neuro-progenitors, we suppressed TRIM3 in NHNP using TRIM3 sh-RNA. TRIM3 sh-RNA successfully reduced TRIM3 protein (Fig. 3A), but did not strongly induce total NICD expression, as noted in Drosophila (Supplementary Fig. 4B). However, we found that suppression of TRIM3 substantially increased NICD nuclear accumulation, similar to the effects in Drosophila (Supplementary Fig. 4C,D). Since loss of Drosophila Brat induces stemness in Drosophila neuroblasts, we determined whether NHNP stemness and differentiation had any effect on human TRIM3. We FACS sorted NHNP cells using stem cell marker CD133 and found that CD133-positive cells have low TRIM3 compared to CD133 negative cells (Fig: 3B). We also differentiated the NHNP cells for 10 days and found that differentiated NHNP cells had more TRIM3 than undifferentiated cells (Fig. 3B). Our data clearly indicated that TRIM3 expression is inversely related to stemness.
Figure 3. TRIM3, the mammalian ortholog of Brat attenuates NOTCH1 signal and reduces stemness in glioma cells.
(A) TRIM3 expression in NHNP. TRIM3 sh-RNA suppresses the TRIM3 expression in NHNP cells. (B) CD133 positive NHNP cells express less TRIM3 than CD133 negative NHNP cells. Differentiated NHNP cells express more TRIM3 and GFAP. (C) GBM derived neurosphere lines N08-74 and 13113 do not express TRIM3, while N08-74 and 13113 that are stably transfected show strong expression of TRIM3 protein. Expression of TRIM3 completely suppresses NOTCH1 signaling target HES1. (D) Flow cytometry data shows expression of TRIM3 in N08-74 and 13113 neurospheres significantly reduces the population of cells expressing CD133. NICD = Notch Intracellular Domain (Active Notch). R = 3 replicates. Scale bar: 50 um. VC = Vector Control. bratIR is brat-RNAi.
We went on to use patient-derived GBM neurosphere cultures (N08-74 and 13113), which lack TRIM3 protein, in order to study the effects of TRIM3 restoration on Notch signaling and stemness (Fig: 3C). To restore TRIM3 expression, we relied on stable transduction process that faithfully expressed the protein (Fig: 3C). We also investigated the cellular localization of TRIM3 and demonstrated that it was almost entirely cytosolic (Supplementary Fig: 4E). Using our TRIM3 over-expressing neurosphere lines, we demonstrated that TRIM3 strongly suppressed the expression of the NOTCH1 target HES1 (Fig: 3C), indicating that TRIM3 normally inhibits NOTCH1 signaling in mammalian systems much like that Brat regulates Notch in Drosophila. Since human NOTCH1 is a fundamental driver of self-renewal and stem cell maintenance (31,32), we next reasoned that TRIM3 would reduce the stem cell component in human GBMs. Using flow cytometry on N08-74 and 13113 GBM neurospheres, we found that TRIM3 restoration was associated with a marked reduction in the percentage of CD133+ glioma stem cells as compared to neurospheres lacking TRIM3 (Fig: 3D-F). These findings indicate that TRIM3 suppresses NOTCH1 signaling and stemness in human gliomas.
TRIM3 reduces nuclear NICD levels in human GBM-derived neurospheres
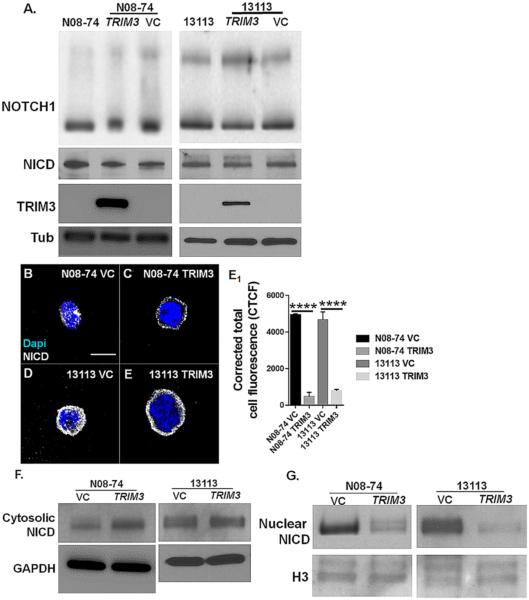
Many cancers show activation of NOTCH1 signaling, similar to our finding of NOTCH1 upregulation in GBM neurospheres lacking TRIM3 (14,33-36). Interestingly, while NOTCH1 activity was clearly increased, our western blots for full-length NOTCH1 and cleaved active NOTCH1 (NICD) that used whole cell (total) protein extracts revealed no substantial differences between their protein levels in TRIM3-expressing N08-74 and 13113 as compared to their TRIM3-null counterparts (Fig: 4A). We hypothesized that NICD is expressed but fails to enter the nucleus to initiate transcription. Using an antibody that recognizes NICD, but not full length NOTCH1, we performed immunocytochemistry and demonstrated reduced levels of NICD in the nucleus, but not the cytoplasm of N08-74 and 13113 upon TRIM3 expression (Fig: 4B-E1). Western blots of nuclear and cytosolic extracts also demonstrated that TRIM3 expression was associated with reduced expression of NICD in the nucleus (Fig: 4G) but not the cytoplasm (Fig: 4F). These results suggested that TRIM3 might suppress the nuclear transportation of NICD.
Figure 4. TRIM3 restricts active NICD transport to nucleus in glioma stem cells.
(A) Western blots using total protein extract from N08-74 (control, TRIM3 expressed and Vector Control) and 13113 (control, TRIM3 expressed and Vector Control) shows that full-length NOTCH1 and active NOTCH1 (NICD) remain unchanged in their expression levels following the expression of TRIM3. (B,C,D,E,E1) Immunocytochemistry data shows TRIM3 expression enhances the nuclear localization of NICD in N08-74 glioma cells. (F,G) Western blot of NICD from cytosolic and nuclear extract supports the immunocytochemical data, showing that TRIM3 expression reduces NICD localization within the nucleus. R = 3 replicates. Scale bar: 5 um (B,C,D,E). VC = Vector Control.
TRIM3 attenuates Importin β1 translocation into nucleus in N08-74 neurospheres
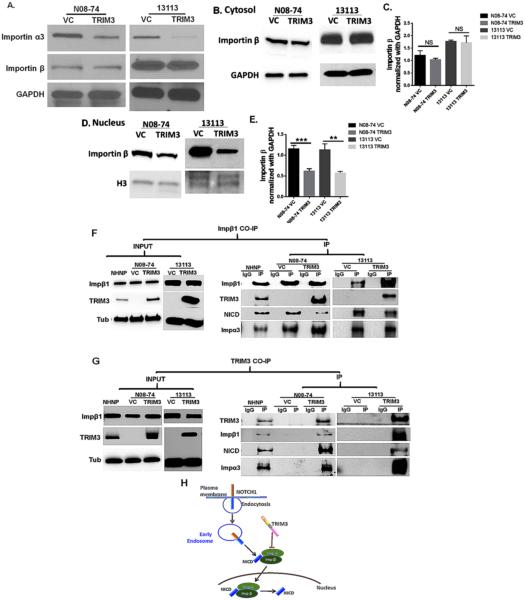
Transport of NICD to the nucleus is a critical step for activation of NOTCH1 signaling. Cleavage of NOTCH1 to generate active NICD occurs in the early endosome and/or in multivesicular bodies (37). After formation, NICD is normally transported into the nucleus by the nuclear transporter Importin complex, composed of Importin α and β. Previous studies indicate that among several types of Importin α and β, NICD nuclear transport depends mostly on Importin α3, α4, α7 and Importin β1 (16,18). Since we observed that NICD localized in the cytoplasm rather than the nucleus after TRIM3 expression, we reasoned that nuclear transportation might be suppressed. To test this, we performed a western blot for Importin α3 and Importin β1 in GBM neurospheres that differentially express TRIM3. We found that total (nuclear and cytoplasmic) Importin α3 expression was reduced following TRIM3 expression, but that total Importin β1 expression remained stable (Fig: 5A). To further investigate the cyto-nuclear translocation of Importins, we examined Importin β1 levels in cytosolic and nuclear extract. We found lower Importin β1 inside the nucleus in TRIM3 expressing cells, with only modest to no reduction of cytosolic levels (Fig: 5B,C,D,E). We confirmed these results using immunofluorescence, which showed greater nuclear accumulation of Importin β1 in GBM cells lacking TRIM3 (Supplementary Fig: 5A-D).
Figure 5. TRIM3 suppresses Importin expression and nuclear translocation affecting active Notch transport into nucleus.
(A) TRIM3 expression in N08-74 and 13113 reduces Importin α3 protein expression, but not Importin β1. (B,C) Cytoplasmic extract shows no significant difference in Importin β1, whereas nuclear extract shows clear reduction in Importin β1 when TRIM3 is expressed (D,E). (F) Co-immunoprecipitation using an antibody against endogenous Importin β1 followed by western blot analysis of input and immunoprecipitation of Importin β1, TRIM3, NICD and Importin α3. The input shows equal protein levels in each lane. We ran the Importin β1 immunoprecipitation of NHNP, N08-74 and 13113 as well as their IgG control. Importin β1 binds with Importin α3, TRIM3 and NICD. In N08-74 and 13113 lines, which don’t express TRIM3, the Importin β1 transport complex does not include TRIM3. (G) Reverse co-immunoprecipitation using antibody against endogenous TRIM3 shows western blot results of Importin β1, TRIM3, NICD and Importin α3. The input shows equal amounts of protein in each lane. The Immunoprecipitation of NHNP, N08-74 and 13113 was loaded along with their IgG immunoprecipitation controls. TRIM3 binds with Importin β1, Importin α3 and NICD. (IgG = immunoprecipitation with control IgG, IP = Immunoprecitation). (H) Model of TRIM3 regulation of NOTCH1 transport through Importin α and β. The presence of TRIM3 protein restricts the entry of Importin α and β into nucleus, thus attenuating active NICD nuclear transport. R = 3 replicates. * P<0.05. Statistics: non-parametric t-test.
To explore whether TRIM3 directly binds with Importin β1 to retain it in the cytosol, we performed co-immunoprecipitation of endogenous Importin β1. Western blot analysis of immunoprecipitated Importin β1 revealed that it strongly binds with TRIM3 in those neurospheres that express TRIM3, but not those that don’t (N08-74 and 13113). In NHNP cells, which normally express TRIM3, binding of TRIM3 and Importin β1 was only modest (Fig: 5F). The reverse co-immunoprecipitation that used a TRIM3 antibody showed that endogenous TRIM3 also binds with Importin β1, providing addition evidence that TRIM3 directly interacts with critical nuclear transporters (Fig: 5G). Since Importin α3 was noted in both the Importin β1 and TRIM3 immunoprecipitations, it is likely that TRIM3 binds to the Importin transport complex. An earlier study has shown that Importin β1 is capable of binding with NICD in oligodendrocyte precursor cells (16). In accordance with this, here we confirmed that in NHNP and in GBM neurospheres, NICD binds with endogenous Importin β1 (Fig: 5F). NICD was also detected in endogenous TRIM3 immunoprecipitation (Fig: 5G), establishing that NICD is a part of the TRIM3, Importin and NICD complex. We also performed immunocytochemistry and found NICD and Importin β1 staining overlapped (Supplementary Fig: 5E-H). Together, these data indicate that TRIM3 attenuates the nuclear transport of NICD by binding to Importin nuclear transport complex and retaining it in the cytoplasm; TRIM3 loss leads to enhanced NICD transport and NOTCH1 signal.
DISCUSSION
We developed a novel adult Drosophila brain tumor model by targeting brat-RNAi to neuroblasts in order to investigate the mechanisms related to disrupted asymmetric cellular division in stem cells (38). The penetrance of the Insc-GAL4 driven brat-RNAi was comparable to that of the Brat hypomorphic allele K06028, as determined by the size of the involved brain and the high number of actively dividing neuroblasts. One of the greatest advantages of using brat-RNAi transgene is that it can be directed to a specific cell type, such as inscuteable positive neuroblasts, so that the other cell types are not affected. While the adult fly brain is normally terminally differentiated and does not contain neuroblasts, the brat-RNAi model creates a phenotype dominated by proliferative neuroblastic stem cells, with a small percentage showing glial differentiation (39). We also observed the expression of stem cell markers Miranda and Deadpan in these proliferating neuroblasts. In our model, we were able to establish that Drosophila Brat attenuates Notch signaling and that this is accomplished at least in part by suppressing nuclear transport of NICD. As opposed to the brat mutants that develop a lethal brain tumor phenotype at the larval stage and cannot be followed longitudinally, tumor-bearing adults in the brat-RNAi model allows the investigation of the temporal evolution of mechanistic events that correspond to functional deterioration and fatal progression.
Notch is well recognized as a critical regulator of cell proliferation and self-renewal signaling pathways and its activation is oncogenic in the proper context (1). During asymmetric division of stem cells, Notch localizes to the apical side to promote stemness, while Brat is a fate determinant that localizes to the basal side to drive differentiation (1). Similar to Notch pathway mutants, loss of Brat also generates tumorous growth (8) by enabling enhanced self-renewal of neuroblasts. In the current study, we observed a concrete link between Brat and Notch signaling in the adult tumor model. brat-RNAi increased the accumulation of active Notch (NICD) in neuroblasts and also led to enhanced expression of Cut, a Notch target. Genetic interaction studies in this model confirmed that suppressing the Notch pathway using Mastermind mutant MamH could reduce tumor burden in brat-RNAi flies. These observations both re-established the role of Notch in neuroblast self-renewal and pointed to a critical contribution of Brat on the Notch signal. We also observed that brat-RNAi increases Musashi protein expression (Supplementary Fig: 6A-C) which, in turn, causes Numb suppression and NICD accumulation.
A recent observation in larval neuroblasts concluded that Brat and Numb regulate parallel pathways (27) rather than intersecting as we have shown in the adult fly model. The differences could be related to the effects of brat-RNAi at the adult stage as opposed to the larval stage of development. Our observation also supports the previously established Numb-independent regulation of the Notch pathway in Drosophila (28). In addition to demonstrating the regulation of Notch signaling by Brat in Drosophila, we also demonstrated that nuclear localization of NICD is suppressed by Brat, potentially providing a mechanism.
Our studies of explanted cultures of human glioblastoma indicated that the findings in Drosophila could be extended to the human disease. The human ortholog of Drosophila Brat, TRIM3, was also found to suppress NOTCH1 signaling. Unlike findings in the fly, we did not observe this strong regulation of NICD expression by TRIM3. However, analogous to Drosophila, TRIM3 was shown to attenuate the nuclear localization of NICD and this process depended on the Importin complex. Previous studies in human cancers have suggested that ligand-independent activation of NOTCH1 intracellular domain (NICD) may play an important role and that NICD localization inside the nucleus is a necessity for neoplastic transformation (40). After NICD is formed in early endosomes and multivesicular bodies (15,19), nuclear localization is accomplished by its shuttling through the Nuclear Pore Complex (NPC). Cyto-nuclear transportation of cargo proteins over 40kD, such as NICD, through the NPC is generally accomplished by the Kryopherins or Importins. Proteins with an NLS (Nuclear Localization Signal) bind with Importin α and this complex binds with Importin β to initiate the multistep process of nuclear localization (41). Previous studies in Drosophila and mammalian systems have demonstrated that NICD nuclear transport depends on the Importin complex (16-18). Here, we observed that TRIM3 bound directly to cytoplasmic Importin β1, retaining both the Importin nuclear transport complex and NICD within the cytoplasm. Our experiments also demonstrated that this retention of NICD in the cytoplasm by TRIM3 was strongly associated with attenuated stem cell properties.
Based on our findings, we have suggested a novel mechanism by which Brat/TRIM3 regulates Notch signaling to maintain a balance of stem cell proliferation and differentiation that is conserved across species (Fig. 5H). In Drosophila, suppression of Brat leads to both NICD expression and its nuclear localization, while in human samples, it seems that the regulation of NICD by TRIM3 occurs primarily at the stage of nuclear transport. Our previous studies, as well as those of others, have indicated the negative regulation of p21 by TRIM3, a finding that could be explained by the effects of Notch signaling, since others have shown a direct regulation of p21 by Notch (5,42,43). Beyond this role, the regulation of nuclear transport machinery by Brat/TRIM3 could imply a far greater impact on neoplastic disease, since nuclear transport is essential for numerous functions critical to proper cell function. TRIM is a large family with many members and here we provide evidence that at least one of its members, TRIM3, can directly bind and regulate nuclear transport in both normal and neoplastic stem cells. This regulation also seems to be evolutionarily conserved between adult Drosophila and human brain tumor models, providing rationale to pursue these mechanisms further.
Supplementary Material
Acknowledgements
We extend thanks to Dr. Barry Yedvobnick and Dr. Cheng-Yu Lee for providing us with stocks and advice. We also thank Dr. Jorgen Knoblich for his generous gift of Brat antibody and Dr. Hideyuki Okano for providing us with the dMusashi antibody. We thank Dr. Cheng-Yu Lee for his helpful comments on the manuscript.
Research reported in this publication was supported in part by the Integrated Cellular Imaging Shared Resource of Winship Cancer Institute of Emory University, FACS and flow cytometry by the Emory Children’s Pediatric Research Center and NIH/NCI under award number P30CA138292. This work was supported by US Public Health Service National Institutes of Health (NIH) grants R01CA149107 and the Georgia Research Alliance (DJB).
Footnotes
Conflict of Interest Statement: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Mukherjee S, Kong J, Brat DJ. Cancer Stem Cell Division: When the Rules of Asymmetry Are Broken. Stem cells and development. 2014 doi: 10.1089/scd.2014.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer research. 2004;64(19):7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 3.Johannessen TC, Wang J, Skaftnesmo KO, Sakariassen PO, Enger PO, Petersen K, et al. Highly infiltrative brain tumours show reduced chemosensitivity associated with a stem cell-like phenotype. Neuropathology and applied neurobiology. 2009;35(4):380–93. doi: 10.1111/j.1365-2990.2008.01008.x. [DOI] [PubMed] [Google Scholar]
- 4.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer research. 2003;63(18):5821–8. [PubMed] [Google Scholar]
- 5.Chen G, Kong J, Tucker-Burden C, Anand M, Rong Y, Rahman F, et al. Human Brat ortholog TRIM3 is a tumor suppressor that regulates asymmetric cell division in glioblastoma. Cancer research. 2014;74(16):4536–48. doi: 10.1158/0008-5472.CAN-13-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nature genetics. 2005;37(10):1125–9. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- 7.Arama E, Dickman D, Kimchie Z, Shearn A, Lev Z. Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene. 2000;19(33):3706–16. doi: 10.1038/sj.onc.1203706. [DOI] [PubMed] [Google Scholar]
- 8.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nature reviews Molecular cell biology. 2010;11(12):849–60. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Current topics in developmental biology. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 10.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 11.Boulay JL, Miserez AR, Zweifel C, Sivasankaran B, Kana V, Ghaffari A, et al. Loss of NOTCH2 positively predicts survival in subgroups of human glial brain tumors. PloS one. 2007;2(6):e576. doi: 10.1371/journal.pone.0000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31(4):557–68. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 13.Nagao M, Sugimori M, Nakafuku M. Cross talk between notch and growth factor/cytokine signaling pathways in neural stem cells. Molecular and cellular biology. 2007;27(11):3982–94. doi: 10.1128/MCB.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stockhausen MT, Kristoffersen K, Poulsen HS. The functional role of Notch signaling in human gliomas. Neuro-oncology. 2010;12(2):199–211. doi: 10.1093/neuonc/nop022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baron M. Endocytic routes to Notch activation. Seminars in cell & developmental biology. 2012;23(4):437–42. doi: 10.1016/j.semcdb.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Nakahara J, Kanekura K, Nawa M, Aiso S, Suzuki N. Abnormal expression of TIP30 and arrested nucleocytoplasmic transport within oligodendrocyte precursor cells in multiple sclerosis. The Journal of clinical investigation. 2009;119(1):169–81. doi: 10.1172/JCI35440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sachan N, Mishra AK, Mutsuddi M, Mukherjee A. The Drosophila importin-alpha3 is required for nuclear import of notch in vivo and it displays synergistic effects with notch receptor on cell proliferation. PloS one. 2013;8(7):e68247. doi: 10.1371/journal.pone.0068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huenniger K, Kramer A, Soom M, Chang I, Kohler M, Depping R, et al. Notch1 signaling is mediated by importins alpha 3, 4, and 7. Cellular and molecular life sciences : CMLS. 2010;67(18):3187–96. doi: 10.1007/s00018-010-0378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Y, Denef N, Schupbach T. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Developmental cell. 2009;17(3):387–402. doi: 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yedvobnick B, Kumar A, Chaudhury P, Opraseuth J, Mortimer N, Bhat KM. Differential effects of Drosophila mastermind on asymmetric cell fate specification and neuroblast formation. Genetics. 2004;166(3):1281–9. doi: 10.1534/genetics.166.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bian XL, Rosas-Acosta G, Wu YC, Wilson VG. Nuclear import of bovine papillomavirus type 1 E1 protein is mediated by multiple alpha importins and is negatively regulated by phosphorylation near a nuclear localization signal. Journal of virology. 2007;81(6):2899–908. doi: 10.1128/JVI.01850-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402(6761):544–7. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- 23.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro-oncology. 2013;15(Suppl 2):ii1–56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heimans JJ, Taphoorn MJ. Impact of brain tumour treatment on quality of life. Journal of neurology. 2002;249(8):955–60. doi: 10.1007/s00415-002-0839-5. [DOI] [PubMed] [Google Scholar]
- 25.Osoba D, Brada M, Prados MD, Yung WK. Effect of disease burden on health-related quality of life in patients with malignant gliomas. Neuro-oncology. 2000;2(4):221–8. doi: 10.1093/neuonc/2.4.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Developmental cell. 2008;14(4):535–46. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komori H, Xiao Q, McCartney BM, Lee CY. Brain tumor specifies intermediate progenitor cell identity by attenuating beta-catenin/Armadillo activity. Development. 2014;141(1):51–62. doi: 10.1242/dev.099382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couturier L, Vodovar N, Schweisguth F. Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nature cell biology. 2012;14(2):131–9. doi: 10.1038/ncb2419. [DOI] [PubMed] [Google Scholar]
- 29.Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435(7042):693–6. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Raheja R, Yeh N, Ciznadija D, Pedraza AM, Ozawa T, et al. TRIM3, a tumor suppressor linked to regulation of p21(Waf1/Cip1.) Oncogene. 2014;33(3):308–15. doi: 10.1038/onc.2012.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development. 2013;140(4):689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 32.Chapouton P, Skupien P, Hesl B, Coolen M, Moore JC, Madelaine R, et al. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(23):7961–74. doi: 10.1523/JNEUROSCI.6170-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallahan D, Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. Journal of virology. 1987;61(1):66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122(7):2251–9. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 35.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nature medicine. 2002;8(9):979–86. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 36.Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(14):6414–8. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kandachar V, Roegiers F. Endocytosis and control of Notch signaling. Current opinion in cell biology. 2012;24(4):534–40. doi: 10.1016/j.ceb.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133(14):2639–48. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- 39.Homem CC, Steinmann V, Burkard TR, Jais A, Esterbauer H, Knoblich JA. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 2014;158(4):874–88. doi: 10.1016/j.cell.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 40.Jeffries S, Capobianco AJ. Neoplastic transformation by Notch requires nuclear localization. Molecular and cellular biology. 2000;20(11):3928–41. doi: 10.1128/mcb.20.11.3928-3941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matamales M, Girault JA. Signaling from the cytoplasm to the nucleus in striatal medium-sized spiny neurons. Frontiers in neuroanatomy. 2011;5:37. doi: 10.3389/fnana.2011.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson A, Radtke F. Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett. 2006;580(12):2860–8. doi: 10.1016/j.febslet.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 43.Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 2005;19(12):1485–95. doi: 10.1101/gad.341405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.