Abstract
VHL-deficient clear cell renal cell carcinomas (ccRCC), the most common form of kidney cancer, express transcripts derived from the novel human endogenous retrovirus HERV-E (named CT-RCC HERV-E). In this study, we define a transcript encoding the entire envelope gene of HERV-E as expressed selectively in ccRCC tumors, as distinct from normal kidney tissues or other tumor types. Sequence analysis of this envelope transcript revealed long open reading frames encoding putative surface and transmembrane envelope proteins. Retroviral envelopes are known to be capable of eliciting immunity in humans. Accordingly, we found that HLA-A*0201-restricted peptides predicted to be products of the CT-RCC HERV-E envelope transcript stimulated CD8+ T cells which could recognize HLA-A*0201-positive HERV-E-expressing kidney tumor cells. Overall, our results offer evidence of unique HERV-E envelope peptides presented on the surface of ccRCC cells, offering potentially useful tumor-restricted targets for T cell-based immunotherapy of kidney cancer.
Introduction
Recently, we isolated and expanded a CD8+ T cell (CTL) clone from the blood of a patient with regressing renal cell carcinoma following an allogeneic hematopoietic stem cell transplant (HSCT) that killed patient tumor cells in vitro (1). This CTL clone was found to have tumor specific cytotoxicity, recognizing an HLA-A11-restricted 10-mer peptide named CT-RCC-1. The transcripts encoding this antigen, CT-RCC-8 and CT-RCC-9 were found to be derived from a novel human endogenous retrovirus type E (named CT-RCC HERV-E) located on chromosome 6q. Both transcripts represent splice variants that share a common region (CR) sequence encoded in the retroviral 5’LTR that is spliced to non-shared regions derived from the protease and polymerase genes, respectively (1). Remarkably, these transcripts were found to be selectively expressed in the clear cell variant of renal cell carcinoma (ccRCC), with no expression observed in normal tissues or any other type of tumor cells (2).
Transcriptional up-regulation of CT-RCC HERV-E in ccRCC appears to occur as a consequence of inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene. Inactivation of VHL with subsequent stabilization of hypoxia-inducible transcription factors (HIF) has been shown to occur in the vast majority of ccRCC tumors. We found that HIF-2α serves as transcriptional factor for CT-RCC HERV-E by binding with a HIF response element (HRE) localized in the proviral 5’LTR (2). Remarkably, this LTR was found to be hypomethylated only in CT-RCC HERV-E-expressing ccRCC cells while other non ccRCC tumors and normal tissues possessed a hypermethylated LTR preventing proviral expression (2).
We have now discovered that a transcript encoding for the entire envelope gene (env) of the CT-RCC HERV-E provirus are also selectively expressed in most ccRCC tumors, but not in normal tissues. This transcript contains sequences which are only found in the loci of the proviral env region on chromosome 6q with expression detected by RT-PCR, Northern blot, and sequence analysis. The proviral env had concurrent expression with the previously identified CT-RCC-8 and -9 transcripts and was found to be exclusively expressed in ccRCC cells, being undetectable in any other tumors or normal tissues. By analyzing the encoding capacities of the CT-RCC-Env transcript, we found long open reading frames (ORFs) that potentially could be translated to produce partial surface (SU) and transmembrane (TM) envelope proteins.
Previously investigators have shown that some HERV-K and HERV-H proviruses express envelope genes that encode highly immunogenic antigens expressed on some tumor cells. Importantly, these tumors can be killed by cytotoxic T cells recognizing these antigens (3, 4). Consistent with these observations, we found three HLA-A*0201- restricted peptides predicted to be derived from proteins encoded by the newly discovered CT-RCC-Env were immunogenic in vitro, stimulating CD8+ T cells that recognized HERV-E expressing ccRCC tumor cells that were HLA-A*0201 positive. These data suggest antigens derived from this newly discovered tumor-restricted HERV-E envelope, could represent excellent targets for T cell based immunotherapy for kidney cancer.
Materials and Methods
Tumor samples and cell lines
The human ccRCC cell lines were established from surgically resected tumors procured at the NIH by the Urologic Oncology Branch of the NCI, the Surgery Branch of the NCI or the Hematology Branch of the NHLBI on IRB approved protocols 97-C-0147, 97-H-0196 and 04-H-0012 as described previously (1). Cell line characterization and authentication were performed by HLA typing and phenotype analysis including the effects of differentiation after 20 passages ex vivo. Once a cell line was authenticated, it was expanded to create a “master stock” of cell aliquots for cryopreservation. Other non-ccRCC cell lines used in experiments were obtained from ATCC. Fresh (primary) ccRCC tumors analyzed in this study were procured by the Urologic Oncology Branch, NCI from nephrectomy samples obtained from 23 patients with VHL disease (2).
RNA isolation, synthesis of cDNA and RT-PCR analysis
Total cellular RNAs were extracted using the RNeasy Mini RNA Purification kit (Qiagen). cDNAs were synthesized using the RT2 First Strand kit (Qiagen). The RT2 SYBR Green ROX qPCR mastermix (Qiagen) was used for quantitative real-time RT-PCR (qRT-PCR). The following primers specific for CT-RCC HERV-E envelope gene were designed for qRT-PCR analysis: forward primer 5’-GGAGTAATAACAGTATTAGGAACCTGCT-3’ and reverse primer 5’-CTTGTGCTGAACTATTTTGGTGAATT-3’. For detection of env gene expression in human normal tissues, human 48 Tissue RapidScan gene expression cDNA panel (Origene) was used as a template.
RNA probe preparation and Northern blotting
Total RNA extracted from ccRCC cell lines expressing CT-RCC HERV-E was used for cDNA synthesis using oligo-dT primer and RT2 First Strand kit (Qiagen). PCR was done using next pair of primers specific for CT-RCC HERV-E env encoding region: 5’-GGAATGACCAACCTCATGTGTC-3’, reverse 5’-GAAGCCTAGGAGCTCATCACCTG-3’. The resulting PCR product was cloned into the pCR4-TOPO vector (Invitrogen). Next, a sequence-verified insert was used as a template for RNA probe preparation. An RNA probe was prepared by in vitro transcription of a Not1-linearized pCR4-TOPO vector carrying the env gene sequence using a DIG Northern starter kit (Roche). For Northern blot analysis, 10 µg of RNA samples were subjected to electrophoresis on a 2% formaldehyde agarose gel in 1 X MOPS buffer and then transferred to a positively charged nylon membrane (Roche). The membrane was hybridized with DIG-labeled RNA probe according to instructions provided by the supplier.
Full-length mRNA sequence identification by RACE
Total RNA was extracted from a ccRCC cell line with a high level of CT-RCC HERV-E expression and then was reverse transcribed using GeneRacer kit (Invitrogen). For 5’RACE and 3’RACE the appropriate adaptor primers were supplied with the GeneRacer kit. Two nested PCR reactions were done using the appropriate GeneRacer nest primers and CT-RCC HERV-E env gene specific primers with the following sequences: forward 5’-GATGGGATGTGGTTCTCTCAGTCG-3’ and reverse 5’-CCAGTATAGTCTAGCTGGAGCCGT-3’. Nested PCR products were then analyzed using TOPO TA cloning kit for sequencing (Invitrogen). The sequencing was performed using BigDye Terminator Cycle Sequencing kit (Applied Biosystems).
Dendritic cells preparation
Peripheral blood mononuclear cells (PBMC) from healthy HLA- A*0201-positive human donors were collected by leukapheresis, washed and cryopreserved in aliquots. Monocyte isolation using negative selection was performed with the MACS Monocyte Isolation Kit II as described by manufacture (Miltenyi Biotec). To generate dendritic cells (DCs), monocytes were plated at 600 – 800K cells/well of a 6-well tissue culture plate in X-VIVO 20 medium supplemented with 10% human serum AB, 1 X penicillin/streptomycin/L-glutamine and 1 X GlutaMAX (Gibco). After one hour incubation at 37°C, non-adherent cells and media were removed and replaced with 2 ml/well of fresh medium with 800 U/ml GMCSF and 1000 U/ml IL-4 (Pepro Tech). After 3 or 4 days of culture at 37°C, cells were matured using medium supplemented with a cytokine cocktail of 800 IU/ml GM-CSF, 1000 IU/ml IL-4, 10 ng/ml TNFα, 10 ng/ml IL-1β, 10 ng/ml IL-6 (Pepro Tech) and 1 µg/ml PGE2 (Sigma), and incubated for another 3 days at 37°C. Mature DCs were then peptide-pulsed as below and used for T cell stimulation.
CTL stimulation
With the computer prediction program BIMAS (http://www-bimas.cit.nih.gov/molbio/hla_bind/), peptides displaying HLA-A*0201-binding motifs were predicted to be derived from the CT-RCC-Env ORF sequences (Table 1). BLASTP analysis was used to confirm that peptides sequences are unique to the CT-RCC HERV-E env-derived putative proteins. HLA-A*0201-restricted Flu and/or CMV peptides were used as a positive control and an HLA-A*0201-restricted MART-1 melanoma-associated antigen as a negative control, respectively (Table 1). The peptides were provided by the NIH Tetramer Facility. Peptides were added to DCs in concentration of 1 ng/mL in a small volume (200 ul) and incubated for 2 hours at 37°C in the conical tubes. DCs were then washed once by media and plated at 50K cells/well of a 96-well culture plate. Primary PBMC cells were added at a responder to stimulator (T/DC) ratios of 5:1. Supplemental IL-7, IL-12, and IL-15 (Pepro Tech) at concentration of 10 ng/ml were added and incubation at 37°C was performed. After 2 days of culturing, 10 U/ml IL-2 and fresh IL-7, IL-12, and IL-15 at concentration of 10 ng/ml were added and the culture was incubated for 3–4 additional days. A second T cell stimulation was performed in a 24-well plate this time using PBMC from the same donor as antigen presenting cells (APCs) with approximately 200K cells per well following the same procedure of culturing.
Table 1.
CT-RCC HERV-E and control peptides used to stimulate T cells.
| Name of protein | Peptide name | Peptide sequencea | Score (BIMAS)b |
|---|---|---|---|
| CT-RCC HERV-E CR | CR1 | 2-FLHKTSVREV | 147 |
| CT-RCC HERV-E CR | CR2 | 11-VLSATIPAT | 29 |
| CT-RCC HERV-E Env | SU1 | 137-SLNITSCYV | 382 |
| CT-RCC HERV-E Env | SU2 | 150-TMGDQWPWEA | 110 |
| CT-RCC HERV-E Env | TM1 | 53-LLLQIMRSFV | 2406 |
| CT-RCC HERV-E Env | TM2 | 45-LLLPCLLPLL | 309 |
| CMV pp65 | CMV | 495-NLVPMVATV | 160 |
| Influenza matrix protein | FLU | 58-GILGFVFTL | 551 |
Position of the starting amino acid in the protein is indicated.
Predicted binding scores to HLA-A*0201 using BIMAS computer assisted analysis.
Enzyme-linked immunosorbent assay (ELISA) test
After two stimulations with peptide pulsed APCs, 100 µl (approximately 100K cells) of each bulk culture were tested for peptide specificity measuring IFNγ secretion by ELISA. Four aliquots from each stimulated culture were examined: two of four aliquots were resuspended with 200 µl of plain media alone and other two aliquots were resuspended with media supplemented with the respective peptide at a concentration of 1 µg/ml. After overnight incubation at 37°C, 20 µl of the supernatant was removed and diluted 10 times. Then 100 µl of the diluted supernatant was assessed for cytokine release using the ELISA following the manufacture’s protocol (R&D Systems).
Intracellular INFγ-secretion assay
To analyze bulk cultures for the presence of peptide-specific CTL, aliquots of T cells were harvested 7 days after the second peptide stimulation and analyzed by an intracellular IFNγ-secretion assay. About 200K T cells were added into a 96-well plate, washed and resuspended in 200 µl new media containing the following: 1 µg/ml corresponding peptide used for T cell stimulation, 1 µl/ml Golgi Plug, 1 µl/ml anti-CD28, and 1 µl/ml anti-CD49d (BD Biosciences). PMA and ionomycin were used to assess for T cell reactivity potential. To analyze bulk cultures for the presence of CTLs recognizing CT-RCC HERV-E expression on the surface of ccRCC cell lines, three different ccRCC cell lines were used as targets: two were HLA-A*0201+ ccRCC cell lines including one that expressed the CT-RCC HERV-E and one that lacked HERV-E expression, and third ccRCC line expressed HERV-E but did not express HLA-A*0201 allele. T cells and RCC cells were co-cultured in a ratio 1:10. The cells were incubated for 5–6 hours at 37°C, then washed and surface stained for CD3, CD56, CD4, and CD8 (BD Biosciences) in FACS buffer for 15 minutes. The unbound antibodies were washed by FACS buffer, then cells were resuspended in 100ul of cytoperm reagent (BD Biosciences) and incubated at 4°C in covered foil plates for 20 minutes. Cells were then washed 2 times with PERM/Wash buffer (BD Biosciences), resuspended in 50 µl master mix containing 2 µl PE- IFNγ Ab (BD Biosciences), and incubated for 15 minutes at 4°C. Cells were washed and then analyzed on a flow cytometer. The gating strategy and flow data analysis are available as Supplemental Material (Supplementary Fig. S1).
To determine CTL cultures specific for CT-RCC HERV-E-derived peptides recognition we used the ratio of percentage of IFNγ positive CD8+ T cells in response to pulsing with HERV-E-originated peptides and the corresponding percentage in response to MART-1 peptide. To find CTL cultures with HERV-E antigen-specific recognition of ccRCC cells we calculated two ratios; ratios of percentages of IFNγ positive CD8+ T cells recognizing ccRCC cells that co-expressed HLA-A*0201and HERV-E and percentages of IFNγ positive CD8+ T cells recognizing either HERV-E+/HLA-A*0201-negative ccRCC cells or HERV-E-negative/HLA-A*0201+ cells. Ratios greater than or equal to 2 were indicated that a CTL bulk culture is specific for HERV-E peptide and kidney cancer cell, respectively.
Results
Detecting expression of CT-RCC HERV-E env gene in ccRCC
CT-RCC HERV-E located at chromosome 6q consists of a full-length, approximately 8.4 Kb, proviral structure organized into gag, pol, and env genes interspersed between two LTRs. This provirus carries termination codons and frame shifts in all genes, preventing its expression in the form of an infectious virus. In previous studies, we reported selective expression of two CT-RCC HERV-E transcripts (CT-RCC-8 and CT-RCC-9) in the clear cell subtype of kidney cancer (1, 2). CT-RCC-8 and CT-RCC-9 represent splice variants containing a 375 bp common region originating from the 5’LTR and unique, 1780 bp and 203 bp non shared regions, derived from the pro and pol genes, respectively.
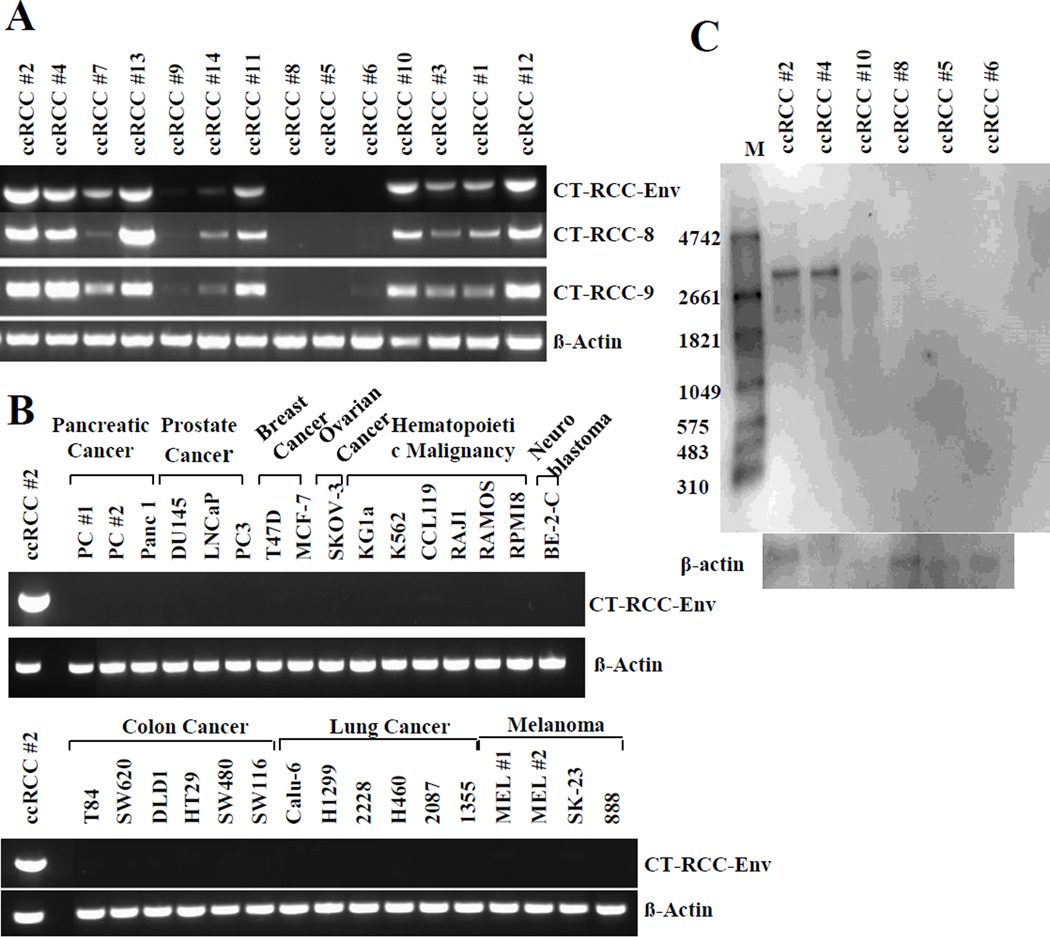
Sequence analysis of CT-RCC HERV-E revealed long ORFs in the env region, suggesting the possibility that components of this provirus’s envelope might also be expressed in kidney cancer cells. To investigate further, we designed primers specific for the CT-RCC HERV-E envelope sequence and performed RT-PCR analysis of all ccRCC cell lines analyzed in our prior studies (1, 2). In the majority of ccRCC cell lines analyzed, expression of the HERV-E envelope was observed (Fig. 1A). On an mRNA level, expression patterns of the env correlated with the expression of the previously identified CT-RCC transcripts. Similar to CT-RCC-8 and CT-RCC-9, no expression of the CT-RCC HERV-E env was detected in any of the non-RCC tumor cell lines analyzed (Fig. 1B).
Figure 1.
Expression of CT-RCC HERV-E envelope transcripts in different cancer cell lines. A, RT-PCR showing expression of three CT-RCC HERV-E transcripts including the Env in the majority of ccRCC cell lines. B, RT-PCR showing lack of expression of the CT-RCC-Env in multiple cancer cell lines other than ccRCC. C, Northern blot analysis of HERV-E env mRNA expression in ccRCC cell lines using a CT-RCC-Env-specific probe. An approximately 3 Kb band was identified in three ccRCC cell lines that were positive for env expression by RT-PCR (ccRCC lines # 2, 4, and 10) but not in three other ccRCC cell lines (ccRCC lines # 5, 6, and 8) that did not express the CT-RCC HERV-E env by RT-PCR. M, RNA marker.
To assess for env gene expression in normal tissues, a cDNA panel from 48 human tissues (RapidScan gene expression cDNA panel, Origene) was analyzed by qRT-PCR and compared to a CT-RCC HERV-E expressing RCC cell line. This analysis showed CT-RCC HERV-E env transcripts were absent in all normal human tissues including normal kidney tissue (Supplementary Table S1).
Identification and sequencing of the full-length env transcript
Northern blot was used to further analyze the expression of CT-RCC HERV-E env mRNA in ccRCC cell lines. A total of six cell lines, including three cell lines that expressed the CT-RCC HERV-E env and three cell lines that were negative for HERV-E expression by RT-PCR were used in the Northern blot analysis. An RNA probe derived from the pCR4-TOPO vector encoding a unique part of HERV-E env genetic loci on chromosome 6q was designed and used for hybridization. RNA samples obtained from the three CT-RCC HERV-E expressing ccRCC cell lines showed positive signals with the RNA probe, demonstrating the presence of an approximately 3 Kb in length env mRNA (Fig. 1C). In contrast, no signal in the Northern blot analysis was detected from the RNA of the three ccRCC cell lines that previously were shown not to express CT-RCC HERV-E sequences by RT-PCR (Fig. 1A).
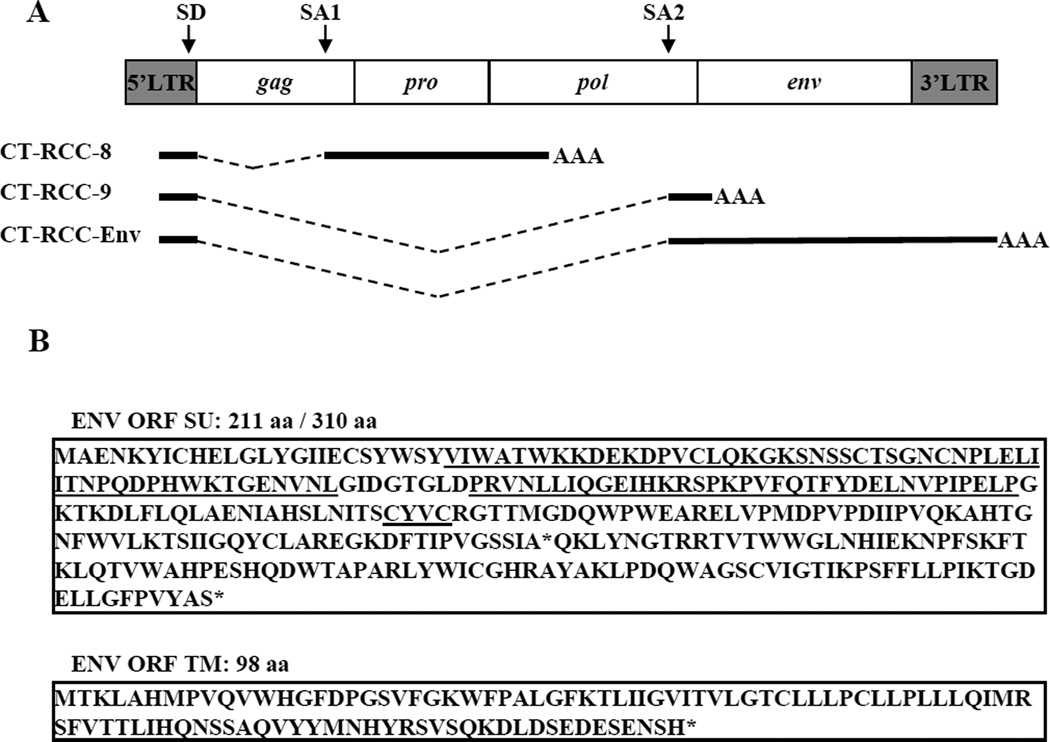
To identify the sequence of env transcript, we used a RACE approach as described in Material and Methods. The cloning and sequencing of PCR products obtained after nested 5’ and 3’ RACE amplification revealed a 3,150 bp long sequence, consistent with the size of env transcript estimated by Northern blot analysis. We named this new transcript CT-RCC-Env (Genbank accession number JQ733905). Comparison of this sequence with the HERV-E genomic locus demonstrated that the CT-RCC-Env transcript is also a spliced variant, sharing the same 375 bp sequence from the proviral 5’LTR as the previously identified CT-RCC-8 and CT-RCC-9 transcripts (Fig. 2A). The non-shared spliced region of the CT-RCC-Env mRNA contained the 3’end of the pol gene along with the full-length env gene sequence. Subsequently, we found that the CT-RCC-Env shared both the 375 bp common region and the 203 bp long pol sequence previously identified in the CT-RCC-9 transcript (Fig. 2A).
Figure 2.
Schematic representation of CT-RCC HERV-E provirus and its three known transcripts. A, A shared 375 bp common region originating from the 5’LTR is spliced into three unique regions of the HERV-E genome. Location of splice donor (SD) and splice acceptor (SA1 and SA2) sites are shown. B, Amino acid sequences predicted to be translated from the common region and envelope ORFs. The ITAM motif is bolded. Motifs RBD, PRR, and CXXC shown in their respective order are underlined. Stop codons are indicated by asterisks (*). SU, the surface protein. TM, the transmembrane protein. CR, common region.
Thus, we identified a new spliced transcript originating from CT-RCC HERV-E, encoding the complete env gene, and having the same splice donor (SD) site located in the 3’end of the 5’LTR as the previously identified CT-RCC-8 and CT-RCC-9 transcripts.
Detection of the CT-RCC-Env in fresh isolated ccRCC tumors
In our previous work, we showed a direct correlation of expression of the CT-RCC HERV-E transcripts with inactivation of the VHL tumor suppressor gene (2). In that analysis, tumors were obtained from nephrectomy samples from patients with VHL disease where specific mutations in the VHL gene had been defined by sequencing. Analysis by qRT-PCR showed 20/23 of these freshly isolated tumors expressed the CT-RCC-8 transcript at high levels (defined as greater than 100 copies relative to GAPDH X 105). In the current study, the same 23 samples were analyzed by qRT-PCR for expression of the newly discovered CT-RCC-Env. Remarkably, we observed CT-RCC-Env expression at variable levels in all 23 freshly isolated tumor samples (Table 2). Since the CT-RCC-Env sequence contains the full component of the CT-RCC-9 sequence, a comparison of expression levels of these two transcripts could not be performed by qRT-PCR.
Table 2.
qRT-PCR results showing expression of CT-RCC-8 and CT-RCC-Env transcripts in tumors obtained from nephrectomy samples from patients with VHL inactivated ccRCC. Copies of CT-RCC transcripts relative to GAPDH ×105 are shown. Control non-tumor normal kidney tissue expression is also shown.
| case number | CT-RCC-8 | CT-RCC-Env |
|---|---|---|
| UT10-0560 | 21537.9 | 31311.31 |
| UT10-0585 | 22636.49 | 24291.61 |
| UT08-0084 | 25090.4 | 21778.52 |
| UT11-0067 | 14840.24 | 14087.66 |
| UT09-0784 | 4226.13 | 6329.35 |
| UT10-0401 | 1854.73 | 3601.29 |
| UT11-0074 | 2729.47 | 3093.17 |
| UT10-0465 | 1852.51 | 2987.65 |
| UT11-0071 | 2033.87 | 2483.89 |
| UT08-0373 | 1929.31 | 2366.46 |
| UT10-0765 | 1534.18 | 1881.42 |
| UT09-0294 | 423.95 | 1176.58 |
| UT09-0320 | 1118.56 | 1002.54 |
| UT10-0927 | 246.98 | 1025.2 |
| UT09-0419 | 191.39 | 567 |
| UT09-0540 | 531 | 488 |
| UT09-0318 | 11.66 | 437.47 |
| UT10-0134 | 132.23 | 398.98 |
| UT09-0433 | 81.54 | 356.02 |
| UT09-0416 | 105.99 | 317.86 |
| UT08-0115 | 247.95 | 291.63 |
| UT09-0335 | 56.06 | 215.74 |
| UT10-0419 | 129.52 | 195.981 |
| Normal kidney | 0.21 | 0.23 |
Prediction of proteins encoded by CT-RCC HERV-E envelope transcripts
We next studied for proteins that could possibly be produced by CT-RCC-Env transcripts. We searched for ORFs starting with ATG in three + frames and then compared all translated products searching against protein databases. We limited this analysis to ORFs encoding sequences with high similarity to previously described retroviral proteins. We found three potentially translatable ORFs that demonstrated high homology to the surface (SU) and transmembrane (TM) subunits of a retroviral envelope. The ORF in the reading frame + 3 containing 402 bp (nts 639–1041) would be predicted to encode a 134 aa sequence that is homologous to the N-terminus of SU proteins (data not shown). The other two ORFs (636 bp long and 297 bp long) were observed to be in the reading frame +1, in the same frame as ORF in the 5’LTR which was previously found to encode the CT-RCC-1 antigen that elicited T cell immunity in vivo in a patient with kidney cancer regression (1). Translation of these two envelope ORFs encodes putative partial 211aa long SU and 98 aa TM proteins, respectively (Fig. 2B). Interestingly, suppression of first termination codon in SU ORF would enlarge the 211 aa long predicted SU protein to 310 aa (Fig. 2B). This putative SU protein retained the canonical active site sequence motifs: a receptor-binding domain (RBD) along with a downstream proline-rich region (PRR) and a CXXC motif, which is known to be highly conserved across a broad range of retroviral envelope proteins. Moreover, this protein was also found to contain an ITAM-signaling motif. This unique motif has the canonical sequence D/Ex0–2YxxL/Ix6–8YxxL/I, where×represents any amino acid.
In summary, the detection of CT-RCC HERV-E envelope transcripts containing long conserved ORFs suggests this provirus may have retained the capacity to produce partial envelope proteins in CT-RCC HERV-E expressing tumors.
Peptides derived from the CT-RCC HERV-E-Env stimulate peptide specific CD8+ T cells that recognize HERV-E expressing ccRCC cells
We next evaluated whether peptides predicted to be derived from the CT-RCC HERV-E-Env could expand antigen-specific CTLs and whether these CTL cultures would recognize ccRCC cell lines that expressed this provirus. DCs were generated from the monocytes of seven different healthy donors who were HLA-A*0201+, then were pulsed with one of six envelope peptides and then were used as APCs to stimulate autologous T cells in vitro. Although the ability to stimulate antigen-specific CTL varied from donor to donor, bulk cultures that responded to the CR1, SU1, and/or TM1 env-derived peptides could be generated from PBMCs of 6 of 7 donors (Table 3). T cell cultures generated from the PBMC of donor 5 did not show clear peptide-specific responses assessed by measuring IFNγ secretion by ELISA (peptide to control ratios of IFNγ secretion were < 2), although there was a trend towards recognition of the SU1 and TM1 peptides (peptide to control ratios of IFNγ secretion were 1.4 and 1.6, respectively). In contrast to the CR1, SU1, and TM1 peptides, antigen-specific T cell recognition of the CR2, SU2 and TM2 peptides could not be induced from the PBMC of any of the 7 donors (data not shown).
Table 3.
Peptide-specific responses of T cells assessed by measuring IFNγ secretion by ELISA.
| DONOR | CR1 PEPTIDE | SU1 PEPTIDE | TM1 PEPTIDE | |||
|---|---|---|---|---|---|---|
| CTL + peptide |
CTL alone | CTL + peptide |
CTL alone | CTL + peptide |
CTL alone | |
| Donor 1 | 491.3 ± 83.6 | 238.9 ± 21.7 | 124.3 ± 5.6 | 115.8 ± 9.2 | 288.2 ± 5.6 | 212.9 ± 42.9 |
| Donor 2 | 678.2 ± 29.0 | 96.7 ± 7.9 | 500.6 ± 12.3 | 479.0 ± 16.6 | 1030 ± 104.4 | 193 ± 2.39 |
| Donor 3 | 322.4 ±2.9 | 174.9 ± 35.2 | 940.9 ± 46.2 | 350.3 ± 23.9 | 608.2 ± 49.5 | 417.8 ± 11.1 |
| Donor 4 | 514.8 ± 5.98 | 295.9 ± 45.7 | 371.5 ± 14.6 | 261.09 ± 37.6 | 1182 ± 120.5 | 237.3 ± 29.1 |
| Donor 5 | 193.3 ± 24.9 | 166.8 ± 18.7 | 390.7 ± 3.8 | 280.2 ± 6.6 | 805.3 ± 1.2 | 496.2 ± 35.6 |
| Donor 6 | 156.6 ± 6.1 | 69.3 ± 38.6 | 128.7 ± 23.7 | 36.2 ± 1.1 | 167.4 ± 39.0 | 52.8 ± 13.5 |
| Donor 7 | 1940.5 ± 12.7 | 576.7 ± 91.5 | 652.6 ± 211.4 | 339.6 ± 38.6 | 358.2 ± 59.2 | 151.9 ± 25.4 |
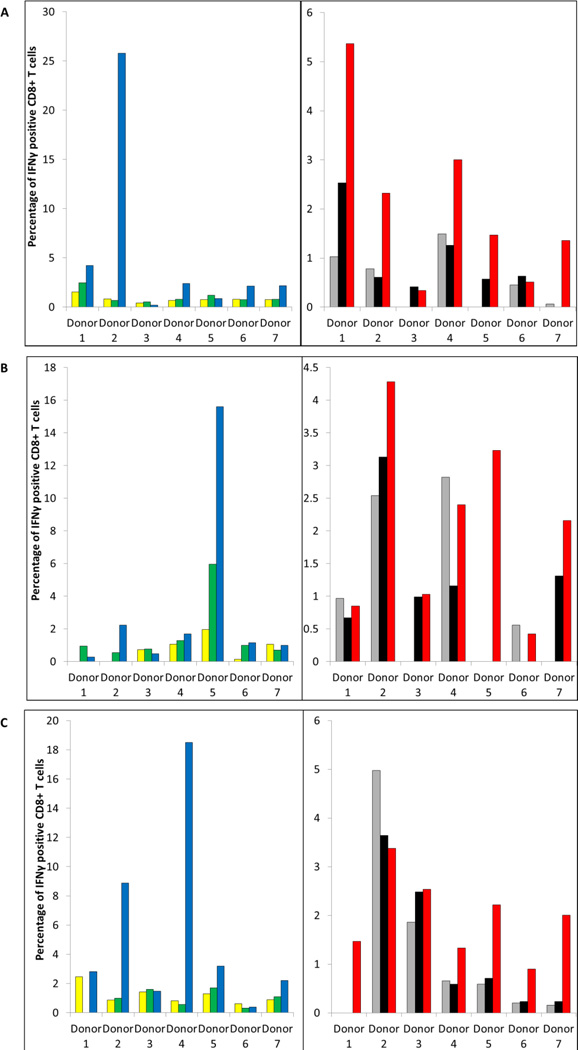
Next, we performed intracellular cytokine staining for IFNγ on CTL cultures stimulated with the CR1, SU1, and TM1 peptides, which our ELISA experiments demonstrated were capable of inducing peptide-specific recognition. Recognition of these peptides was confirmed to reside predominantly in CD8+ T cell populations contained within these bulk cultures (Fig. 3; Supplementary Fig. S1). Further, intracellular cytokine staining for IFNγ demonstrated that these peptide specific CD8+ T cells also recognized ccRCC cells that co-expressed HLA-A*0201and the HERV-E provirus but not ccRCC cell lines that were either HLA-A*0201-negative or HERV-E-negative (Fig. 3; Table 4).
Figure 3.
CT-RCC HERV-E envelope derived peptides stimulate peptide specific CD8+ T cells that recognize HERV-E expressing ccRCC cells. Flow cytometry data are shown for CTL bulk cultures stimulated two times with the HERV-E-env peptides: A, CR1, B, SU1, and C, TM1. Axis X, numeration of healthy donors. Axis γ, the percentage of IFNγ positive CD8+ T cells following pulsing with control peptide MART-1 (green), pulsing with corresponding HERV-E env-derived peptide (blue), or after co-culturing with HERV-E+/HLA-A*0201- negative ccRCC line (grey), HLA-A*0201+/HERV-E-negative ccRCC line (black), and HLA-A*0201+/HERV-E+ ccRCC line (red). Yellow bars show the percentage of IFNγ positive CD8+ T cells alone.
Table 4.
The total number of donors and CTL bulk cultures demonstrating antigen-specific recognition of CT-RCC HERV-E-derived peptides and CT-RCC HERV-E expressing ccRCC cells.
| HLA-A2-restricted peptide |
Number of donors analyzed |
Number of donors with CTL recognizing peptide |
Number of donors with CTL recognizing HERV-E expressing ccRCC cells |
|---|---|---|---|
| CR1 | 7 | 4 | 5 |
| SU1 | 7 | 2 | 1 |
| TM1 | 7 | 4 | 4 |
Taken altogether, these data show that three HLA-A*0201-restricted peptides derived from the putative proteins of CT-RCC HERV-E-Env could be used to expand CD8+ peptide-reactive CTLs that recognized endogenously expressed HERV-E-derived antigens on the surface of HERV-E+/HLA-A*0201-positive ccRCC cells.
Discussion
Although thousands of endogenous retroviral elements have been identified in the human genome, most have accumulated mutations or insertions/deletions and lack uninterrupted ORFs to code full-length proteins. Furthermore, promoter methylation renders the vast majority of HERV’s transcriptionally inactive. Nevertheless, an increasing number of transcripts and protein products derived from HERVs have been detected in a number of human malignancies including, but not limited to melanoma (5–7), breast cancer (4, 8), prostate cancer (9), gastrointestinal cancer (10), ovarian cancer (11–12), leukemias (13–14) and renal cell carcinoma (1, 2). Recently, we identified two transcripts (CT-RCC-8 and -9) derived from a HERV-E located on chromosome 6q that were expressed in ccRCC encoding an antigen which elicited T cell mediated anti-tumor immunity. Here, we present data for the first time showing this HERV-E also expresses a transcript that encodes the entire envelope gene. Similar to CT-RCC-8 and -9, this envelope-derived transcript was also found to be selectively expressed in the clear cell variant of RCC, both in fresh tumors at early stages and in established tumor cell lines. All three CT-RCC HERV-E transcripts were found to represent spliced variants sharing the same SD site (Fig. 2A). The frequency and pattern of RNA splicing were previously shown to vary among different types of HERVs, although an SD site in the leader region seems to be ubiquitously used in spliced subgenomic mRNAs (15). The splice acceptor (SA) site for CT-RCC-8 mRNA is found in the gag region (SA1) while the SA sites for CT-RCC-Env and CT-RCC-9 are identical and are located in the terminal part of the pol gene (SA2; Fig. 2A). Jern et al. also demonstrated that the SA sites for the env transcripts are typically located in the region between the pol and env genes or within the terminal portion of pol, which is similar to the SA2 site (15). A study of spliced HERV-H env transcripts in human T cell leukemia cell lines and lymphocytes showed splice sites in the proximity of a protease region (similar to SA1 for the CT-RCC-8) were also used in the presence of a functional envelope SA site and contribute to the alternative splicing of a number of different subgenomic mRNAs (13).
Although most HERVs have extensive deletions and mutations, some have retained ORFs coding for functional proteins. The maintenance of any HERV ORF over a significant evolutionary time period suggests a biological function. Indeed, the ERVWE1 HERV-W env locus on chromosome 7q21.2 has retained an uninterrupted ORF that encodes the envelope derived Syncytin-1 protein which has a functional role in placental morphogenesis (16). Importantly, some defects in HERV ORFs may be overcome by at least three compensatory mechanisms: complementation, termination suppression, and translational frame shifting. Roebke et al. recently showed that Xq22.3 HERV-W env is capable of producing N-terminally truncated HERV-W protein ex vivo (named N-Trenv). Remarkably, reversion of one stop codon was found to result in the expression of a full-length HERV-W Env protein from this locus (17). We found long ORFs in the newly discovered CT-RCC-Env that potentially could be translated to produce partial SU and TM envelope proteins (Fig. 2B). Furthermore, in the event of termination suppression, it is possible that a longer 310 aa SU sequence could be translated. Analysis of a putative SU protein encoded by the CT-RCC-Env transcript, revealed the presence of canonical sequence motifs of retroviral envelopes including an ITAM-motif (Fig. 2B). The majority of proteins containing ITAMs are transmembrane proteins that exist as a component of immunoreceptor complexes expressed on host cells. However, a number of oncogenic retroviruses have been shown to encode Env proteins that possess ITAM signaling motifs which may activate pathways leading to cellular transformation. For example, the Tax protein of HTLV-1promotes cellular proliferation by activating the expression of a number of cellular genes (18), and the Env protein of JSRV activates signaling pathways including the MEK/extracellular signal-regulated kinase and Akt protein kinase cascades (19). Likewise, the Env ITAM-containing protein from MMTV was shown to activate Src and Syk tyrosine kinases, both of which have transforming capabilities (20). Although unproven, the presence of an ITAM-motif suggests HERV-E expression potentially could lead to the production of env-derived proteins with biological functions.
The capacity of CT-RCC transcripts to produce full or partial length proteins which are antigenic, capable of inducing a cellular or humoral immune response in the host remains an active area of investigation. In our previous work, we identified an HLA-A11-restricted 10-mer peptide derived from the common region of CT-RCC HERV-E transcripts that was the target of a RCC specific CTL response in a patient that had sustained regression of cancer after an allogeneic stem cell transplant (1). In this work we show that peptides predicted to be derived from the newly discovered CT-RCC- ENV that have high binding affinity for HLA-A*0201 are immunogenic in vitro, stimulating antigen specific CD8+ T cells that recognize HERV-E-expressing ccRCC cells. Artificial pulsing of CTLs with peptides likely account for the higher percentages of IFNγ positive CD8+ T cells that reacted to peptide pulsed cells compared to ccRCC tumor target that endogenously expressed these HERV-E env peptides at lower levels (Fig. 3). Notably, some exceptions were observed, for instance CR1-stimulated cells from PBMC of donor 1 that contained CD8+ T cells with specificity to HERV-E+/HLA-A*0201 ccRCC cells had similar reactivity to peptide-pulsed cells (Fig. 3A). Regardless, the observation that these CTL contained cells that responded to HERV-E+/HLA-A*0201 positive ccRCC cell lines provide evidence that env-derived peptides are endogenously processed and presented on the surface of ccRCC cells and can serve as targets for CD8+ T cells.
While it would have been interesting to conduct similar experiments using T cells obtained from patients with metastatic RCC, we did not have access to high numbers of patient monocytes that would be needed to generate dendritic cells to conduct these experiments. However, when PBMCs collected from 4 patients with metastatic clear cell RCC that were HLA-A*0201 positive were pulsed with CT-RCC ENV-derived peptides, antigen specific T cells were detectable in the range of 0.07– 1.36% in CD8+ T cells using intracellular staining for IFNγ (data not shown). Remarkably, these populations were not detectable in the PBMC of HLA identical siblings from all 4 of these patients, suggesting CT-RCC-ENV reactive CTL should be expandable from the blood of patients with metastatic RCC.
There is a growing number of reports suggesting antigens derived from the envelope region of HERVs are highly immunogenic. For instance, investigators have recently observed cytotoxic T cells specific to antigens derived from the envelope of a HERV-K in patients with HERV-K-expressing breast cancer and ovarian cancer (4, 12). Further, HLA-A*0201- restricted peptides derived from a putative 273 aa protein encoded in the HERV-H locus on chromosome Xp22 were shown to generate CTL from peripheral blood T cells that induced tumor-specific lysis of cancer cell lines that endogenously expressed the HERV-H Xp22.3 (3). Remarkably, this provirus was found to be specifically expressed in gastrointestinal cancer, but not in other cancers or normal tissues (10). These findings are even more remarkable when one considers the recent observation that an antigenic peptide was discovered to be translated from a small alternative ORF in a retroviral element where the main ORF encoding for the retroviral protein was absent as a consequence of mutations (21). In this context, an HLA-A*0201-restricted peptide was found to be translated from the smallest possible ORF of the HERV-K-MEL env transcript, an ORF which remarkably is no longer than the peptide coding sequence itself (21). The observation that this peptide was expressed in most melanomas, as well as the observation that naturally occurring CTL responses directed against this epitope could be detected in patients with melanoma suggests a potential use of this peptide for therapeutic cancer vaccination (21).
In contrast to most HERV-derived transcripts and antigens discovered in cancers to date, CT-RCC HERV-E transcripts including the newly discovered env transcript described herein appear to have expression restricted to clear cell kidney tumors, which occurs as a consequence of VHL inactivation. Importantly, as VHL inactivation is a cancer inciting event, expression of this HERV-E provirus occurs in fresh tumors even at the earliest stages of disease (2). Recent work by Rooney et al. compared the results of cytolytic activity of local immune infiltrates with associated genetic changes across 18 tumor types using a comprehensive analysis of high-dimensional data sets such as The Cancer Genome Atlas (TCGA). In this study, they also mapped TCGA RNA-Seq data to the annotation of expressed HERVs and assessed associations with cytolytic activity in different tumors (22). Remarkably, they found independent evidence of elevated expression of CT-RCC HERV-E provirus which was restricted to clear cell kidney cancer but was undetectable in normal tissues (in this work, CT-RCC HERV-E was named as ERVE-4 according to revised nomenclature for HERV loci, ref. 23). They also demonstrated an association of cytolytic activity in ccRCC with expression of this HERV-E suggesting this provirus produces tumor-specific peptide epitopes (22).
T cell immunotherapy for cancer is a rapidly growing field. Recent clinical trials have shown that patients with melanoma and other malignancies can now benefit from adoptive T cell transfer. Despite great progress with new treatments such as targeted inhibitors and inhibitors of immune check points (anti-CTLA-4 and anti-PD-1 monoclonal antibodies), metastatic RCC is generally lethal with mean survival being less than a year. Therefore, more effective therapies are needed for patients with this disease. Recently, PD-L1 molecule which is expressed at high level in various cancer types including RCC was chosen as a target for anticancer immunotherapy. Minami et al. identified new HLA-A24+ restricted PD-L1 peptides that could be used for peptide-based anticancer vaccines for metastatic RCC patients (24). One of the limitations to implementing this approach is expression of PD-L1 on non-cancer cells, with the risk that PD-L1-recognizing CTLs could attack normal cells. Therefore, the kidney cancer reactive TCRs that target antigens which are not expressed on normal tissues still needs further development. In our work, we show for the first time that the CT-RCC HERV-E envelope-encoding transcript is expressed in the majority of ccRCC tumors but not in normal tissues. Peptide antigens predicted to be derived from putative envelope proteins appear to be immunogenic, stimulating CD8+ T cells in vitro that recognize CT-RCC HERV-E-expressing ccRCC tumors. These data suggests antigens derived from this newly discovered HERV-E envelope could represent excellent targets for T cell based immunotherapy for kidney cancer.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by the intramural research program of NIH, National Heart, Lung, and Blood Institute, Hematology Branch.
We acknowledge ACKC (Action to Cure Kidney Cancer) for their support of the ACKC fellows who conducted this research and The Dean R. O’Neill and Edward Rancic Memorial Fellowship for generous contributions supporting this research.
Footnotes
No potential conflicts of interest were disclosed.
References
- 1.Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy J, et al. Regression of kidney cancer following allogeneic stem-cell transplantation associated with T-cells recognizing a HERV-E antigen. J Clin Invest. 2008;118:1099–1109. doi: 10.1172/JCI34409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherkasova E, Malinzak E, Rao S, Takahashi Y, Senchenko V, Kudryavtseva A, et al. Inactivation of the von Hippel-Lindau tumor suppressor leads to selective expression of a human endogenous retrovirus in kidney cancer. Oncogene. 2011;30:4697–4706. doi: 10.1038/onc.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullins C, Linnebacher M. Endogenous retrovirus sequences as a novel class of tumor-specific antigens: an example of HERV-H env encoding strong CTL epitopes. Cancer Immunol Immunother. 2012;61:1093–1100. doi: 10.1007/s00262-011-1183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang-Johanning F, Radvanyi L, Rycaj K, Plummer J, Yan P, Sastry K, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serafino A, Balestrieri E, Pierimarchi P, Matteucci C, Moroni G, Oricchio E, et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res. 2009;315:849–862. doi: 10.1016/j.yexcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 7.Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003;63:8735–8741. [PubMed] [Google Scholar]
- 8.Wang-Johanning F, Frost A, Jian B, Epp L, Lu D, Johanning G. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 9.Wang-Johanning F, Frost A, Jian B, Azuero R, Lu D, Chen D, et al. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98:187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 10.Wentzensen N, Coy J, Knaebel H, Linnebacher M, Wilz B, Gebert J, et al. Expression of an endogenous retroviral sequence from the HERV-H group in gastrointestinal cancers. Int J Cancer. 2007;121:1417–1423. doi: 10.1002/ijc.22826. [DOI] [PubMed] [Google Scholar]
- 11.Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen D, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 12.Rycaj K, Plummer J, Yin B, Li M, Garza J, Radvanyi L, et al. Cytotoxicity of human endogenous retrovirus K specific T cells toward autologous ovarian cancer cells. Clin Cancer Res. 2014;21:471–483. doi: 10.1158/1078-0432.CCR-14-0388. [DOI] [PubMed] [Google Scholar]
- 13.Lindeskog M, Blomberg J. Spliced human endogenous retroviral HERV-H env transcripts in T-cell leukaemia cell lines and normal leukocytes: alternative splicing pattern of HERV-H transcripts. J Gen Virol. 1997;78:2575–2585. doi: 10.1099/0022-1317-78-10-2575. [DOI] [PubMed] [Google Scholar]
- 14.Boyd M, Maclean N, Oscier D. Detection of retrovirus in patients with myeloproliferative disease. Lancet. 1989;1:814–817. doi: 10.1016/s0140-6736(89)92273-3. [DOI] [PubMed] [Google Scholar]
- 15.Jern P, Sperber G, Ahlsen G, Blomberg J. Sequence variability, gene structure, and expression of full-length human endogenous retrovirus H. J Virol. 2005;79:6325–6337. doi: 10.1128/JVI.79.10.6325-6337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blond J, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roebke C, Wahl S, Laufer G, Stadelmann C, Sauter M, Mueller-Lantzsch N, et al. An N-terminally truncated envelope protein encoded by a human endogenous retrovirus W locus on chromosome Xq22.3. Oncogene. 2010;7:69–83. doi: 10.1186/1742-4690-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grassmann R, Aboud M, Jeang K. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene 2. 2005;4:5976–5985. doi: 10.1038/sj.onc.1208978. [DOI] [PubMed] [Google Scholar]
- 19.Liu S, Miller A. Oncogenic transformation by the jaagsiekte sheep retrovirus envelope protein. Oncogene. 2007;26:789–801. doi: 10.1038/sj.onc.1209850. [DOI] [PubMed] [Google Scholar]
- 20.Katz E, Lareef M, Rassa J, Grande S, King L, Russo J, et al. MMTV Env encodes an ITAM responsible for transformation of mammary epithelial cells in three-dimensional culture. J Exp Med. 2005;201:431–439. doi: 10.1084/jem.20041471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiavetti F, Thonnard J, Colau D, Boon T, Coulie P. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62:5510–5516. [PubMed] [Google Scholar]
- 22.Rooney M, Shukla S, Wu C, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolitic activity. Cell. 2014;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer J, Blomberg J, Seal R. A revised nomenclature for transcribed human endogenous retroviral loci. Mob DNA. 2011;2:1–8. doi: 10.1186/1759-8753-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minami Ta, Minami To, Shimizu N, Yamamoto Y, De Velasko M, Nozawa M, et al. Identification of programmed Death Ligand-1 derived peptides capable of inducing cancer-reactive cytotoxic T lymphocytes from HLA-A24+ patients with renal cell carcinoma. J Immunother. 2015;38:285–291. doi: 10.1097/CJI.0000000000000090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.