Abstract
The liver has a remarkable regeneration capacity, and, after surgical removal of its mass, the remaining tissue undergoes rapid regeneration through compensatory growth of its constituent cells. Although hepatocytes synchronously proliferate under the control of various signaling molecules from neighboring cells, there have been few detailed analyses on how biliary cells regenerate for their cell population after liver resection. The present study was undertaken to clarify how biliary cells regenerate after partial hepatectomy of mice through extensive analyses of their cell cycle progression and gene expression using immunohistochemical and RT-PCR techniques. When expression of PCNA, Ki67 antigen, topoisomerase IIα and phosphorylated histone H3, which are cell cycle markers, was immunohistochemically examined during liver regeneration, hepatocytes had a peak of the S phase and M phase at 48–72 h after resection. By contrast, biliary epithelial cells had much lower proliferative activity than that of hepatocytes, and their peak of the S phase was delayed. Mitotic figures were rarely detectable in biliary cells. RT-PCR analyses of gene expression of biliary markers such as Spp1 (osteopontin), Epcam and Hnf1b demonstrated that they were upregulated during liver regeneration. Periportal hepatocytes expressed some of biliary markers, including Spp1 mRNA and protein. Some periportal hepatocytes had downregulated expression of HNF4α and HNF1α. Gene expression of Notch signaling molecules responsible for cell fate decision of hepatoblasts to biliary cells during development was upregulated during liver regeneration. Notch signaling may be involved in biliary regeneration.
Keywords: biliary epithelial cells, liver regeneration, mouse, Notch, osteopontin
Introduction
While the liver is an internal organ that is responsible for the metabolism and storage of nutrition, it has a remarkable regeneration capacity. After surgical removal of 70% of its mass, the remaining tissue undergoes rapid regeneration which completes, usually within 10 days after surgery, through compensatory growth of each hepatic constituent cell, including hepatocytes [7, 30]. Hepatocytes rapidly and synchronously exit the G0 phase and enter the cell cycle in response to resection. It is proposed that there is an initial activation of the TNFα cascade in Kupffer cells, which stimulates multiple diverse growth factor and metabolic pathways in hepatocytes [2, 3, 5, 13, 18, 25, 37]. Biliary cell proliferation occurs in a late phase in case of rats, compared with that of hepatocytes [10, 22]. Although biliary epithelial cells may restore their cell population through proliferation of their own population during liver regeneration, there have been few detailed analyses on their cell cycle progression. By contrast, in rodent models of liver injury using some drugs such as 2-acetylaminofluorene or D-galactosamine, liver stem-like or progenitor-like cells, which are known as oval cells, extensively proliferate and are postulated to generate both cell populations of both hepatocytes and biliary epithelial cells [6, 21]. Liver stem-like or progenitor-like cells may locate in the canals of Hering (bile ductules) [6, 21]. Recent cell labeling studies, in which hepatocytes and biliary epithelial cells are genetically labeled, gave controversial results for their origin, showing that oval cells are derived from hepatocytes, or that they originate from biliary epithelial cells or ductular cells [9, 23, 28, 31]. It may be intriguing to examine whether hepatocytes can generate biliary cells or not during liver regeneration after resection. Upregulation of Notch signaling can induce adult mature hepatocytes to give rise to biliary epithelial cells as demonstrated in biliary development at fetal stages [24, 35, 38]. During biliary development, the induction may include Jag1/Notch2 signaling; the Jag1 signal of portal mesenchyme cells is received through the Notch2 receptor of periportal biliary progenitors [24, 35]. The Notch signaling may activate transcription of the HES (hairy and enhancer of split)/HEY (HES-related with YRPW motif) family member genes, including HES1, which encode bHLH/orange domain transcriptional repressors [17, 20]. TNFα and FGF signaling also play decisive roles in the oval cell reaction [14, 16, 19, 34]. However, it remains to be revealed which signaling operates in biliary cell proliferation after resection of liver pieces.
In the present study, we immunohistochemically examined the cell cycle of biliary epithelial cells, and expression of biliary markers during regeneration after partial hepatectomy using mice. We found that biliary epithelial cells had much lower proliferation activity than that of hepatocytes, and that biliary gene expression, including osteopontin and cytokeratins expression, was detectable in periportal hepatocytes during liver regeneration.
Materials and Methods
Animals
C57BL/6J strain male mice (8 week old; CLEA Japan, Tokyo) were used. Animals anesthetized with isoflurane (Wako Pure Chemical Industries, Osaka, Japan) underwent 70% partial hepatectomy (PH) according to methods described by Higgins and Anderson [11], and Mitchell and Willenbring [26]. Sham operations were also carried out for control experiments (SH). At least three animals for each time point except for liver samples at 0 h and 336 h after partial hepatectomy (PH0 and PH336), and ten sections for each animal were examined. All animal experiments were carried out in compliance with the “Guide for Care and Use of Laboratory Animals” of Shizuoka University.
Histology and immunohistochemistry
For histology and immunohistochemistry, liver tissues were fixed in a cold mixture of 95% ethanol and acetic acid (99:1 v/v) overnight, and embedded in paraffin. Paraffin sections were cut at 6 µm.
When a peroxidase-labeled secondary antibody was used, endogenous peroxidase activity in dewaxed sections was blocked by treatment with PBS containing 3% H2O2 for 10 min before incubation with the primary antibody. The antigenicities of HNF4α, HNF1α, SOX9, topoisomerase IIα, Ki67 and cytokeratin no. 19 (CK19) on paraffin sections were retrieved by TE (10mM Tris, 1mM EDTA [ethylenediaminetetraacetic acid], 0.05% [w/v] Tween 20, pH 9.0) treatment at 95°C for 10 min after dewaxing. In case of mouse monoclonal anti-proliferating cell nuclear antigen (PCNA) antibody (Dako Japan, Tokyo, Japan), sections were blocked for endogenous mouse IgG with M.O.M. Kit (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s instructions. For Ep-CAM immunoshistochemistry, frozen sections were used.
Hydrated sections were incubated with the primary antibodies listed in Table 1 overnight at 4°C. The primary antibodies were diluted in 5% normal donkey serum (Jackson ImmunoResearch Lab., West Grove, PA, USA). After thorough washing with PBS containing 0.1% Tween (PBS/T), sections were incubated with a species-specific peroxidase-labeled anti-mouse, rat, goat or rabbit IgG antibody (Jackson ImmunoResearch) for 2 h. After thorough washing, sections were stained with 3, 3′-diaminobenzidine (DAB), and then with hematoxylin. For immunofluorescence, sections were incubated with a species-specific fluorochrome-labeled secondary antibody (Jackson ImmunoResearch) diluted in PBS/T for 2 h at room temperature, washed again, and mounted in buffered glycerol containing p-phenylenediamine [15]. In some immunofluorescence experiments, nuclei were stained with 4’, 6-diamidine-2’-phenylindole dihydrochloride (DAPI). Double immunofluorescent analyses were carried out for osteopontin and carbamoylphosphate synthase I (CPSI), osteopontin and HNF4α, HNF4α and CPSI, HNF1α and CPSI, and CK19 and CPSI using a species-specific different fluorochrome-labeled secondary antibody. Control incubations were carried out in 5% normal donkey serum in place of the primary antibodies. The specificities of the antibodies against transcription factors from Santa Cruz Technology were checked by preabsorption experiments with antigenic peptides.
Table 1. Primary antibodies used in immunohistochemistry.
| Antibody | Source | Dilution |
|---|---|---|
| Goat anti-human HNF1α antibody | Santa Cruz Biotechnology, Santa Cruz, CA, USA | 1:100* |
| Goat anti-human HNF4α antibody | Santa Cruz Biotechnology, Santa Cruz, CA, USA | 1:100* |
| Goat anti-mouse osteopontin antibody | R&D Systems, Minneapolis, MN, USA | 1:100 |
| Mouse anti-proliferating cell nuclear antigen antibody clone PC10 | Dako Japan, Tokyo, Japan | 1:1000 |
| Rabbit anti-calf keratin antiserum | Dako, Carpinteria, CA, USA | 1:200 |
| Rabbit anti-HES1 antibody (D6P2U) | Cell Signaling Technology Japan, Tokyo, Japan | 1:100 |
| Rabbit anti-human HNF4α antibody | Santa Cruz Biotechnology, Santa Cruz, CA, USA | 1:500* |
| Rabbit anti-human phospho-histone H3 (Ser10) antiboby | Cell Signaling Technology Japan, Tokyo, Japan | 1:100 |
| Rabbit anti-human SOX9 antibody | Millipore Corporation, Temecula, CA, USA | 1:500* |
| Rabbit anti-human topoisomerase IIα antibody [EP1102Y] | abcam, Tokyo, Japan | 1:100* |
| Rabbit anti-mouse carbamoylphosphate synthase I (CPSI) antibody | Nitou et al., 2002 | 1:500 |
| Rabbit anti-mouse Ki67 antibody | Novus Biologicals, Littleton, CO, USA | 1:500* |
| Rat anti-mouse cytokeratin no.19 antibody | Developmental Studies Hybridoma Bank, Iowa City, IA, USA | 1:100* |
| Rat anti-mouse Ep-CAM antibody | Developmental Studies Hybridoma Bank, Iowa City, IA, USA | 1:200** |
*Antigen retrieval treatment (TE treatment for 10 min) was carried out in paraffin sections. **Frozen sections fixed in MEMFA were used.
Immunohistochemical detection of nuclear localization of PCNA, Ki67, topoisomerase IIα and phosphorylated histone H3 (P.H3) was used as markers for late G1-M phase, late G1-M phase, S phase and M phase of the cell cycle, respectively [27, 36].
Dewaxed and dehydrated sections were incubated with fluorescein isothionate-labeled soybean agglutinin (SBA) or Dolichos biflorus agglutinin (DBA) (Vector Laboratories, Burlingame, CA) for 30 min [33]. After thorough washing in PBS, the sections were observed using a fluorescent microscope.
Hematoxylin-eosin (H-E) staining was carried out for demonstration of histology and mitoses.
RT-PCR
Total RNA was extracted from regenerating livers using IsogenII (Nippon Gene, Tokyo, Japan). Complementary DNA was synthesized from total RNA (2 µg) in 20 µl of reaction mixture containing 2.5 µM oligo dT primer, 0.25 mM dNTP, 2 U/µl RNase inhibitor, and 10 U/µl PrimeScriptR II Reverse Transcriptase (Takara Bio Inc., Otsu, Japan), according to the manufacturer’s instructions.
PCR reaction was conducted in 20 µl of the reaction mixture, using Ex-Taq DNA polymerase (Takara Bio Inc.; 0.025 U/µl). Primers listed in Table 2 were used at 0.5 µM. After various dilutions of template cDNA, we optimized the concentration for each primer. In these concentrations, amplification by PCR did not reach a plateau and could be used for semi-quantitative analysis. PCR cycles were as follows: initial denaturation at 94°C for 1.5 min, followed by 20–36 cycles at 94°C for 30 sec, at 60°C for 30 sec, at 72°C for 1 min, and final extension at 72°C for 10 min. PCR products were separated by 1% agarose gel electrophoresis.
Table 2. Primers used in RT-PCR analysis.
| Symbol | Description | Accession No. | Sequence | PCR product length (bp) |
|---|---|---|---|---|
| Actb | β-actin | NM_007393.3 | F: 5’-GACGGCCAGGTCATCACTAT-3’ | 337 |
| R: 5’-ACATCTGCTGGAAGGTGGAC-3’ | ||||
| Afp | α-fetoprotein (AFP) | NM_007423.4 | F: 5’-GGAGGCTATGCATCACCAGT-3’ | 165 |
| R: 5’-GTTCAGGCTTTTGCTTCACC-3’ | ||||
| Alb | Albumin | NM_009654.3 | F: 5’-GTAGTGGATCCCTGGTGGAA-3’ | 179 |
| R: 5’-CTTGTGCTTCACCAGCTCAG-3’ | ||||
| Cps1 | Carbamoyl phosphate synthase I | NM_001080809.1 | F: 5’-GCATTCATACCGCCTTCCTA-3’ | 117 |
| R: 5’-GCTCAGCAACACCAAGGAAT-3’ | ||||
| Epcam | Epithelial cell adhesion molecule (Ep-CAM) | NM_008532.2 | F: 5’-GGGTGAGATCCACAGAGAGC -3’ | 401 |
| R: 5’-GGGCAGCCTTAATCACAAAA-3’ | ||||
| Fgf7 | Fibroblast growth factor 7 | NM_008008.4 | F: 5’-TTGACAAACGAGGCAAAGTG -3’ | 289 |
| R: 5’-TTGACAGGAATCCCCTTTTG-3’ | ||||
| Fgfr2b | Fibroblast growth factor receptor 2 isoform IIIb (FGFR2b) | NM_201601.2 | F: 5’-GAGATCATCGCCTGCTCCATCC -3’ | 132 |
| R: 5’-TTGTTACTGCTGTTCCTGCTCCC-3’ | ||||
| Hnf1b | HNF1β | NM_009330.2 | F: 5’-TCCAACTTGGTCACGGAGGTC-3’ | 181 |
| R: 5’-ACATCTTGTTGGGTGGAGAGGAG-3’ | ||||
| Jag1 | Jagged1 | NM_013822.5 | F: 5’-AGTAAACGGGATGGAAACAG-3’ | 583 |
| R: 5’-GCGGTGCCCTCAAACTCT-3’ | ||||
| Krt19 | Cytokeratin 19 | NM_008471.2 | F: 5’-AAACCTCAATGATCGTCTCGCC-3’ | 198 |
| R: 5’-TCTTGGAGTTGTCAATGGTGGC-3’ | ||||
| Notch2 | Notch2 | NM_010928.2 | F: 5’-ACTCCTCCTCAGGCAGAACA-3’ | 660 |
| R: 5’-TTTGGCCGCTTCATAACTTC-3’ | ||||
| Sox9 | SOX9 | NM_011448.4 | F: 5’-CAGCCCCTTCAACCTTCCTC-3’ | 82 |
| R: 5’-AGTTCTGATGGTCAGCGTAGTCG-3’ | ||||
| Spp1 | Osteopontin | NM_001204203.1 | F: 5’-GCAGTCTTCTGCGGCAGGCA-3’ | 494 |
| R: 5’-ACCTCGGCCGTTGGGGACAT-3’ | ||||
| Tnfrsf12a | Tumor necrosis factor receptor superfamily, member 12a (Fn14) | NM_013749.2 | F: 5’-GACCACACAGCGACTTCTGC -3’ | 258 |
| R: 5’-GAATGAATGGACGACGAGTG-3’ | ||||
| Tnfsf12 | Tumor necrosis factor (ligand) superfamily, member 12 (TWEAK) | NM_011614.3 | F: 5’- CCCCTACTTATCCCTGACTCC-3’ | 299 |
| R: 5’-CCCCTTCCCACAATCTTCA-3’ |
F, forward primer; R, reverse primer.
In situ hybridization
cDNA coding for partial sequences of mouse Spp1 mRNA was cloned by RT-PCR. The primers used were designed based on the sequence of mouse gene (NCBI Accession Number, NM_001204203.1; sequence 45~538 [size of RNA probe, 494 bases]). Both sense and antisense digoxigenin-labeled riboprobes were synthesized from plasmids containing its cDNA by using a DIG RNA labeling kit (Roche Diagnostics, Mannheim, Germany). Liver tissues for in situ hybridization were fixed using MEMFA (3.7% formaldehyde, 100mM MOPS [3-morpholinopropanesulfonic acid], 2mM EGTA [O,O’-bis (2-aminoethyl) ethyleneglycol-N,N,N’,N’-tetraacetic acid], 1mM MgSO4 [pH7.4]), and then frozen sections were cut. In situ hybridization on frozen sections was carried out according to Akai et al. [1] with some modifications, which included changing the hybridization temperature from 70 to 65°C. The proteinase K concentration was 2 µg/ml, and the length of the proteinase K treatment was modified according to the size of the tissue.
Results
Cell cycle progression during liver regeneration
Many PCNA-, Ki67-, and topoisomerase IIα-positive hepatocyte nuclei were detected at 48 and 72 h after partial hepatectomy (Figs. 1A-E; Supplementary Figs. 1A-L). Stages with the highest proportion of hepatocytes with positive nuclei for each cell cycle marker in all hepatocytes were at 48 h for PCNA and topoisomerase IIα, and at 72 h for Ki67 (Figs. 1A-C). Mitotic figures of hepatocytes and P.H3-positive staining of hepatocyte nuclei, including their mitotic figures, were most often observed at 48 h, and gradually decreased during liver regeneration (Figs. 1D and E; Supplementary Figs. 1M-P).
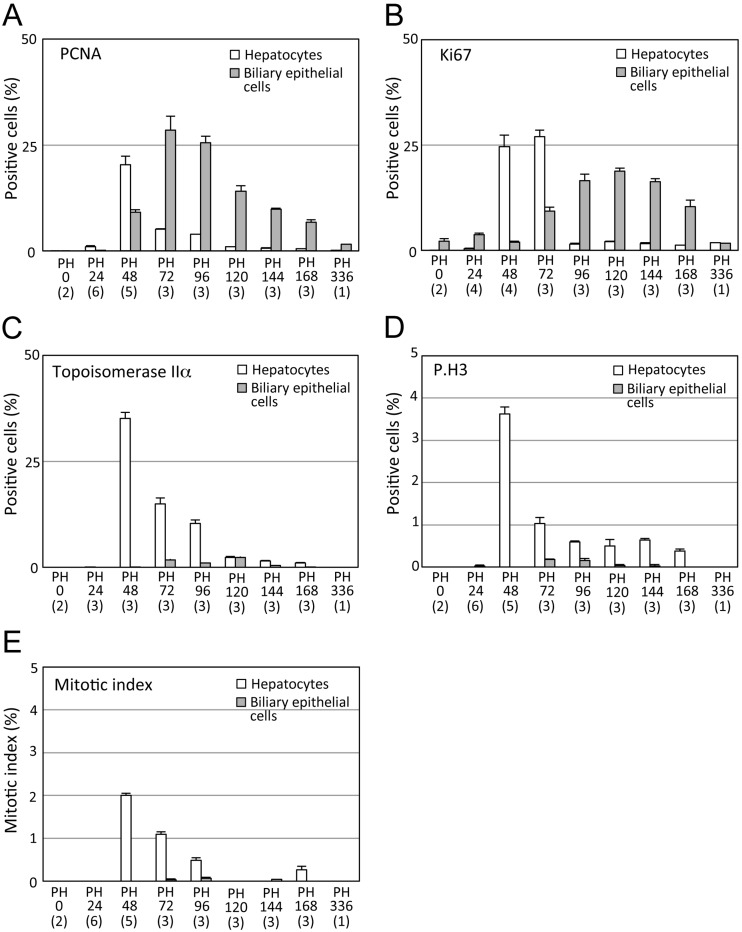
Fig. 1.
Cell cycle progression during mouse liver regeneration. A, Percentage of hepatocytes and biliary epithelial cells with PCNA-positive nuclei. B, Percentage of hepatocytes and biliary epithelial cells with Ki67-positive nuclei. C, Percentage of hepatocytes and biliary epithelial cells with topoisomerase IIα-positive nuclei. D, Percentage of hepatocytes and biliary epithelial cells with P.H3-positive nuclei. E, Mitotic index of hepatocytes and biliary epithelial cells. Over 1,000 cells per liver were counted. Data are shown as mean +/− standard deviation. While most hepatocytes semi-synchronously enter the S or M phase at PH48 and PH72, biliary epithelial cells have delayed and slow cell cycle progression (A-E). The number in parentheses is the number of animals examined.
The positive immunoreaction of biliary epithelial cell nuclei with anti-PCNA, Ki67 and topoisomerase IIα antibodies commenced at various stages; Ki67-, PCNA- and topoisomerase IIα-positive biliary cell nuclei appeared at 0, 48 and 72 h after liver resection, respectively (Figs. 1A-C; Supplementary Figs. 1A-L). Stages with the highest proportion of biliary cells with positive nuclei for PCNA and Ki67 markers in all biliary epithelial cells were at 72 and 120 h, respectively. That for topoisomerase IIα-positive biliary cells was between 72 and 120 h. The proportion of topoisomerase IIα−positive biliary cells was significantly much smaller than those of hepatocytes (Figs. 1A-C). Biliary epithelial cells had few mitotic figures in H-E stained slides, and P.H3 immunohistochemistry also supported the data (Figs. 1D and E; Supplementary Figs. 1M-P).
Immunoreaction of hepatocytes and biliary epithelial cells for each cell cycle marker in livers of sham operations was similar to that at PH0 (Supplementary Figs. 1Q-T).
Gene expression of hepatocyte and biliary markers
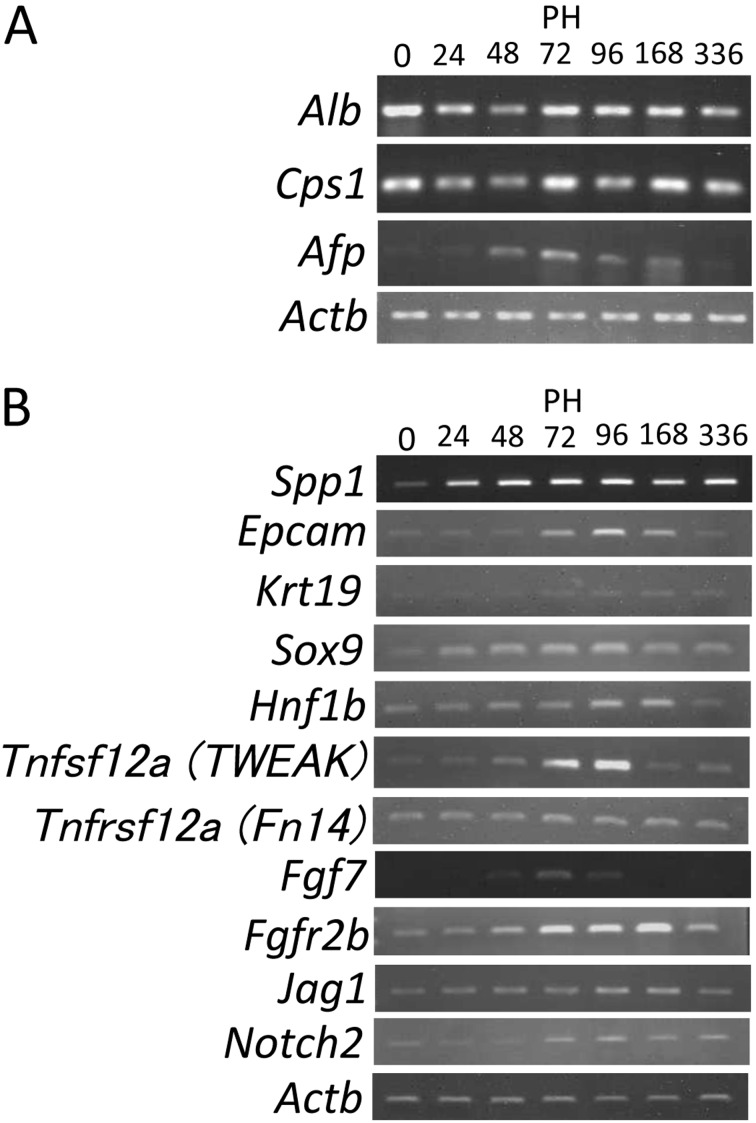
When Spp1, Sox9, Hnf1b, Krt19, and Epcam mRNAs of biliary markers were examined during liver regeneration using RT-PCR, they were upregulated between 48 and 168 h after liver resection (Fig. 2B). Hepatocyte markers (Cps1 and Alb mRNAs) did not significantly change their expression during liver regeneration (Fig. 2A). Expression of Afp mRNA was transiently upregulated between 48 and 168 h after liver resection.
Fig. 2.
RT-PCR analyses of expression of hepatocyte and biliary markers during liver regeneration. A, Expression of Alb, Cps1, and Afp mRNAs. Afp mRNA is transiently upregulated at PH48-168 during liver regeneration. B, Expression of Spp1, Epcam, Krt19, Sox9, Hnf1b, Tweak, Fn14, Fgf7, Fgfr2b, Jag1 and Notch2 mRNAs. Biliary markers such as Spp1, Epcam, Krt19, Sox9, Hnf1b mRNAs, and mRNAs of signaling molecules for oval cell reactions or controling biliary differentiation (Tnfsf12a [TWEAK], Tnfrsf12a [Fn14], Fgf7, Fgfr2b, Jag1 and Notch2) are slightly or moderately upregulated at PH72-168 during liver regeneration. The numbers in parentheses denote cycles of each PCR.
Expression of osteopontin and its mRNA in periportal hepatocytes
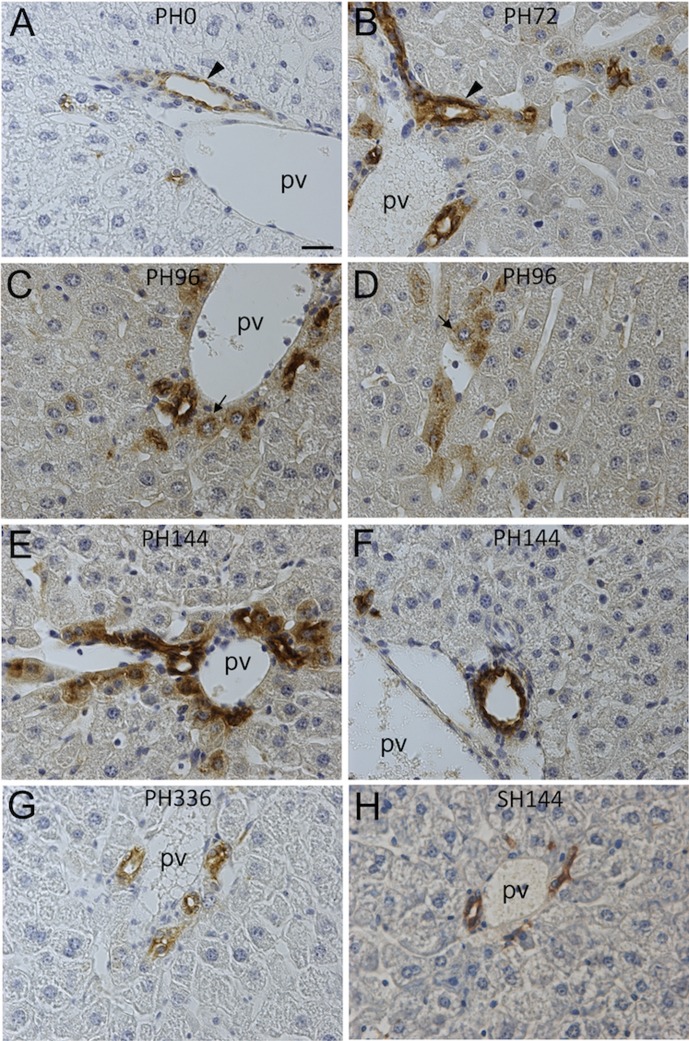
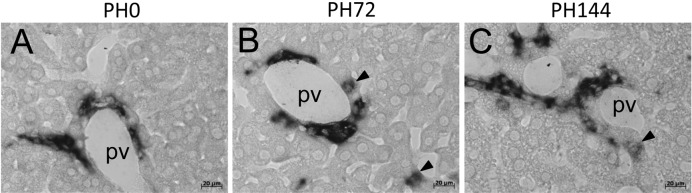
Whereas osteopontin expression was immunohistochemically detectable in biliary epithelial cells and ductular cells, it was absent in all hepatocytes of normal livers and SH livers (Figs. 3A and H). Osteopontin protein expression started to be detectable in periportal hepatocytes at 72 h after liver resection in addition to periportal biliary cells (Fig. 3B). At 96–168 h, the osteopontin immunostaining in periportal hepatocytes was very remarkable, although some portal veins did not have osteopontin-positive hepatocytes (Figs. 3C-F). At 336 h, its expression was immunohistochemically returned to a normal level (Fig. 3G). In situ hybridization analyses of Spp1 mRNAs demonstrated that some periportal hepatocytes expressed Spp1 at 72 and 144 h during liver regeneration (Figs. 4B and C). Spp1 mRNA was expressed only in biliary epithelial cells in normal liver (Fig. 4A). Spp1 mRNA and its protein were not detectable in nonperiportal hepatocytes, including pericentral ones, throughout liver regeneration (data not shown).
Fig. 3.
Immunohistochemical detection of osteopontin expression during liver regeneration. After immunohistochemistry of osteopontin, sections were counterstained with hematoxylin. A, liver section at PH0. B, liver section at PH72. C, D, liver section at PH96. E, F, liver section at PH144. G, liver section at PH336. H, liver section at SH144. Only biliary epithelial cells express osteopontin protein at PH0 and SH144 (A, H). At PH72, periportal hepatocytes become positive for osteopontin in addition to biliary epithelial cells (arrowhead) (B). Positive staining of osteopontin in periportal hepatocytes is conspicuous at PH96 and PH144 (C, E). Periportal hepatocytes (arrow) are positive around small portal veins, which are not accompanied by bile ductules (D). Portal area where osteopontin expression of periportal hepatocytes is not remarkable is observed at PH144 (F). Periportal hepatocytes express osteopontin at PH336 similarly to those of PH0 livers (G). pv, portal vein. Bar indicates 20 µm.
Fig. 4.
In situ hybridization analyses of Spp1 expression during liver regeneration. A, liver section at PH0. B, liver section at PH72. C, liver section at PH144. Spp1 mRNA is expressed only in biliary epithelial cells at PH0 (A), but is also expressed in some periportal hepatocytes at PH72 (B) and PH144 (C) (arrowheads). pv, portal vein. Bars indicate 20 µm.
Expression of other biliary markers in periportal hepatoctyes
Periportal hepatocytes were positively immunostained for polyclonal anti-cytokeratin antibody in addition to biliary cells during liver regeneration, whereas the antibody marked only biliary cells in normal liver (Supplementary Figs. 2A-C). This antibody reacted with almost all hepatocytes at 48 h after partial hepatectomy (Supplementary Fig. 2B). For CK19 immunostaining, hepatocytes, including periportal ones, were negative, and only biliary epithelial cells and ductular cells were labeled throughout liver regeneration (Supplementary Figs. 2D-F). Ep-CAM immunostaining also exhibited similar reactivity to that of CK19, and reacted only with biliary cells, but not with hepatocytes (Supplementary Figs. 2G-I). The anti-SOX9 antibody reacted only with nuclei of biliary cells in normal liver, but also bound to nuclei of many hepatocytes at 72 h (Supplementary Figs. 2J and K). The positive immunoreaction was gradually confined to periportal hepatocytes during liver regeneration (Supplementary Fig. 2L). Fluorescein isothionate-labeled DBA and SBA reacted with some biliary cells, especially on their apical side, in normal liver. This staining pattern did not change for biliary epithelial cells throughout liver regeneration (data not shown).
Double immunofluorescent analyses of expression for biliary and hepatocyte markers in periportal hepatoctyes
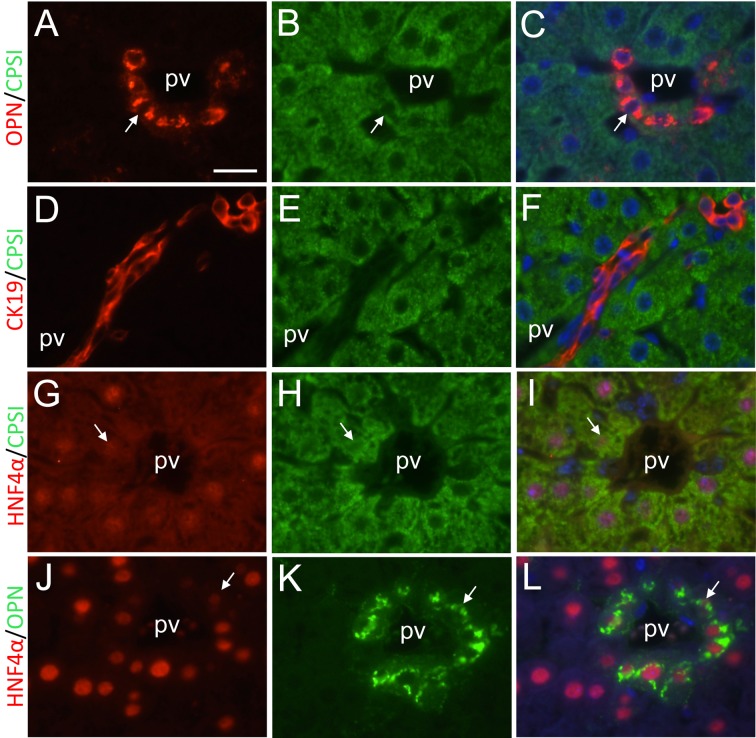
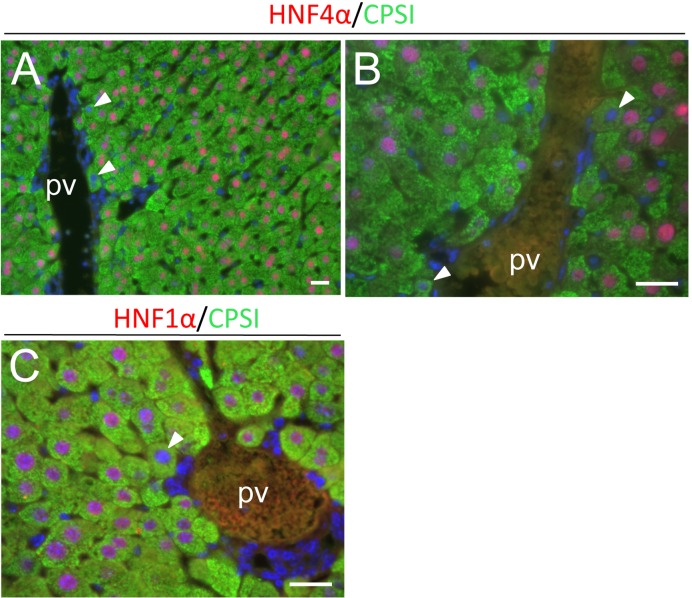
Double immunofluorescent analyses demonstrated that CPSI- and HNF4α-positive hepatocytes expressed osteopontin in periportal regions, and that CK19 expression was confined to only biliary cells, which did not express CPSI, during liver regeneration (Figs. 5A-L). HNF4α expression was downregulated in some periportal hepatocytes at 144 h (Figs. 5G and I). At 72 h, negative or very weak HNF4α and HNF1α staining was also noted in some periportal hepatocytes (Figs. 6A-C). In these hepatocytes, CPSI expression was not downregulated. Downregulation of HNF4α and HNF1α expression was not detected in nonperiportal hepatocytes, including pericentral hepatocytes, during liver regeneration.
Fig. 5.
Double immunofluorescent analyses of expression of biliary and hepatocyte marker proteins. A, B, C, Osteopontin immunostaining, CPSI immunostaining and their double immunostaining at PH168, respectively. D, E, F, CK19 immunostaining, CPSI immunostaining and their double immunostaining at PH168, respectively. G, H, I, HNF4α immunostaining, CPSI immunostaining and their double immunostaining at PH144, respectively. J, K, L, HNF4α immunostaining, osteopontin immunostaining and their double immunostaining at PH168, respectively. Arrows indicate osteopontin- and CPSI-positive periportal hepatocytes (A-C), HNF4α-weakly positive and CPSI-positive periportal hepatocytes (G-I), and HNF4α-weakly positive and osteopontin-positive periportal hepatocytes (J-L). CK19-positive signals are restricted in biliary epithelial cells, and not expressed in CPSI-positive periportal hepatoctyes (D-F). pv, portal vein. Bar indicates 20 µm.
Fig. 6.
Double immunofluorescent analyses of expression of CPSI (green) and HNF4α or HNF1α (red) during liver regeneration (PH72). Nuclei were stained with DAPI (blue). Some periportal hepatocytes have HNF4α- or HNF1α-negative or very weakly positive nuclei at this time point (arrowheads)(A-C). pv, portal vein. Bars indicate 20 µm.
Gene expression for biliary signaling
Gene expression for biliary signaling such as FGF and Notch was examined using RT-PCR. Jag1 and Notch2 expression was transiently upregulated after 48 or 72 h after liver resection (Fig. 2B). Fgf7, Fgfr2b, Tnfsf12 and Tnfrsf12a, mRNAs for FGF7, its receptor FGFr2b, TWEAK (tumor necrosis factor-like weak inducer of apoptosis) and its receptor Fn14 (FGF-inducible 14), respectively, were also slightly or moderately upregulated between 48 and 168 h during liver regeneration (Fig. 2B).
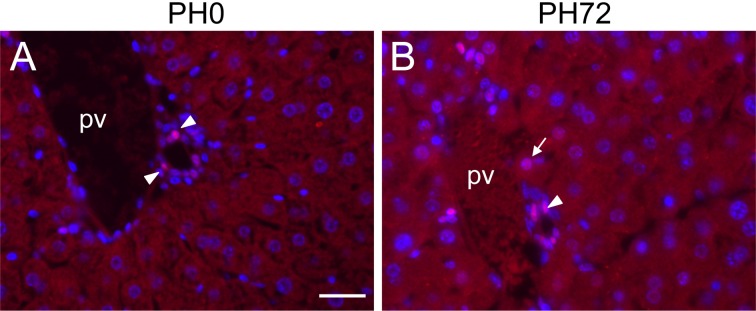
To demonstrate active Notch signaling during liver regeneration, nuclear localization of HES1 protein was immunohistochemically examined. As a result, some periportal hepatocytes had positive nuclear staining of HES1 in addition to nuclei of biliary epithelial cells at 72 h after liver resection (Fig. 7B). At PH0, nuclear immunostaining was detectable only in biliary epithelial cells, but not in hepatocytes (Fig. 7A). Nuclei of nonperiportal hepatocytes were not reactive with our HES1 antibody throughout liver regeneration.
Fig. 7.
Immunohistochemical detection of nuclear localization of HES1 protein (red) in periportal hepatocytes during liver regeneration. Nuclei were stained with DAPI (blue). A, liver section at PH0. B, liver section at PH72. Nuclear localization of HES1 protein is detectable only in biliary epithelial cells (arrowheads) at PH0 (A), but a periportal hepatocyte having weakly HES1-positive nucleus (arrow) is observed in addition to biliary cells with moderately positive nuclei at PH72 (B). pv, portal vein. Bar indicates 20 µm.
Discussion
Our immunohistochemical analyses of several cell cycle markers, including PCNA, Ki67, topoisomerase IIα and P.H3, and histological analyses of mitotic index demonstrated that the cell cycle of hepatocytes was semi-synchronously progressed during mouse liver regeneration, and that the first peaks of the S phase and M phase in hepatocytes were approximately at 48–72 h in our liver resection protocol. By contrast, biliary epithelial cells had much poorer cell cycle progression than hepatocytes did. Immunohistochemical detection of topoisomerase IIα and P.H3 proteins and mitotic index data showed that a very low proportion of biliary epithelial cells is proliferating during liver regeneration, although both anti-PCNA and Ki67 antibodies reacted with nuclei of many biliary epithelial cells. PCNA may also be involved in DNA repair, suggesting that PCNA can be expressed by cells that are not proliferating [27, 36]. Ki67 protein, which is thought to be exclusively expressed in proliferating cells, may be associated with ribosomal RNA transcription in quiescent and proliferating cells [4]. These may be the reasons why many biliary epithelial cells expressed both PCNA and Ki67 proteins in their nuclei during liver regeneration. In any event, our data for the cell cycle of hepatocytes and biliary epithelial cells during mouse liver regeneration agree with data of the rat in the paper by Grisham [10], in which 3H-thymidine incorporation was used for cell cycle evaluation.
The present data for the cell cycle progression of hepatocytes indicate that hepatocytes may restore their original cell population or whole mass mainly through cell proliferation after liver resection. In contrast, our data for biliary epithelial cells suggest that their proliferation, which was poor during liver regeneration, may not account for whole restoration of their population. Although the biliary duct system can restore its original volume or length through cell elongation and cell arrangement after liver resection, remarkable morphological changes in bile ducts did not occur during liver regeneration (data not shown).
It is of note that expression of osteopontin and its mRNA, which is biliary markers and was undetectable in hepatocytes of normal mouse liver, was upregulated in periportal hepatocytes during liver regeneration. Furthermore, the present study, for the first time, demonstrated that some periportal hepatocytes had remarkably downregulated HNF4α and HNF1α expression, and nuclear localization of HES1 protein during liver regeneration, which suggests active Notch signaling, although their number was small. From these data, it is possible that biliary epithelial cells partially restore their population from transdifferentiation of periportal hepatocytes. Nishikawa et al. [29] indicated that hepatocytes could generate biliary epithelial cells when they are cultured in vitro. Yanger et al. [38] have shown that upregulated Notch signaling in adult hepatocytes induces biliary differentiation. It has been recently demonstrated that hepatocytes can generate biliary cells in several mouse models of chronic liver injury using Cre-ERT2-reporter systems for genetic cell labeling [28, 31]. During liver development, periportal hepatoblasts, one of liver progenitor cells, may give rise to biliary cells under the influence of portal mesenchymal cells [12, 24, 32].
On the other hand, we indicated that both CK19 and Ep-CAM proteins, markers of biliary epithelial cells, were expressed only in biliary epithelial cells during liver regeneration, but not in periportal hepatocytes coexpressing osteopontin and mature hepatocyte markers such as CPSI. Downregulation of CPSI in periportal hepatocytes was not immunohistochemically detected at 72 and 144/168 h. If the transdifferentiation of periportal hepatocytes into biliary cells can occur, periportal hepatocytes coexpressing CPSI and CK19 or Ep-CAM are supposed to be detected. However, we did not observe such periportal hepatocytes in our immunohistochemical analyses, suggesting that the transdifferentiation does not happen. Thus, detailed cell lineage analyses using Cre-ERT2-reporter systems for genetic cell labeling are required for biliary regeneration after partial hepatectomy.
Font-Burgada et al. [8] have recently shown that normal periportal hepatocytes express osteopontin, which is not consistent with our data. The difference may be due to those of antibodies used or sensitivities for immunohistochemical detection. Although osteopontin-positive preiportal cells appearing after partial hepatectomy, which we showed, can be originated from “periportal hybrid cells” expressing hepatocyte markers and low amounts of SOX9 and other bile-duct-enriched genes, observed by Font-Burgada et al. [8], our data indicated that proliferation activities in hepatocytes were similar in three zones of the hepatic lobule, implying that special expansion of “periportal hybrid cells” does not occur during liver regeneration.
When gene expression for biliary signaling such as Jag1-Notch2 signaling, which works during fetal biliary development [12, 24, 35], was examined using RT-PCR in the present study, this signaling was transiently upregulated at 72–168 h during regeneration after liver resection. This result suggests that Jag1-Notch2 signaling act in biliary regeneration after liver resection.
It is also intriguing that mRNAs for FGF7-FGFR2b signaling were upregulated in liver regeneration after resection as demonstrated in the present study, which may act in oval cell reactions during liver regeneration caused by some chemicals [34]. TWEAK ligand and its receptor Fn14 mRNAs, and AFP mRNA were also slightly or moderately upregulated in our liver regeneration experiments. These data suggest that molecular mechanisms underlying regeneration in injured livers such as FGF7-FGFR2b and TWEAK signaling can also operate in cellular signaling during liver regeneration after partial hepatectomy.
Supplementary Material
Acknowledgments
We thank Professor Emeritus Takeo Mizuno of the University of Tokyo and Prof. Nelson Fausto of the University of Washington for their interest in our study and encouragement. The authors also thank Dr. T. Koike for his discussion, and Messrs. Y. Akahori, T. Ueno, Y. Akai and Y. Uchida, and Miss T. Abo for their technical assistance. This work was supported in part by Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (JSPS) (Grant Number 25440153) and by Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government (Grant Number 26119706).
References
- 1.Akai Y., Oitate T., Koike T., Shiojiri N.2014. Impaired hepatocyte maturation, abnormal expression of biliary transcription factors and liver fibrosis in C/EBPα (Cebpa)-knockout mice. Histol. Histopathol. 29: 107–125. [DOI] [PubMed] [Google Scholar]
- 2.Anders R.A., Subudhi S.K., Wang J., Pfeffer K., Fu Y.X.2005. Contribution of the lymphotoxin beta receptor to liver regeneration. J. Immunol. 175: 1295–1300. doi: 10.4049/jimmunol.175.2.1295 [DOI] [PubMed] [Google Scholar]
- 3.Berasain C., García-Trevijano E.R., Castillo J., Erroba E., Lee D.C., Prieto J., Avila M.A.2005. Amphiregulin: an early trigger of liver regeneration in mice. Gastroenterology 128: 424–432. doi: 10.1053/j.gastro.2004.11.006 [DOI] [PubMed] [Google Scholar]
- 4.Bullwinkel J., Baron-Lühr B., Lüdemann A., Wohlenberg C., Gerdes J., Scholzen T.2006. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J. Cell Physiol. 206: 624–635. doi: 10.1002/jcp.20494 [DOI] [PubMed] [Google Scholar]
- 5.Cressman D.E., Greenbaum L.E., DeAngelis R.A., Ciliberto G., Furth E.E., Poli V., Taub R.1996. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science 274: 1379–1383. doi: 10.1126/science.274.5291.1379 [DOI] [PubMed] [Google Scholar]
- 6.Evarts R.P., Nagy P., Marsden E., Thorgeirsson S.S.1987. A precursor-product relationship exists between oval cells and hepatocytes in rat liver. Carcinogenesis 8: 1737–1740. doi: 10.1093/carcin/8.11.1737 [DOI] [PubMed] [Google Scholar]
- 7.Fausto N., Campbell J.S., Riehle K.J.2006. Liver regeneration. Hepatology 43:(Suppl 1): S45–S53. doi: 10.1002/hep.20969 [DOI] [PubMed] [Google Scholar]
- 8.Font-Burgada J., Shalapour S., Ramaswamy S., Hsueh B., Rossell D., Umemura A., Taniguchi K., Nakagawa H., Valasek M.A., Ye L., Kopp J.L., Sander M., Carter H., Deisseroth K., Verma I.M., Karin M.2015. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell 162: 766–779. doi: 10.1016/j.cell.2015.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuyama K., Kawaguchi Y., Akiyama H., Horiguchi M., Kodama S., Kuhara T., Hosokawa S., Elbahrawy A., Soeda T., Koizumi M., Masui T., Kawaguchi M., Takaori K., Doi R., Nishi E., Kakinoki R., Deng J.M., Behringer R.R., Nakamura T., Uemoto S.2011. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 43: 34–41. doi: 10.1038/ng.722 [DOI] [PubMed] [Google Scholar]
- 10.Grisham J.W.1962. A morphologic study of deoxyribonucleic acid synthesis and cell proliferation in regenerating rat liver; autoradiography with thymidine-H3. Cancer Res. 22: 842–849. [PubMed] [Google Scholar]
- 11.Higgins G.M., Anderson R.M.1931. Experimental pathology of the liver: I. Restoration of the liver of the white rat following partial surgical removal. Arch. Pathol. (Chic) 12: 186–202. [Google Scholar]
- 12.Hofmann J.J., Zovein A.C., Koh H., Radtke F., Weinmaster G., Iruela-Arispe M.L.2010. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development 137: 4061–4072. doi: 10.1242/dev.052118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huh C.G., Factor V.M., Sánchez A., Uchida K., Conner E.A., Thorgeirsson S.S.2004. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc. Natl. Acad. Sci. USA 101: 4477–4482. doi: 10.1073/pnas.0306068101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakubowski A., Ambrose C., Parr M., Lincecum J.M., Wang M.Z., Zheng T.S., Browning B., Michaelson J.S., Baetscher M., Wang B., Bissell D.M., Burkly L.C.2005. TWEAK induces liver progenitor cell proliferation. J. Clin. Invest. 115: 2330–2340. doi: 10.1172/JCI23486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson G.D., Nogueira Araujo G.M.1981. A simple method of reducing the fading of immunofluorescence during microscopy. J. Immunol. Methods 43: 349–350. doi: 10.1016/0022-1759(81)90183-6 [DOI] [PubMed] [Google Scholar]
- 16.Karaca G., Swiderska-Syn M., Xie G., Syn W.K., Krüger L., Machado M.V., Garman K., Choi S.S., Michelotti G.A., Burkly L.C., Ochoa B., Diehl A.M.2014. TWEAK/Fn14 signaling is required for liver regeneration after partial hepatectomy in mice. PLoS ONE 9: e83987. doi: 10.1371/journal.pone.0083987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh M., Katoh M.2007. Notch signaling in gastrointestinal tract (review) (review). Int. J. Oncol. 30: 247–251. [PubMed] [Google Scholar]
- 18.Kiso S., Kawata S., Tamura S., Higashiyama S., Ito N., Tsushima H., Taniguchi N., Matsuzawa Y.1995. Role of heparin-binding epidermal growth factor-like growth factor as a hepatotrophic factor in rat liver regeneration after partial hepatectomy. Hepatology 22: 1584–1590. [PubMed] [Google Scholar]
- 19.Knight B., Yeoh G.C., Husk K.L., Ly T., Abraham L.J., Yu C., Rhim J.A., Fausto N.2000. Impaired preneoplastic changes and liver tumor formation in tumor necrosis factor receptor type 1 knockout mice. J. Exp. Med. 192: 1809–1818. doi: 10.1084/jem.192.12.1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kodama Y., Hijikata M., Kageyama R., Shimotohno K., Chiba T.2004. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology 127: 1775–1786. doi: 10.1053/j.gastro.2004.09.004 [DOI] [PubMed] [Google Scholar]
- 21.Lemire J.M., Shiojiri N., Fausto N.1991. Oval cell proliferation and the origin of small hepatocytes in liver injury induced by D-galactosamine. Am. J. Pathol. 139: 535–552. [PMC free article] [PubMed] [Google Scholar]
- 22.Lesage G., Glaser S.S., Gubba S., Robertson W.E., Phinizy J.L., Lasater J., Rodgers R.E., Alpini G.1996. Regrowth of the rat biliary tree after 70% partial hepatectomy is coupled to increased secretin-induced ductal secretion. Gastroenterology 111: 1633–1644. doi: 10.1016/S0016-5085(96)70027-6 [DOI] [PubMed] [Google Scholar]
- 23.Malato Y., Naqvi S., Schürmann N., Ng R., Wang B., Zape J., Kay M.A., Grimm D., Willenbring H.2011. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Invest. 121: 4850–4860. doi: 10.1172/JCI59261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCright B., Lozier J., Gridley T.2002. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development 129: 1075–1082. [DOI] [PubMed] [Google Scholar]
- 25.Mead J.E., Fausto N.1989. Transforming growth factor alpha may be a physiological regulator of liver regeneration by means of an autocrine mechanism. Proc. Natl. Acad. Sci. USA 86: 1558–1562. doi: 10.1073/pnas.86.5.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell C., Willenbring H.2008. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 3: 1167–1170. doi: 10.1038/nprot.2008.80 [DOI] [PubMed] [Google Scholar]
- 27.Muskhelishvili L., Latendresse J.R., Kodell R.L., Henderson E.B.2003. Evaluation of cell proliferation in rat tissues with BrdU, PCNA, Ki-67(MIB-5) immunohistochemistry and in situ hybridization for histone mRNA. J. Histochem. Cytochem. 51: 1681–1688. doi: 10.1177/002215540305101212 [DOI] [PubMed] [Google Scholar]
- 28.Nagahama Y., Sone M., Chen X., Okada Y., Yamamoto M., Xin B., Matsuo Y., Komatsu M., Suzuki A., Enomoto K., Nishikawa Y.2014. Contributions of hepatocytes and bile ductular cells in ductular reactions and remodeling of the biliary system after chronic liver injury. Am. J. Pathol. 184: 3001–3012. doi: 10.1016/j.ajpath.2014.07.005 [DOI] [PubMed] [Google Scholar]
- 29.Nishikawa Y., Tokusashi Y., Kadohama T., Nishimori H., Ogawa K.1996. Hepatocytic cells form bile duct-like structures within a three-dimensional collagen gel matrix. Exp. Cell Res. 223: 357–371. doi: 10.1006/excr.1996.0091 [DOI] [PubMed] [Google Scholar]
- 30.Riehle K.J., Dan Y.Y., Campbell J.S., Fausto N.2011. New concepts in liver regeneration. J. Gastroenterol. Hepatol. 26:(Suppl 1): 203–212. doi: 10.1111/j.1440-1746.2010.06539.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekiya S., Suzuki A.2014. Hepatocytes, rather than cholangiocytes, can be the major source of primitive ductules in the chronically injured mouse liver. Am. J. Pathol. 184: 1468–1478. doi: 10.1016/j.ajpath.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 32.Shiojiri N.1997. Development and differentiation of bile ducts in the mammalian liver. Microsc. Res. Tech. 39: 328–335. doi: [DOI] [PubMed] [Google Scholar]
- 33.Shiojiri N., Nagai Y.1992. Preferential differentiation of the bile ducts along the portal vein in the development of mouse liver. Anat. Embryol. (Berl.) 185: 17–24. doi: 10.1007/BF00213597 [DOI] [PubMed] [Google Scholar]
- 34.Takase H.M., Itoh T., Ino S., Wang T., Koji T., Akira S., Takikawa Y., Miyajima A.2013. FGF7 is a functional niche signal required for stimulation of adult liver progenitor cells that support liver regeneration. Genes Dev. 27: 169–181. doi: 10.1101/gad.204776.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanimizu N., Miyajima A.2004. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. J. Cell Sci. 117: 3165–3174. doi: 10.1242/jcs.01169 [DOI] [PubMed] [Google Scholar]
- 36.Tzankov A., Zimpfer A., Went P., Maurer R., Pileri S.A., Geley S., Dirnhofer S.2005. Aberrant expression of cell cycle regulators in Hodgkin and Reed-Sternberg cells of classical Hodgkin’s lymphoma. Mod. Pathol. 18: 90–96. doi: 10.1038/modpathol.3800276 [DOI] [PubMed] [Google Scholar]
- 37.Yamada Y., Kirillova I., Peschon J.J., Fausto N.1997. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc. Natl. Acad. Sci. USA 94: 1441–1446. doi: 10.1073/pnas.94.4.1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanger K., Zong Y., Maggs L.R., Shapira S.N., Maddipati R., Aiello N.M., Thung S.N., Wells R.G., Greenbaum L.E., Stanger B.Z.2013. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 27: 719–724. doi: 10.1101/gad.207803.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.