Abstract
Background
One of the most prevalent congenital birth defects is cleft palate. The palatal skeleton is derived from the cranial neural crest and platelet-derived growth factors (Pdgf) are critical in palatogenesis. Of the two Pdgf receptors, pdgfra is required for neural crest migration and palatogenesis. However, the role pdgfrb plays in the neural crest, or whether pdgfra and pdgfrb interact during palatogenesis is unclear.
Results
We find that pdgfrb is dispensable for craniofacial development in zebrafish. However, the palatal defect in pdgfra;pdgfrb double mutants is significantly more severe than in pdgfra single mutants. Data in mouse suggest this interaction is conserved and that neural crest requires both genes. In zebrafish, pdgfra and pdgfrb are both expressed by neural crest within the pharyngeal arches and pharmacological analyses demonstrate Pdgf signaling is required at these times. While neither proliferation nor cell death appears affected, time-lapsed confocal analysis of pdgfra;pdgfrb mutants shows a failure of proper neural crest condensation during palatogenesis.
Conclusions
We provide data showing that pdgfra and pdgfrb interact during palatogenesis in both zebrafish and mouse. In zebrafish, this interaction affects proper condensation of maxillary neural crest cells, revealing a previously unknown interaction between Pdgfra and Pdgfrb during palate formation.
Keywords: pdgfra, pdgfrb, palatogenesis, cranial neural crest
Introduction
Human palatal clefting is a highly prevalent congenital disorder, affecting nearly 1 in 700 children (Dixon et al., 2011). Studies involving model organisms have provided information on both genetic and morphogenetic processes that control palate formation. In mouse and zebrafish, early palate formation is highly conserved and involves migration of cranial neural crest cells (CNCC) from two distinct regions of the first pharyngeal arch. The frontonasal prominence forms from neural crest migrating around the top of the eye and the maxillary prominence forms from neural crest condensing upon the oral ectoderm and under the eye (Eberhart et al., 2006; Kimmel and Eberhart, 2008; Osumi-Yamashita et al., 1994; Swartz et al., 2011; Wada et al., 2005). Reciprocal signaling between the maxillary neural crest and the oral ectoderm then drive the formation of the palate (Eberhart et al., 2006; Swartz et al., 2011; Wada et al., 2005). Numerous signaling pathways are involved in this process, including the platelet-derived growth factor (Pdgf) pathway (Eberhart et al., 2008; Soriano, 1997; Tallquist and Soriano, 2003).
There are two Pdgf receptors, alpha and beta, and four ligands in human and mouse, and six ligands in zebrafish (Betsholtz et al., 2001; Eberhart et al., 2008; Tallquist and Kazlauskas, 2004). Ligands and receptors can homo- or heterodimerize, leading to phosphorylation of intracellular receptor tyrosine residues (Tallquist and Kazlauskas, 2004). This phosphorylation activates several downstream effector pathways that promote cell migration, proliferation, and/or survival (Klinghoffer et al., 2002; Tallquist and Kazlauskas, 2004). Pdgf signaling is important throughout development in numerous tissues including the gonads, lung, intestine, skin, central nervous system, skeleton, vasculature, and both cardiac and cranial neural crest (Andrae et al., 2008; Hoch and Soriano, 2003; Smith and Tallquist, 2010).
Multiple studies have focused on the role Pdgfra plays in the cranial neural crest. Pdgfra is expressed in both premigratory and migratory neural crest cell populations in mouse and zebrafish (Eberhart et al., 2008; Liu et al., 2002; Schatteman et al., 1992). In mouse, conditional loss of Pdgfra in the neural crest leads to numerous cranial defects including cleft palate, a shortened skull, absent hyoid bone, and incomplete ossification of the basosphenoid, presphenoid, and alisphenoid bones (He and Soriano 2013; He and Soriano 2015; Tallquist et al., 2003). A hypomorphic mutant of pdgfra in zebrafish has cleft palate as well, which is attributed to disrupted migration of the frontonasal CNCC (Eberhart et al., 2008). Neural crest cell migration is also defective in Pdgfra mutant mice (He and Soriano, 2013; Vasudevan and Soriano, 2014), highlighting the conserved function of this gene across species. While pdgfra is strongly implicated in palatogenesis, the role of pdgfrb in craniofacial development is less understood.
Pdgfrb knockout mice exhibit kidney, heart, and hematological defects (Mellgren et al., 2008; Soriano, 1994), but no overt craniofacial abnormalities. Mutations specific to the PI3K-domains of both Pdgfra and Pdgfrb result in craniofacial defects as severe as a Pdgfra null (Klinghoffer et al., 2002). Furthermore, combined loss of Pdgfra and Pdgfrb in heart epicardium or aortic arch NCC results in a more severe and penetrant phenotype (Richarte et al, 2007; Smith et al., 2011). Collectively, these results suggest that, in certain tissues, PDGF receptors have cooperative functions.
Here, we sought to investigate the roles of the PDGF receptors during palatogenesis. While loss of pdgfrb does not cause any overt craniofacial defects, pdgfra hypomorphic mutant zebrafish have cleft palate. Double pdgfra;pdgfrb mutants show an exacerbated palate defect, compared to hypomorphic pdgfra mutants. Data in mouse reveals a conservation of this interaction and demonstrate that neural crest cells autonomously require both receptors. Using a broad Pdgf Inhibitor, we show that craniofacial development is most sensitive to Pdgf activity from the initiation of neural crest migration, at 10 hours post fertilization (hpf), to the formation of the maxillary domain, at 30 hpf. We observe that neural crest cells express both pdgfra and pdgfrb at the onset of palatogenesis in zebrafish. While neither cell proliferation nor cell death appears altered in the maxillary domain of pdgfra;pdgfrb mutants, CNCC fail to condense appropriately, revealing a previously unknown compensatory role of Pdgf receptors in palatogenesis.
Results
Pdgfrb partially compensates for loss of Pdgfra during craniofacial development
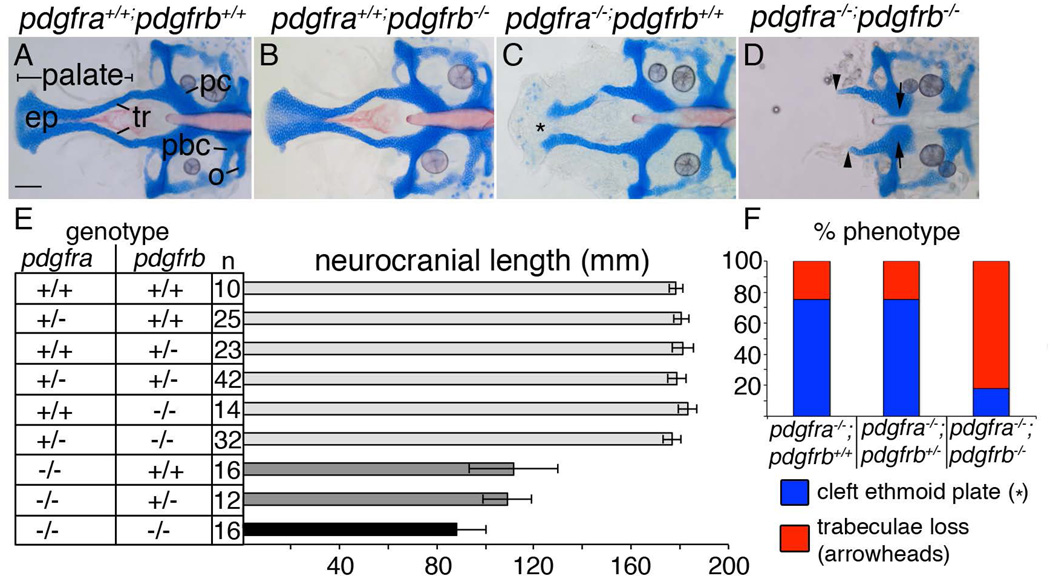
Previous work suggested that Pdgfra and Pdgfrb might interact during craniofacial development. To investigate this interaction, we generated pdgfra;pdgfrb double mutants, using our hypomorphic pdgfra mutant and a putative null pdgfrb mutant (Eberhart et al., 2008; Kok et al., 2014). While pdgfrb-mutant embryos develop normal craniofacial skeletons (Fig. 1B, compared to Fig. 1A), pdgfra mutant embryos exhibit cleft palate where the ethmoid plate fails to fuse, resulting in shorter neurocranial lengths (Fig. 1C, asterisk, Fig 1E; Eberhart et al., 2008; McCarthy et al., 2013). Viscerocranial elements are quantifiably smaller as well (data not shown; McCarthy et al., 2013).
Figure 1.
pdgfra and pdgfrb interact during craniofacial development. (A–D) 5 dpf flatmounted zebrafish neurocrania. Anterior is to the left. (A) Wild type and (B) pdgfrb mutants display normal craniofacial skeletons, while (C) pdgfra mutants display a shortened neurocrania and cleft palate (asterisk). (D) pdgfra;pdgfrb double mutants lose the palatal skeleton (arrowheads) and a region of the parachordals adjacent to the posterior basicapsular commissure (arrows). (E) Bar chart depicting neurocranial length measured from the anterior ethmoid plate to the posterior occipital arch of the neurocranium. Similarly shaded bars are not significantly different from one another (ANOVA, p=<0.05). (F) Bar chart depicting percent of phenotypes observed per genotype listed showing higher percentage of pdgfra−/−;pdgfrb−/− mutants displaying trabeculae loss, blue bar depicts cleft ethmoid plate (asterisk in C) and red bar depicts loss of trabeculae (arrowheads in D). ep=ethmoid plate, o=occipital arch, pbc=posterior basicapsular commissure, pc= parachordals, tr=trabeculae. scale bar= 20 µm.
While loss of a single allele of pdgfrb has no effect on the phenotype of pdgfra mutants, analysis of double pdgfra;pdgfrb mutants reveals a significantly reduced neurocranium, measured from the anterior ethmoid plate to the posterior occipital arch (Fig. 1D, arrowheads, 1E, analysis from 3 different clutches), compared to all other genotypes. Neurocranial width and symplectic length are also reduced in double pdgfra;pdgfrb mutants compared to all other genotypes (data not shown). Other defects observed in double mutants include cartilage loss between the polar cartilages and the posterior basicapsular commissure, and loss of notochord ossification (Fig. 1D, arrows). Furthermore, double pdgfra;pdgfrb mutants display a higher percentage of trabeculae loss phenotypes compared to either pdgfra−/−;pdgfrb+/+ or pdgfra−/−;pdgfrb+/− genotypes (Fig. 1F). Together, these data show that loss of both Pdgf receptors results in a synergistic interaction affecting the craniofacial skeleton, including the palate.
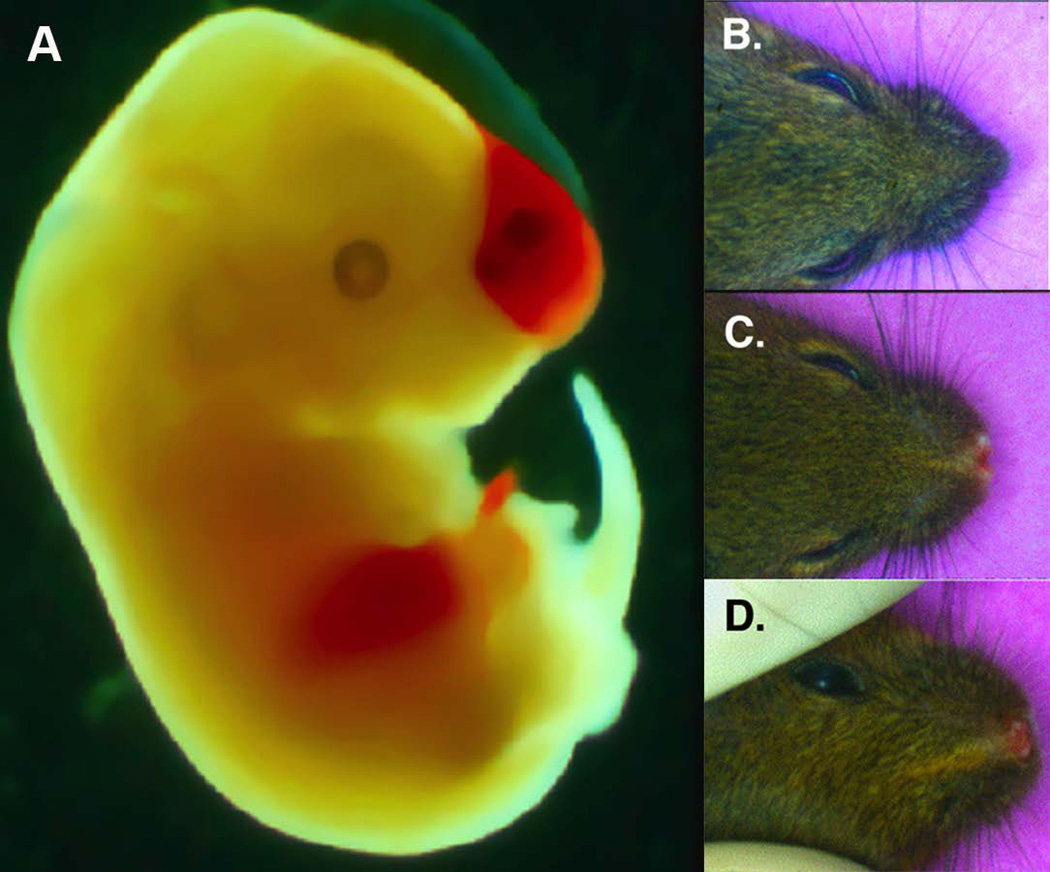
Because previous reports show conservation of Pdgfra function across species (Eberhart et al., 2006; Tallquist et al., 2003; Vasudevan and Soriano, 2014), we next investigated Pdgf receptor genetic interactions in mice. We surveyed mouse embryos and pups possessing hypomorphic Pdgf receptors (Pdgfrαfl, PdgfrβF3, materials and methods and Tallquist et al., 2000, respectively) and/or null alleles for gross cranial facial malformations. At E14.5 compound mutant embryos exhibit a blood filled blister in the frontal nasal region (Fig. 2A, n=7/10).
Figure 2. Genetic interactions between PDGFRα and PDGFRβ in craniofacial development.
(A) E14.5 PDGFRα+/−; PDGFRβF3/F3 with hemorrhage and blistering. (B–D) P28 PDGFR mutant animals (B) PDGFRβF3/F3 hypomorph C) PDGFRαfl/+; PDGFRβF3/F3 (D) PDGFRαfl/fl; PDGFRβF3/F3.
It was noted many years ago that mice heterozygous for the Pdgfrα Ph allele have wider and shorter frontonasal cavities (Gruneberg and Truslove, 1960). We subsequently found this phenotype was exacerbated when PDGFRβ signaling was disrupted. We observed midline facial defects in 50% of mice bearing Pdgf receptor hypomorphic alleles (Fig. 2C–D compared to 2B, n=27/54). The craniofacial bones in the mutant had a significant reduction in the crown length resulting in a stunted snout, as well as a defect in the fusion of the nasal cartilage (data not shown). On a few occasions we observed cleft palates in mice of the genotype shown in Figure 2D. Mutation in at least one allele of Pdgfrα was required for this phenotype as mice homozygous for either Pdgfrβ hypomorphic or null alleles exhibited no observable cranial facial defects (Fig. 2B and 3D). These data demonstrate that loss of PDGFRβ signaling in a sensitized background (reduced signaling through PDGFRα) results in craniofacial defects and thus suggests a supporting role for Pdgfrb in craniofacial development.
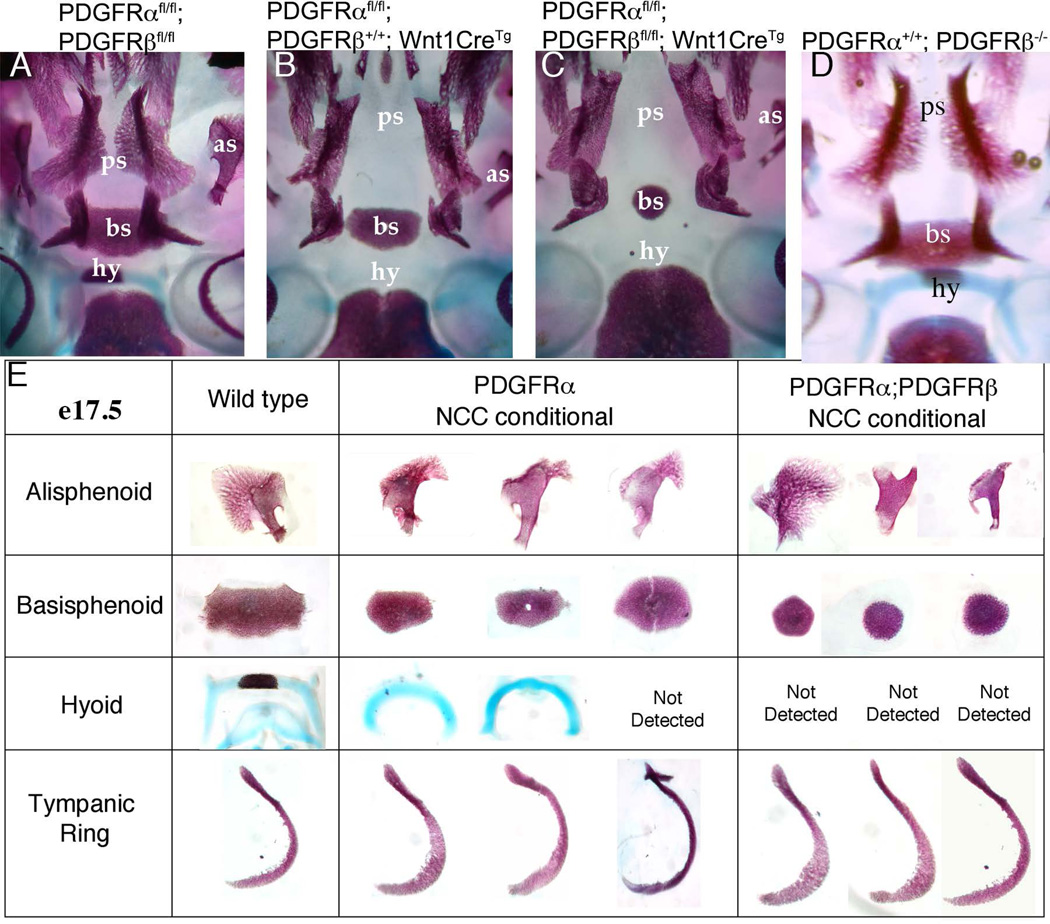
Figure 3.
Loss of PDGFRβ in NCC increased cranial bone malformatons. (A–C) A ventral view of the mouse skull derived from E17.5 embryos of the indicated genotype. Anterior is up. (E) Individual cranial skeletal elements dissected from the indicated genotypes. In Pdgfrafl/fl;Pdgfrbfl/fl;Wnt1:Cre+ embryos, the alisphenoid and basisphenoid were reduced, while the hyoid was undetectable. as=alisphenoid, bs=basisphenoid, hy= hyoid, ps=palatal shelf.
To determine if these genetic interactions of Pdgf receptors were solely a result of disrupted signaling in NCC, we examined cranial bone preparations from E16.5–E18.5 Pdgfra;Pdgfrb NCC conditional (Pdgfrαfl/fl;Pdgfrβfl/fl; Wnt1CreTg) embryos to assess the frequency of cranial bone phenotypes. Previously, we have shown that Wnt1CreTg recombination of Pdgf receptor loxP-flanked alleles lead to efficient reduction in receptor expression in first branchial arch NCC derivatives (Richarte et al., 2007). We found that deletion of both Pdgf receptors in NCC resulted in increased severity of malformation of NCC-derived bones including the hyoid arch, basisphenoid, and alisphenoid bones. The disrupted bones were the same bones affected by loss of Pdgfra in NCC, but bone formation was even more rudimentary than observed in Pdgfrα NCC mutants. The most prominent craniofacial defect observed in Pdgfra NCC conditional mutants is the midline frontonasal clefting, where the palatine bone and nasal cartilage fail to fuse (Fig. 3B). The cleft palate defect occurs with 100% penetrance in the Pdgfra NCC conditional (Tallquist et al., 2003), and no additional defects could be detected with the loss of the Pdgfrb in the NCC lineage. In addition to previously noted abnormalities in the alisphenoid, basisphenoid, and pterygoid bones (He et al., 2013) Pdgfra NCC conditional embryos show delayed development of the hyoid bone. When Pdgfrβ is additionally deleted, the hyoid bone fails to be stained by either by Alizarin red or Alcian Blue (Fig. 3C and E). In control embryos, the basisphenoid is a very dense structure that bridges the width between the two sphenoid bones. In the Pdgfra NCC conditional, the basisphenoid spans the width between the sphenoid bones, but no longer overlaps with the sphenoid bones. In the double NCC conditional embryos, the basisphenoid is significantly reduced (Fig. 3B–E). This spectrum of defects was consistently seen in Pdgfrα;Pdgfrβ NCC mutants at all stages examined from E16.5-P0 (n>10). These data show that a genetic interaction between Pdgfra and Pdgfrb in craniofacial development is conserved across vertebrates.
Pdgf activity is required between 10 and 30 hours post fertilization for proper craniofacial development
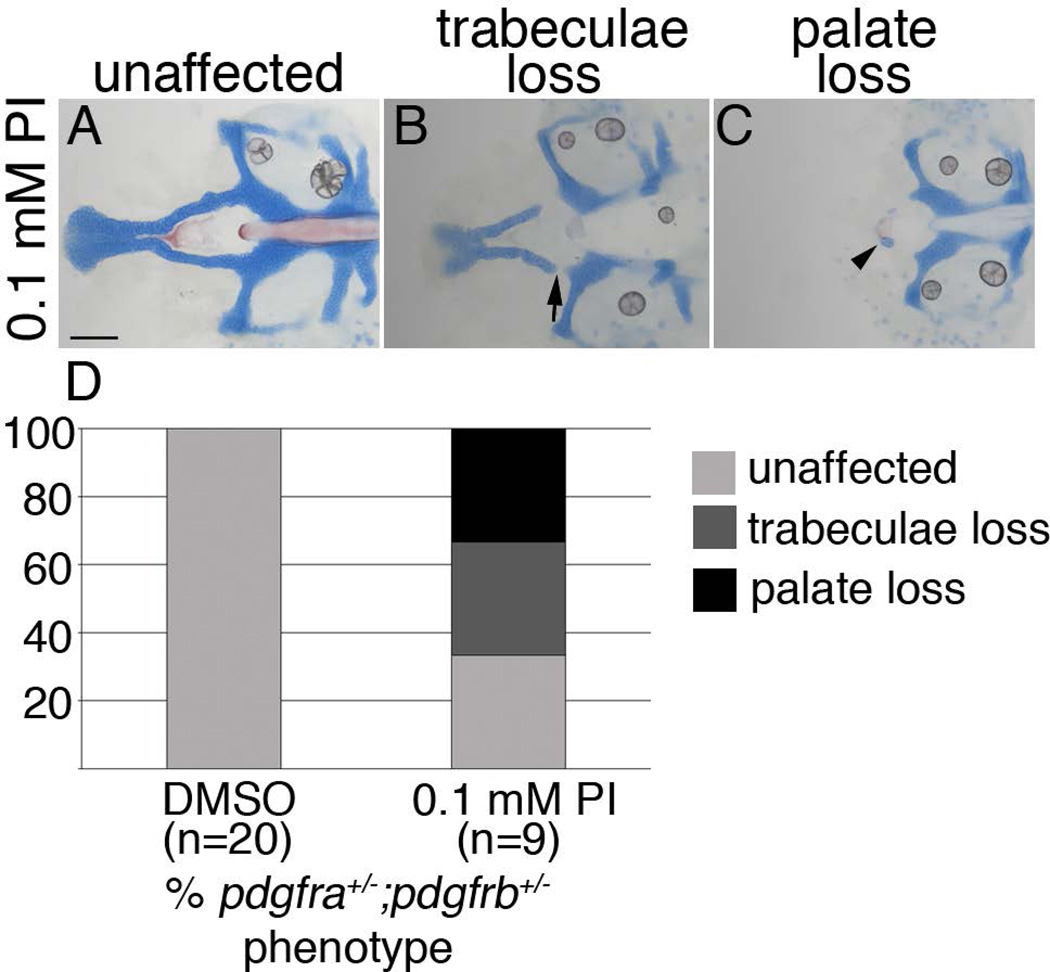
To determine when craniofacial development requires Pdgf function, we utilized PDGF Inhibitor V (Calbiochem), which broadly inhibits Pdgf activity. In an initial dose response analysis, three concentrations of inhibitor were tested on wild-type fish, 1, 1.5, and 2 µM, from 10 hpf, or just prior to neural crest migration, to 30 hpf, when most neural crest cells have condensed within the pharyngeal arches and palatogenesis has begun. A concentration of 1 µM did not cause any gross craniofacial or vascular defects, even though concentrations of 0.25 µM had previously been shown to cause vascular defects, on a different zebrafish genetic background (Wiens et al., 2010). We found that treating wild type embryos with 1.5 mM Pdgf Inhibitor V from 10 to 30 hpf caused complete loss of the palate in 100% of treated embryos, with embryos treated after 30 hpf showing no overt craniofacial defects (data not shown). To guard against potential off target effects of the inhibitor, we next tested suboptimal concentrations of the inhibitor on embryos obtained from incrosses of double carrier fish. We found that a concentration of 0.1 µM caused few defects to wild-type embryos (n=5). However, embryos heterozygous at both loci had a near complete loss of the palate in 33% of cases (n=3) and trabeculae loss in 33% of cases (n=3; Fig. 4). Three of five wild-type clutch mates were normal; the remaining two had trabeculae loss. While not observed in our analyses in Figure 1, here we did find a single untreated pdgfra+/−;pdgfrb−/− zebrafish with cleft palate, providing further evidence for the requirement of Pdgfrb when Pdgfra function is attenuated. Collectively, these data suggest that Pdgf function is required as CNCCs migrate to and condense within the pharyngeal arches.
Figure 4.
Craniofacial development is sensitive to Pdgf signaling loss between 10 and 30 hpf (A–C) 5 dpf flatmounted pdgfra+/−;pdgfrb+/− zebrafish neurocrania, exposed to 0.1 mM Pdgf Inhibitor V (PI) from 10 to 30 hpf, anterior to the left. (D) Bar chart depicting percentage of pdgfra+/−;pdgfrb+/− embryos displaying unaffected (light grey bar, A), trabeculae loss (dark grey bar, arrow in B), or palate loss (black bar, arrowhead in C) craniofacial defects in DMSO or 0.1 mM Pdgf Inhibitor V (PI). scale bar= 20 µm.
Neural crest cells express pdgfra and pdgfrb dynamically as they condense within the pharyngeal arches
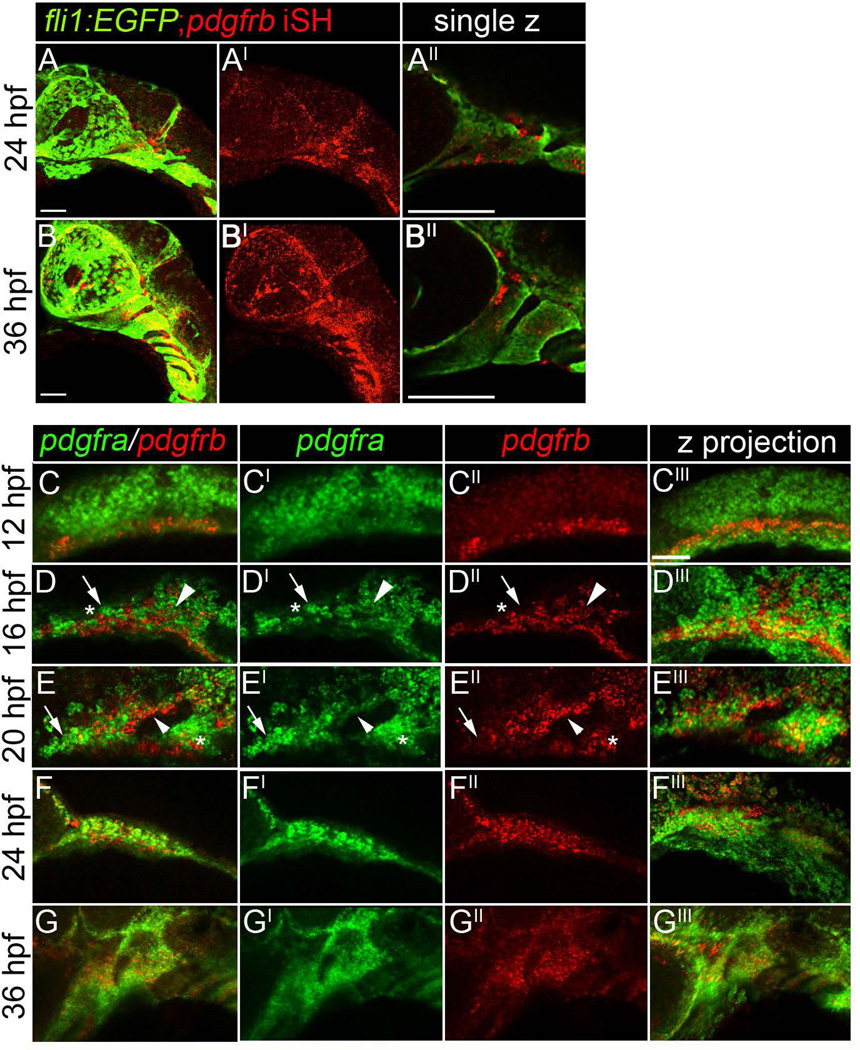
Both Pdgf receptors function in proper craniofacial development between 10 and 30 hpf. CNCC express pdgfra during this time window (Eberhart et al., 2008). It was unclear, however, where pdgfrb was expressed at these times. To confirm that pdgfrb was expressed in the neural crest of zebrafish, we used fluorescence in situ of pdgfrb in a fli1:EGFP transgenic, which labels the neural crest and vasculature, to look for co-localization. Indeed, at both 24 and 36 hpf, pdgfrb expression occurs in the neural crest and the vasculature (Fig. 5A–A’, 5B–B’; Wiens et al., 2010). Analysis of single z-slices demonstrates that neural crest cells within the pharyngeal arches express pdgfrb (Fig. 5A”, B”) These data suggest that neural crest cells express both pdgfra and pdgfrb as CNCC populate the pharyngeal arches.
Figure 5.
Neural crest cells express both pdgfra and pdgfrb. (A–A”) A 24 hpf fli1:EGFP transgenic embryo labeled for pdgfrb mRNA via fluorescence in situ hybridization, anterior is to the left. (B–B”) A 36 hpf fli1:EGFP transgenic embryo and labeled for pdgfrb mRNA via fluorescence in situ hybridization, anterior is to the left. (C–C’”) A 12, (D–D’”) 16, (E–E’”) 20, (F–F’”) 24, (G–G’”) 36 hpf embryo stained for both pdgfra and pdgfrb mRNA via fluorescence in situ hybridization. The neural crest expresses pdgfra (Eberhart et al., 2008), while pdgfrb is restricted medial and ventral to pdgfra expression at 12 hpf. At 16 hpf, cells singly positive for either pdgfra or pdgfrb are observed within the crest stream (D–D”, arrow and asterisk, respectively). We also observe cells doubly positive within the stream (Arrowhead). More extensive co-expression of pdgfra and pdgfrb is observed at 20, 24, and 36 hours post fertilization (arrowheads in E–E”). Scale bar= 20 µm.
To characterize the relative expression of both pdgfra and pdgfrb, we used double fluorescent in situ analysis. At 12 hpf, we detect little to no overlap between pdgfra and pdgfrb (Fig. 5C–C’”). From earlier studies, pdgfra at these time points is expressed by migrating neural crest (Eberhart et al., 2008), while pdgfrb expression occurred ventral and medial to pdgfra positive cells. At 16 hpf, we observe the first overlap in expression of both receptors (Fig. 5D–D’”, arrowhead). However, we also observed cells singly positive for pdgfra or pdgfrb (Fig. 5D–D”, arrow and asterisk, respectively). While singly positive cells remain, co-expression appears common by 20 hpf, although the relative intensity of staining varies, suggesting that the relative levels of the two receptors may vary across cells (Fig. 5E–E”, small arrowhead; Eberhart et al., 2008; Wiens et al., 2010). Both receptors remain expressed in the pharyngeal arch until at least 36 hpf (Fig. 5G–G’”).
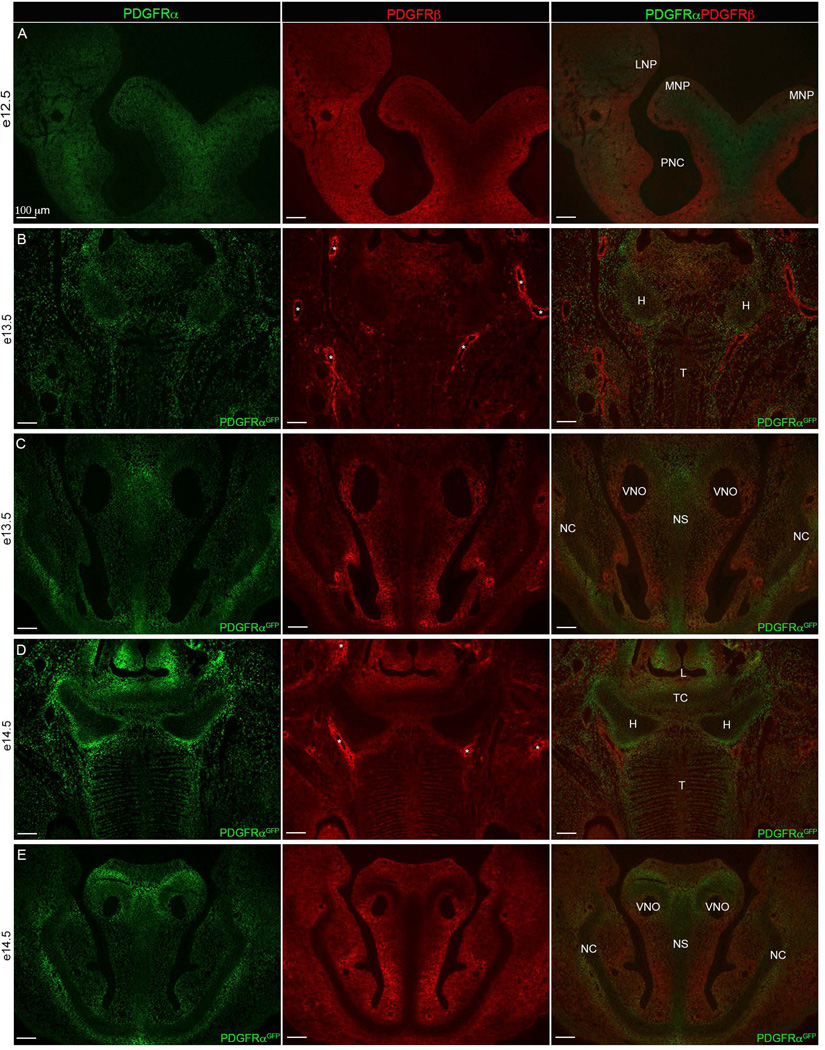
We also determined Pdgf receptor expression patterns in the mouse, detecting Pdgf receptors using either antibodies to PDGFRα and PDGFRβ or fluorescence detection of GFP in PdgfrαGFP embryos. In the developing head mesenchyme at e12.5, the two receptors were initially diffusely localized in similar regions (Fig. 6A), similar to what we observed in zebrafish. At later time points, e13.5 and e14.5, we observed a largely non-overlapping pattern of expression between PDGFRα and PDGFRβ. PDGFRβ was detected in the perichondrial layer surrounding both the hyoid bone and nasal cartilage (Fig. 6B–E). PDGFRα was continuous throughout the hyoid bone but noticeably higher in the perichondrial layer. Cartilage making up the nasal capsule, nasal septum, and surrounding the vomeronasal organs were also devoid of Pdgfrb expression while Pdgfra expression appeared to be fairly universal throughout. Thus, the expression of both Pdgf receptors is highly dynamic in the neural crest during craniofacial development. More detailed expression characterizations along with promoter analyses in both species will help determine the overall conservation of Pdgf receptor expression.
Figure 6. Pdgfrα and Pdgfrβ craniofacial expression.
(A) e12.5 (B–C) e13.5 and (D–E) e14.5. (A) PDGFRα (green) and PDGFRβ (red) protein expression in 10 µm transverse sections. (B–E) fluorescence detection of nuclear PDGFRαGFP transgene (green) and PDGFRβ protein (red) through (B, D) tongue and hyoid region and (C, E) frontonasal region. Overlap shows that the hyoid and frontonasal cartilages are devoid of Pdgfrβ expression while Pdgfrα expression is present. Asterisks (*) indicate blood vessels surrounded by Pdgfrβ expression exists. LNP=Lateral nasal process, MNP=medial nasal process, PMN= Primitive nasal capsule, H=Hyoid, T=Tongue, L= Laryngeal Aditus, TC= Thyroid Cartilage, VNO=Vomeronasal organ (Jacobson’s organ), NS=Nasal Septum, NC=Nasal Capsule.
Condensation of the maxillary domain fails in pdgfra;pdgfrb mutants
Palatogenesis involves two populations of neural crest, the frontonasal CNCC and the maxillary CNCC. Frontonasal CNCC migrate over the eye to reach the oral ectoderm and form the medial ethmoid plate (Eberhart et al., 2008; Wada et al., 2005). The maxillary CNCC first migrate to and form a tightly compacted group of cells (or condense) under the eye, upon the oral ectoderm, forming the upper jaw, lateral ethmoid plate and trabeculae (Eberhart et al., 2006; Swartz et al., 2011; Wada et al., 2005). In pdgfra mutants, frontonasal CNCC fail to migrate to the oral ectoderm, explaining the clefting of the ethmoid plate (Eberhart et al., 2008). The phenotype of pdgfra;pdgfrb double mutants and our inhibitor analyses would suggest that there is an additional defect to these processes in the maxillary CNCC of double mutants. To analyze the morphogenesis of maxillary neural crest in zebrafish, we performed time-lapsed confocal analyses in fli:EGFP transgenics.
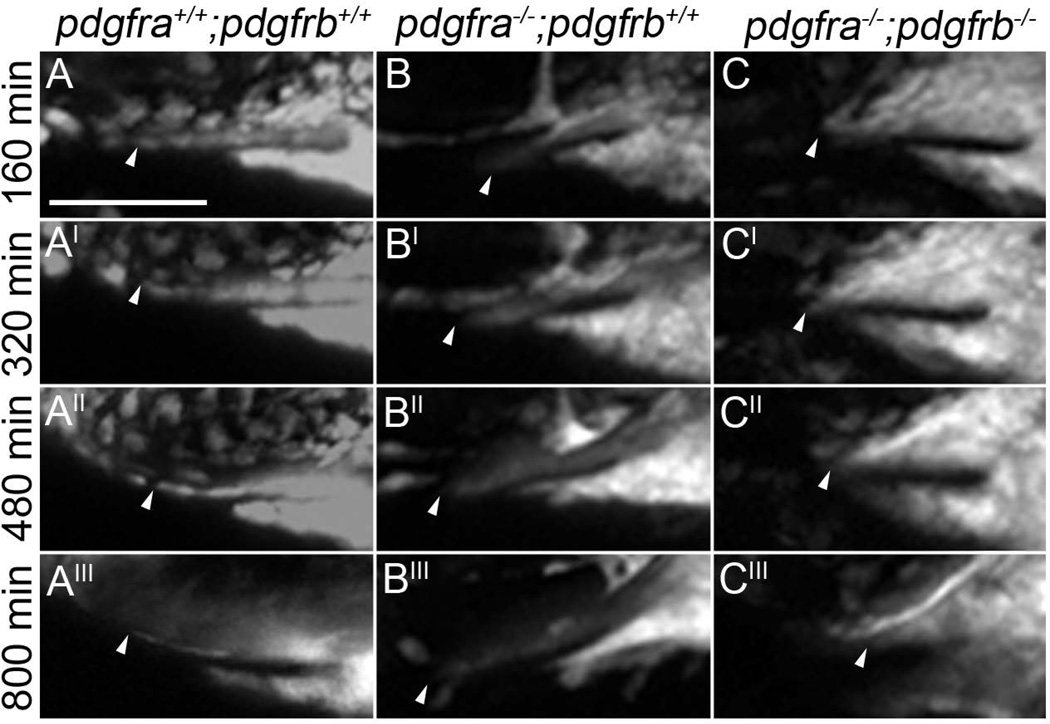
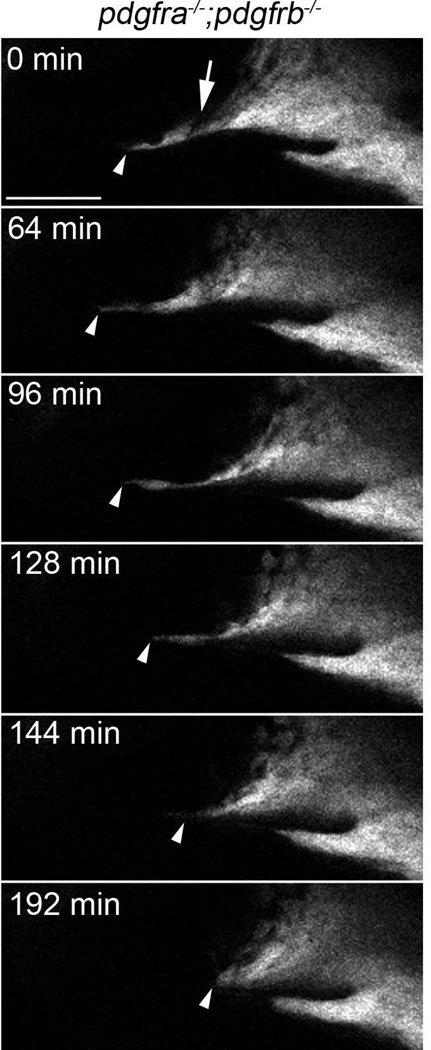
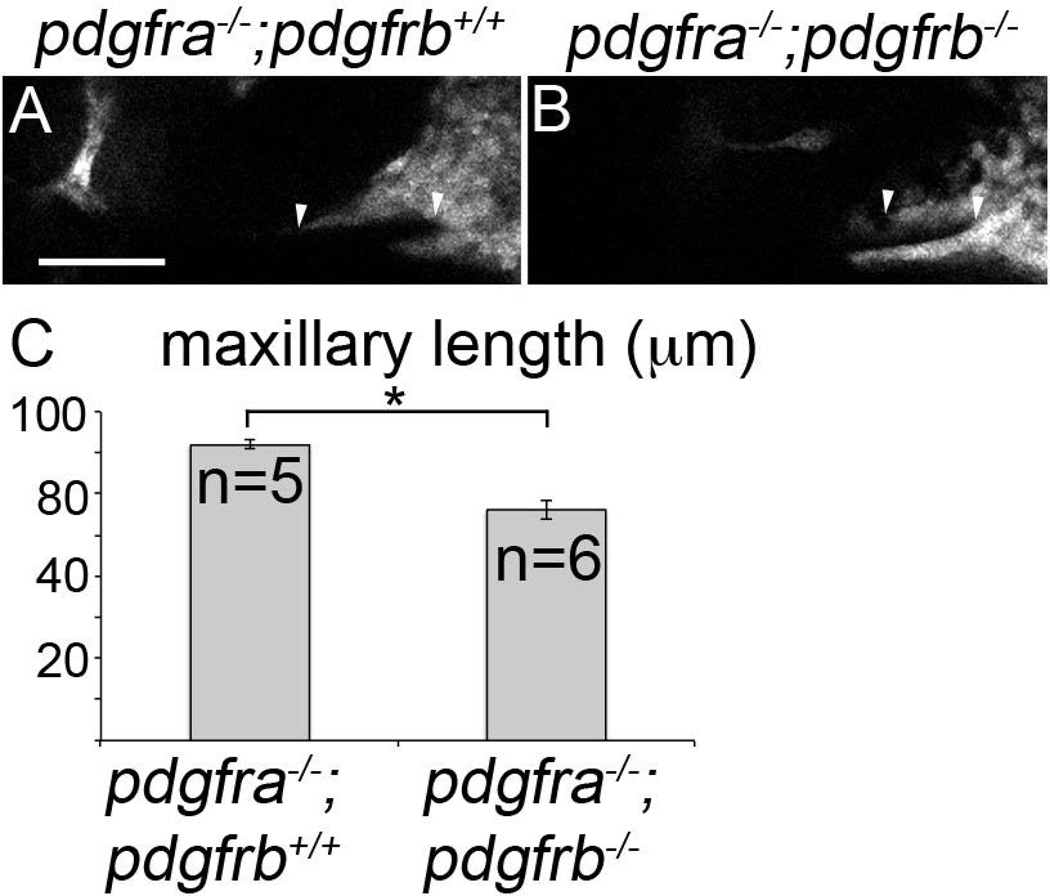
In wild type, maxillary neural crest cells have migrated to and condensed below the eye, meeting with the frontonasal mass by 24 hpf (Fig 7A–A’”, arrowheads denote most anterior maxillary cells in all images, Supplemental movie S1). Likewise, maxillary cells migrate to and condense below the eye in pdgfra mutants (Fig 7B–B’”, Supplemental movie S2). However, these cells do not connect with the mislocalized frontonasal CNCC (Eberhart et al., 2008). In contrast, maxillary cells fail to condense under the eye in pdgfra;pdgfrb mutants (Fig 7C–C’”, Supplemental movie S3). Early during maxillary neural crest formation, we did find that neural crest cells moved anteriorly and under the eye in pdgfra;pdgfrb double mutants, but then retracted posteriorly again (Fig 8, arrowhead). Furthermore, we found that gaps in the maxillary condensation were apparent in these double mutants (Fig. 8, arrow). This may suggest that loss of both Pdgf receptors results in overall failure of the morphogenesis of the anterior maxillary neural crest. We quantified the maxillary condensation by measuring the length of the oral ectoderm that had neural crest cells upon it, in lateral view. We found that that pdgfra−/−;pdgfrb−/− mutants display statistically significant reductions in the length of the condensation at 30 hpf (Fig. 9B compared to 9A, Fig 9C; students t-test, *p=0.0136). Because condensation is required for skeletogenesis, these data suggest that the exacerbated phenotype observed in pdgfra;pdgfrb mutants is, at least partly, caused by a failure of proper maxillary neural crest condensation during palatogenesis.
Figure 7.
Maxillary CNCC fail to condense properly in pdgfra−/−;pdgfrb−/− embryos beginning at 24 hpf. (A–A’”, B–B’”, C–C’”) Stills of confocal movies of pdgfra+/+;pdgfrb+/+, pdgfra−/−;pdgfrb+/+ and pdgfra−/−;pdgfrb−/− embryos, respectively. Anterior is to the left. Arrowheads point to the anterior most maxillary domain. In a pdgfra−/−;pdgfrb+/+ embryo (B–B’”), the maxillary CNCC condensation progress anteriorly throughout craniofacial morphogenesis, while in pdgfra−/−;pdgfrb−/− embryo (C–C’”), maxillary domain CNCC fails to extend, compared to pdgfra+/+;pdgfrb+/+ (A–A’”). Scale bar= 50 µm.
Figure 8.
Maxillary neural crest cells fail to extend anteriorly in pdgfra−/−;pdgfrb−/− mutants. Stills of supplemental movie S3 showing maxillary neural crest extending anteriorly in pdgfra−/−;pdgfrb−/− mutants early, then retracting posteriorly later (arrowhead). A gap in the condensation is also present at the beginning of the movie (arrow). Scale bar= 50 µm.
Figure 9.
Maxillary domain CNCC length is reduced in pdgfra;pdgfrb mutants at 30 hpf. (A–B) 30 hpf single z-stack confocal images of fli1:EGFP transgenic embryos with indicated genotypes above the panels, anterior is to the left. EGFP-labeling is depicted as grey. The maxillary condensation extends further anteriorly in (A) pdgfra−/−;pdgfrb+/+ embryos compared to (B) pdgfra−/−;pdgfrb−/− embryos (arrowhead denotes length measured). (C) Bar chart depicting maxillary neural crest length (Students t-test *p=0.0136). scale bar= 50 µm.
Proliferation and cell death are not significantly altered in the maxillary domain of pdgfra;pdgfrb mutants
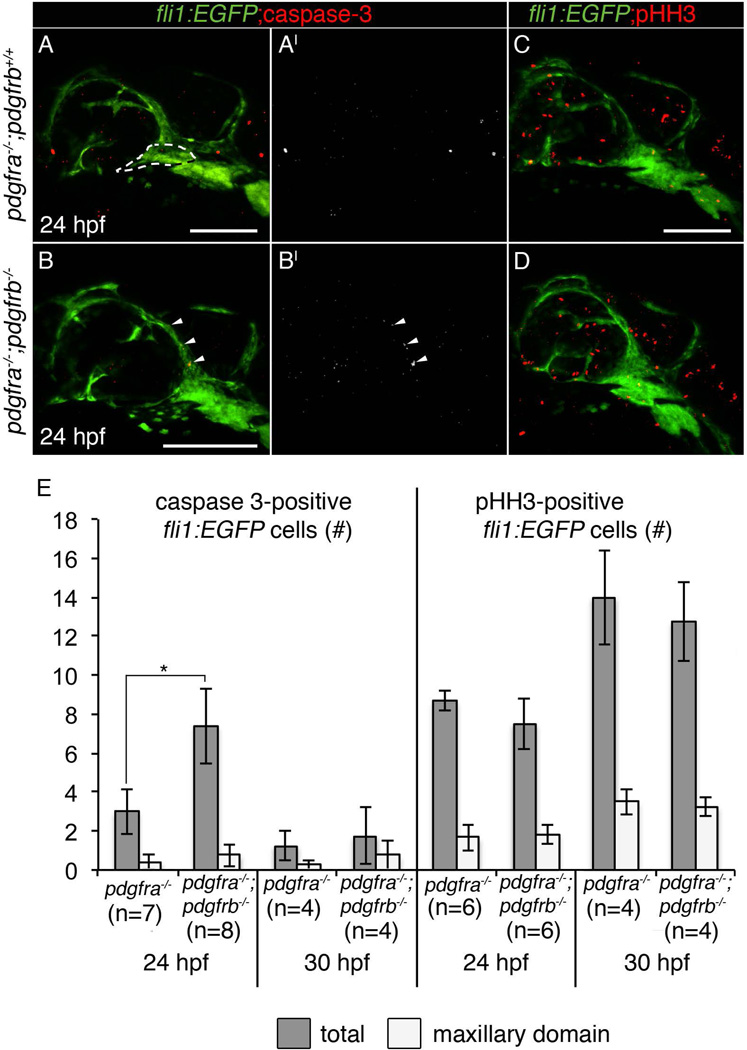
Loss of both pdgfra and pdgfrb led to more severe palate phenotypes compared to either loss of pdgfra or pdgfrb alone. Cell death is not observed in pdgfra single mutants (Eberhart et al., 2008; McCarthy et al., 2013). However, ethanol treatment causes increased neural crest cell death in pdgfra mutants (McCarthy et al., 2013), demonstrating that in certain contexts Pdgf signaling is critical for neural crest cell survival. Therefore, we analyzed cell death in fli1:EGFP transgenics using an antibody to active caspase-3 (Sigma). We chose two time points within the window when Pdgf inhibition disrupts palatal development, 24 and 30 hpf. Loss of pdgfrb alone does not cause any increase in cell death compared to wild type siblings at 24 hpf (neural crest cell death: pdgfrb−/− mean=1.29, pdgfrb+/+ mean=2.5, p=0.175). In the absence of both receptors, we see an increase in total cell death at 24 hpf (Fig. 10B–B’, 10A–A’, 10E *p=0.021, arrowheads), however, cell death located in the maxillary domain is not significantly different (Fig. 10E, see panel A for outline of maxillary domain area). Cell death occurred primarily above the eye in pdgfra;pdgfrb mutant embryos (Fig. 10B–B’, arrowheads), and this is an area that does not contribute to the palate in zebrafish (Eberhart et al., 2006; Swartz et al., 2011). At 30 hpf, cell death was not increased in pdgfra;pdgfrb mutants compared to pdgfra mutants alone (Fig. 10E). Therefore, at least at the times analyzed, loss of Pdgf signaling does not elevate maxillary neural crest cell death.
Figure 10.
Neither cell death nor proliferation is significantly affected in the maxillary domain of pdgfra;pdgfrb mutants. (A–A’, B–B’, C–D) 24 hpf fli1:EGFP zebrafish embryo with corresponding genotypes listed left of panels, stained for active caspase 3 (A and B in red; A’ and B’ in grey), and phospho-histone H3 (pHH3) in (C–D). Anterior is to the left. (B–B’) pdgfra−/−;pdgfrb−/− embryos show increased total cell death at 24 hpf, but not in the maxillary domain crest (arrowheads show cell death above the eye, maxillary domain is outlined in panel A). (E) Bar chart representing either total (grey bars) or maxillary domain (light bars) number of active caspase 3-positive or pHH3-positive fli1:EGFP cells in corresponding genotypes (Students T-test, p=<0.05). scale bar= 100 µm.
To quantify cell proliferation, we again utilized the neural crest-labeling fli:EGFP transgenic and stained for the proliferation marker pHH3 (Sigma). However, when compared to single pdgfra mutants, we found no change in cell proliferation in pdgfra;pdgfrb mutants, at either 24 or 30 hpf (Fig 10C–E). Together, these data suggest that, apoptosis and loss of proliferation within the maxillary domain are not altered during times in which the condensation of neural crest cells is disrupted. However, we cannot rule out an involvement of apoptosis or proliferation in the overall phenotype.
Discussion
Here, we show that Pdgfra and Pdgfrb interact during craniofacial development. In both mouse and zebrafish, Pdgfrb appears dispensable for craniofacial development. However, in both species, a requirement for Pdgfrb in craniofacial development is revealed when combined with loss of Pdgfra. In zebrafish, we go on to show that loss of Pdgf function during a time window when both receptors are expressed in the neural crest is critical for proper craniofacial development. Finally, we find that the maxillary CNCC fails to properly condense in pdgfra;pdgfrb mutants.
Despite the fact that Pdgfra and Pdgfrb are homologs with nearly identical signaling capabilities, their function in development has diverged extensively throughout evolution (Gu and Gu, 2003). However, there are now a handful of examples where these two genes interact during development. Pdgfc mediates the survival response of retinal ganglia cells following eye-injury and this response requires both Pdgfra and Pdgfrb function (Tang et al., 2010). Proper aortic arch and ventricular septum development requires both Pdgfra and Pdgfrb (Richarte et al., 2007), populations of cardiac neural crest cells express the receptors, and Pdgfra;Pdgfrb deficient neural crest cells have a more penetrant aortic arch phenotype than the single mutant neural crest cells (Richarte et al., 2007). More recently, epithelial-to-mesenchymal transition of epicardial cells has also been shown to require the function of both receptors (Smith et al., 2011). Additionally, while loss of PI3K signaling through Pdgfrb results in normal craniofacial development, the combined loss of PI3K signaling through both Pdgf receptors results in craniofacial defects as severe as a Pdgfra single mutant (Klinghoffer et al., 2002). Our results provide a novel example where the requirement of one receptor is only revealed when function of the other receptor is impaired. The mechanism of this apparent compensatory mechanism remains to be revealed.
Pdgf signaling and the maintenance of cell behaviors
We previously showed that pdgfra protects against ethanol-induced neural crest cell death (McCarthy et al., 2013). We hypothesized that ethanol may broadly impact Pdgf signaling, and thus, we predicted that pdgfra;pdgfrb double mutants would phenocopy ethanol-treated pdgfra single mutants. While pdgfra;pdgfrb mutants had an elevation in cell death at 24 hpf, this cell death was limited primarily to an area above the eye, that does not contribute to the palate (Eberhart et al., 2006; Swartz et al., 2011). Because fli:EGFP transgenics mark both neural crest and vasculature, and Pdgfrb has a known role in mural cell biology (Wiens et al., 2010; Mellgren et al., 2008; Soriano, 1994), this cell death may be vessel-associated. Our results here would suggest that the elevated cell death in ethanol-treated pdgfra mutants (McCarthy et al., 2013) is not simply a consequence of broadly inhibiting the Pdgf signaling pathway.
Recently, Pdgfra has been shown to be involved in cell proliferation in mouse (He and Soriano, 2015). While we did see a trend for decreased cell proliferation in pdgfra;pdgfrb fish, the effect was not significant. This may be due to the low amount of cell proliferation occurring in zebrafish at the time points examined, or slight differences in the roles of Pdgf signaling during development across species. Future studies would aim at understanding the full breadth of neural crest proliferative dynamics in pdgfra;pdgfrb zebrafish, for example, by utilizing the cell cycle transgenic fucci (Sugiyama et al., 2014). While it is possible that neural crest cell death and/or proliferation may be affected at time points not examined, our data suggest that during the time in which neural crest cell condensation is disrupted neither cell death nor proliferation are significantly altered in pdgfra;pdgfrb mutants. Similarly, neither cell death nor proliferation was found to be altered in the cardiac neural crest in Pdgfra;Pdgfrb mouse mutants (Richarte et al., 2007). Thus, these data implicate that the Pdgfra;Pdgfrb interaction involves cell movements.
The maxillary CNCC is a highly dynamic population of neural crest that, upon condensation on the oral ectoderm, undergo numerous cell rearrangements to form the palate (Dougherty et al., 2012; Eberhart et al., 2008; Swartz et al., 2011; Wada et al., 2005). Using zebrafish, we were able to show a novel role of Pdgf signaling in maxillary CNCC condensation. Our in situ data shows that while pdgfra is expressed very early in neural crest migration (Eberhart et al., 2008), pdgfrb only becomes expressed in the neural crest populating the pharyngeal arches beginning at 16 hpf. Our inhibitor analysis demonstrated that craniofacial development required Pdgf signaling as maxillary neural crest cells condense upon the oral ectoderm. Using time-lapse confocal microscopy, we showed that while the frontonasal-migrating neural crest fails in pdgfra mutants, the maxillary CNCC was still able to condense anteriorly towards the area normally populated by frontonasal CNCC. This condensation completely fails in pdgfra;pdgfrb mutants, and we see no movement of the maxillary CNCC anteriorly at any time point during this process. How this condensation fails is of ongoing interest, but a similar loss of condensation occurs in smoothened (smo) mutant zebrafish (Eberhart et al., 2006). Smo function is required in the oral ectoderm for condensation (Eberhart et al., 2006) and several Pdgfs are expressed in the oral ectoderm (Eberhart et al., 2008; our unpublished results). It will also be of great interest to determine if Pdgf signaling functions downstream of Shh to stabilize neural crest cells upon the roof of the oral ectoderm.
In other systems where Pdgfra and Pdgfrb interact, cells rely on these signaling pathways to promote proper cell movements. For example, in the epicardium, Pdgfra and Pdgfrb are required for proper migration and epithelial to mesenchymal transition of these cells (Smith et al., 2011). Epicardial conditional Pdgfra;Pdgfrb mutant mice show decreased Sox9 expression and overexpressing Sox9 resulted in restored epicardial migration, epithelial-to-mesenchymal transition gene profiles, and actin reorganization dynamics (Smith et al., 2011). Loss of these receptors in cardiac neural crest, likewise, causes cell migration defects (Richarte et al., 2007). It may be possible that Sox9 loss is a mechanism by which cardiac and maxillary neural crest fail to migrate and condense, respectively. Loss of sox9 in zebrafish causes chondrocyte-stacking defects, as well as disrupted craniofacial cartilage formation (Yan et al., 2002). An important future question is whether sox9 is regulated by Pdgf signaling and whether the loss of sox9 explains the condensation defect in pdgfra;pdgfrb mutants.
Evolutionary conservation of Pdgf signaling during craniofacial development
CNCC express Pdgfra in fish, amphibian, avian, and mammalian species (Ho et al, 1994; Liu et al 2002; Marcelle et al., 1992; Orr-Urtreger and Lonai, 1992; Schatteman et al., 1992). Loss of Pdgfra signaling causes midfacial clefting in mouse and zebrafish (Eberhart et al., 2008; Soriano et al., 1997; Tallquist and Soriano, 2003). In both species, Pdgfra function is required autonomously in the neural crest cells that generate the craniofacial skeleton (Eberhart et al., 2008; Tallquist and Soriano, 2003). Also consistent across zebrafish and mouse is the requirement for Pdgfra signaling in the migration of frontonasal CNCC (Eberhart et al., 2008; He and Soriano, 2013). Collectively, these results suggest that Pdgfra function is conserved across vertebrate species
The cytoplasmic signaling necessary for craniofacial development also appears conserved. In mouse, mutant analysis has demonstrated that craniofacial development relies predominantly upon the PI3K effector of Pdgfra (Klinghoffer et al., 2002). In zebrafish, elevating PI3K signaling, by knocking down the negative regulator Ptena, rescues craniofacial development in pdgfra mutants (McCarthy et al., 2013).
Our current analyses demonstrate that a genetic interaction between Pdgfra and Pdgfrb in craniofacial development is conserved between mouse and zebrafish. While Pdgfrb appears completely dispensable for craniofacial development in the presence of Pdgfra, loss of or attenuated Pdgfra function reveals a requirement for Pdgfrb. In both mouse and zebrafish, Pdgfra;Pdgfrb double mutants have more extensive craniofacial defects than either Pdgfra or Pdgfrb mutants alone. In mouse, Pdgfra;Pdgfrb double PI3K effector domain mutants recapitulate the Pdgfra neural crest conditional mutant phenotype, suggesting that PI3K is also the major effector for Pdgfrb during craniofacial development (Klinghoffer et al., 2002). With the highly conserved nature of this pathway in mind, the functional data we provide here in zebrafish will likely provide valuable insight into future studies in the mammalian system.
We now know that Pdgfra interacts with a multitude of genes (Klinghoffer et al., 2002; Schmal et al., 2006), including Pdgfrb. Recent studies have linked PDGFRA to isolated cleft palate in human (Rattanasopha et al., 2012), and due to the prevalent nature of this defect, the interactions between PDGFRA and other genes may help explain this variability. Understanding the full breadth of these interactions will be vital in understanding the etiology of cleft lip and cleft lip/cleft palate diseases.
Experimental Procedures
Zebrafish care and use
All embryos were raised and cared for using established protocols with IACUC approval. Lines used in this study include pdgfrab1059(Eberhart et al., 2008), pdgfrbum148(Kok et al., 2014), and Tg(fli1:EGFP)y1(Lawson and Weinstein, 2002) called fli1:EGFP in the text. Embryos were treated with 0.1, 1, 1.5, and 2 µM Pdgf Inhibitor V (Calbiochem) from a 10mM stock in DMSO diluted in embryo medium.
Mice
All mouse protocols were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center and the University of Hawaii and conform to NIH guidelines for care and use of laboratory animals. Mice were maintained on a mixed C57/BL6J X 129SV background. Mouse lines include: PdgfraGFP (Hamilton et al., 2003); Pdgfra− (Soriano, 1994); PdgfrbF3 (Tallquist et al., 2000); Pdgfrafl (Tallquist 2003); Pdgfrbfl (Richarte et al., 2007); and Wnt1-CreTg (Danielian et al., 1998). It should be noted that mice containing Pdgfrαfl allele exhibit hypomorphic phenotypes. For example, mice with one null allele and one floxed allele are not viable due to a range of phenotypes that including spina bifida and cleft palate. We hypothesize this occurs because the conditional allele still possesses the neomyocin resistance cassette which reportedly can disrupt transcript expression (Meyers et al., 1998).
Murine skeletal bone preparations
Staining for ossified bone (Alizarin Red; Sigma) and glycosaminoglycans of cartilage (Alcian blue; Sigma) was accomplished according to previously described methods (Hogan, 1994). The bone preparations were stored in 100% glycerol for preservation. After whole cranial images were photographed, individual bones were dissected and compared.
Immunohistochemistry and In situ hybridization
Embryos were fixed and processed as described previously (Maves et al., 2002; McCarthy et al., 2013) using anti-active caspase 3 (Promega) and anti-phospho histone H3 (Sigma) primary antibodies with AlexaFluor 568 secondary antibodies. Fluorescent in situ hybridization was performed as described previously (Jowett and Yan, 1996). A students T-test was performed for statistical analysis. All bar charts were made using Microsoft Excel 2011.
Zebrafish cartilage staining
5 day postfertilization embryos were stained with Alcian Blue and Alizarin Red for cartilage and bone (Walker and Kimmel, 2007), then flat mounted (Kimmel et al., 1998). Images were taken with a Zeiss Axio Imager-AI scope and measurements of neurocranial and viscerocranial elements were performed as described (McCarthy et al., 2013). We used ANOVA and Tukey-Kramer post-hoc test for all statistical analysis.
Confocal microscopy and figure processing
Confocal z-stacks were collected on a Zeiss LSM 710 using Zeb software. All images were processed in Adobe Photoshop CS. In figure 8, images were quantified using Zeb software and students T-test was performed for statistical analysis.
Supplementary Material
Bullet Points.
pdgfra and pdgfrb genetically interact in palatogenesis.
Neural crest conditional loss of both Pdgfra;Pdgfrb in mouse causes exacerbated craniofacial defects compared to either mutant alone.
pdgfra and pdgfrb are co-expressed in the neural crest beginning at 20 hpf, when maxillary neural crest begin to condense upon the oral ectoderm.
In zebrafish, maxillary neural crest condensation is defective in pdgfra;pdgfrb double mutants.
Acknowledgments
We would like to thank Nathan Lawson for contributing the pdgfrbum148 zebrafish line. We would also like to thank Anna Percy for the maintenance and care of all zebrafish lines. This work was supported by NIH/NIDCR grant R01DE020884 and NIH/NIAAA grant U24AA014811 (Riley PI) to JKE; NIH/NIAAA F31AA020731 to NM; F31GM73417-01 to AMR; and from the March of Dimes Birth Defects Foundation (5-FY2003-132), The Chun Foundation, and NIH/NHLBI grant (HL074257) to MDT; NIH/NIAAA K99AA023560 to CBL.
Footnotes
No competing interests to disclose.
References
- Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. The origins of the neural crest. Part I: embryonic induction. Mech Dev. 1997;69:3–11. doi: 10.1016/s0925-4773(97)00132-9. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Betsholtz C, Karlsson L, Lindahl P. Developmental roles of platelet-derived growth factors. Bioessays. 2001;23:494–507. doi: 10.1002/bies.1069. [DOI] [PubMed] [Google Scholar]
- Crump JG, Swartz ME, Eberhart JK, Kimmel CB. Moz-dependent Hox expression controls segment-specific fate maps of skeletal precursors in the face. Development. 2006;133:2661–2669. doi: 10.1242/dev.02435. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty M, Kamel G, Shubinets V, Hickey G, Grimaldi M, Liao EC. Embryonic fate map of first pharyngeal arch structures in the sox10: kaede zebrafish transgenic model. J Craniofac Surg. 2012;23:1333–1337. doi: 10.1097/SCS.0b013e318260f20b. [DOI] [PubMed] [Google Scholar]
- Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, Postlethwait JH. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart JK, Swartz ME, Crump JG, Kimmel CB. Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development. 2006;133:1069–1077. doi: 10.1242/dev.02281. [DOI] [PubMed] [Google Scholar]
- Gruneberg H, Truslove GM. Two closely linked genes in the mouse. Genet., Res. 1960;1:69–90. [Google Scholar]
- Gu J, Gu X. Natural history and functional divergence of protein tyrosine kinases. Gene. 2003;317:49–57. doi: 10.1016/s0378-1119(03)00696-6. [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol. 2003;23(11):4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Soriano P. A critical role for PDGFRalpha signaling in medial nasal process development. PLoS Genet. 2013;9:e1003851. doi: 10.1371/journal.pgen.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Soriano P. Sox10ER(T2) CreER(T2) mice enable tracing of distinct neural crest cell populations. Dev Dyn. 2015;244:1394–1403. doi: 10.1002/dvdy.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Symes K, Yordan C, Gudas LJ, Mercola M. Localization of PDGF A and PDGFR alpha mRNA in Xenopus embryos suggests signalling from neural ectoderm and pharyngeal endoderm to neural crest cells. Mech Dev. 1994;48:165–174. doi: 10.1016/0925-4773(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Hoch RV, Soriano P. Roles of PDGF in animal development. Development. 2003;130:4769–4784. doi: 10.1242/dev.00721. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costanini F, Lacy E. Manipulating the Mouse Embryo: A laboratory manual. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. (1994) [Google Scholar]
- Jowett T, Yan YL. Double fluorescent in situ hybridization to zebrafish embryos. Trends Genet. 1996;12:387–389. doi: 10.1016/s0168-9525(96)90091-8. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Eberhart JK. The midline, oral ectoderm, and the arch-0 problem. Integr Comp Biol. 2008;48:668–680. doi: 10.1093/icb/icn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Kruze G, Ullmann B, BreMiller RA, Larison KD, Snyder HC. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol. 1998;203:245–263. doi: 10.1006/dbio.1998.9016. [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Hamilton TG, Hoch R, Soriano P. An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development. Dev Cell. 2002;2:103–113. doi: 10.1016/s1534-5807(01)00103-4. [DOI] [PubMed] [Google Scholar]
- Knight RD, Schilling TF. Cranial neural crest and development of the head skeleton. Adv Exp Med Biol. 2006;589:120–133. doi: 10.1007/978-0-387-46954-6_7. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2014;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Liu L, Korzh V, Balasubramaniyan NV, Ekker M, Ge R. Platelet-derived growth factor A (pdgf-a) expression during zebrafish embryonic development. Dev Genes Evol. 2002;212:298–301. doi: 10.1007/s00427-002-0234-3. [DOI] [PubMed] [Google Scholar]
- Marcelle C, Eichmann A. Molecular cloning of a family of protein kinase genes expressed in the avian embryo. Oncogene. 1992;7:2479–2487. [PubMed] [Google Scholar]
- Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- McCarthy N, Wetherill L, Lovely CB, Swartz ME, Foroud TM, Eberhart JK. Pdgfra protects against ethanol-induced craniofacial defects in a zebrafish model of FASD. Development. 2013;140:3254–3265. doi: 10.1242/dev.094938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellgren AM, Smith CL, Olsen GS, Eskiocak B, Zhou B, Kazi MN, Ruiz FR, Pu WT, Tallquist MD. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ Res. 2008;103:1393–1401. doi: 10.1161/CIRCRESAHA.108.176768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Gen. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Lonai P. Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development. 1992;115:1045–1058. doi: 10.1242/dev.115.4.1045. [DOI] [PubMed] [Google Scholar]
- Osumi-Yamashita N, Ninomiya Y, Doi H, Eto K. The contribution of both forebrain and midbrain crest cells to the mesenchyme in the frontonasal mass of mouse embryos. Dev Biol. 1994;164:409–419. doi: 10.1006/dbio.1994.1211. [DOI] [PubMed] [Google Scholar]
- Rattanasopha S, Tongkobpetch S, Srichomthong C, Siriwan P, Suphapeetiporn K, Shotelersuk V. PDGFRa mutations in humans with isolated cleft palate. Eur J Hum Genet. 2012;20:1058–1062. doi: 10.1038/ejhg.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richarte AM, Mead HB, Tallquist MD. Cooperation between the PDGF receptors in cardiac neural crest cell migration. Dev Biol. 2007;306:785–796. doi: 10.1016/j.ydbio.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatteman GC, Morrison-Graham K, van Koppen A, Weston JA, Bowen-Pope DF. Regulation and role of PDGF receptor alpha-subunit expression during embryogenesis. Development. 1992;115:123–131. doi: 10.1242/dev.115.1.123. [DOI] [PubMed] [Google Scholar]
- Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res. 2011;108:e15–e26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Tallquist MD. PDGF function in diverse neural crest cell populations. Cell Adh Migr. 2010;4:561–566. doi: 10.4161/cam.4.4.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 1994;8:1888–1896. doi: 10.1101/gad.8.16.1888. [DOI] [PubMed] [Google Scholar]
- Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- Stanier P, Moore GE. Genetics of cleft lip and palate: syndromic genes contribute to the incidence of non-syndromic clefts. Hum Mol Genet. 2004;13(Spec No 1):R73–R81. doi: 10.1093/hmg/ddh052. [DOI] [PubMed] [Google Scholar]
- Sugiyama M, Saitou T, Kurokawa H, Sakaue-Sawano A, Imamura T, Miyawaki A, Iimura T. Live imaging-based model selection reveals periodic regulation of the stochastic G1/S phase transition in vertebrate axial development. PLoS Comput Biol. 2014;10:e1003957. doi: 10.1371/journal.pcbi.1003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz ME, Sheehan-Rooney K, Dixon MJ, Eberhart JK. Examination of a palatogenic gene program in zebrafish. Dev Dyn. 2011;240:2204–2220. doi: 10.1002/dvdy.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Klinghoffer RA, Heuchel R, Mueting-Nelsen PF, Corrin PD, Heldin CH, Johnson RJ, Soriano P. Retention of PDGFR-beta function in mice in the absence of phosphatidylinositol 3’-kinase and phospholipase Cgamma signaling pathways. Genes Dev. 2000 Dec 15;14(24):3179–3190. doi: 10.1101/gad.844700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development. 2003;130:507–518. doi: 10.1242/dev.00241. [DOI] [PubMed] [Google Scholar]
- Tallquist M, Kazlauskas A. PDGF signaling in cells and mice. Cytokine Growth Factor Rev. 2004;15:205–213. doi: 10.1016/j.cytogfr.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Tang Z, Arjunan P, Lee C, Li Y, Kumar A, Hou X, Wang B, Wardega P, Zhang F, Dong L, Zhang Y, Zhang SZ, Ding H, Fariss RN, Becker KG, Lennartsson J, Nagai N, Cao Y, Li X. Survival effect of PDGF-CC rescues neurons from apoptosis in both brain and retina by regulating GSK3beta phosphorylation. J Exp Med. 2010;207:867–880. doi: 10.1084/jem.20091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Arjunan P, Lee C, Li Y, Kumar A, Hou X, Wang B, Wardega P, Zhang F, Dong L, Zhang Y, Zhang SZ, Ding H, Fariss RN, Becker KG, Lennartsson J, Nagai N, Cao Y, Li X. Survival effect of PDGF-CC rescues neurons from apoptosis in both brain and retina by regulating GSK3beta phosphorylation. J Exp Med. 2010;207:867–880. doi: 10.1084/jem.20091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA. Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A. 2010;152A:2984–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan HN, Soriano P. SRF regulates craniofacial development through selective recruitment of MRTF cofactors by PDGF signaling. Dev Cell. 2014;31:332–344. doi: 10.1016/j.devcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, Schilling TF. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 2005;132:3977–3988. doi: 10.1242/dev.01943. [DOI] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Wiens KM, Lee HL, Shimada H, Metcalf AE, Chao MY, Lien CL. Platelet-derived growth factor receptor beta is critical for zebrafish intersegmental vessel formation. PLoS One. 2010;5:e11324. doi: 10.1371/journal.pone.0011324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat Rev Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, Chung BC, Westerfield M, Haffter P, Hopkins N, Kimmel C, Postlethwait JH. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129:5065–5079. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.