Abstract
The current study comprehensively examined the association between common variants in the Na+-coupled bicarbonate transporter (NCBT) genes and blood pressure (BP) responses to dietary sodium intervention. A 7-day low-sodium followed by a 7-day high-sodium dietary intervention was conducted among 1906 Han participants from rural areas of northern China. Nine BP measurements were obtained at baseline and each intervention using a random-zero sphygmomanometer. A mixed-effect model was used to assess the additive associations of 76 common variants in five NCBT genes, including SLC4A4, SLC4A5, SLC4A7, SLC4A8 and SLC4A10, with salt-sensitivity phenotypes. The Bonferroni method was used to adjust for multiple testing. SLC4A4 marker rs4254735 was significantly associated with diastolic BP (DBP) response to low-sodium intervention (P=5.05×10−4), with mean (95% confidence interval [CI]) response of −2.91 (−3.21, −2.61) and −0.40 (−1.84, 1.05) mmHg for genotype AA and AG, respectively. In addition, BP responses to high-sodium intervention significantly increased with the number of minor C alleles of SLC4A4 marker rs10022637. Mean systolic BP (SBP) responses among those with genotypes TT, CT, and CC were 4.62 (4.29, 4.99), 5.94 (5.31, 6.58) and 6.00 (3.57, 8.43) mmHg (P=1.14×10−4); mean DBP responses were 1.72 (1.41, 2.03), 3.22 (2.52, 3.92) and 3.94 (1.88, 5.99) mmHg (P=2.26×10−5), and mean arterial pressure responses were 2.69 (2.40, 2.97), 4.13 (3.57, 4.70) and 4.61 (2.51, 6.71) mmHg (P=2.07×10−6), respectively. Briefly, the present study indicated that common variants in the SLC4A4 gene might contribute to the variation of BP responses to dietary sodium intake in Han Chinese population.
INTRODUTION
Hypertension is a complex disorder influenced by genetic and environmental factors and their interactions.1 High salt intake is one of the most important environ- mental risk factors for elevated blood pressure (BP).2, 3 However, BP responses to dietary sodium intake vary among individuals, a phenomenon known as salt sensitivity.4 BP salt sensitivity has been associated with increased risks for hypertension, cardiovascular disease, and premature death.5, 6 Substantial evidences have suggested that genetic factors play an important role in determining BP salt sensitivity.7, 8 Previous reports from the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) Collaborative Research Group have identified many biological candidate genes which are apparently related to salt-sensitivity of BP. In general, most biological pathways involved in BP regulation (such as apelin system,9 renin-angiotensin-aldosterone system,10 epithelial sodium channel,11 kallikrein-kinin system,12 and endothelin system13) are related to salt-sensitivity of BP. However, the genomic mechanisms underlying BP salt sensitivity are far from being completely elucidated. Thus, identification of novel genetic variants for salt sensitivity will help to elucidate the potential interactions between genetic factors and dietary sodium intake on the regulation of BP.
The solute carrier 4 (SLC4) membrane transporter proteins involve in kidney acid-base regulation, intracellular pH maintaining and the balance of cation composition.14, 15 There are currently five mammalian Na+-coupled bicarbonate (HCO3−) transporter (NCBT) genes in the SLC4 family: SLC4A4 (encodes NBCe1), SLC4A5 (encodes NBCe2), SLC4A7 (encodes NBCn1), SLC4A8 (encodes NDCBE) and SLC4A10 (encodes NBCn2/NCBE).16, 17 NCBTs transport Na+ and HCO3− across the basolateral membranes of many cell types, including renal tubule cells, into the interstitial fluid and ultimately into the circulation, and the transport activities of NCBTs are different. Disruptions of NCBT genes in the kidney lead to severe electrolyte, acid-base disorders and cause BP deregulation.18, 19
Previous genome-wide association studies (GWASs) and candidate gene association studies have identified SLC4A4,20 SLC4A5,21, 22 and SLC4A723, 24 as susceptibility genes of hypertension. Single nucleotide polymorphisms (SNPs) of the SLC4A5 gene were also identified to be associated with salt-sensitive hypertension in White participants.25 These findings were concurrently reinforced by the experimental studies which showed that the Slc4a5 knockout mice were hypertensive,26 and the Slc4a7 knockout mice were mildly hypertensive at rest, and displayed altered vascular functions.27 SLC4A8 and SLC4A10 were also reported to be involved in the process of Na+ re-absorption.28, 29 To the best of our knowledge, none studies have reported associations of variants in the NCBT genes with BP response to dietary sodium intake in Chinese population.
The current study aimed to comprehensively investigate the association between common variants in five NCBT genes and BP responses to dietary sodium intake in the GenSalt study.
MATERIALS AND METHODS
Study population
All study subjects were participants of the GenSalt study, a family-based dietary feeding study examining gene-dietary sodium and potassium interaction on BP among a rural Han population in north China where habitual salt intake was high.8 Potential probands and their families were identified through a community-based BP screening carried out among persons aged 18–60 years in the study villages. Those with a mean systolic BP (SBP) between 130–160 mmHg and/or a mean diastolic BP (DBP) between 85–100 mmHg and no use of antihypertensive medications, as well as their spouses, siblings and offspring were recruited for the dietary intervention study. Individuals with stage-2 hypertension, current or recent use of antihypertensive medications, secondary hypertension, history of clinical cardiovascular disease, diabetes, chronic kidney disease, along with pregnant women, heavy alcohol users and those currently on a low-sodium diet were excluded from the study. More details of the study population and methods for the GenSalt study have been presented elsewhere.8, 30 Among 1906 participants eligible for the dietary intervention, 1871 (98.2%) and 1860 (97.6%) completed the low-sodium intervention and high-sodium intervention, respectively, were included in the current analysis.
Written informed consents were obtained from all participants after detailed explanation of the study. The study had been approved by the Institutional Review Board at all the participating institutions.
Dietary intervention
The dietary intervention included low-sodium and high-sodium feeding among probands and their siblings and offspring. In the first 3-day baseline observation, the study participants consumed their usual diet, then they received a 7-day low-sodium (3 g of sodium chloride or 51.3 mmol of sodium), followed by a 7-day high-sodium (18 g of sodium chloride or 307.8 mmol of sodium) dietary intervention. During both intervention phases, dietary potassium intake remained unchanged. All foods were cooked without salt, and pre-packaged salt was added to the study participants’ meal, as specified in the protocol. To ensure study participants’ compliance to the intervention program, they were instructed to avoid consuming any foods or beverages that were not provided by the study. In addition, three urinary excretions (one 24-hour and two overnight) were collected on 3-day baseline examination and on days 5, 6, and 7 of each intervention phase to confirm participants’ compliance with the dietary sodium intervention. The results from the 24-hour urinary excretions of sodium and potassium showed excellent compliance with the study diet: The mean (standard deviation) of 24-hour urinary excretions of sodium and potassium were 242.4 (66.7) mmol and 36.9 (9.6) mmol at baseline, 47.5 (16.0) mmol and 31.4 (7.7) mmol during the low-sodium intervention, 244.3 (37.7) mmol and 35.7 (7.5) mmol during the high-sodium intervention, respectively.
Phenotype measurement
During the 3 days of baseline examination, trained staff collected information on family structure, demographic characteristics, personal and family medical history and lifestyle risk factors using a standard questionnaire. BP was measured with the participants in the sitting position after 5 minutes of rest, and participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 minutes before their BP measurements. Three BP measurements were obtained each morning of the 3-day baseline observation and on days 5, 6, and 7 of each intervention period by the trained and certified observers using random-zero sphygmomanometer according to a standard protocol.10 All BP observers were blinded to the participants’ dietary intervention. In addition, body weight, height, and waist circumference were measured twice in light indoor clothing without shoes during the baseline examination. Body mass index (BMI) was calculated as kilograms per meters squared (kg/m2).
BP levels at baseline and during the intervention were calculated as the mean of 9 measurements from each period. Mean arterial pressure (MAP) was calculated by the formula: MAP=DBP+[(SBP−DBP)/3]. Mean BP response to low-sodium intervention was calculated as mean BP during low-sodium intervention minus the mean BP at baseline, and mean BP response to high-sodium intervention was calculated as mean BP during high-sodium intervention minus that during low-sodium intervention.
SNP selection and genotyping
SNPs located within the five NCBT genes (including about 5kb flanking regions each upstream and downstream of the genes) were genotyped among all participants using chip-based hybridization assays Affymetrix 6.0 platform (Affymetrix, Santa Clara, CA). SNPs were excluded if they had a call rate less than 95%, or significantly deviated from Hardy-Weinberg equilibrium (HWE) after adjustment for multiple comparisons (false discovery rate [FDR], P<0.05), or had a minor allele frequency (MAF) less than 1%. Within the five NCBT genes, 133 SNPs met the quality control criteria, and 76 SNPs were tagged (r2<0.9) using Haploview software (version4.2, http://www.broad.mit.edu/mpg/haploview) for inclusion in the current analysis. Detailed characteristics of the NCBT genes including the gene symbol, physical position, number of tag SNPs and encoded protein were presented in Table 1. Quality control information on the tagged 76 SNPs was listed in Supplemental Table 1.
Table 1.
Characteristics of five NCBT genes
| Gene symbol | Locus | Physical position (±5 kb)a | No. of tag SNPs | Encoded protein |
|---|---|---|---|---|
| SLC4A4 | 4q13.3 | chr4:72,048,003-72,442,804 | 42 | electrogenic sodium bicarbonate cotransporter 1,NBCe1 |
| SLC4A5 | 2p13.1 | chr2:74,443,561-74,547,152 | 13 | electrogenic sodium bicarbonate cotransporter 2, NBCe2 |
| SLC4A7 | 3p24.1 | chr3:27,409,212-27,503,245 | 4 | sodium bicarbonate cotransporter 3, NBC3/ NBCn1 |
| SLC4A8 | 12q13.13 | chr12:51,890,612-51,914,547 | 5 | electroneutral sodium bicarbonate exchanger 1,NDCBE |
| SLC4A10 | 2q24.2 | chr2:162,475,845-162,846,786 | 12 | sodium-driven chloride bicarbonate exchanger, NCBE/ NBCn2 |
Abbreviations: SNP, single-nucleotide polymorphism;
Genomic locations are based on NCBI Build 37 of the genome.
Statistical analysis
Baseline characteristics and BP responses were summarized as mean±standard deviations for continuous variables and as percentages for categorical variables. Deviation from HWE was tested by χ2 test. Additive associations between SNPs and BP responses to each sodium intervention were examined using a mixed-effect linear model (Proc Mixed procedure) using SAS software (version9.3; SAS Institute, Cary, NC). A sandwich estimator was used to account for correlations of individuals within families. Age, gender and baseline BMI were adjusted in multivariable analyses. To correct for multiple testing of the 76 variants, Bonferroni method was used with the threshold of 6.58×10−4 (0.05/76).
RESULTS
Characteristics of the GenSalt participants and BP responses to sodium interventions are shown in Table 2. On average, participants were 38.7 years of age, with BMI of 23.3 kg/m2. Approximately 52.9% of the study participants were male. BP levels significantly decreased from baseline to low-sodium dietary intervention and increased in response to the high-sodium dietary intervention (all P<0.0001). In the low-sodium intervention phase, average SBP, DBP and MAP responses were −5.5, −2.8, and −3.7 mmHg, respectively. And SBP, DBP and MAP increased by 4.9, 1.9, and 2.9 mmHg, respectively, during high-sodium intervention. BP responses to dietary intervention were normally distributed and varied widely among study participants.
Table 2.
Characteristics of 1,906 GenSalt dietary intervention participants
| Variables | Mean ± SD or No.(percentage) | Median (Inter-quartile Range) |
|---|---|---|
| Age, years | 38.7 ± 9.6 | 39.0 (33.0, 46.0) |
| Male, No.(%) | 1010 (52.9) | - |
| BMI, kg·m2 | 23.3 ± 3.2 | 22.9 (21.1, 25.2) |
| Baseline BP, mm Hg | ||
| Systolic | 116.9 ± 14.2 | 115.8 (106.4, 127.1) |
| Diastolic | 73.7 ± 10.3 | 73.3 (66.7, 80.7) |
| Mean arterial pressure | 88.1 ± 10.9 | 87.7 (80.0, 95.4) |
| BP during low salt, mm Hg | ||
| Systolic | 111.4 ± 12.2 | 110.0 (102.7, 119.3) |
| Diastolic | 71.0 ± 9.7 | 70.7 (64.2, 77.3) |
| Mean arterial pressure | 84.5 ± 9.7 | 84.1 (77.4, 90.6) |
| BP during high salt, mm Hg | ||
| Systolic | 116.3 ± 13.6 | 114.4 (106.7, 124.6) |
| Diastolic | 72.9 ± 10.3 | 72.4 (66.0, 79.1) |
| Mean arterial pressure | 87.4 ± 10.6 | 86.7 (79.6, 94.3) |
| BP response to low-sodium, mmHg | ||
| Systolic | −5.5 ± 7.0* | −4.4 (−8.9, −1.3) |
| Diastolic | −2.8 ± 5.5* | −2.7 (−5.6, 0.4) |
| Mean arterial pressure | −3.7 ± 5.3* | −3.3 (−6.6, −0.6) |
| BP response to high-sodium, mm Hg | ||
| Systolic | 4.9 ± 6.0* | 4.7 (0.6, 8.2) |
| Diastolic | 1.9 ± 5.4* | 1.8 (−1.6, 5.3) |
| Mean arterial pressure | 2.9 ± 5.0* | 2.7 (−0.4, 5.9) |
Abbreviations: BMI, body mass index; BP: blood pressure.
P-value<0.0001 when compared to no BP change during sodium interventions.
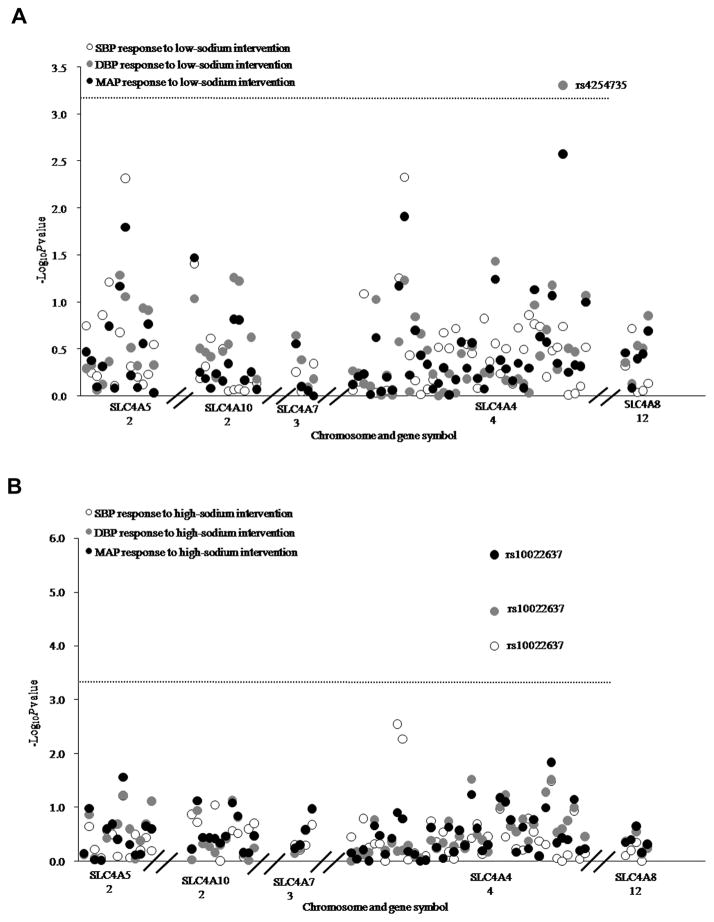
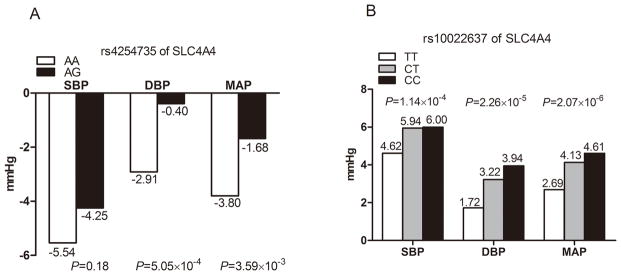
Figure 1A presents the association of each SNP with absolute SBP, DBP and MAP responses to low-sodium intervention. After adjustment for multiple testing, SLC4A4 marker rs4254735 was significantly associated with DBP response to low-sodium intake (P=5.05×10−4). Mean DBP response to low-sodium intervention according to SLC4A4 rs4254735 was shown in Figure 2A. Despite the low MAF of the G allele (2%), participants with heterozygous AG genotype had a decreased response to the low-sodium intervention compared to those with AA genotype. Mean DBP response (95%CI) was −2.91 (−3.21, −2.61) mmHg among those with AA genotype, and −0.40 (−1.84, 1.05) mmHg among those with AG genotype, respectively.
Figure 1.
−Log10P values for the associations of 76 tag SNPs in NCBT genes with blood pressure responses to low- sodium (A) and high-sodium (B) interventions. Labeled SNP was significant after Bonferroni correction. The horizontal dashed lines indicated the Bonferroni corrected significant level (P=6.58×10−4).
Figure 2.
Blood pressure responses to low-sodium intervention among participants with different genotypes of rs4254735 (A) and high-sodium intervention with different genotypes of rs10022637 (B).
As shown in Figure 1B, SLC4A4 marker rs10022637 was significantly associated with the absolute SBP, DBP and MAP responses to high-sodium intervention (P=1.14×10−4, 2.26×10−5 and 2.07×10−6, respectively). Data in Figure 2B showed that BP responses to high-sodium intervention increased with the number of minor C alleles of rs10022637. For example, mean SBP response to high-sodium intervention among participants with rs10022637 genotypes TT, CT, and CC were 4.62 (4.30, 4.95), 5.94 (5.31, 6.58) and 6.00 (3.60, 8.40) mmHg, respectively. Exact P values for all SNPs association tests were shown in Supplemental Table 2.
DISCUSSION
As the first study to examine the association between the NCBT genes and salt-sensitivity of BP among a large sample of Han Chinese population, we identified a significant association between SLC4A4 marker rs4254735 and DBP response to low-sodium intervention. In addition, SLC4A4 marker rs10022637 was significantly associated with SBP, DBP and MAP responses to high-sodium intervention. The minor C allele of rs10022637 was associated with dose-dependent increase in BP responses to high-sodium intervention. These results indicated that the NCBT genes might be mechanistically involved in BP salt sensitivity and their genetic variants contributed to the variation of this complex phenotype. And the novel genetic variants identified in our findings that determine an individual’s susceptibility to dietary sodium intervention is crucial for the identification of individuals who will benefit the most from a low-sodium diet in Han Chinese population. Moreover, our findings will help in the understanding of the genetic mechanisms underlying hypertension.
In mammals, there are five NCBTs encoded by different genes.18 The electrogenic NBCe1 is encoded by the SLC4A4 gene on chromosome 4q13.3. It has a Na+:HCO3− stoichiometry of 1:3 and mediates the efflux of sodium/bicarbonate at the basolateral membrane of proximal tubule cells essential for bicarbonate absorption of the proximal tubule in the kidney.31 This gene has been reported to be associated with decreased body weight/size, hematocrit, and abnormal ion homeostasis in mice,32 and the expression of SLC4A4 gene was found to be increased in the renal cortex of spontaneously hypertensive rats.31 In a previous report, Yang and colleagues identified SLC4A4 as a hypertension susceptible gene in Han Chinese with a genome-wide gene-based association study.20 They found that SLC4A4 gene was differentially expressed in 12 matched hypertension patients and control pairs.20 The markers rs4254735 and rs10022637 identified in the current study were both located in the intronic region which might possibly play regulatory roles in altering the expression of the SLC4A4 gene. In addition, we used the web tool RegulomeDB,33 a database that annotates SNPs with known and predicted regulatory elements in the intergenic regions of the Human genome, to speculate the functional implication of these 2 significant SNPs. Little evidence showed that rs4254735 and its highly correlated SNPs were causally associated with the regulation of SLC4A4 expression, so the function of rs4254735 and other correlated SNPs need to be further investigated. Moreover, no information on SNP rs10022637 was provided in the RegulomeDB database. However, SNP rs4130912, which was highly correlated with rs10022637 (r2=0.94, D′=1 in Hapmap CHB), had strong binding evidences and possibly a functional SNP for the regulation of SLC4A4 expression.33 To fully interpret these associations, further dissections of the regions surrounding these SNPs and functional studies are warranted in the future. We didn’t detect any GG homozygote for rs4254735 in our participants, even so, this study represented the first examination of rs4254735, and highlighted the need for replication of this result in other populations. In addition, our work provided important information for future studies of unique SLC4A4 variants in relation to BP salt sensitivity.
A previous study conducted in 185 Caucasians demonstrated that two SLC4A5 SNPs rs7571842 and rs10177833 were associated with BP salt-sensitivity.25 However, rs10177833 was not genotyped in our study, and rs7571842 was not significantly associated with BP responses to dietary sodium intervention in the present study (Supplemental Table 2). Several possibilities may explain the discrepancies between studies. Our study was conducted in Han Chinese population, where linkage disequilibrium (LD) structure may be different from that of Western population. For example, rs7571842 and rs10177833 were weakly correlated (r2=0.43, D′=0.72) in Han Chinese population, but they were strongly correlated (r2=0.85, D′=0.97) in the European population. If the identified variant is in LD with the true causal variant, it may not be replicated in other populations with a distinct ethnic background. Furthermore, the same genetic variants may have different BP effects due to their interactions with genetic and environmental factors that are unique to certain populations. Moreover, the effects of other potential factors, such as different characteristics of subjects, sodium-intervention design, and lifestyle factors also might make the inconsistency of results between Han Chinese and Europeans.
Several strengths of this study should be noted. To our knowledge, this is the first report of a comprehensive analysis of associations between common variants in the NCBT genes and BP responses to dietary sodium intervention. In the GenSalt study, stringent quality control procedures were employed during the dietary intervention, phenotype data collection, genotyping, and data analysis. Measurement error for BP phenotype was reduced by nine BP measurements measured by a random-zero sphygmomanometer. The large and homogenous Han Chinese population in this study makes the analysis robust to population stratification. However, although the Affymetrix 6.0 genotyping platform generally provides good coverage of common genetic variants, some important rare, low-frequency, and structural variants may be missed in the current study.
In summary, we provided the first evidence that common variants in the NCBT genes were associated with BP responses to dietary sodium intervention in the Han Chinese population. The findings reported here suggest that dietary sodium intervention might be particularly effective in lowering BP among individuals with specific variants of SLC4A4 gene. Still, replications of these results in other populations are needed, and further functional studies are critically important for identifying true causal variants.
Supplementary Material
What is known about the topic
Blood pressure (BP) responses to dietary sodium intake vary among individuals, a phenomenon known as salt sensitivity, and genetic factors play important roles in determining BP salt sensitivity.
The Na+-coupled bicarbonate (HCO3−) transporter (NCBT) genes play important roles in blood pressure regulation through acid-base regulation, intracellular pH maintaining and the balance of cation composition in the kidney.
Emerging evidences have linked mammalian Na+-coupled bicarbonate (HCO3−) transporter (NCBT) genes to hypertension or salt-sensitive hypertension in White participants.
What this study adds
SLC4A4 marker rs4254735 was significantly associated with diastolic BP response to low-sodium intervention. BP responses to high-sodium intervention significantly increased with the number of minor C alleles of SLC4A4 marker rs10022637 in Han Chinese population.
The present study indicated that common variants of SLC4A4 gene may contribute to the variation of BP response to dietary sodium intake in the Han Chinese population.
Acknowledgments
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by research grants (U01HL072507, R01HL087263 and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, USA. This study is also funded by a research grant (2012AA02A516) from the High-Tech Research and Development Program of China (863 Plan) from the Ministry of Science and Technology of China.
Footnotes
CONFLICT OF INTEREST
The authors declared no conflict of interest.
Supplementary Information accompanies this paper on the Journal of Human Hypertension website (http://www.nature.com/jhh)
References
- 1.Whelton PK, He J, Appel LJ, Cutler JA, Havas S, Kotchen TA, et al. Primary prevention of hypertension: clinical and public health advisory from The National High Blood Pressure Education Program. Jama. 2002;288:1882–1888. doi: 10.1001/jama.288.15.1882. [DOI] [PubMed] [Google Scholar]
- 2.Mozaffarian D, Fahimi S, Singh GM, Micha R, Khatibzadeh S, Engell RE, et al. Global sodium consumption and death from cardiovascular causes. The New England journal of medicine. 2014;371:624–634. doi: 10.1056/NEJMoa1304127. [DOI] [PubMed] [Google Scholar]
- 3.He J, Whelton PK. What is the role of dietary sodium and potassium in hypertension and target organ injury? The American journal of the medical sciences. 1999;317:152–159. doi: 10.1097/00000441-199903000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Cowley AW., Jr Genetic and nongenetic determinants of salt sensitivity and blood pressure. The American journal of clinical nutrition. 1997;65:587s–593s. doi: 10.1093/ajcn/65.2.587S. [DOI] [PubMed] [Google Scholar]
- 5.Morimoto A, Uzu T, Fujii T, Nishimura M, Kuroda S, Nakamura S, et al. Sodium sensitivity and cardiovascular events in patients with essential hypertension. Lancet (London, England) 1997;350:1734–1737. doi: 10.1016/S0140-6736(97)05189-1. [DOI] [PubMed] [Google Scholar]
- 6.Weinberger MH, Fineberg NS, Fineberg SE, Weinberger M. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension. 2001;37:429–432. doi: 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- 7.Beeks E, Kessels AG, Kroon AA, van der Klauw MM, de Leeuw PW. Genetic predisposition to salt-sensitivity: a systematic review. Journal of hypertension. 2004;22:1243–1249. doi: 10.1097/01.hjh.0000125443.28861.0d. [DOI] [PubMed] [Google Scholar]
- 8.Gu D, Rice T, Wang S, Yang W, Gu C, Chen CS, et al. Heritability of blood pressure responses to dietary sodium and potassium intake in a Chinese population. Hypertension. 2007;50:116–122. doi: 10.1161/HYPERTENSIONAHA.107.088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Q, Hixson JE, Rao DC, Gu D, Jaquish CE, Rice T, et al. Genetic variants in the apelin system and blood pressure responses to dietary sodium interventions: a family-based association study. Journal of hypertension. 2010;28:756–763. doi: 10.1097/HJH.0b013e3283370d32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu D, Kelly TN, Hixson JE, Chen J, Liu D, Chen JC, et al. Genetic variants in the renin-angiotensin-aldosterone system and salt sensitivity of blood pressure. Journal of hypertension. 2010;28:1210–1220. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Q, Gu D, Hixson JE, Liu DP, Rao DC, Jaquish CE, et al. Common variants in epithelial sodium channel genes contribute to salt sensitivity of blood pressure: The GenSalt study. Circulation Cardiovascular genetics. 2011;4:375–380. doi: 10.1161/CIRCGENETICS.110.958629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu D, Zhao Q, Kelly TN, Hixson JE, Rao DC, Cao J, et al. The role of the kallikrein-kinin system genes in the salt sensitivity of blood pressure: the GenSalt Study. American journal of epidemiology. 2012;176 (Suppl 7):S72–80. doi: 10.1093/aje/kws277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Defago MD, Gu D, Hixson JE, Shimmin LC, Rice TK, Gu CC, et al. Common genetic variants in the endothelial system predict blood pressure response to sodium intake: the GenSalt study. American journal of hypertension. 2013;26:643–656. doi: 10.1093/ajh/hps099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romero MF, Chen AP, Parker MD, Boron WF. The SLC4 family of bicarbonate (HCO(3)(−)) transporters. Molecular aspects of medicine. 2013;34:159–182. doi: 10.1016/j.mam.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero MF. Molecular pathophysiology of SLC4 bicarbonate transporters. Current opinion in nephrology and hypertension. 2005;14:495–501. doi: 10.1097/01.mnh.0000168333.01831.2c. [DOI] [PubMed] [Google Scholar]
- 16.Parker MD, Boron WF. The divergence, actions, roles, and relatives of sodium-coupled bicarbonate transporters. Physiological reviews. 2013;93:803–959. doi: 10.1152/physrev.00023.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pushkin A, Kurtz I. SLC4 base (HCO3−, CO3 2−) transporters: classification, function, structure, genetic diseases, and knockout models. American journal of physiology Renal physiology. 2006;290:F580–599. doi: 10.1152/ajprenal.00252.2005. [DOI] [PubMed] [Google Scholar]
- 18.Aalkjaer C, Boedtkjer E, Choi I, Lee S. Cation-coupled bicarbonate transporters. Comprehensive Physiology. 2014;4:1605–1637. doi: 10.1002/cphy.c130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boedtkjer E, Aalkjaer C. Disturbed acid-base transport: an emerging cause of hypertension. Frontiers in physiology. 2013;4:388. doi: 10.3389/fphys.2013.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang HC, Liang YJ, Chen JW, Chiang KM, Chung CM, Ho HY, et al. Identification of IGF1, SLC4A4, WWOX, and SFMBT1 as hypertension susceptibility genes in Han Chinese with a genome-wide gene-based association study. PloS one. 2012;7:e32907. doi: 10.1371/journal.pone.0032907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor JY, Wu CY, Darling D, Sun YV, Kardia SL, Jackson JS. Gene-environment effects of SLC4A5 and skin color on blood pressure among African American women. Ethnicity & disease. 2012;22:155–161. [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt SC, Xin Y, Wu LL, Cawthon RM, Coon H, Hasstedt SJ, et al. Sodium bicarbonate cotransporter polymorphisms are associated with baseline and 10-year follow-up blood pressures. Hypertension. 2006;47:532–536. doi: 10.1161/01.HYP.0000196949.26088.3c. [DOI] [PubMed] [Google Scholar]
- 23.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Wang L, Lin X, Huang J, Charles Gu C, He M, et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Human molecular genetics. 2015;24:865–874. doi: 10.1093/hmg/ddu478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carey RM, Schoeffel CD, Gildea JJ, Jones JE, McGrath HE, Gordon LN, et al. Salt sensitivity of blood pressure is associated with polymorphisms in the sodium-bicarbonate cotransporter. Hypertension. 2012;60:1359–1366. doi: 10.1161/HYPERTENSIONAHA.112.196071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groger N, Vitzthum H, Frohlich H, Kruger M, Ehmke H, Braun T, et al. Targeted mutation of SLC4A5 induces arterial hypertension and renal metabolic acidosis. Human molecular genetics. 2012;21:1025–1036. doi: 10.1093/hmg/ddr533. [DOI] [PubMed] [Google Scholar]
- 27.Boedtkjer E, Praetorius J, Matchkov VV, Stankevicius E, Mogensen S, Fuchtbauer AC, et al. Disruption of Na+,HCO(3)(−) cotransporter NBCn1 (slc4a7) inhibits NO-mediated vasorelaxation, smooth muscle Ca(2)(+) sensitivity, and hypertension development in mice. Circulation. 2011;124:1819–1829. doi: 10.1161/CIRCULATIONAHA.110.015974. [DOI] [PubMed] [Google Scholar]
- 28.Leviel F, Hubner CA, Houillier P, Morla L, El Moghrabi S, Brideau G, et al. The Na+-dependent chloride-bicarbonate exchanger SLC4A8 mediates an electroneutral Na+ reabsorption process in the renal cortical collecting ducts of mice. The Journal of clinical investigation. 2010;120:1627–1635. doi: 10.1172/JCI40145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilgen G, Huebner AK, Tanimoto N, Sothilingam V, Seide C, Garcia Garrido M, et al. Lack of the sodium-driven chloride bicarbonate exchanger NCBE impairs visual function in the mouse retina. PloS one. 2012;7:e46155. doi: 10.1371/journal.pone.0046155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.GenSalt: rationale, design methods and baseline characteristics of study participants. Journal of human hypertension. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlowski A, Ciancio MC, Caldiz CI, De Giusti VC, Aiello EA. Reduced sarcolemmal expression and function of the NBCe1 isoform of the Na(+)-HCO(3)(−) cotransporter in hypertrophied cardiomyocytes of spontaneously hypertensive rats: role of the renin-angiotensin system. Cardiovascular research. 2014;101:211–219. doi: 10.1093/cvr/cvt255. [DOI] [PubMed] [Google Scholar]
- 32.Gawenis LR, Bradford EM, Prasad V, Lorenz JN, Simpson JE, Clarke LL, et al. Colonic anion secretory defects and metabolic acidosis in mice lacking the NBC1 Na+/HCO3− cotransporter. The Journal of biological chemistry. 2007;282:9042–9052. doi: 10.1074/jbc.M607041200. [DOI] [PubMed] [Google Scholar]
- 33.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.