Abstract
While immunotherapeutic strategies are emerging as adjunctive treatments for cancer, sensitive methods of monitoring the immune response after treatment remain to be established. We used a novel next generation sequencing (NGS) approach to determine whether quantitative assessments of tumor infiltrating lymphocyte (TIL) content and the degree of overlap of T cell receptor (TCR) sequences in brain tumors and peripheral blood were predictors of immune response and overall survival in glioblastoma (GBM) patients treated with autologous tumor lysate-pulsed dendritic cell (DC) immunotherapy. A significant correlation was found between a higher estimated TIL content and increased time to progression (TTP) and overall survival (OS). In addition, we were able to assess the proportion of shared TCR sequences between tumor and peripheral blood at time points before and after therapy, and found the level of TCR overlap to correlate with survival outcomes. Higher degrees of overlap, or the development of an increased overlap following immunotherapy, correlated with improved clinical outcome, and may provide insights into the successful, antigen-specific immune response.
Keywords: cancer, immunotherapy, glioblastoma, TCR sequencing, DC vaccination
INTRODUCTION
Glioblastoma (GBM) is both the most common and lethal adult primary solid CNS malignancy (1). The median overall survival ranges from 12–18 months when conventional therapy is utilized (2). The poor outcome of this disease highlights the need for additional treatment modalities. Autologous tumor lysate-pulsed dendritic cell vaccine therapy may offer a high degree of tumor specificity for highly heterogenous tumors, such as GBM, and has demonstrated a tolerable safety and toxicity profile in Phase I and II trials (3-6).
While DC vaccine strategies have shown great promise, there are limitations with this treatment modality. Distinct subsets of patients in early clinical trials appear to derive dramatic benefit and long-term survival from the therapy, whereas in others, the course of the disease appears unaltered (7). We theorize that the action of the autologous tumor lysate DC vaccine is to prime and/or boost an immune response against individualized, tumor-specific, mutated or overexpressed tumor-specific antigens. T lymphocyte clones specific for these antigens would then invade the tumor and mediate its destruction. However, the precise identity of these targets are unknown and likely differ from tumor to tumor (and possibly spatially and temporally within the same patient), as evidenced by studies examining the antigen targets of expanded populations of tumor-infiltrating lymphocytes in other malignancies (8, 9).
Reliable biomarkers, imaging modalities and/or peripheral blood immune monitoring assays that can predict and characterize the immune and clinical responses generated by therapies, such as the DC vaccine, are currently still in development. Previously, our group demonstrated that intracellular cytokine signaling (specifically that of STAT1 and STAT5 T cells) correlated with survival and also that regulatory T-cell population shifts were associated with clinical outcome (10, 11).
The presence, quantity and phenotype of tumor infiltrating lymphocytes (TILs) have been examined in various malignancies, including glioma, and found to correlate with prognosis and/or grade (12-14). In the context of dendritic cell vaccination, we previously demonstrated that patients with a mesenchymal gene expression tumor subtype possessed an increased TIL infiltration measured by non-quantitative IHC, which correlated with longer survival after DC vaccination compared to patients with other gene expression signatures (7).
In the present study, we used next generation deep sequencing of the TCRVβ CDR3 region to 1) quantitatively measure TIL content in tumors and 2) to compare the array of TCR sequences present (TCR repertoire) within a tumor to those in the peripheral blood of the same patient, and correlate these factors with survival after DC vaccination. This was performed in an attempt to generate a predictive model by which to assess which patients would potentially benefit, and are currently benefitting, from DC vaccination.
Tracking TCR repertoire shifts in the systemic circulation and tumor after treatment is a means of detecting treatment associated immune responsiveness without the need to know the specificity of the receptors (15). Examination of the TCR repertoire has previously been used to characterize the immune response to systemic, nonspecific immunotherapies, such as CTLA-4 and PD-1 blockade (16-18). Based on this work, we hypothesized that a quantitative method to evaluate TIL content in glioblastomas could be utilized to predict survival independently of tumor subtypes assessed by gene expression analysis. We also utilized this NGS data to identify and track T cell populations found in the tumor with those in the systemic circulation (19), and applied this technique to glioblastoma patients treated with autologous tumor lysate-pulsed DC vaccination.
METHODS
Patient characteristics from DC vaccination Trials
This study focused on 15 glioblastoma multiforme (GBM) patients from Phase I and Phase II DC vaccination trials at UCLA (9 newly diagnosed, 6 recurrent GBM) from whom adequate residual fresh-frozen tumor tissue for TIL analysis was available and for whom >2-year survival data was available. Patients were treated with autologous tumor lysate-pulsed dendritic cell (DC) vaccination between 2003 and 2012. Patients provided informed consent for this study, approved by UCLA Medical Institutional Review Board (IRB) prior to treatment. The clinical characteristics and pathology biomarkers are outlined in Supplementary Table 1.
Preparation of Autologous Dendritic Cells (DC)
Monocyte-derived DCs were established from adherent peripheral blood mononuclear cells (PBMC) obtained through leukapheresis, as we have recently described (5). All ex vivo DC preparations were performed in the UCLA-Jonsson Comprehensive Cancer Center GMP facility under sterile and monitored conditions. Briefly, adherent peripheral blood mononuclear cells PBMCs were obtained from patients via leukapheresis and transformed into monocyte-derived dendritic cells by culture in RPMI media supplemented with 10% autologous serum, 500 U/mL of IL-4 and 500 U/mL GM-CSF for 7 days. DCs were collected by vigorous rinsing and washing with a 0.9% NaCl solution. Flow cytometry was utilized in order to analyze the purity and activation status of the DC population using CD83, CD86, and HLA-DR (release criteria required >30% large cell gate being CD86+ and HLA-DR+). DCs were pulsed (co-cultured) with autologous tumor lysate overnight prior to injection.
Treatment Schema
Baseline brain MRI scans were performed one month prior to initiation of immunotherapy and then bimonthly after the completion of the experimental treatment. Each vaccine dose was administered as an intradermal (i.d.) injection in the arm region below patient's axilla. Patients were monitored for two hours after immunization at UCLA’s Clinical and Translational Research Center (CTRC). Patients received three i.d. injections at biweekly intervals. TTP was calculated from the date of the first DC vaccination to the date at which MR imaging confirmed tumor progression. OS was calculated from the date of the first DC vaccination to the date of death from any cause.
IHC Staining
Multiplex immunohistochemistry was preformed on patient FFPE tumor samples. Briefly, tissue was deparaffinized in xylene, followed by incubation in decreasing concentrations of ethanol. Heat-induced epitope retrieval was then conducted; tissue was submerged in a pH 9.0 Tris-EDTA solution while incubated in a pressure cooker. After cooling, tissue was blocked for one hour (with Dako antibody diluent) and stained at room temperature with an antibody to CD8 (Dako, clone C8/144B). Slides were then washed and stained with the TSA Plus Fluorescein System, respectively. Slides were mounted using a VectaShield HardSet Mounding Medium with DAPI. Three images per section were taken using a Perkin Elmer Vectra 2 Automated Quantitative Pathology imaging system, using the 20x objective. An automated image analysis software program (Inform) was used to quantify the density of CD8 T cells/frame.
High-throughput TCRβ sequencing
After surgical resection, residual tumor not required for clinical diagnosis was collected from each patient for the experimental studies herein. Blood was washed and removed from the specimen; necrotic areas were subsequently removed. Prior to DNA isolation, quality control H&E staining was conducted to insure the presence of >20% tumor within the tissue. Genomic DNA was extracted from patient tumor samples, both pre-DC vaccination, and when available, post DC-vaccination. Genomic DNA was also isolated from patient PBMC samples collected prior to DC vaccination, 2 weeks following the 3rd DC vaccination, and at later timepoints if, and when tumor progression occurred. gDNA extraction was accomplished using Qiagen DNeasy Blood extraction Kit (Qiagen, Gaithersburg, MD, USA). For each sample, TCRB CDR3 regions were amplified and sequenced from 2 μg of genomic DNA, or if there was less than 2 μg, from all-available extracted DNA. Amplification and sequencing of TCRB CDR3 regions (defined according to the IMGT collaboration (20)), were carried out using the ImmunoSEQ platform at Adaptive Biotechnologies (Seattle, WA, USA). Briefly, a multiplexed PCR method was employed using a mixture of 60 forward primers specific to TCR Vβ gene segments and 13 reverse primers specific to TCR Jβ gene segments. Reads of 87 bp were obtained using the Illumina HiSeq System. Raw HiSeq sequence data were preprocessed to remove errors in the primary sequence of each read, and to compress the data. A nearest neighbor algorithm was used to collapse the data into unique sequences by merging closely related sequences, to remove both PCR and sequencing errors (21-23). Samples in which >100 unique TCRs were identified within tumors were used in this study. The estimated TIL content was calculated as previously described (23). Briefly, we quantified the amount of DNA input into the assay and converted this to the equivalent total number of nucleated cells, assuming approximately 6.4 pg genomic DNA per diploid cell. ImmunoSEQ then amplifies and sequences the molecules with rearranged TCR beta chains. Because the ImmunoSEQ assay includes analysis of DJ rearrangements, most T cells produce two sequences, one of which is a complete VDJ rearrangement and the other either a VDJ or an incomplete DJ rearrangement. To estimate the number of starting templates with TCR® rearrangements that were in the sample, the average number of sequence reads for each starting template was measured. Synthetic control templates are also spiked at limiting dilution into each sample such that each template may be present at most as a single copy. One then can compute the average number of reads for each sequenced spiked synthetic template. The total number of T cells sequenced is then derived as the total number of sequencing reads divided by this computed average reads per template (fold-coverage), further divided by two, because there are two alleles per T cell. The total fraction of T cells in the mixture is this number of T cells divided by the total number of input cells as determined by DNA input quantity.
The overlap of TCR sequences was calculated as previously published (19, 24). Briefly, to assess the similarity of TCRB repertoires between tumor and PBMC samples from each patient, we used the metric of TCRB repertoire overlap. For two samples, a and b, we identified the n TCRB CDR3 sequences present in both samples, along with the sequencing read counts C of each sequence in each sample. TCRB repertoire overlap between samples was defined as the sum of the sequencing reads from shared TCRB sequences divided by the total number of sequencing reads observed in both samples, and ranges from 0 to 1. To transform this overlap metric into a distance metric, we calculated [(1/TCR Overlap)−1]. This distance metric is higher for less-related TCRB repertoires and ranges from 0 to infinity. This calculation roughly corresponds to the number of sequencing reads required (in excess of one) to expect one read from a TCRB rearrangement shared between the two samples.
RESULTS
Baseline TIL density correlates with survival after vaccination
Based on our earlier studies that correlated the clinical benefits of an autologous dendritic cell vaccine with the presence of TILs and the mesenchymal gene expression signature of glioblastoma (7), we hypothesized that the quantitative assessment of pretreatment TIL levels might be able to predict survival outcomes in our patient population without the need for gene expression profiling analysis.
To test this question, we performed next generation sequencing of the rearranged TCRVβ gene repertoire found in 15 patients for whom we had residual fresh-frozen tumor after harvesting for autologous tumor lysate as part of the vaccine preparation (specimen from resection prior to vaccination) and from the same patients if they recurred after DC vaccination and underwent another resection (5 such pairs were available). Supplementary Table S1 outlines the clinical characteristics of these patients.
Next generation TCRVβ gene sequencing was also performed on each patient’s PBMC obtained at the same time points (pre/post DC vaccination and at tumor progression) so that we had matched TCRVβ sequencing data from each patient’s tumor(s) and PBMC. Quantitative assessment of the tumor infiltrating T-cell content in the pre-vaccine specimens from all 15 patients was performed. Supplementary Table S2 lists the resulting values for TIL content, clonality, and TCR overlap derived from TCR sequencing analysis. As a control, multiplex immunofluorescent staining for T cells was performed (Supplementary Fig. S1). This staining confirmed that the vast majority of T cells were found within the tumor parenchyma itself, and not contaminants within the tumor blood vessels. We then correlated the estimated TIL content with TTP and OS in our patient population. The estimated TIL count of glioblastomas, prior to DC vaccination, was the most significant predictor of outcome in our patient population. Cox regression analyses demonstrated that higher estimated TIL content in the pretreatment tumor was a strong predictor of longer TTP (HR=0.252, 95% CI=0.066-0.965, p=0.0442) and OS (HR=0.277, 95% CI=0.078–0.978, p=0.0461, Supplementary Table S3) in GBM patients who received DC vaccination.
In order to assess whether the prognostic role of TILs may be independent of DC vaccination, we performed TCR sequencing on an additional six GBM patients who did not receive immunotherapy (3 newly diagnosed and 3 recurrent GBM samples). To account for spatial TIL heterogeneity within the tumors, we also collected three different pieces of tumor for genomic DNA analysis. In these patients who did not receive immunotherapy, the estimated TIL content did not significantly correlate with TTP (HR=1.258; p=0.835) nor OS (HR=1.260; p=0.837; Supplementary Figure 2), suggesting that the prognostic benefit of TIL is specifically relevant for patients who subsequently go on to receive immunotherapy.
We also dichotomized patients into high and low estimated TIL content groups, based on the top quartile versus the lower three quartiles. Patients in the top TIL quartile group had significantly longer TTP and OS compared to the lower three TIL quartiles (p = 0.0088 and p = 0.0038) (Fig. 1A and B). No patients with estimated TIL content below the top quartile survived past 30 months. All long-term survivors (>5 year OS) in this patient population had an estimated TIL content in the top quartile. To corroborate the findings from our NGS-based estimated TIL content, we performed quantitative immunofluorescent IHC on representative tumor samples for which we had FFPE sections. These data were consistent with the estimated TIL content derived from TCR sequencing (Fig. 1C).
Figure 1. Elevated estimated TIL content in pretreatment tumor effectively segregates the survival of glioblastoma patients after DC vaccination.
The TTP and OS was compared between patients whose estimated TIL content was dichotomized into the top quartile (Red) or the lower 3 quartiles (orange). The estimated TIL content was dichotomized at 0.787. A log-rank test was performed to compare the TTP (A) and the OS (B). TTP; **p<0.0087. OS; **p<0.0038. (C) Representative immunofluorescent staining of CD8 TIL from patients in the top quartile (17-232, 1-708) and lower 3 quartiles (21-828, 6-815). Quantitation of CD8 was performed using inForm image analysis software.
TCR overlap between tumor and blood are indicative of responses to vaccination
Although we showed that pretreatment TIL content could predict the clinical outcome for patients receiving DC vaccination, there remained a group of patient outliers who did not possess high pretreatment TIL content but may still have potentially benefitted from DC vaccination, given the survival differences with similar patients historically. In this sub-group of patients, we theorized that although the initial estimated TIL content might be low, the DC vaccination may have induced tumor-specific T-cell responses. By using the TCR sequences obtained from relapsed tumor (post-DC vaccination) we could potentially monitor the maintenance, gain, or loss of specific TCRs over the course of immunotherapy.
To test this hypothesis, we used NGS in order to sequence the CDR3 region of rearranged TCRβ genes from five patients for whom we had obtained both primary and relapsed tumor samples, and peripheral blood samples obtained before treatment, after treatment, and following tumor progression. The frequency of the top 25 clones in tumor biopsies at pretreatment and the frequency of these clones in pretreatment and post-treatment blood samples is outlined in Supplementary Fig. S3. We found little overlap between the TCRVβ sequences identified in different patients.
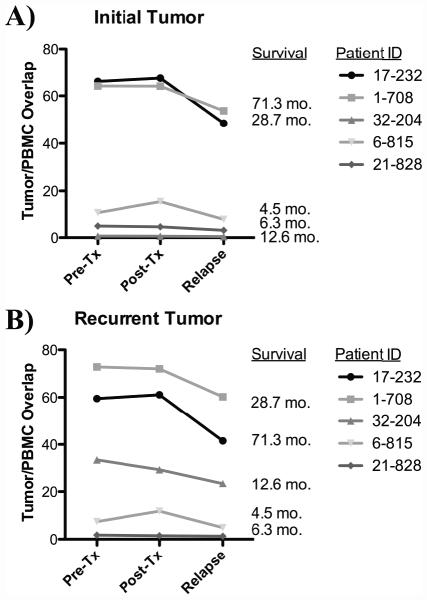
We then evaluated whether the subset of unique TCR sequences found within each patient’s tumor tissue could be detected in the peripheral blood. We defined this TCR overlap as the percentage of unique TCRβ chain sequences identified within each tumor that was also present within the peripheral blood. By restricting our analysis just to the discrete TCR repertoire present within the tumor, we could then just compare the overlap of these unique TCR sequences with those in the systemic circulation (blood), and correlated this overlap index with survival. The two patients (1-708 and 17-232) who displayed the highest overlap between pretreatment tumor and pretreatment peripheral blood TCR sequences (Figures 2A and 2B) had the longest overall survival (28.7 months and 71.3 months, respectively). Furthermore, the high degree of TCR overlap persisted over time, including after DC vaccination, to when the tumor eventually relapsed and another surgical resection was warranted. Of note, patient 17-232 did have a tumor recurrence, but the actual cause of death was from a deep vein thrombosis and the associated pulmonary embolism, and not from tumor progression. Thus, these data suggest to us that this patient was continuing to mount an antitumor T-cell response, even at recurrence.
Figure 2: TCR Overlap between pretreatment and post-treatment peripheral blood and/or tumors are associated with survival. /B> Correlation between pretreatment peripheral blood TCR sequences (x-axis) and post-treatment peripheral blood TCR sequences (y-axis) overlaid with presence of T-cells within the TIL populations.
: TCRs present only in peripheral blood samples; Blue: TCRs present only in pretreatment TILs and peripheral blood; Green: TCRs present only in post-treatment TILs; Orange: persistent TCRs present in both sets of TILs and peripheral blood. (A-B, High-High TCR Overlap Group) (C, Low-High TCR Overlap Group) (D-E, Low-Low TCR Overlap Group). A) Patient 1-708 (overall survival= 28.7 months); B) Patient 17-232 (overall survival= 71.3 months); C) Patient 32-204 (overall survival= 12.6 months); D) Patient 6-815 (overall survival= 6.3 months); E) Patient 21-828 (overall survival= 4.5 months).
In contrast, patients 6-815 and 21-828 exhibited short survival (4.5 and 6.3 months, respectively), and demonstrated the smallest TCR overlap between pretreatment tumor and the pretreatment peripheral blood. This minimal degree of TCR overlap remained unchanged, even after DC vaccination and upon tumor relapse (Fig. 2D and E). Perhaps of most interest, patient 32-204 had low pretreatment tumor to pretreatment peripheral blood TCR overlap, but subsequently developed a 200-fold increase in TCR overlap between post-treatment tumor and post-treatment peripheral blood after DC vaccination (Fig. 2C). This patient exhibited an intermediate survival of 12.6 months, falling between the patients (17-232 and 1-708) who had persistently high TCR overlap with extended survival, and patients (6-815 and 21-828) who had persistently low TCR overlap and shorter survival. The pattern of this overlap between the initial tumor (prior to DC vaccination) and circulating T cells, and the recurrent tumor (after DC vaccination), is outlined in Fig. 3, and suggests that the overlap of the TCR repertoire between the tumor and blood may be a sensitive method to evaluate anti-tumor T cell responses after active DC vaccination.
Figure 3. Dynamic monitoring of tumor-infiltrating TCR sequences in glioblastoma patients treated with autologous tumor lysate-pulsed DC vaccination.
/B> Assessment of the TCR overlap between the initial tumor (prior to DC vaccination) and PBMC taken at time points pre-vaccine, post-vaccine, and at relapse (B) Assessment of the TCR overlap between the recurrent tumor (after DC vaccination) and PBMC taken at time points pre-vaccine, post-vaccine, and at relapse.
DISCUSSION
Although therapies targeting defined epitopes are conducive to well-established immune monitoring assays, many of the newest agents to receive FDA approval are not targeted to a specific antigen and are patient-specific. Since it is unclear what antigen/targets each patient’s antitumor T cell response will be directed against, new assays that can track T-cell populations without the prior knowledge of their specificity will become critical. New clinical trials should incorporate pre-surgical/surgical designs that include the collection of tumor tissue for laboratory analyses (if possible before and after immunotherapy). Herein, we describe a new approach utilizing multiple analytes from a single TCR deep-sequencing assay to monitor and predict response to DC vaccination. We determined that the estimated TIL content of pretreatment tumor directly correlated with survival and was predictive of which patients would benefit from the therapy. The fraction of TCR sequences within tumors that can also be found systemically also correlated with clinical outcome and provided insight into the determinants of successful, antigen-specific, antitumor immune response.
We evaluated this novel technology on specimens from patients who had previously been enrolled in clinical trials of autologous tumor lysate dendritic cell vaccines for the treatment of glioblastoma. As is often the case when working with such material, our study was limited in the number of patients with fully available sets of blood and tumor specimens. Part of our study required comparing initial tumors to recurrent tumors from the same patient and as such the number of initial-recurrent tumor pairs was even more limited. Our patient cohort was heterogeneous, and included patients who were both newly diagnosed and had recurrent GBM and somewhat varied by molecular status (although none bore an isocitrate dehydrogenase mutation). However, even with the small number and heterogeneous group of patients, we were able to successfully demonstrate that TCR sequencing data can help distinguish patients that will benefit and are benefitting from immunotherapy with the DC vaccine.
We showed that an increased estimated TIL content prior to therapy was associated with prolonged TTP and OS. All very long-term survivors in this trial had estimated TIL content in the top quartile. The correlations between estimated TIL content and TTP and OS argues that estimated TIL content in the initial tumor specimen may be used to select patients for DC vaccination and may be applicable to other immunotherapies as well. In contrast, TIL content was not associated with survival in patients who were not treated with immunotherapy. Thus, our data suggests that the use of TCR sequencing to predict such survival indices may be directly relevant only for patients undergoing immunotherapy treatments such as DC Vax. We also did not find a direct relationship between increased TCR clonality and survival in these patients, suggesting that a diverse, polyclonal T-cell response may be critical for effective antitumor immune responses.
Although TIL content was the strongest predictor of long-term survival after DC vaccination, a subset of patients did not possess high-pretreatment TIL content, but still appeared to benefit. As DC vaccination is administered systemically, we compared tumor to peripheral blood TCR overlap over the course of DC vaccination therapy to monitor the immune response and predict clinical outcome. Questions remain regarding whether the systemic immune response reflects that of the tumor environment, but we believe that overlap between the TCRs found within the peripheral blood compared to the tumor would indicate an immune environment supportive of an antitumor immune response and would be important for survival in our cohort of patients.
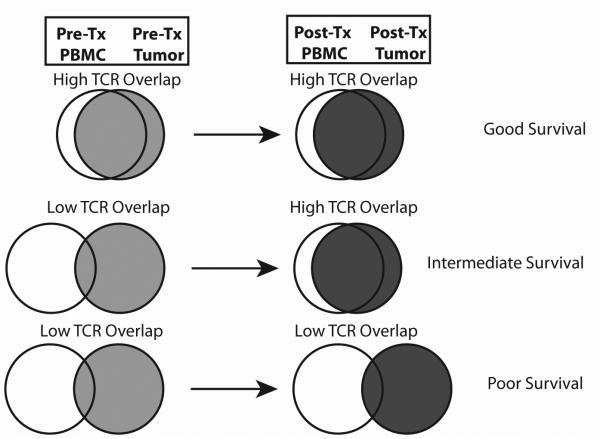
Analysis of the data suggested that DC vaccination patients could be categorized into 3 response groups (Fig. 4): 1) High-high overlap 2) low-high overlap, and 3) low-low overlap responders. High-high overlap responders possessed an elevated overlap between the TCR repertoire in the tumor and peripheral blood before DC vaccination therapy. At recurrence, these patients continued to demonstrate this high degree of overlap. The maintenance or antitumor effect of these co-occurring T cells may be boosted by the DC vaccination, which may have otherwise been rendered anergic by the immunosuppressive effects of the tumor. Low-low overlap responders were those who demonstrated a low degree of TCR overlap between the tumor and peripheral blood before and after DC vaccination, suggesting both a poor initial immune response and a poor response to therapy. In these patients the course of the disease remained unaltered and did not demonstrate extended survival. While purely speculative, patients in this group may have fewer somatic mutations, may be less antigenic, or more immunosuppressive tumors. Low-high overlap responders had an initially low degree of TCR overlap between tumor and peripheral blood, but the overlap increased after DC vaccination. Such a situation might suggest that the DC vaccination actually generated the antitumor immune response de novo, where there was none to begin with. This group contrasts with the high-high overlap group, which may have only needed a boost from the intradermal vaccine to re-invigorate the pre-existing anti-tumor T cell response. This speculative model will need to be tested in prospectively designed clinical trials for validation.
Figure 4. Development of a TCR overlap model that demonstates how to track the evolution of tumor-infiltrating TCRs after DC vaccination in glioblastoma patients.
The quantity and diversity of TCR transcripts are determined from glioblastoma tumor samples and peripheral blood. The TCR overlap was defined as the percentage of unique TCR beta chain sequences identified in each tumor that were also identified in the peripheral blood (before treatment, after treatment, and at relapse). We formulated a model depicted in the Venn diagrams. (Top) Patients who have a high TCR overlap with the blood from the initial tumor and relapsed tumor are predicted to have long survival outcomes. (Middle) Patients whose initial tumor sample have low TCR overlap with the blood, but develop a high TCR overlap in the relapsed tumor after DC vaccination, are predicted to have intermediate survival. (Bottom) Patients who have a low TCR overlap with both the initial and relapsed tumor and blood are predicted to have short survival outcomes.
In conclusion, estimated TIL content may be used as a predictor of clinical response and way to select patients for DC vaccination therapy. Furthermore, monitoring of the systemic and tumor environment TCR overlap is a promising approach to monitor immune response to DC vaccination. Ongoing studies in the lab are attempting to identify unique TCR clones from paired alpha and beta chains of ex vivo-expanded TIL, and coordinating this effort with tumor cell sequencing and additional peptide-synthesis and screening in order to aid antigen discovery. However, next-generation sequencing of TCRs can provide an accurate estimate of the redundancy of the TIL repertoire and will give us specific and clinically relevant information without needing to identify these targets.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by NIH/NCI grants R21-CA186004 and R01CA154256 (RMP), RO1 CA125244 (LML), R25 NS079198 (RGE, RMP and LML), NIH/NCATS UCLA CTSI Grant Number UL1TR000124 (RMP, LML, MH), the Musella Foundation for Brain Tumor Research (RMP), and Adaptive Biotechnologies (EY, CS, HSR).
Funding
This work was supported in part by NIH/NCI grants R21-CA186004 and R01CA154256 (RMP), RO1 CA125244 (LML), R25 NS079198 (RMP, RGE, and LML), NIH/NCATS UCLA CTSI Grant Number UL1TR000124 (RMP, LML, MH), the Musella Foundation for Brain Tumor Research (RMP), and Adaptive Biotechnologies (EY, CS, HSR).
Abbreviations
- TCR
T cell receptor
- CNS
central nervous system
- DC
dendritic cell
- qPCR
quantitative polymerase chain reaction
- OS
overall survival
- TTP
time to tumor progression
- TIL
tumor infiltrating lymphocyte
- HR
hazard ratio
- GBM
glioblastoma
Footnotes
Conflict of Interest
No potential conflicts of interest to be disclosed.
REFERENCES
- 1.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro Oncol. 2013;15(Suppl 2):ii1–56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp RMW, van den Bent MJ, Weller M, Fisher B, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Okada HKP, Ueda R, Hoji A, Kohanbash G, et al. Induction of CD8+ T-cell responses against novel glioma-associated antigen peptides and clinical activity by vaccinations with {alpha}-type 1 polarized dendritic cells and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in patients with recurrent malignant glioma. J Clin Oncol. 2011;29:330–6. doi: 10.1200/JCO.2010.30.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phuphanich S, Wheeler CJ, Rudnick JD, Mazer M, Wang H, Nuno MA, et al. Phase I trial of a multi-epitope-pulsed dendritic cell vaccine for patients with newly diagnosed glioblastoma. Cancer Immunol Immunother. 2012 doi: 10.1007/s00262-012-1319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prins RM, Wang X, Soto H, Young E, Lisiero DN, Fong B, et al. Comparison of glioma-associated antigen peptide-loaded versus autologous tumor lysate-loaded dendritic cell vaccination in malignant glioma patients. J Immunother. 2013;36:152–7. doi: 10.1097/CJI.0b013e3182811ae4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampson JH, Archer GE, Mitchell DA, Heimberger AB, Herndon JE, 2nd, Lally-Goss D, et al. An epidermal growth factor receptor variant III-targeted vaccine is safe and immunogenic in patients with glioblastoma multiforme. Mol Cancer Ther. 2009;8:2773–9. doi: 10.1158/1535-7163.MCT-09-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prins RMSH, Konkankit V, Odesa SK, Eskin A, et al. Gene expression profile correlates with T-cell infiltration and relative survival in glioblastoma patients vaccinated with dendritic cell immunotherapy. Clin Cancer Res. >2011;17:1603–1615. doi: 10.1158/1078-0432.CCR-10-2563. 17:: 1603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kvistborg P, Shu CJ, Heemskerk B, Fankhauser M, Thrue CA, Toebes M, et al. TIL therapy broadens the tumor-reactive CD8(+) T cell compartment in melanoma patients. Oncoimmunology. 2012;1:409–18. doi: 10.4161/onci.18851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013 doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everson RJ, Wang X, Safaee M, Scharnwber R, Lisiero DN, Soto H, Liau L, Prins RM. Cytokine Responsiveness of CD8+ T cells is a reproducible biomarker for the clinical efficacy of dendritic cell vaccination in glioblastoma patients. 2014:2–11. doi: 10.1186/2051-1426-2-10. R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong B, Jin R, Wang X, Safaee M, Lisiero DN, Yang I, et al. Monitoring of regulatory T cell frequencies and expression of CTLA-4 on T cells, before and after DC vaccination, can predict survival in GBM patients. PloS one. 2012;7:e32614. doi: 10.1371/journal.pone.0032614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 13.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer. 2011;105:93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang I, Tihan T, Han SJ, Wrensch MR, Wiencke J, Sughrue ME, et al. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2010;17:1381–5. doi: 10.1016/j.jocn.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emerson RO, Sherwood AM, Rieder MJ, Guenthoer J, Williamson DW, Carlson CS, et al. High-throughput sequencing of T cell receptors reveals a homogeneous repertoire of tumor-infiltrating lymphocytes in ovarian cancer. J Pathol. 2013 doi: 10.1002/path.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robert L, Tsoi J, Wang X, Emerson R, Homet B, Chodon T, et al. CTLA4 blockade broadens the peripheral T-cell receptor repertoire. Clin Cancer Res. 2014;20:2424–32. doi: 10.1158/1078-0432.CCR-13-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robert L, Harview C, Emerson R, Wang X, Mok S, Homet B, et al. Distinct immunological mechanisms of CTLA-4 and PD-1 blockade revealed by analyzing TCR usage in blood lymphocytes. Oncoimmunology. 2014;3:e29244. doi: 10.4161/onci.29244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajor DL, Xu X, Torigian DA, Mick R, Garcia LR, Richman LP, et al. Immune activation and a 9-year ongoing complete remission following CD40 antibody therapy and metastasectomy in a patient with metastatic melanoma. Cancer immunology research. 2014;2:1051–8. doi: 10.1158/2326-6066.CIR-14-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emerson RO, Sherwood AM, Rieder MJ, Guenthoer J, Williamson DW, Carlson CS, et al. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. The Journal of pathology. 2013;231:433–40. doi: 10.1002/path.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yousfi Monod M, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004;20(Suppl 1):i379–85. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
- 21.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 22.Robins HS, Ericson NG, Guenthoer J, O'Briant KC, Tewari M, Drescher CW, et al. Digital genomic quantification of tumor-infiltrating lymphocytes. Science translational medicine. 2013;5:214ra169. doi: 10.1126/scitranslmed.3007247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu D, Emerson RO, Sherwood A, Loh ML, Angiolillo A, Howie B, et al. Detection of minimal residual disease in B lymphoblastic leukemia by high-throughput sequencing of IGH. Clin Cancer Res. 2014;20:4540–8. doi: 10.1158/1078-0432.CCR-13-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cha E, Klinger M, Hou Y, Cummings C, Ribas A, Faham M, et al. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Science translational medicine. 2014;6:238ra70. doi: 10.1126/scitranslmed.3008211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.