Abstract
Current therapies for high-grade gliomas extend survival only modestly. The glioma microenvironment, including glioma-associated microglia/macrophages (GAMs), is a potential therapeutic target. The microglia/macrophage cytokine CSF1 and its receptor CSF1R are overexpressed in human high-grade gliomas. To determine if the other known CSF1R ligand IL-34 is expressed in gliomas, we examined expression array data of human high-grade gliomas and performed RT-PCR on glioblastoma sphere-forming cell lines (GSCs). Expression microarray analyses indicated that CSF1, but not IL-34, is frequently overexpressed in human tumors. We found that while GSCs did express CSF1, most GSC lines did not express detectable levels of IL-34 mRNA. We therefore studied the impact of modulating CSF1 levels on gliomagenesis in the context of the GFAP-V12Ha-ras-IRESLacZ (Ras*) model. Csf1 deficiency deterred glioma formation in the Ras* model while CSF1 transgenic overexpression decreased the survival of Ras* mice and promoted the formation of high-grade gliomas. Conversely, CSF1 overexpression increased GAM density, but did not impact GAM polarization state. Regardless of CSF1 expression status, most GAMs were negative for the M2 polarization markers ARG1 and CD206; when present, ARG1+ and CD206+ cells were found in regions of peripheral immune cell invasion. Therefore, our findings indicate that CSF1 signaling is oncogenic during gliomagenesis through a mechanism distinct from modulating GAM polarization status.
Keywords: CSF1, IL-34, glioma, macrophages, microglia
INTRODUCTION
High-grade gliomas are associated with a grim prognosis despite treatment with surgery, temozolomide, radiation therapy and anti-angiogenic therapy (1). Cells in the tumor microenvironment such as glioma-associated microglia/macrophages (GAMs) are being investigated as targets for therapy (2). Microglia are the resident macrophages of the central nervous system (CNS) and comprise the primary line of innate immune response in the brain and spinal cord (3). Gliomas are associated with microglial/macrophage infiltration (4). Increased expression of microglia/macrophage related genes has been associated with the shortest survival outcome in adult human glioblastoma (grade IV glioma, GBM) (5). However, in contrast, another study found a positive correlation between longer survival in high-grade astrocytomas and the expression of genes associated with microglia/macrophages (6). Therefore, the role of GAMs in glioma formation/progression remains unclear.
CSF1 is a cytokine for microglia and macrophages (7). Csf1 was previously identified as a candidate glioma oncogene in Sleeping Beauty-induced high-grade astrocytomas (8). CSF1 and its receptor CSF1R are over-expressed in human GBMs (8–10), and in one study CSF1 expression levels were found to be correlated with glioma grade (9). In human gliomas, CSF1 was found to be expressed by GFAP+ cells (8) and cultured human glioblastoma sphere-forming cells (GCSs) have also been shown to express CSF1 by ELISA (11); indicating that CSF1 is produced by the tumor cells themselves. Studies using cell lines or established tumors have indicated that CSF1/CSF1R signaling functions to regulate monocyte/microglial migration/invasion or to influence GAM polarization state (11–15). However, the role of CSF1 signaling during de novo glioma formation within an unperturbed CNS has not been previously investigated.
Activated microglia/macrophages have been categorized as either M1 polarized (‘classically activated’) which express cytokines and enzymes that are pro-inflammatory, or M2 polarized (‘alternatively activated’) which express cytokines and enzymes that have anti-inflammatory and pro-tumor actions (16). Some studies report human GBMs to be associated with primarily M2 polarized GAMs (9,17). A study also found that GAMs in intracranial allografts of murine GL261 glioma cells express markers of M2 polarization by immunohistochemistry (IHC) (18). However, although distinctions in microglia/macrophage M1 vs. M2 macrophage populations have been made in the context of gliomas and other CNS pathologies, it is also possible that polarization states may not be entirely dichotomous (16,19). In support of this for GAMs, studies analyzing the mRNA expression profile of GAMs isolated from GBMs or from intracranial allografts of GL261 glioma cells found GAMs not to be predominantly M1 or M2 polarized (20,21). Thus, further studies of GAM phenotypes and the signaling molecules regulating polarization states are essential to better understand the function(s) of this cell population.
In this study we utilized mining of expression array datasets from high-grade gliomas and RT-PCR analysis of expression of CSF1R ligands in human GSC lines to determine that CSF1 is more commonly over-expressed than IL-34 in human high-grade gliomas. We therefore utilized an in vivo autochthonous mouse model to study the role of CSF1 signaling on glioma formation and GAMs within an unperturbed CNS. Our studies support an oncogenic role for CSF1 signaling during gliomagenesis independent from regulation of GAM polarization state.
MATERIALS AND METHODS
See supplementary materials and methods for additional details.
Cell culture
GSC lines (22), human cortex fetal neural stem cells (NSCs, a gift from Dr. Clive Svendsen) (23), and normal human astrocytes (NHAs) (24) have been previously described. The U87, A172 and T98G glioma cell lines were maintained in growth medium (DMEM + 10% fetal bovine serum + antibiotics) at 37°C with 5% CO2, were purchased from the American Type Culture Collection and used within 6 months of receipt.
Mice
Work was reviewed and approved by the University of Wisconsin-Madison Institutional Animal Care and Use Committee. The Ras* transgene and Csf1op allele were found to be linked. Therefore, to generate Ras*; Csf1op/op and Ras*; Csf1op/+ mice, a recombinant chromosome harboring both Ras* and Csf1op was generated and then crossed to Csf1op/+ mice to generate experimental Ras*; Csf1op/op (n=49) and control Ras*; Csf1op/+ (n=43) mice, which were aged until moribund or to an endpoint of 270 days. Csf1op/+ mice have been shown to be phenotypically identical to wild-type Csf1+/+ mice (25). GFAP-tTA mice (26) express the “TET-OFF” version of the tetracycline transactivator (tTA) in the GFAP compartment. CSF1 over-expressing (CSF1 OE mice) were generated by crossing TRE-CSF1 (secreted form)-IRES-EGFP mice (referred to hereafter as TRE-CSF1 mice) to GFAP-tTA mice (27). GFAP-V12Ha-ras-IRESLacZ (referred to hereafter as Ras*) mice (28) were generously provided by the Guha laboratory. Ras* mice were crossed to TRE-CSF1; GFAP-tTA mice to obtain Ras*; TRE-CSF1; GFAP-tTA mice (experimental cohort, referred to as Ras*; CSF1 OE mice, n = 35) as well as Ras* Control (n=40) mice. TRE-CSF1 only mice and GFAP-tTA only mice exhibited no difference in Csf1, Iba1, or Cd11b levels in the brain (27), therefore the Ras* Control cohort consisted of a combination of Ras*; TRE-CSF1 mice and Ras*; GFAP-tTA mice. Ras*; CSF1 OE mice and Ras* Control mice were aged until moribund or to a pre-determined endpoint of 120 days. Ras*; Ccr2RFP/+; CSF1 OE mice were generated by first crossing Ccr2RFP/RFP mice (29) to TRE-CSF1; GFAP-tTA mice and then crossing the resulting Ccr2RFP/+; TRE-CSF1; GFAP-tTA mice to Ras* mice. The resulting Ras*; Ccr2RFP/+; CSF1 OE mice were then aged until moribund.
Tissue isolation
To monitor GFP or RFP expression, some mice were perfused with phosphate buffered saline (PBS) followed by 4% paraformaldehyde (PFA) solution in 100mM phosphate buffer (pH 7.4) under pentobarbital anesthesia. Brains were fixed overnight in 4% PFA, cryoprotected by submerging in sucrose solution, embedded in OCT and cryosectioned. For other mice, following CO2 asphyxiation, tissues were fixed overnight in 10% neutral buffered formalin at 4°C prior to paraffin embedding.
IHC/Immunofluorescence (IF)
IHC or IF was performed following standard protocols as previously described (27).
RNA isolation and RT-PCR
RNA was isolated from frozen cell pellets by TRIZOL (Invitrogen) followed by generation of first strand cDNA and RT-PCR as described previously (27). RT-PCR primer sequences are listed in Supplemental Materials and Methods.
Pathological analyses
World Health Organization grading criteria was employed which defines low-grade astrocytomas as lacking significant mitotic activity, necrosis or endothelial proliferation (30). Tumors with any of these features were classified as high-grade. Brains with either no significant pathology or pathologies other than gliomas such as increased glial cellularity, ischemia, and inflammation were classified as “No tumor”.
Microglial imaging
For determining microglial density, gliomas were categorized as small, medium or large if they comprised <50%, about 50%, or >50% of a brain hemisphere, respectively. 8, 16 and 24 random field confocal images were acquired for small, medium and large gliomas, respectively, and the average % area occupied by IBA1 staining per field was determined using ImageJ software by a blinded observer. Normal brains for comparison were obtained from mice similar in age to the median age of the mice with gliomas that were examined. For M1/M2 polarization studies, confocal images were acquired at a low magnification (20X objective lens) and the presence or absence of co-localized staining was confirmed by acquiring confocal images at high magnification (60X objective lens).
Sequencing
DNA from gliomas and normal brain sections identified in FFPE samples was extracted using the QIAampDNA FFPE Tissue Kit (Qiagen). Following PCR amplification with the Csf1op sequencing amplification primers (Supplemental Materials and Methods), purified PCR products were then subjected to Sanger sequencing using the reverse Csf1op amplification primer to detect the Csf1op mutation.
Statistical analysis
Statistical analysis was performed as indicated using Prism software (GraphPad). Error bars represent standard error.
RESULTS
CSF1 and not IL-34 is the predominantly expressed ligand of CSF1R in human high-grade gliomas
Although CSF1 and IL-34 are both CSF1R ligands, they have been shown to activate biologically distinct signaling in vitro (31,32). Therefore, to identify the relevant CSF1R ligand(s) in human high-grade gliomas, mRNA expression array data at www.oncomine.org was mined. Dataset analysis revealed that CSF1 expression is up-regulated in gliomas compared to normal brain in most studies of high-grade human gliomas (GBM and Anaplastic Astrocytoma (AA)) while IL-34 expression is down-regulated or unchanged (Table 1).
Table 1. CSF1 and IL-34 expression changes in human GBM and AA compared to normal brain in datasets in www.oncomine.org.
CSF1 is up-regulated significantly in GBMs or AAs in 4 of 6 datasets and IL-34 is down-regulated significantly in 3 of 4 datasets. White rows and grey rows denote statistically significant and non-significant differences in expression, respectively.
| CSF1 | |||
|---|---|---|---|
| Dataset | Comparison | p value | Fold change |
| Sun et al | GBM vs. normal | 7.52E-13 | 2.4 |
| Sun et al | AA vs. normal | 1.46E-05 | 2.758 |
| TCGA | Brain GBM vs. normal | 4.40E-05 | 1.38 |
| Murat et al | GBM vs. normal | 0.043 | 1.307 |
| Lee et al | GBM vs. normal | 0.126 | 2.021 |
| Shai et al | GBM vs. normal | 0.226 | 1.04 |
| IL-34 | |||
| Dataset | Comparison | p value | Fold change |
| Sun et al | GBM vs. normal | 3.75E-17 | −2.308 |
| Sun et al | AA vs. normal | 2.48E-04 | −1.998 |
| Murat et al | GBM vs. normal | 0.05 | −1.213 |
| Lee et al | GBM vs. normal | 0.108 | −1.55 |
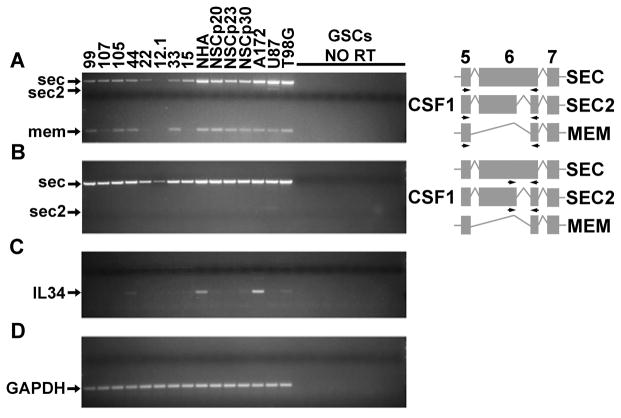
CSF1 mRNA undergoes alternative splicing resulting in the expression of CSF1 isoforms that are primarily secreted or membrane bound. Membrane and secreted CSF1 can have biologically distinct activities as determined by in vivo studies in mice (33,34). Therefore, RT-PCR was used to examine the expression of CSF1 splice variants and IL-34 in human GSCs, as well as human glioma cell lines. All of the GSC and glioma cell lines tested express the mRNA encoding a secreted CSF1 isoform (SEC) (Figure 1A, B). Several GSCs also express the mRNA encoding the membrane-bound CSF1 isoform (Figure 1A). A second secreted CSF1 isoform (SEC 2) that is annotated in the human genome but not in the mouse genome was weakly detected only in U87 cells (Figure 1A, B). IL-34 expression, however, was not detected in most of the GSC lines queried, and among the glioma cell lines was only robustly detectable in the A172 cell line (Figure 1C). Therefore, mining of GBM and AA expression array datasets revealed that CSF1 but not IL-34 is up-regulated in human high-grade gliomas, a result which is supported by RT-PCR analyses of GSCs and glioma cells lines.
Figure 1.
RT-PCR indicates that CSF1 is the primary CSF1R ligand expressed by human GSC lines (99, 107, 105, 44, 22, 12.1, 33, 15) and glioma cell lines (A172, U87, T98G). Expression was also examined in immortalized NHAs and three separate passages (p20, p23, p30) of human NSCs. Samples omitting reverse transcriptase (NO RT) are shown to demonstrate no amplification from genomic DNA from each of the 8 GSC lines. For CSF1 RT-PCRs, the three alternative splicing events in exon 6 leading to the formation of transcripts encoding two different secreted (SEC and SEC2) and membrane-bound (MEM) isoforms are shown, along with the location of the primer pairs utilized. A) RT-PCR with a primer pair that detects all three CSF1 splice variants. B) RT-PCR with a primer pair that specifically detects both secreted CSF1 isoforms. C) RT-PCR for IL-34. D) RT-PCR for GAPDH demonstrates the presence of cDNA for all RT samples.
CSF1 deficiency deters glioma formation in vivo
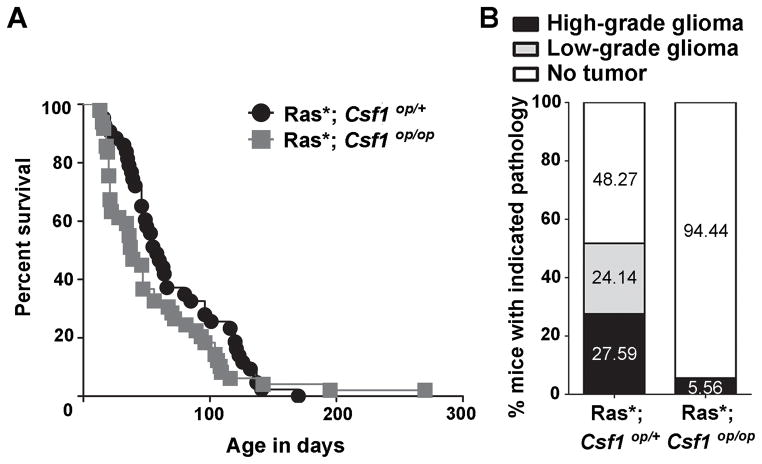
To determine if CSF1 is required for glioma formation in vivo, mice deficient for functional CSF1 (Csf1op/op) (35), were crossed to GFAP-V12Ha-ras-IRESLacZ (Ras*) mice. In Ras* transgenic mice, expression of constitutively active Ras is driven by the GFAP promoter, which results in the formation of low- and high-grade astrocytomas (28). An experimental cohort of Ras*; Csf1op/op mice and a littermate control cohort of Ras*; Csf1op/+ mice were generated and no significant difference in survival between the two cohorts (p = 0.14) was observed (Figure 2A). Brains of mice from both Ras*; Csf1op/op and Ras*; Csf1op/+ genotypes were examined for pathology (Figure 2B, Supplemental Figure S1A). Despite expressing the Ras* transgene (Supplemental Figure S1B), Ras*; Csf1op/op mice displayed a significant reduction in glioma formation, with only one glioma detected in the brains analyzed (5.56%). Despite having pathological features that classified it as high-grade, the tumor was of limited volume (Supplemental Figure S1A). GAMs were detected in the glioma, however they appeared to be reduced in density as compared to high-grade gliomas from the Ras*; Csf1op/+ cohort (Supplemental Figure S1C). Furthermore, the mutation causing the Csf1op allele was maintained in the tumor (Supplemental Figure S1D), indicating that the tumor did not form due to reversion of the frame-shift mutation that causes the allele. In contrast, gliomas were detected in 51.73% of the control Ras*; Csf1op/+ brains analyzed. Therefore, CSF1 deficiency does not impact time to a moribund state in Ras* mice, but significantly deters glioma formation in vivo.
Figure 2.
Csf1 loss deters glioma formation in the Ras* glioma model in vivo. A) Kaplan-Meier survival curve depicting time to a moribund state of Ras*; Csf1op/op and littermate control Ras*; Csf1op/+ mice. Log-rank analysis reveals no significant difference in time to a moribund state (p = 0.14) B) Stacked bars representing the percentage of mice harboring the indicated brain pathologies for analyzed Ras*; Csf1op/op (n=18) and littermate control Ras*; Csf1op/+ (n=29) cohorts. Chi-square analysis reveals a significant difference (p = 0.0046) between the two groups.
Secreted CSF1 over-expression promotes the formation of high-grade gliomas with increased GAM density in vivo
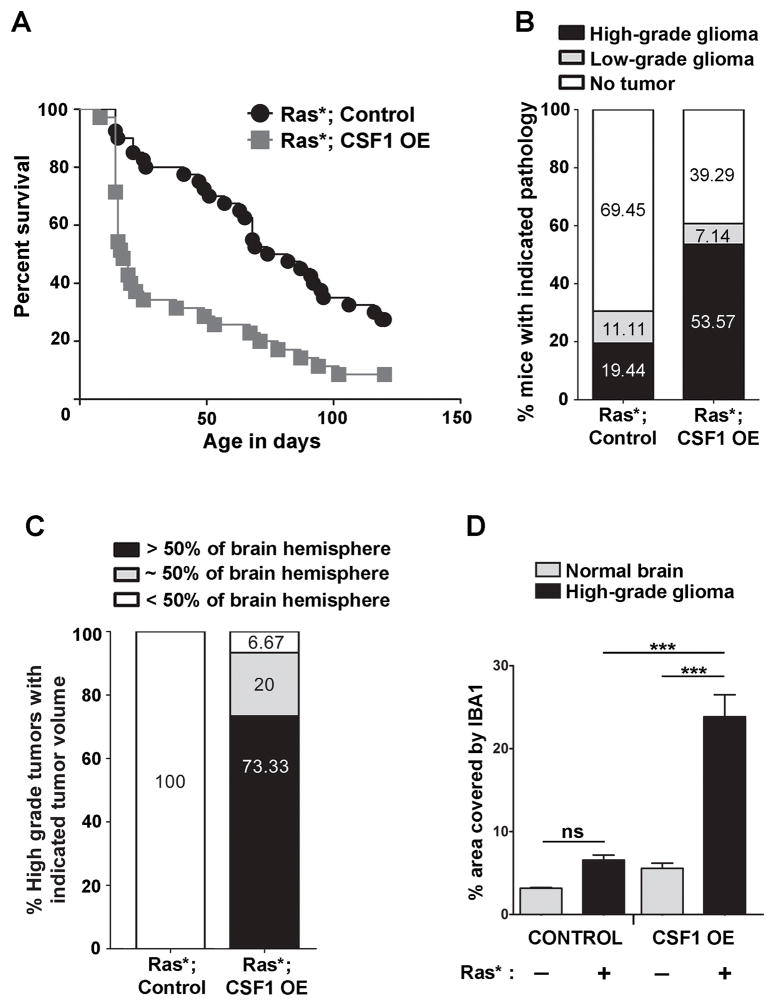
Since CSF1 over-expression has been associated with high-grade gliomas (8,9,11) and expression of the mRNA encoding the secreted isoform was observed in all GSC and glioma cell lines, the impact of over-expression of secreted CSF1 upon glioma formation in vivo was examined. TRE-CSF1 mice were previously generated, which when crossed with GFAP-tTA mice results in CSF1 over-expression in the CNS (27). TRE-CSF1; GFAP-tTA mice will hereafter be referred to as CSF1 over-expressing mice (CSF1 OE mice). By crossing CSF1 OE mice with Ras* mice, Ras*; CSF1 OE mice and littermate Ras* Control mice were generated. Ras*; CSF1 OE mice had a significant reduction in time to a moribund state compared to the Ras* Control cohort (p = 0.0002) (Figure 3A). Brains of mice from both Ras*; CSF1 OE and Ras* Control mice were examined for pathology (Figure 3B, Supplemental Figure S2A). Ras*; CSF1 OE mice display significantly increased high-grade glioma formation (53.57%) as compared to Ras* Control mice (19.44%) (p = 0.017). Low-grade gliomas were detected in 7.14% of Ras*; CSF1 OE mice and 11.11% of Ras* Control mice. In addition, the high-grade gliomas in Ras*; CSF1 OE mice were significantly larger than those from the Ras* Control cohort (p <.001) (Figure 3C).
Figure 3.
Secreted CSF1 over-expression promotes the formation of high-grade gliomas with increased GAM density in vivo. A) Kaplan-Meier survival curve depicting time to a moribund state of Ras*; CSF1 OE (grey squares) and littermate Ras* Control (black circles) cohorts. Log-rank analysis reveals a significant difference in time to a moribund state (p = 0.0002). B) Stacked bars representing the percentage of mice harboring the indicated brain pathologies for Ras*; CSF1 OE (n = 28) and Ras* Control (n = 36) mice. Chi-square analysis reveals a significant difference (p = 0.017) between the two groups. C) Stacked bars indicating the percentage of high-grade gliomas from the Ras*; CSF1 OE (n=15) and Ras* Control (n=7) groups with the indicated proportion of tumor area in the brain sections analyzed. Chi-square analysis reveals a significant difference in tumor area between the two groups (p <.001). D) Left- Quantification of microglial density (average % area covered by IBA1 per 40× field) in normal brains (grey bars) vs. GAM density in high-grade gliomas (black bars) from Control mice without or with the Ras* transgene, respectively. Right – Quantification of microglial density in normal brains (grey bars) vs. GAM density in high-grade gliomas (black bars) from CSF1 OE mice without or with the Ras* transgene, respectively. n = 3 for all groups except Ras* Control high-grade, where n = 4. ANOVA followed by Tukey’s post-hoc tests for the indicated comparisons were performed. ***p<0.001, ns = non-significant.
To determine if CSF1 over-expression in Ras*; CSF1 OE gliomas is associated with a concomitant change in GAM density compared to Ras* Control gliomas, immunofluorescence for IBA1 (a pan microglia/macrophage marker) was performed (Supplemental Figure S3B). GAM density as measured by % area covered by IBA1 was significantly increased in high-grade Ras*; CSF1 OE gliomas compared to high-grade gliomas in Ras* Control mice (p < 0.001) (Figure 3D). GAM densities were also significantly increased in Ras*; CSF1 OE high-grade gliomas compared to microglial density in CSF1 OE normal brains lacking the Ras* transgene (p < 0.001) (Figure 3D). As a second method for determining GAM density, the percentage of cells within tumors that were IBA1+ was also calculated. Using this method, Ras*; CSF1 OE high-grade gliomas were also found to have increased GAMs compared to high-grade gliomas in Ras* Control mice (p< 0.03, Supplemental Figure S2C). Studies of the limited number of low-grade tumors available from Ras*; CSF1 OE mice (n = 2) indicate that GAMs density appears higher in Ras*; CSF1 OE low-grade gliomas as compared to Ras* Control (n = 3) low-grade gliomas (Supplemental Figure S2D), however the number of tumors available precluded statistical analysis. Therefore, CSF1 over-expression promotes the development of aggressive high-grade tumors in vivo with increased GAM density.
The majority of GAMs in Ras* gliomas do not express markers of M1 or M2 polarization, regardless of CSF1 over-expression status
Given that CSF1/CSF1R signaling has been associated with promoting macrophage M2 phenotypes (12,15), GAM polarization phenotypes were investigated in Ras*; CSF1 OE and Ras* Control gliomas. Immunofluorescence based approaches were utilized so that only GAMs and not microglia from any adjacent normal brain would be analyzed. Gliomas were co-stained for IBA1 along with markers for M2 (ARG1 and CD206) or M1 (iNOS and CD16/CD32) polarization states, respectively.
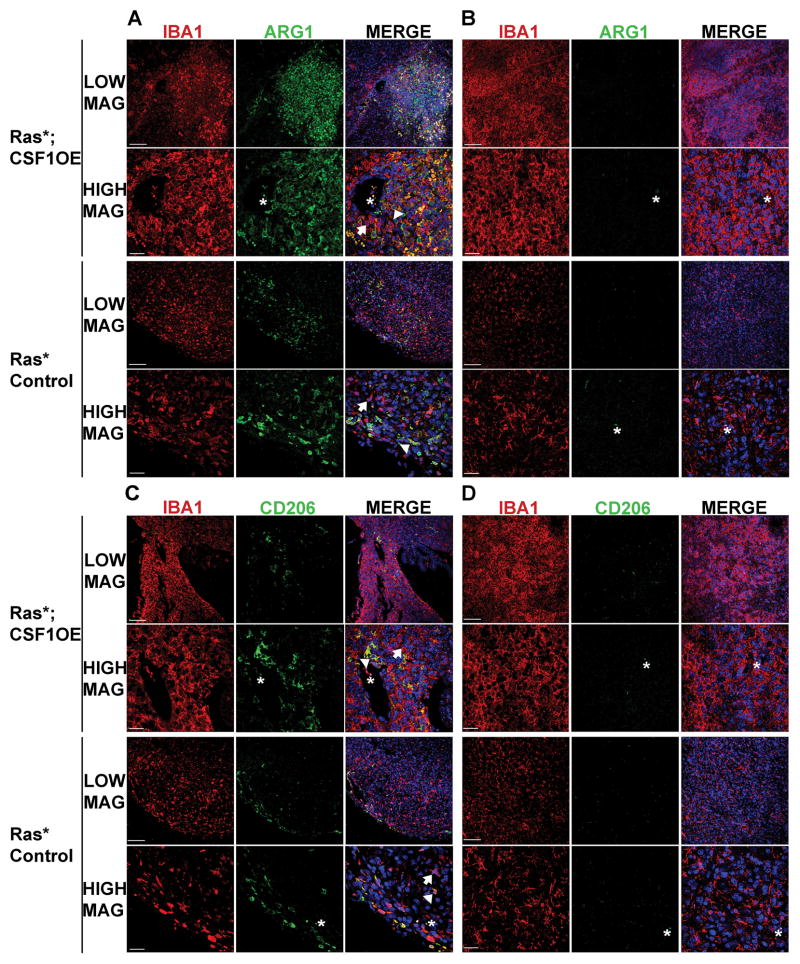
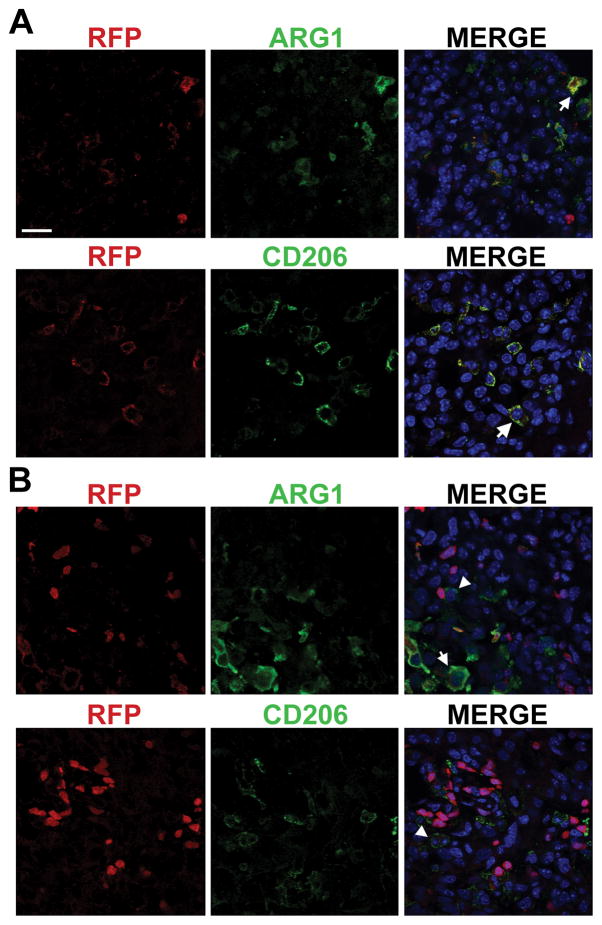
Although ARG1+ GAMs were detected in both Ras*; CSF1 OE and Ras* Control high-grade gliomas, they usually appeared in clusters in smaller, defined areas of the tumor, such as in or adjoining sites of hemorrhage, blood vessels, and the meninges (Figure 4A). However, the majority of GAMs in both Ras*; CSF1 OE and Ras* Control high-grade gliomas were ARG1− (Figure 4B). Similar results were found for CD206 (Figure 4C, D). To better characterize the cell population(s) expressing M2 markers, high-grade gliomas were identified in Ras*; Ccr2RFP/+; CSF1 OE mice. In Ccr2RFP/+ mice, CCR2 and therefore RFP, is expressed in circulating monocytes but not brain resident microglia. Consequently, RFP can be used to mark macrophages invading from the periphery (29). ARG1+ RFP+ and CD206+ RFP+ cells were observed in tumors (Figure 5A). In some tumor areas, ARG1+ or CD206+ cells were located in proximity to RFP+ cells, but expressed either low or undetectable levels of RFP (Figure 5B). This indicates that M2 polarized GAMs are found in tumor regions where immune cells are invading from the periphery, and that at least a subset of M2 polarized GAMs are invading macrophages.
Figure 4.
The majority of GAMs in Ras* high-grade gliomas do not express the M2 polarization markers ARG1 or CD206, regardless of CSF1 over-expression status. LOW MAG and HIGH MAG denote images that were acquired for the indicated genotype at lower magnification and higher magnification, respectively. Immunofluorescence staining for the GAM marker IBA1 (red), ARG1 or CD206 (green) and DAPI (blue) was performed. A) Representative images of a Ras*; CSF1 OE high-grade glioma and a Ras* Control high-grade glioma depicting the presence of IBA1+ GAMs that are also ARG1+. B) Representative images of a Ras*; CSF1 OE high-grade glioma and a Ras* Control high-grade glioma depicting the presence of IBA1+ GAMs, but no ARG1+ cells. C) Representative images of a Ras*; CSF1 OE high-grade glioma and a Ras* Control high-grade glioma depicting the presence of IBA1+ GAMs that are also CD206+. D) Representative images of a Ras*; CSF1 OE high-grade glioma and a Ras* Control high-grade glioma depicting the presence of IBA1+ GAMs, but no CD206+ cells. Arrowheads indicate examples of ARG1+ IBA1+ or CD206+ IBA1+ GAMs, arrows indicate examples of ARG1− IBA1+ or CD206− IBA1+ GAMs and asterisks indicate examples of auto-fluorescence of red blood cells visible in blood vessels and regions of hemorrhage. Scale bar for LOW MAG images = 100μm, scale bar for HIGH MAG images = 30μm.
Figure 5.
M2 GAMs are localized in areas of invasion of peripheral immune cells. Immunofluorescence staining for ARG1 or CD206 is in green, RFP fluorescence (a marker for Ccr2 expression) in red and DAPI in blue. A) Representative images of ARG1+ RFP+ and CD206+ RFP+ cells found in gliomas. B) Representative images including ARG1+ or CD206+ cells that are adjacent to RFP+ cells but themselves express low to undetectable levels of RFP. Arrows indicate examples of ARG1+ RFP+ or CD206+ RFP+ cells and arrowheads indicate examples of ARG1+ or CD206+ cells that do not appear to express RFP. Scale bar = 20 μm.
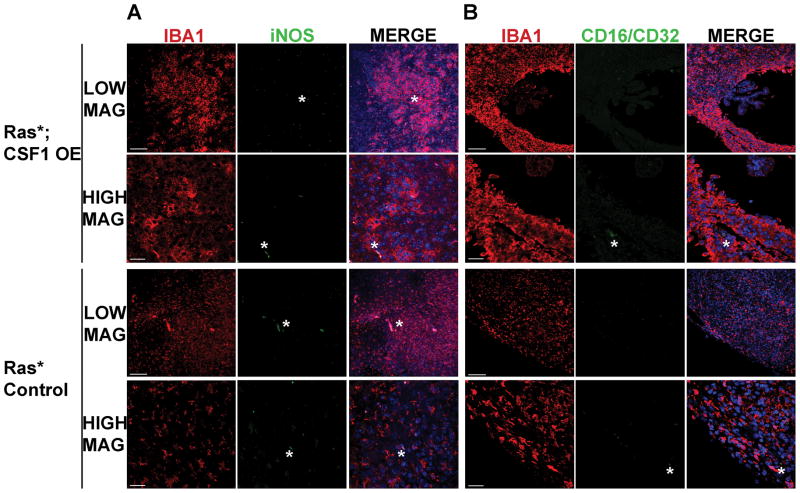
In low-grade tumors, ARG1 expression was not detected in GAMs from either Ras*; CSF1 OE (n=2) or Ras* Control (n=3) mice (Supplemental Figure S3A), and CD206 was detected in a limited number of GAMs in only one Ras*; CSF1 OE low-grade glioma (Supplemental Figure S3B, C). Conversely, the M1 markers iNOS and CD16/CD32 were not detected in GAMs in any high- (Figure 6A–D) or low-grade (Supplemental Figure S4) gliomas from either Ras*; CSF1 OE or Ras* Control mice. Positive staining for iNOS and CD16/CD32 in IBA1+ cells was detected in other tissues, indicating that the antibodies used are effective (Supplemental Figure S5). Therefore, our results indicate that the majority of GAMs are not positive for ARG1 or CD206, two commonly utilized markers of the M2 phenotype, and that, when present, at least a subset of M2 polarized GAMs have a peripheral origin. Additionally, in vivo transgenic CSF1 over-expression does not impact the M2 polarization status of GAMs in Ras* gliomas.
Figure 6.
GAMs in Ras* high-grade gliomas do not express the M1 polarization markers iNOS or CD16/CD32. Immunofluorescence staining for IBA1 (red), iNOS in (A) or CD16/32 in (B) (green) and DAPI (blue) was performed. LOW MAG and HIGH MAG denote images that were acquired for the indicated genotype at lower magnification and higher magnification, respectively. Representative images showing that no IBA1+ GAMs in high-grade gliomas were found to be positive for iNOS (A) or CD16/32 (B). Asterisks indicate examples of auto-fluorescence of red blood cells. Scale bar for LOW MAG images = 100μm, scale bar for HIGH MAG images = 30μm.
DISCUSSION
The increased expression of CSF1 and its receptor CSF1R in human high-grade gliomas suggest an oncogenic role for the CSF1/CSF1R signaling axis in gliomagenesis. Although most studies using cell lines or established tumors have supported a role for CSF1 signaling in promoting glioma phenotypes (8,11,13) other studies have not (36,37). Therefore, we studied the role of CSF1 during gliomagenesis in an autochthonous glioma model that allowed us to investigate the effects of modulating CSF1 levels on de novo gliomagenesis in vivo in an intact immune environment. The GFAP-Ras* model was chosen for our studies because glioma formation in this model is driven by a single transgene and allows for tumor development to occur in the presence of an intact immune environment without injections of cells or viruses that may artificially breach the blood brain barrier and potentially cause an immune response. Although RAS mutations are rare in human high-grade gliomas, dysregulation of RAS signaling pathways through other mechanisms is thought to commonly occur (38). High-grade gliomas in both GFAP-Ras* Control and GFAP-Ras*; CSF1 OE mice express high levels of OLIG2 and low levels of CD44 (Supplemental Figure S6), a characteristic of the proneural subtype of human high-grade gliomas (39).
Although Csf1 and Il-34 have both been shown to be important for regulating microglial numbers (40–42), our RT-PCR data and mining of microarray expression data indicate that of the two, CSF1 is the CSF1R ligand that is robustly expressed in high-grade gliomas. However, our studies cannot rule out the possibility that IL-34 has a role in the context of low-grade gliomas, during early stages of glioma development, or in a limited number of high-grade tumors. Of the CSF1 splice variants, the transcript encoding secreted CSF1 was expressed in all GSCs and glioma cells lines; therefore we focused our over-expression studies on the secreted isoform. However, the transcript encoding membrane bound CSF1 was also detected in many of the GSCs and glioma cell lines; therefore it may have a role in gliomagenesis as well.
Csf1 deficiency was found to deter glioma formation in the Ras* model, but not to prolong the survival of Ras* mice. Of the Ras*; Csf1op/op moribund mice that did not have frank tumors, most (81.25%) had non-tumor brain pathologies including increased glial cellularity, ischemia or inflammation. Similar non-tumor brain pathologies were also observed in 92.9% of Ras*; Csf1op/+ mice that did not develop frank tumors. These non-tumor brain pathologies likely contributed to their moribund state. In a breast cancer study, Csf1 deficiency was found not to affect tumor incidence or growth, but delayed the development of invasive metastatic carcinomas (43). Additionally, in a pancreatic neuroendocrine tumor model, Csf1 deficiency was shown to reduce tumor burden, however tumors in Csf1 deficient mice were similar in volume to those in controls (44). In contrast, in the Ras* glioma model, Csf1 appears to have an impact on both tumor initiation and tumor growth, as only one glioma of limited tumor volume was observed in the Ras*; Csf1op/op cohort. Our data also indicate that although Csf1 deficiency deters glioma formation in the Ras* model, rarely there might be other oncogenic signaling mechanisms that substitute for CSF1 signaling during tumor development.
GAMs and normal microglia express CSF1R, and CSF1R signaling has been shown to induce proliferation, differentiation and chemotaxis of microglia and macrophages (45–47). However, there is also evidence that CSF1R is expressed in tumor cells themselves in a subset of human gliomas (10,48). In high-grade gliomas from Ras*; CSF1-OE mice, CSF1R expression extensively co-localizes with IBA1 (Supplemental figure S7), therefore we focused our analyses on the impact of CSF1 over-expression on GAMs. However, our results do not definitively rule out an autocrine role for CSF1 signaling in gliomagenesis. Further studies utilizing genetic models will be required to fully address this possibility.
Our results indicate that one mechanism by which CSF1 over-expression promotes gliomagenesis is by increasing GAM density. Moreover, the percentage of cells within high-grade tumors in Ras*; CSF1 OE mice that are GAMs (average 18.7%, Supplemental Figure S2C) is in line with GAM measurements utilizing IBA1 in human high-grade gliomas (49). Our in vivo results are also consistent with previous findings that CSF1R inhibition reduces GAM numbers in GL261 glioma allografts (13). Studies utilizing the murine GL261 glioma cell line indicated that a signaling loop exists where glioma cells attract microglia through CSF1R signaling while microglia promote glioma cell invasion (13). In the current study, the observation that gliomas from CSF1 OE mice were large, infiltrative tumors is also supportive of a role for CSF1 signaling in promoting glioma invasion in the context of de novo gliomagenesis. Therefore, our data supports a model whereby increased CSF1 expression during de novo gliomagenesis promotes increases in microglial numbers, which then promote aggressive tumor pathologies such as increased tumor size. This model is also consistent with human glioma studies where reduced survival has been correlated with expression of microglia/macrophage markers (5).
Many studies of GAM polarization phenotypes have focused on mRNA expression studies of pools of isolated cells. Utilizing IHC, we were able to elucidate the expression of M1 and M2 markers in individual cells. In our study, only a minority of GAMs in Ras* high-grade gliomas express the M2 polarization markers ARG1 or CD206. Two observations support that cells expressing M2-polarization markers, when present in gliomas, are peripherally derived: 1) ARG1+ or CD206+ cells are physically located next to potential sites of invasion from the periphery and 2) Some ARG1+ or CD206+ cells express a reporter for Ccr2, a marker which has been used to label invading monocytes/macrophages in CNS disease (29). A few cells expressing M2 markers were adjacent to RFP+ cells but were not themselves RFP+. Our studies cannot distinguish between the possibilities that these RFP− M2 polarized GAMs are either brain-resident microglia recruited to sites of peripheral invasion or that they represent peripheral cells that have down-regulated Ccr2 reporter expression. Our data supports the hypothesis that glioma associated microglia and glioma associated macrophages can exhibit distinct polarization states within a glioma. Differences in phenotypes of microglia and invading macrophages have been observed in a mouse model of experimental autoimmune encephalomyelitis (EAE) (50), and our experiments support a similar phenomenon in glioma. Further studies in glioma models will be required to determine if GAM subsets have differential impacts on gliomagenesis.
In vitro monocyte to macrophage differentiation studies have associated CSF1 with an M2-like polarization state (12,15). In our study, transgenic CSF1 over-expression did not impact the presence of GAMs expressing the M2 markers ARG1 or CD206. This is consistent with our previous in vivo findings in a non-glioma setting indicating that CSF1 over-expression alone does not induce a basally M1 or M2 polarized microglial phenotype (27). An in vivo RCAS glioma study utilizing established gliomas showed that CSF1R inhibition shrank gliomas by decreasing the expression of M2 markers such as Arg1 and Cd206 in GAMs (14). Because global transcription patterns of GAMs and not phenotypes of individual GAMs were examined, it remains to be determined whether the decrease in M2 marker expression observed is due to CSF1R inhibition having differential impacts on GAM subsets.
In summary, we have identified secreted CSF1 to be a relevant CSF1R ligand in gliomas. By utilizing in vivo autochthonous models, we have shown that CSF1 has oncogenic effects during glioma development in vivo. Although CSF1 transgenic over-expression was found to modulate GAM density and glioma volume, it did not impact expression of M1 and M2 markers in GAMs. Since there is conflicting evidence regarding the M2 polarization state of GAMs in high-grade gliomas, it is possible that there is GAM heterogeneity both across and within tumors. Therefore, the Ras* autochthonous glioma model can also be useful in testing the efficacy of CSF1R inhibition based therapeutics in gliomas where the majority of the GAMs are not inherently M2 polarized, as well as the impact of CSF1R inhibition on phenotypically different GAM subsets.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Goldhirsh Foundation (LSC), the University of Wisconsin Graduate School (LSC, ID), NINDS R01NS085364 (LSC), R21CA161704UL1RR025011 (JSK, PAC), NCI HHSN261201000130C (JSK, PAC), P30CA014520 (JSK, PAC), NIHR01NS75995 (JSK, PAC), R01CA158800 (JSK, PAC), the Headrush Brain Tumor Research Professorship award (JSK) and the Roger Loff Memorial “Farming Against Brain Cancer” Fund for GBM Research (JSK, PAC). JSK and PAC also appreciate support from the Wisconsin Partnership Program core grant to the Center for Stem Cell and Regenerative Medicine and the University of Wisconsin (Graduate School, School of Medicine and Public Health and Dept. of Neurological Surgery). The UWCCC experimental pathology core is supported by the University of Wisconsin Carbone Cancer Center Cancer Center Support Grant P30 CA014520.
We thank the UWCCC experimental pathology core and Dr. Ruth Sullivan for histopathology assistance. We thank Dr. Arash Bashirullah’s laboratory for use of the confocal microscope. We thank Brady Strittmatter for technical assistance. We thank Dr. Jyoti Watters, Dr. Albee Messing, members of Dr. Paul Marker’s laboratory, Dr. Delinda Johnson and the UWCCC brain tumor disease oriented working group for providing reagents and for many helpful discussions.
Footnotes
The authors have no conflict of interests to declare.
References
- 1.Alifieris C, Trafalis DT. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol Ther. 2015;152:63–82. doi: 10.1016/j.pharmthera.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 2.da Fonseca AC, Badie B. Microglia and macrophages in malignant gliomas: recent discoveries and implications for promising therapies. Clin Dev Immunol. 2013;2013:264124. doi: 10.1155/2013/264124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol. 2011;11(11):775–87. doi: 10.1038/nri3086. [DOI] [PubMed] [Google Scholar]
- 4.Gutmann DH. Microglia in the tumor microenvironment: taking their TOLL on glioma biology. Neuro Oncol. 2015;17(2):171–3. doi: 10.1093/neuonc/nou346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engler JR, Robinson AE, Smirnov I, Hodgson JG, Berger MS, Gupta N, et al. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PLoS One. 2012;7(8):e43339. doi: 10.1371/journal.pone.0043339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donson AM, Birks DK, Schittone SA, Kleinschmidt-DeMasters BK, Sun DY, Hemenway MF, et al. Increased immune gene expression and immune cell infiltration in high-grade astrocytoma distinguish long-term from short-term survivors. Journal of immunology. 2012;189(4):1920–7. doi: 10.4049/jimmunol.1103373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 2004;14(11):628–38. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Bender AM, Collier LS, Rodriguez FJ, Tieu C, Larson JD, Halder C, et al. Sleeping beauty-mediated somatic mutagenesis implicates CSF1 in the formation of high-grade astrocytomas. Cancer Res. 2010;70(9):3557–65. doi: 10.1158/0008-5472.CAN-09-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komohara Y, Ohnishi K, Kuratsu J, Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J Pathol. 2008;216(1):15–24. doi: 10.1002/path.2370. [DOI] [PubMed] [Google Scholar]
- 10.Komohara Y, Horlad H, Ohnishi K, Fujiwara Y, Bai B, Nakagawa T, et al. Importance of direct macrophage - Tumor cell interaction on progression of human glioma. Cancer Sci. 2012 doi: 10.1111/cas.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu A, Wei J, Kong LY, Wang Y, Priebe W, Qiao W, et al. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12(11):1113–25. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, et al. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(13):4560–5. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coniglio SJ, Eugenin E, Dobrenis K, Stanley ER, West BL, Symons MH, et al. Microglial Stimulation of Glioblastoma Invasion Involves Epidermal Growth Factor Receptor (EGFR) and Colony Stimulating Factor 1 Receptor (CSF-1R) Signaling. Mol Med. 2012;18(1):519–27. doi: 10.2119/molmed.2011.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nature medicine. 2013;19(10):1264–72. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. Journal of immunology. 2006;177(10):7303–11. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 16.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. The Journal of clinical investigation. 2012;122(3):787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prosniak M, Harshyne LA, Andrews DW, Kenyon LC, Bedelbaeva K, Apanasovich TV, et al. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin Cancer Res. 2013;19(14):3776–86. doi: 10.1158/1078-0432.CCR-12-1940. [DOI] [PubMed] [Google Scholar]
- 18.Gabrusiewicz K, Hossain MB, Cortes-Santiago N, Fan X, Kaminska B, Marini FC, et al. Macrophage Ablation Reduces M2-Like Populations and Jeopardizes Tumor Growth in a MAFIA-Based Glioma Model. Neoplasia. 2015;17(4):374–84. doi: 10.1016/j.neo.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Reports. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hattermann K, Sebens S, Helm O, Schmitt AD, Mentlein R, Mehdorn HM, et al. Chemokine expression profile of freshly isolated human glioblastoma-associated macrophages/microglia. Oncol Rep. 2014;32(1):270–6. doi: 10.3892/or.2014.3214. [DOI] [PubMed] [Google Scholar]
- 21.Szulzewsky F, Pelz A, Feng X, Synowitz M, Markovic D, Langmann T, et al. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One. 2015;10(2):e0116644. doi: 10.1371/journal.pone.0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zorniak M, Clark PA, Leeper HE, Tipping MD, Francis DM, Kozak KR, et al. Differential expression of 2′,3′-cyclic-nucleotide 3′-phosphodiesterase and neural lineage markers correlate with glioblastoma xenograft infiltration and patient survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(13):3628–36. doi: 10.1158/1078-0432.CCR-12-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svendsen CN, ter Borg MG, Armstrong RJ, Rosser AE, Chandran S, Ostenfeld T, et al. A new method for the rapid and long term growth of human neural precursor cells. J Neurosci Methods. 1998;85(2):141–52. doi: 10.1016/s0165-0270(98)00126-5. [DOI] [PubMed] [Google Scholar]
- 24.Caldwell MA, He X, Wilkie N, Pollack S, Marshall G, Wafford KA, et al. Growth factors regulate the survival and fate of cells derived from human neurospheres. Nat Biotechnol. 2001;19(5):475–9. doi: 10.1038/88158. [DOI] [PubMed] [Google Scholar]
- 25.Pollard JW, Stanley ER. Pleiotropic roles for CSF-1 in development defined by the mouse mutation osteopetrotic (op) Adv Dev Biochem. 1996;4:153–93. [Google Scholar]
- 26.Wang J, Lin W, Popko B, Campbell IL. Inducible production of interferon-gamma in the developing brain causes cerebellar dysplasia with activation of the Sonic hedgehog pathway. Mol Cell Neurosci. 2004;27(4):489–96. doi: 10.1016/j.mcn.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 27.De I, Nikodemova M, Steffen MD, Sokn E, Maklakova VI, Watters JJ, et al. CSF1 overexpression has pleiotropic effects on microglia in vivo. Glia. 2014;62(12):1955–67. doi: 10.1002/glia.22717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding H, Roncari L, Shannon P, Wu X, Lau N, Karaskova J, et al. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 2001;61(9):3826–36. [PubMed] [Google Scholar]
- 29.Saederup N, Cardona AE, Croft K, Mizutani M, Cotleur AC, Tsou CL, et al. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS One. 2010;5(10):e13693. doi: 10.1371/journal.pone.0013693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.WHO. Classification of Tumours of the Central Nervous System. IARC Press; 2007. [Google Scholar]
- 31.Chihara T, Suzu S, Hassan R, Chutiwitoonchai N, Hiyoshi M, Motoyoshi K, et al. IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ. 2010;17(12):1917–27. doi: 10.1038/cdd.2010.60. [DOI] [PubMed] [Google Scholar]
- 32.Yamane F, Nishikawa Y, Matsui K, Asakura M, Iwasaki E, Watanabe K, et al. CSF-1 receptor-mediated differentiation of a new type of monocytic cell with B cell-stimulating activity: its selective dependence on IL-34. J Leukoc Biol. 2014;95(1):19–31. doi: 10.1189/jlb.0613311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai XM, Zong XH, Sylvestre V, Stanley ER. Incomplete restoration of colony-stimulating factor 1 (CSF-1) function in CSF-1-deficient Csf1op/Csf1op mice by transgenic expression of cell surface CSF-1. Blood. 2004;103(3):1114–23. doi: 10.1182/blood-2003-08-2739. [DOI] [PubMed] [Google Scholar]
- 34.Ryan GR, Dai XM, Dominguez MG, Tong W, Chuan F, Chisholm O, et al. Rescue of the colony-stimulating factor 1 (CSF-1)-nullizygous mouse (Csf1(op)/Csf1(op)) phenotype with a CSF-1 transgene and identification of sites of local CSF-1 synthesis. Blood. 2001;98(1):74–84. doi: 10.1182/blood.v98.1.74. [DOI] [PubMed] [Google Scholar]
- 35.Wiktor-Jedrzejczak W, Bartocci A, Ferrante AW, Jr, Ahmed-Ansari A, Sell KW, Pollard JW, et al. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc Natl Acad Sci U S A. 1990;87(12):4828–32. doi: 10.1073/pnas.87.12.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jadus MR, Williams CC, Avina MD, Ly M, Kim S, Liu Y, et al. Macrophages kill T9 glioma tumor cells bearing the membrane isoform of macrophage colony stimulating factor through a phagocytosis-dependent pathway. J Immunol. 1998;160(1):361–8. [PubMed] [Google Scholar]
- 37.Sielska M, Przanowski P, Wylot B, Gabrusiewicz K, Maleszewska M, Kijewska M, et al. Distinct roles of CSF family cytokines in macrophage infiltration and activation in glioma progression and injury response. J Pathol. 2013;230(3):310–21. doi: 10.1002/path.4192. [DOI] [PubMed] [Google Scholar]
- 38.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–46. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondo Y, Duncan ID. Selective reduction in microglia density and function in the white matter of colony-stimulating factor-1-deficient mice. J Neurosci Res. 2009;87(12):2686–95. doi: 10.1002/jnr.22096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13(8):753–60. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, et al. Stroma-Derived Interleukin-34 Controls the Development and Maintenance of Langerhans Cells and the Maintenance of Microglia. Immunity. 2012 doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193(6):727–40. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pyonteck SM, Gadea BB, Wang HW, Gocheva V, Hunter KE, Tang LH, et al. Deficiency of the macrophage growth factor CSF-1 disrupts pancreatic neuroendocrine tumor development. Oncogene. 2012;31(11):1459–67. doi: 10.1038/onc.2011.337. [DOI] [PubMed] [Google Scholar]
- 45.Chaubey S, Jones GE, Shah AM, Cave AC, Wells CM. Nox2 is required for macrophage chemotaxis towards CSF-1. PLoS One. 2013;8(2):e54869. doi: 10.1371/journal.pone.0054869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lelli A, Gervais A, Colin C, Cheret C, Ruiz de Almodovar C, Carmeliet P, et al. The NADPH oxidase Nox2 regulates VEGFR1/CSF-1R-mediated microglial chemotaxis and promotes early postnatal infiltration of phagocytes in the subventricular zone of the mouse cerebral cortex. Glia. 2013;61(9):1542–55. doi: 10.1002/glia.22540. [DOI] [PubMed] [Google Scholar]
- 47.Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Curr Opin Immunol. 2006;18(1):39–48. doi: 10.1016/j.coi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 49.Deininger MH, Seid K, Engel S, Meyermann R, Schluesener HJ. Allograft inflammatory factor-1 defines a distinct subset of infiltrating macrophages/microglial cells in rat and human gliomas. Acta Neuropathol. 2000;100(6):673–80. doi: 10.1007/s004010000233. [DOI] [PubMed] [Google Scholar]
- 50.Yamasaki R, Lu H, Butovsky O, Ohno N, Rietsch AM, Cialic R, et al. Differential roles of microglia and monocytes in the inflamed central nervous system. J Exp Med. 2014;211(8):1533–49. doi: 10.1084/jem.20132477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.