Abstract
Amelogenin is the most abundant enamel protein involved in enamel mineralization. Our goal was to determine whether all three regions of amelogenin (N-terminus, C-terminus, central core) are required for enamel formation. Amelogenin RNA is alternatively spliced, resulting in at least 16 different amelogenin isoforms in mice, with M180 and LRAP expressed most abundantly. Soon after secretion by ameloblasts, M180 is cleaved by MMP20 resulting in C-terminal truncated (CTRNC) amelogenin. We aimed to determine whether the 2 transgenes (Tg), LRAP and CTRNC together, can improve LRAPTg/Amelx−/− and CTRNCTg/Amelx−/− enamel thickness and prism organization, which were not rescued in Amelx−/− enamel. We generated CTRNCTg/LRAPTg/Amelx−/− mice and analyzed developing and mature incisor and molar enamel histologically, by microCT, SEM and microhardness testing. CTRNCTg and LRAPTg overexpression together significantly improved the enamel phenotype of LRAPTg/Amelx−/− and CTRNCTg/Amelx−/− mouse enamel, however enamel microhardness was recovered only when M180Tg was expressed, alone or with LRAPTg. We determined that both LRAP and CTRNC, which together express all three regions of the amelogenin protein (N-terminus, C-terminus and hydrophobic core) contribute to the final enamel thickness and prism organization in mice.
Keywords: enamel, ameloblasts, amelogenin, Mmp20, secretory stage
1. Introduction
Amelogenin proteins make up 90% of the enamel matrix secreted by ameloblasts during the secretory stage and include at least 16 alternative splice products [1]. Amelogenin proteins are cleaved during the secretory stage by matrix metalloproteinase-20 (MMP20) [2], resulting in the accumulation of different amelogenin cleavage products in the developing enamel matrix of between ~5–30 kDa [3–5]. Ablation of the murine amelogenin gene leads to non-prismatic, thin enamel, around 10% the thickness of WT, similar to human AI with an amelogenin mutation [6]. The two most abundant amelogenin splice variants are M180, (mouse protein, 180 amino acids) and LRAP (59 amino acids). M180 and LRAP amelogenin may have different functions and their expression is regulated differently [7], however both splice variants have identical, highly conserved N- and C-termini [8] (Fig. 1). M180 is the largest and most abundant amelogenin splice variant, and is also thought to be the most crucial, rescuing approximately 50% the thickness of knockout enamel [9], and improving enamel prism organization [10, 11]. The LRAP isoform, which is highly expressed in secretory stage enamel, albeit at a lower level than M180, is believed to contribute to enamel structure and thickness in cooperation with full-length M180 [9] but not if expressed alone [12] in Amelx−/− mice. We aimed to determine whether expression of all three regions of amelogenin, when expressed as CTRNCTg and LRAPTg, are involved in enamel formation.
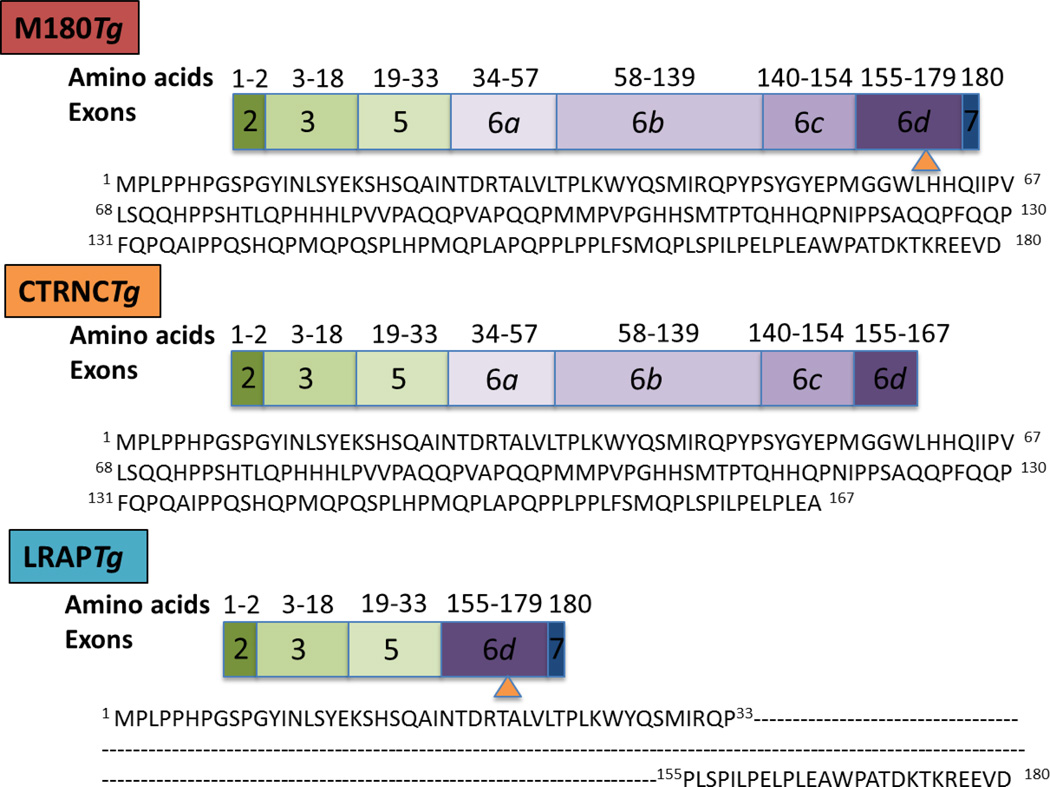
Fig. Schematic of amelogenin transgenes used in this study.
M180Tg and LRAPTg represent the major amelogenin isoforms, while CTRNCTg represents the major amelogenin cleavage product. M180Tg contains exons 2, 3, 5, 6a–6d, 7; CTRNCTg contains exons 2, 3, 5, 6a-part of 6d; LRAPTg contains exons 2, 3, 5, 6d, 7. Amino acid numbers and sequences corresponding to transgene exons are shown. Orange arrowhead indicates the site of the first MMP20 cleavage.
Although overexpression of an M180 transgene with a C-terminus deletion (CTRNCTg) did not rescue Amelx−/− mouse enamel [13], cleaved M180 may interact with LRAP to guide mineralization in vivo since M180 is cleaved at the C-terminus early in the secretory stage by Mmp20. We therefore hypothesize that transgenes for cleaved M180 (CTRNCTg) and LRAP overexpressed together in Amelx−/− mice can improve rescue over either transgene alone, due to a cumulative effect on enamel phenotype from the presence of all three domains of amelogenin.
2. Results
2.1 Endogenous and transgenic expression of amelogenin isoforms and cleavage products
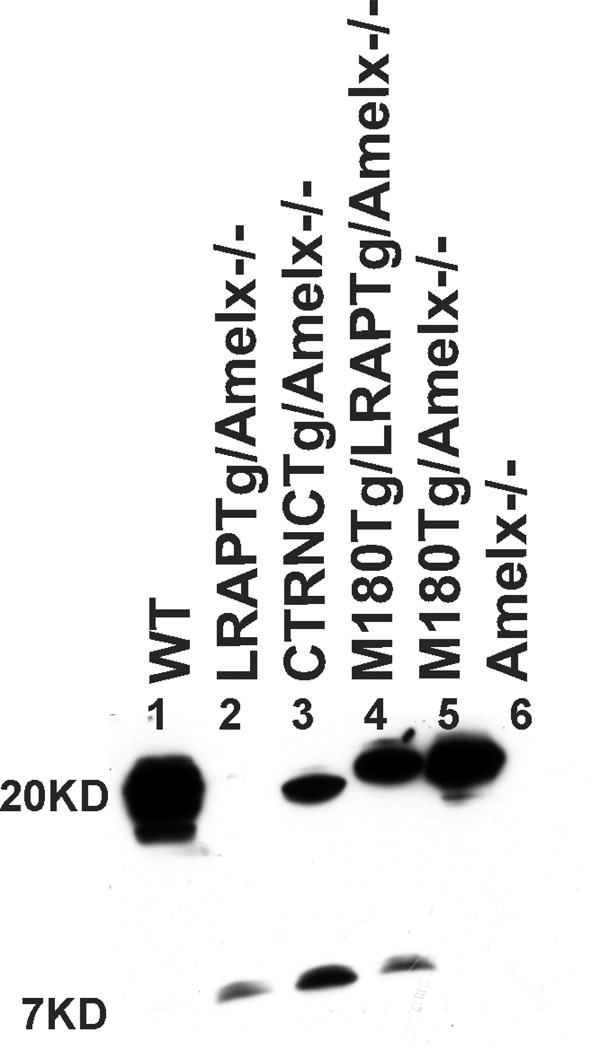
We compared relative protein expression of M180Tg (~20KD) CTRNCTg (~19KD) and LRAPTg (~7KD) transgenes compared to WT amelogenin using Western blot (Fig. 1). Band intensities were normalized relative to WT in multiple immunoblots (not shown). LRAPTg (~7 kD) was expressed at a higher level in LRAPTg/Amelx−/−, M180Tg/LRAPTg/Amelx−/− and CTRNCTg/LRAPTg/Amelx−/− mice (Fig. 2, lanes 2, 3, 4) than endogenous LRAP in WT mice, as the LRAP band was not visible in this immunoblot (Fig. 2, lane 1). Endogenous M180 and truncated M180 (CTRNC) appeared as a large smear around 17 – 20 kD in WT, which also includes additional amelogenin splice variants and cleavage products (Fig. 2, lane 1). The protein size difference between M180Tg and CTRNCTg, which lacks the last 13 amino acids in M180Tg’s C-terminus, is apparent, when comparing CTRNCTg/LRAPTg/Amelx−/− to M180Tg/LRAPTg/Amelx−/− and M180Tg/Amelx−/− of the Western blot (Fig. 2, lanes 3, 4, 5). The CTRNCTg and M180Tg band intensities normalized to WT amelogenin expression of the smear around 17–20 kD indicated similar but slightly lower expression levels of the transgenes (Fig. 2, lanes 3, 4, 5). This is due to the presence of additional endogenous amelogenin cleavage products in the WT developing enamel.
Fig. 2. Amelx protein expression in WT, Amelx−/− and transgenic 5 day-old developing molars.
WT (lane 1) molars had a smear of amelogenin protein between 17–20 kD representing most of the splice variants and cleavage products expressed during the secretory stage. In this immunoblot, the LRAP band (~7 kD) is not visible (although it was at longer exposure times), but other immunoblots of different WT mice show faint bands around 7 kD representing LRAP. Lanes 2–5 are from Tg mice in Amelx−/− backgrounds. Lane 6 is a Amelx−/− molar showing no amelogenin protein. LRAPTg expression in double transgenic/Amelx−/− mice (lanes 3 & 4) is noticeably higher than that in WT molars. M180Tg (lanes 4 & 5) and CTRNCTg (lane 3) expression was slightly less than to endogenous WT levels (lane 1), presumably due to the presence of less cleavage products.
2.2 Truncated M180 (CTRNC) and LRAP transgenes together significantly improve Amelx−/− enamel thickness
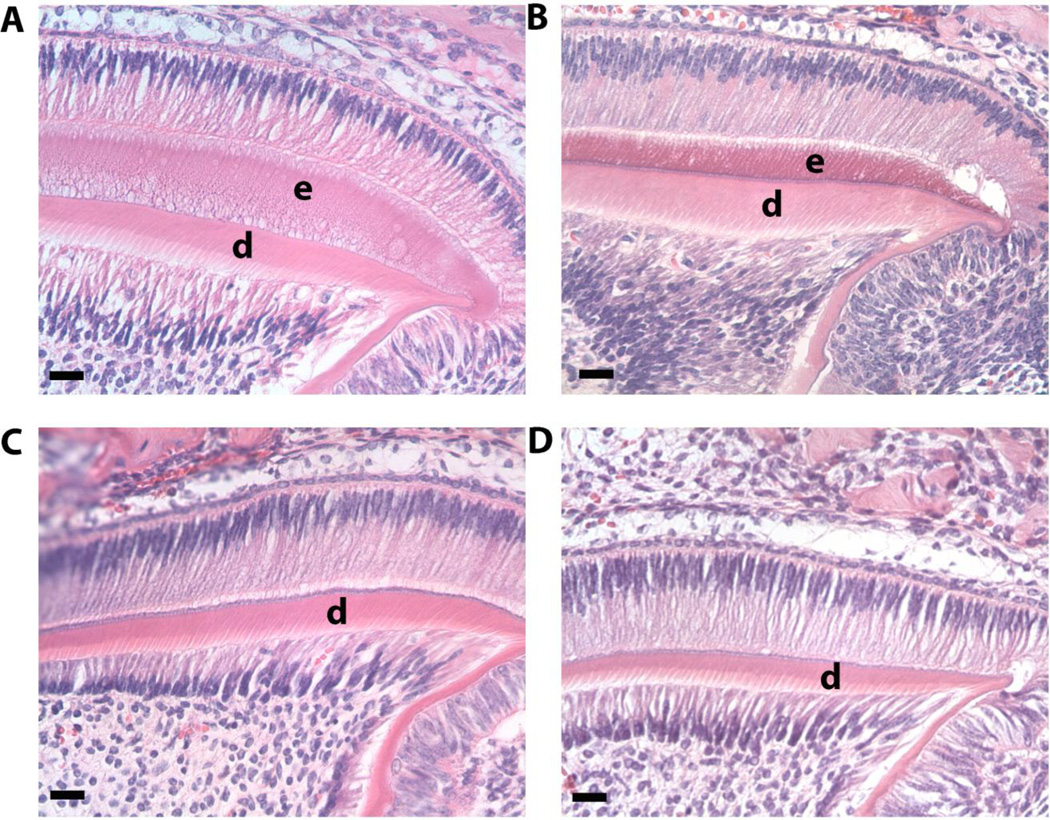
We analyzed secretory stage enamel histologically in developing molars from 5 day-old mice. Although the enamel layer was not as thick as WT (Fig. 3A) CTRNCTg/LRAPTg/Amelx−/− (Fig. 3B) molars have more enamel than both Amelx−/− (Fig. 3D) and LRAPTg/Amelx−/− (Fig. 3C) molars, which had little to no enamel.
Fig. 3. Histological analysis of developing enamel in 5 day-old 1st molars.
Images of H&E stained molars were obtained within the distal cusp of the developing 1st molar at 40× and showed noticeably more enamel in WT (A) and CTRNCTg/LRAPTg/Amelx−/− (B) compared to LRAPTg/Amelx−/− (C) which showed a thin layer of enamel, and Amelx−/− (D), which had no enamel presence. Loss of endogenous amelogenin (B, C, D) and the addition of transgene overexpression (B & C) appeared to have no effect on ameloblasts, odontoblasts or dentin.
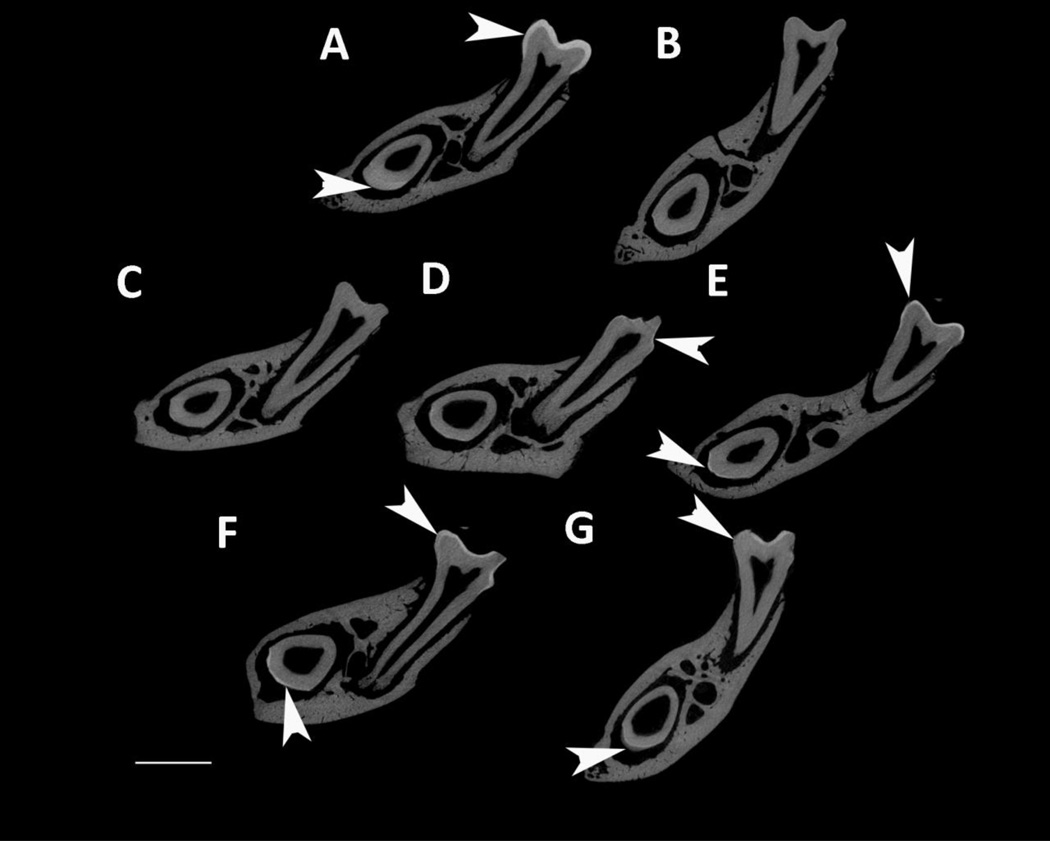
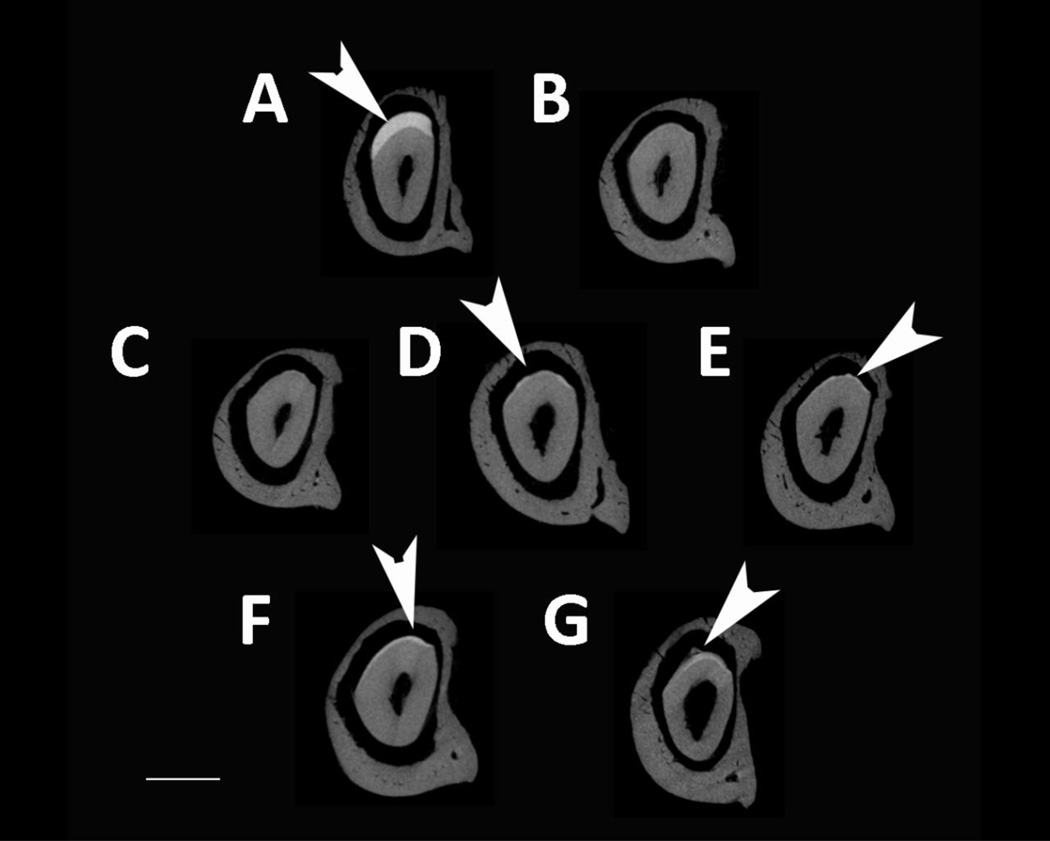
MicroCT was used to analyze enamel mineral in adult mandibles at a location that was sectioned through both the mature first molar and the developing incisor near the beginning of maturation, when the enamel is not fully mineralized in WT mice (Fig. 4). Amelx−/− and LRAPTg/Amelx−/− molar and incisor enamel are mostly not visible, too thin to be detectable by the microCT’s resolution (Fig. 4B & 4C). Thin layers of enamel were visible in CTRNCTg/Amelx−/− molars (Fig. 4D), M180Tg/KO incisors (Fig. 4E, M180Tg/LRAPTg/Amelx−/− incisors (Fig. 4F) and CTRNCTg/LRAPTg/Amelx−/− incisors (Fig. 4G), while thicker layers of mineralized enamel were visible in M180Tg/Amelx−/− molars (Fig 4E), M180Tg/LRAPTg/Amelx−/− (Fig. 4F) molars and CTRNCTg/LRAPTg/Amelx−/− molars (Fig. 4G). Notably, the enamel in CTRNCTg/LRAPTg/Amelx−/− incisors (Fig. 4G) appeared less mineralized than the visible enamel in the other incisors at the same stage.
Fig. 4. MicroCT analysis of adult molar and incisor enamel.
MicroCT scanned sections were selected in the same location developmentally in each mouse mandible: within the distal root of the first molar, which is adjacent to early maturation-stage of the incisor. Arrowheads point to mineralized enamel layers visible in molars and incisors. Little to no enamel was visible in Amelx−/− and LRAPTg/Amelx−/− molars and incisors (B & C), and CTRNCTg/Amelx−/− incisors (D). Thin enamel layers were visible in CTRNCTg/Amelx−/− molars (D), and M180Tg/Amelx−/− (E), M180Tg/LRAPTg/Amelx−/− (F) and CTRNCTg/LRAPTg/Amelx−/− (G) incisors. Thicker enamel layers were visible in WT molars and incisors (A), M180Tg/Amelx−/− (E), M180Tg/LRAPTg/Amelx−/− (F), and CTRNCTg/LRAPTg/Amelx−/− molars (G). Interestingly, incisor enamel in CTRNCTg/LRAPTg/Amelx−/− (G) appeared much less mineralized than that in the other genotypes with visible incisor enamel (A, E, F).
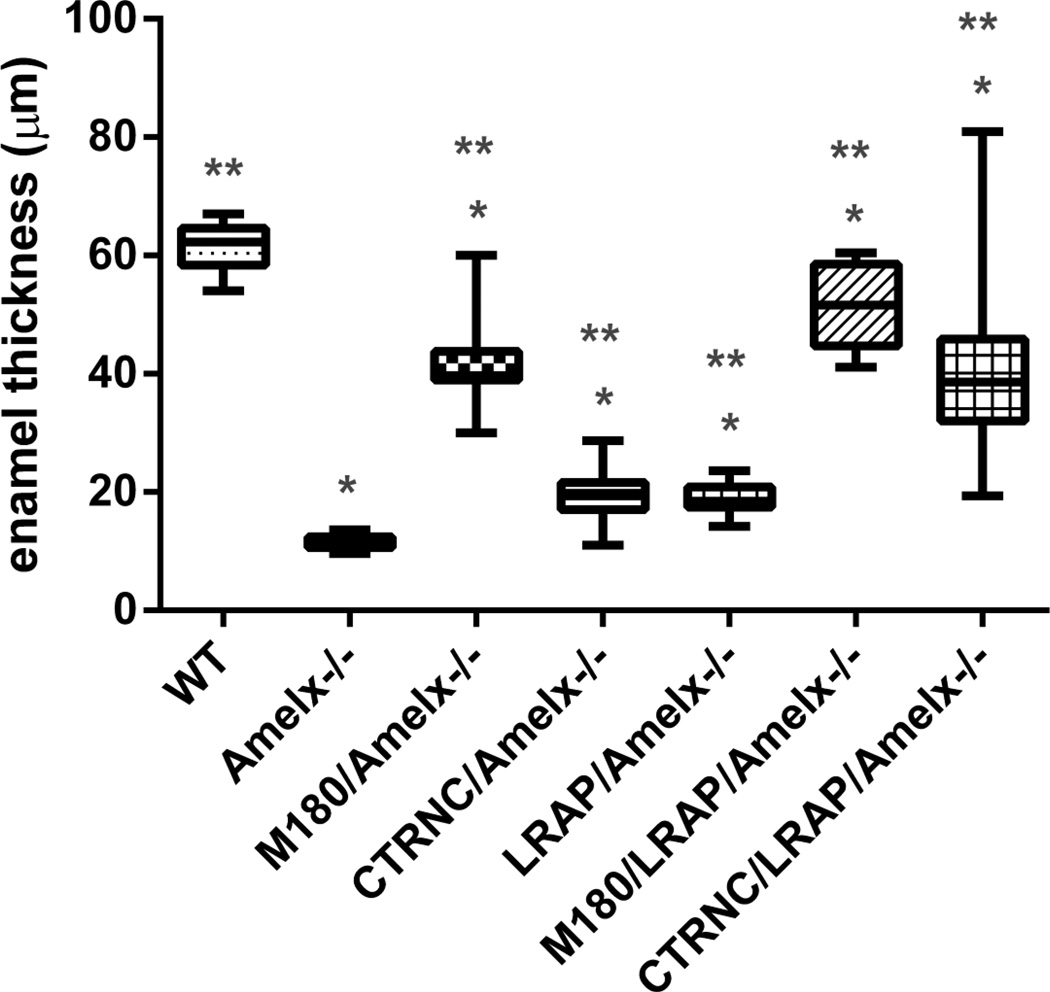
We also examined enamel mineral in incisors from Amelx−/− and transgenic mice by microCT at a later stage of development in the maturation stage, just before eruption, in the last section with alveolar one surrounding the incisor (Fig. 5). In contrast to Amelx−/− (Fig. 5B) and LRAPTg/Amelx−/− (Fig. 5C) incisors in which enamel was not visible, thin layers of mineralized enamel were visible in incisors from mice with the M180Tg with (Fig. 5F) and without (Fig. 5E) LRAPTg, and both CTRNCTg and LRAPTg (Fig. 5G). WT incisor enamel (Fig. 5A) was noticeably thicker than that of any other genotype, presumably due to the lower transgene expression levels in incisors compared to molars [10], leading to less overall rescue in incisors than occurred in molars (Fig. 4).We measured enamel thickness in adult first molars that were imaged by SEM. Molar enamel thickness of CTRNCTg/LRAPTg/Amelx−/− mice improved to 67% the thickness of WT from 19% for Amelx−/−), 32% for CTRNCTg/Amelx−/− and 31% for LRAPTg/Amelx−/− (Fig. 6). M180Tg/LRAPTg/Amelx−/− molar was 83% the thickness of WT and significantly thicker than CTRNCTg/LRAPTg/Amelx−/− molar enamel (Fig. 6). These data are consistent with histological and microCT analyses which indicate significantly more rescue of Amelx−/− enamel thickness with overexpression both CTRNCTg and LRAPTg versus either transgene alone.
Fig. 5. MicroCT analysis of adult incisor maturation stage enamel.
MicroCT scanned sections were selected in the same location developmentally in each mouse mandible: prior to eruption in the maturation-stage of the incisor in the final section that shows the incisors completely surrounded by alveolar bone. Arrows point to mineralized enamel layers visible in incisors. Little to no enamel was visible in Amelx−/−, LRAPTg/Amelx−/−, CTRNCTg/Amelx−/− incisors (B, C, D). Thin mineralized enamel layers were visible in M180Tg/Amelx−/− (E), M180Tg/LRAPTg/Amelx−/− (F) and CTRNCTg/LRAPTg/Amelx−/− (G) incisors. Thicker mineralized enamel layers were only visible in WT incisors (A).
Fig. 6. Molar enamel thickness.
Enamel thickness was measured from SEM images of longitudinal sections through distal first molars. All of the transgenic molar enamel was significantly thicker than Amelx−/− molar enamel (p<0.05), and not as thick as WT enamel (p<0.05). However, mice with M180Tg overexpressed alone or with LRAPTg had thicker enamel than those mice without M180Tg (CTRNCTg/Amelx−/−, LRAPTg/Amelx−/− and CTRNCTg/LRAPTg/Amelx−/−). CTRNCTg/LRAPTg/Amelx−/− mice had thicker molar enamel than both CTRNCTg/Amelx−/− and LRAPTg/Amelx−/− mice (p<0.05). * = significantly different from WT (p<0.05), ** = significantly different from Amelx−/− (p<0.05). Boxes represent a range of values between first and third quartile, the line in the center being the median, while the vertical lines represent the maximum and minimum values.
2.3 CTRNC and LRAP transgenes together improved enamel structure
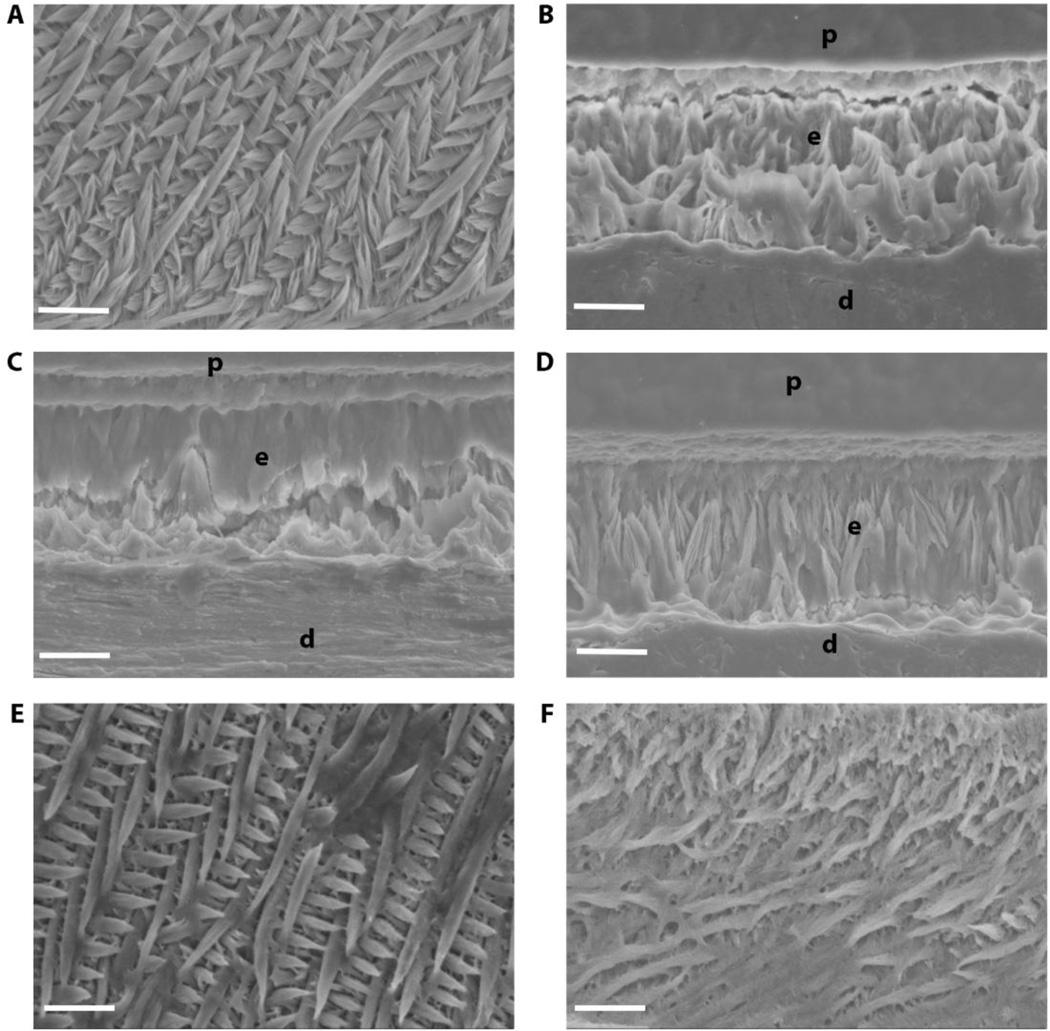
Because we wanted to observe the combined effect of truncated M180 and LRAP transgenes on enamel formation, we used SEM to analyze structure of acid-etched incisor enamel (Fig. 7). WT enamel was highly organized with decussating prisms (Fig. 7A). The thin, non-prismatic Amelx−/− enamel in adult mouse incisors was not significantly affected by the addition of truncated M180 transgene (CTRNCTg) (Fig. 7A, D) or LRAPTg (Fig. 7A, C), when expressed alone. However when these transgenes were expressed together (CTRNCTg/LRAPTg/Amelx−/−) the enamel had more prism organization (Fig. 7F). This suggests an important role for all three domains (N-terminus, central core, C-terminus) of amelogenin, which were represented by the combination of CTRNCTg and LRAPTg. M180Tg/LRAPTg/Amelx−/− enamel was highly organized with decussating enamel prisms and similar structure as WT (Fig. 7E).
Fig. 7. Incisor enamel structure imaged by SEM.
Polished and etched longitudinal sections through mandibular incisor enamel were imaged by scanning electron microscopy in secondary mode at 2000×. While WT (A) exhibits the classic decussating prismatic organization of incisor enamel, Amelx−/− (B), LRAPTg/Amelx−/− (C), and CTRNCTg/Amelx−/− (D) incisor enamel was non-prismatic, with CTRNCTg/Amelx−/− enamel exhibiting slightly more organized mineral. CTRNCTg/LRAPTg/Amelx−/− (F) incisor enamel had noticeably more mineral organization with some decussating prisms, although not as much decussating prisms as appeared in M180Tg/LRAPTg/Amelx−/− incisor enamel (E). p = plastic embedding material, e = enamel, d = dentin
2.4 CTRNC and LRAP transgenes did not improve enamel hardness
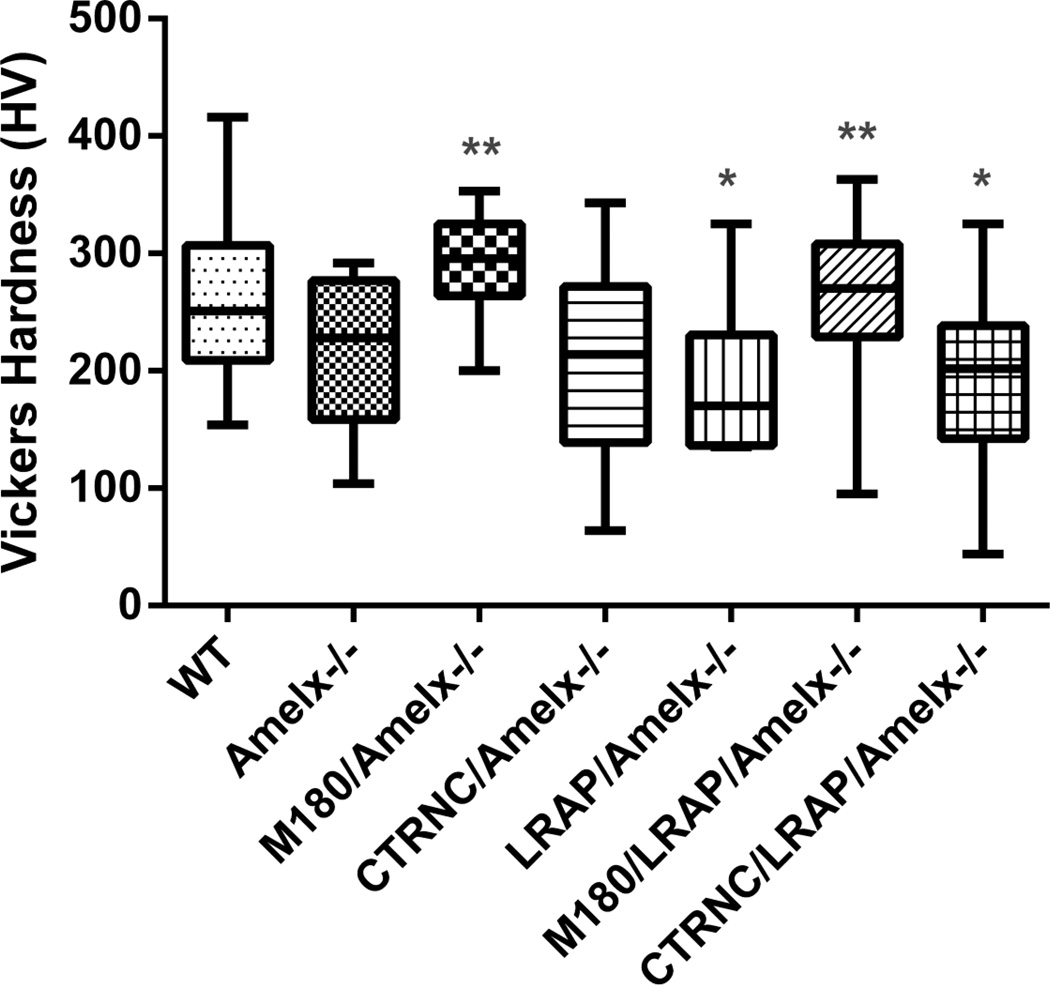
Microhardness measurements of molar enamel showed that CTRNCTg/LRAPTg/Amelx−/− enamel was significantly softer than WT enamel, and enamel of M180/LRAP/Amelx−/−mice was not different from WT (Fig. 8). Notably, M180Tg/LRAPTg/Amelx−/−incisor enamel structure was more organized (Fig. 7E) and thicker (Fig. 6) than CTRNCTg/LRAPTg/Amelx−/−, indicating a crucial role of the C-terminus of M180 in thickness, structure and resulting mechanical properties.
Fig. 8. Vickers Microhardness Testing.
Polished longitudinal sections through mandibular molar enamel were used for microhardness testing. If possible, 20 indents per mouse were obtained in adult molar enamel. M180Tg overexpression either alone or with LRAPTg significantly improved enamel hardness over Amelx−/− or transgenic mice without M180Tg. * = significantly different from WT (p<0.05), ** = significantly different from Amelx−/− (p<0.05). Boxes represent a range of values between first and third quartile, the line in the center being the median, while the vertical lines represent the maximum and minimum values.
3. Discussion
Although LRAP transgene overexpression upregulated wild-type mouse ameloblast differentiation in vivo [14], little is known about the role of the LRAP amelogenin isoform in enamel mineralization during the secretory stage after it is secreted. Like M180, LRAP is cleaved at its C-terminus by MMP20 [15], and while the role of this cleavage is unknown, it is possible that LRAP cleavage products also play a crucial role in enamel formation, albeit at a lower concentration than M180 cleavage products.
Additionally, while many investigators have shown the importance of M180 and its C-terminus both in vivo [10, 11, 13, 16] and in vitro [17–22], little is understood about the role of cleaved M180 without it’s C-terminus (CTRNC) or of the C-terminus itself, although truncated M180 has been suggested to stabilize amorphous calcium phosphate in vitro [23, 24]. Interestingly, the C-terminal amino acids of amelogenin were truncated in two reported amelogenesis imperfecta-causing mutations [25, 26].
We showed that C-terminal cleaved M180 transgene (CTRNCTg) and LRAPTg, when overexpressed together in Amelx−/− mice, have a pronounced effect on both incisor and molar enamel, improving both thickness and prism organization but not mechanical properties. In previous studies, when express separately, CTRNCTg [13] and LRAPTg [12, 27] had very little effect on the Amelx−/− enamel phenotype, although mechanical properties of LRAPTg/Amelx−/− enamel were significantly improved over Amelx−/− [27]. However, when M180Tg was overexpressed in addition to LRAPTg, both enamel thickness and enamel prism organization were considerably improved [9] over both LRAPTg/Amelx−/− [12, 27] and M180Tg/Amelx−/− [10] enamel. Although a recent in vivo study suggests that M180 is sufficient for thickness and mechanical properties [11], the enamel prismatic architecture was not thoroughly examined at high magnification. Taken together, our data along with that from the literature discussed above provide crucial mechanistic insights regarding the role of the most abundant amelogenins expressed during the secretory stage: M180, cleaved M180 (referred to here as CTRNC) and LRAP.
In vivo, CTRNC may interact with LRAP to promote crystallite elongation during the secretory stage, as suggested by the improved enamel thickness of CTRNCTg/LRAPTg/Amelx−/− mice (Fig. 5). Furthermore, as evidenced by the noticeable improvement of enamel structure, CTRNC and LRAP seem to also promote proper crystallite orientation, permitting partial decussating prisms to form (Fig. 7F), however, surprisingly, the enamel hardness was not improved (Fig. 8). When the expression levels of the M180 and LRAP transgenes are similar, the only difference between the LRAPTg/Amelx−/− versus the M180Tg/Amelx−/− mice is the presence of the hydrophobic core domain in the M180Tg/Amelx−/− mice, suggesting an important role for the core domain in crystal elongation and enamel thickness.
Although the improvement in enamel structure was remarkable compared to Amelx−/− (Fig. 7B) and either LRAPTg or CTRNCTg overexpressed separately (Fig. 7C&D), the prism organization of enamel in mice with both M180Tg and LRAPTg was even more strikingly organized (Fig. 7E), appearing to have near normal decussating prisms. Furthermore, the presence of M180Tg alone or with LRAPTg resulted in significantly harder enamel, confirming a crucial role of the C-terminus of M180 in enamel structure, resulting mechanical properties and thickness. As mentioned the importance of the C-terminus of M180 has been reported both in vitro [17–22] and in vivo [13, 16].
In conclusion, we demonstrated in vivo that CTRNC and LRAP transgenes when overexpressed together can considerably improve the enamel phenotype of Amelx−/− mice, although not as drastically as the combination of M180 and LRAP, suggesting important roles for M180, cleaved M180 and LRAP in enamel crystallite formation, elongation and orientation during the secretory stage of enamel formation. Selective ablation of the LRAP splice variant is critically important to developing better understanding of LRAP’s role in enamel formation.
4. Methods
4.1. Generation of CTRNC and LRAP double transgenic mice
To generate CTRNCTg/LRAPTg/Amelx−/− mice, mice overexpressing both CTRNC [13] and LRAP [12] transgenes from the bovine amelogenin promoter [28] were mated with male Amelx−/0 mice [6]. After two generations of mating, both male and female mice were genotyped using PCR primers to detect transgenes [12, 13, 28] as well as amelogenin WT and Amelx−/− [6] DNA. To analyze expression of endogenous and transgenic amelogenin in CTRNCTg/LRAPTg/Amelx−/− male and female mice and controls (WT, Amelx−/−, CTRNCTg/Amelx−/−, M180Tg/Amelx−/−, LRAPTg/Amelx−/−, M180Tg/LRAPTg/Amelx−/−), first molars were harvested from 5 day-old mouse pups and protein was extracted. Equal amounts of protein were loaded in each lane and run on a 4–20% SDS-PAGE gel (BioRad). Membranes were immunoblotted with an antibody against full-length Amelx (FL-191, Santa Cruz Biotechnology, Santa Cruz, CA), with a goat anti-rabbit secondary antibody (Santa Cruz Biotechnology).
4.2. Histology of developing enamel
5 day-old male and female CTRNCTg/LRAPTg/Amelx−/− and control mouse heads were dissected, formalin-fixed, decalcified, paraffin-embedded and sectioned. Sections through developing first molars and incisors with ameloblasts in the secretory stage were carefully chosen, and then deparaffinized in xylene and dehydrated in graded alcohols before H&E staining.
4.3. μCT of adult enamel
Six 6 week-old male and female adult hemi-mandibles were dissected and fixed. μCT was used to compare mineral densities of incisor and molar enamel in specific regions in CTRNCTg/LRAPTg/Amelx−/− mice and controls. Adult mouse incisor and molar enamel were assessed for mineralization levels [29]. Hemimandibles with soft tissues removed were scanned in a μCT-40 (Scanco, Brüttisellen, Switzerland) at 70 kV, 114 mA, and 6 μm resolution. Images were processed with μCT-40 evaluation software and ImageJ [30] was used to orient the incisors and molars to clearly observe enamel mineralization in two locations: 1) Within the distal root of the 1st molar and developing incisor in the early maturation stage, and 2) within the maturation stage incisor, the last section before eruption, with the incisor completely enclosed by bone.
4.4. Structure and enamel thickness of adult enamel
Hemi-mandibles were embedded in plastic and polished longitudinally for scanning electron microscopy (SEM) analysis for enamel prism organization and enamel thickness [29]. Mandibles from 6 week-old male and female mice and controls were dissected, and erupted portions of mandibular incisors and molars were washed and dehydrated with graded alcohol and acetone, and embedded in Embed812 resin (Electron Microscopy Sciences, Hatfield, PA), ground in a sagittal plane, and polished to 0.25 μm with diamond suspensions (Electron Microscopy Sciences), etched with 0.1M phosphoric acid for 15 s, gold coated and imaged with a Zeiss Evo LS 10 SEM. Enamel thicknesses were measured from images taken at 1000× in first molars, and enamel structure was visualized in incisors at 2000×. Both thickness and structure were analyzed at the same location of molars and incisors, respectively, in at least 6 samples per genotype.
4.5. Microhardness of adult enamel
Erupted portions of dissected, 6 week-old male and female mandibular incisors and molars from CTRNCTg/LRAPTg/Amelx−/− and control mice were washed and dehydrated with graded alcohol and acetone, and embedded sagittally in Embed812 resin (Electron Microscopy Sciences). Samples were then ground and polished to 0.25 μm with diamond suspensions (Electron Microscopy Sciences). The polished samples were tested for enamel microhardness on an M400 HI testing machine (Leco, St. Joseph, MI). Testing was performed with load of 10 g for 5 sec with a Vickers tip, 20 indentations per sample on at least 4 teeth per group and averaged.
4.6. Statistical methods
We used ANOVA with the Tukey post hoc test to detect differences (p < 0.05) between groups of teeth analyzed for enamel thickness as measured by SEM, and Vickers microhardness (GraphPad Software, San Diego, CA, USA).
Highlights.
CTRNCTg and LRAPTg overexpression significantly improved the enamel of Amelx−/−.
Enamel microhardness was recovered when M180Tg was expressed alone or with LRAPTg.
All three regions of amelogenin contribute to final enamel thickness and structure.
Acknowledgements
The authors thank Carolyn Gibson for scientific input and generating the LRAP and CTRNC transgenic mouse lines, Ashok Kulkarni and Carolyn Gibson for the Amelx−/− mouse, Jo Buchanan for assistance with image analysis, and Justine Dobeck from the Forsyth Biostructure Core for assistance with histology. These experiments were supported by NIH/NIDCR grant R00DE022624 (MKP).
Abbreviations
- M180
mouse amelogenin, 180 amino acids
- LRAP
leucine rich amelogenin peptide
- CTRNC
C-terminal truncated amelogenin
- Tg
transgene
- Amelx
amelogenin
- Mmp20
matrix metalloproteinase-20
- KO
knockout mouse
- WT
wild-type mouse
- μCT
microcomputed tomography
- SEM
scanning electron microscopy
- AI
amelogenesis imperfecta
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
MKP designed project. YX, AR and MKP performed experiments, collected and analyzed data. MKP wrote the paper. All authors read, contributed to discussion and approved the final manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Bartlett JD, Ball RL, Kawai T, Tye CE, Tsuchiya M, Simmer JP. Origin, splicing, and expression of rodent amelogenin exon 8. J Dent Res. 2006;85:894–899. doi: 10.1177/154405910608501004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, et al. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- 3.Brookes SJ, Robinson C, Kirkham J, Bonass WA. Biochemistry and molecular biology of amelogenin proteins of developing dental enamel. Arch Oral Biol. 1995;40:1–14. doi: 10.1016/0003-9969(94)00135-x. [DOI] [PubMed] [Google Scholar]
- 4.Simmer JP, Fincham AG. Molecular mechanisms of dental enamel formation. Crit Rev Oral Biol Med. 1995;6:84–108. doi: 10.1177/10454411950060020701. [DOI] [PubMed] [Google Scholar]
- 5.Fincham AG, Moradian-Oldak J. Recent advances in amelogenin biochemistry. Connect Tissue Res. 1995;32:119–124. doi: 10.3109/03008209509013713. [DOI] [PubMed] [Google Scholar]
- 6.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 7.Xu L, Harada H, Taniguchi A. The exon 6ABC region of amelogenin mRNA contribute to increased levels of amelogenin mRNA through amelogenin protein-enhanced mRNA stabilization. J Biol Chem. 2006;281:32439–32444. doi: 10.1074/jbc.M605406200. [DOI] [PubMed] [Google Scholar]
- 8.Delgado S, Ishiyama M, Sire JY. Validation of amelogenesis imperfecta inferred from amelogenin evolution. J Dent Res. 2007;86:326–330. doi: 10.1177/154405910708600405. [DOI] [PubMed] [Google Scholar]
- 9.Gibson CW, Li Y, Suggs C, Kuehl MA, Pugach MK, Kulkarni AB, et al. Rescue of the murine amelogenin null phenotype with two amelogenin transgenes. Eur J Oral Sci. 2011;119(Suppl 1):70–74. doi: 10.1111/j.1600-0722.2011.00882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Suggs C, Wright JT, Yuan ZA, Aragon M, Fong H, et al. Partial rescue of the amelogenin null dental enamel phenotype. J Biol Chem. 2008;283:15056–15062. doi: 10.1074/jbc.M707992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snead ML, Zhu DH, Lei Y, Luo W, Bringas PO, Jr, Sucov HM, et al. A simplified genetic design for mammalian enamel. Biomaterials. 2011;32:3151–3157. doi: 10.1016/j.biomaterials.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen E, Yuan ZA, Wright JT, Hong SP, Li Y, Collier PM, et al. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif Tissue Int. 2003;73:487–495. doi: 10.1007/s00223-002-0036-7. [DOI] [PubMed] [Google Scholar]
- 13.Pugach MK, Li Y, Suggs C, Wright JT, Aragon MA, Yuan ZA, et al. The amelogenin C-terminus is required for enamel development. J Dent Res. 2010;89:165–169. doi: 10.1177/0022034509358392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stahl J, Nakano Y, Kim SO, Gibson CW, Le T, DenBesten P. Leucine rich amelogenin peptide alters ameloblast differentiation in vivo. Matrix Biol. 2013;32:432–442. doi: 10.1016/j.matbio.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, et al. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu D, Paine ML, Luo W, Bringas P, Jr, Snead ML. Altering biomineralization by protein design. J Biol Chem. 2006;281:21173–21182. doi: 10.1074/jbc.M510757200. [DOI] [PubMed] [Google Scholar]
- 17.Aoba T, Tanabe T, Moreno EC. Function of amelogenins in porcine enamel mineralization during the secretory stage of amelogenesis. Adv Dent Res. 1987;1:252–260. doi: 10.1177/08959374870010021401. [DOI] [PubMed] [Google Scholar]
- 18.Shaw WJ, Campbell AA, Paine ML, Snead ML. The COOH terminus of the amelogenin, LRAP, is oriented next to the hydroxyapatite surface. J Biol Chem. 2004;279:40263–40266. doi: 10.1074/jbc.C400322200. [DOI] [PubMed] [Google Scholar]
- 19.Moradian-Oldak J, Tan J, Fincham AG. Interaction of amelogenin with hydroxyapatite crystals: an adherence effect through amelogenin molecular self-association. Biopolymers. 1998;46:225–238. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 20.Moradian-Oldak J, Bouropoulos N, Wang L, Gharakhanian N. Analysis of self-assembly and apatite binding properties of amelogenin proteins lacking the hydrophilic C-terminal. Matrix Biol. 2002;21:197–205. doi: 10.1016/s0945-053x(01)00190-1. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Avila OM, Wu S, Cheng Y, Lee R, Khan F, Habelitz S. Self-assembly of amelogenin proteins at the water-oil interface. Eur J Oral Sci. 2011;119(Suppl 1):75–82. doi: 10.1111/j.1600-0722.2011.00907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margolis HC, Beniash E, Fowler CE. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 23.Kwak SY, Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Litman A, et al. Role of 20-kDa amelogenin (P148) phosphorylation in calcium phosphate formation in vitro. J Biol Chem. 2009;284:18972–18979. doi: 10.1074/jbc.M109.020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwak SY, Kim S, Yamakoshi Y, Simmer JP, Beniash E, Margolis HC. Regulation of calcium phosphate formation by native amelogenins in vitro. Connect Tissue Res. 2014;55(Suppl 1):21–24. doi: 10.3109/03008207.2014.923853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kindelan SA, Brook AH, Gangemi L, Lench N, Wong FS, Fearne J, et al. Detection of a novel mutation in X-linked amelogenesis imperfecta. J Dent Res. 2000;79:1978–1982. doi: 10.1177/00220345000790120901. [DOI] [PubMed] [Google Scholar]
- 26.Greene SR, Yuan ZA, Wright JT, Amjad H, Abrams WR, Buchanan JA, et al. A new frameshift mutation encoding a truncated amelogenin leads to X-linked amelogenesis imperfecta. Arch Oral Biol. 2002;47:211–217. doi: 10.1016/s0003-9969(01)00111-x. [DOI] [PubMed] [Google Scholar]
- 27.Gibson CW, Li Y, Daly B, Suggs C, Yuan ZA, Fong H, et al. The leucine-rich amelogenin peptide alters the amelogenin null enamel phenotype. Cells Tissues Organs. 2009;189:169–174. doi: 10.1159/000151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson CW, Yuan ZA, Li Y, Daly B, Suggs C, Aragon MA, et al. Transgenic mice that express normal and mutated amelogenins. J Dent Res. 2007;86:331–335. doi: 10.1177/154405910708600406. [DOI] [PubMed] [Google Scholar]
- 29.Pugach MK, Suggs C, Li Y, Wright JT, Kulkarni AB, Bartlett JD, et al. M180 amelogenin processed by MMP20 is sufficient for decussating murine enamel. J Dent Res. 2013;92:1118–1122. doi: 10.1177/0022034513506444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasband WS. ImageJ, US National Institutes of Health. 1997–2002 [Google Scholar]