Abstract
Phasins are important proteins controlling poly-3-hydroxybutyrate (PHB) granules formation, their number into the cell and stability. The genome sequencing of the endophytic and diazotrophic bacterium Herbaspirillum seropedicae SmR1 revealed two homologous phasin genes. To verify the role of the phasins on PHB accumulation in the parental strain H. seropedicae SmR1, isogenic strains defective in the expression of phaP1, phaP2 or both genes were obtained by gene deletion and characterized in this work. Despite of the high sequence similarity between PhaP1 and PhaP2, PhaP1 is the major phasin in H. seropedicae, since its deletion reduced PHB accumulation by ≈50% in comparison to the parental and ΔphaP2. Upon deletion of phaP1, the expression of phaP2 was sixfold enhanced in the ΔphaP1 strain. The responsive backup expression of phaP2 partially rescued the ΔphaP1 mutant, maintaining about 50% of the parental PHB level. The double mutant ΔphaP1.2 did not accumulate PHB in any growth stage and showed a severe reduction of growth when glucose was the carbon source, a clear demonstration of negative impact in the fitness. The co-occurrence of phaP1 and phaP2 homologous in bacteria relatives of H. seropedicae, including other endophytes, indicates that the mechanism of phasin compensation by phaP2 expression may be operating in other organisms, showing that PHB metabolism is a key factor to adaptation and efficiency of endophytic bacteria.
Keywords: polyhydroxybutyrate, PHB, phasin, Herbaspirillum seropedicae, granule-associated proteins, backup regulation
Introduction
Herbaspirillum seropedicae SmR1 is a diazotrophic β-Proteobacterium that associates beneficially with economically relevant species of Gramineae (Baldani et al., 1986) and produces poly-3-hydroxybutyrate (PHB) granules as means of carbon and energy storage (Catalan et al., 2007; Kadowaki et al., 2011). Therefore, H. seropedicae SmR1 is an important model to study the impact of PHB metabolism on endophytic growth and adaptation. Thirteen genes probably involved in PHB metabolism were identified in the strain SmR1 (Kadowaki et al., 2011; Pedrosa et al., 2011), including four phaC, two phaZ and two phaP genes encoding PHA synthases, PHA depolymerases and phasins, respectively. Although H. seropedicae SmR1 possesses three genes encoding proteins homologous to PHA synthases (Pedrosa et al., 2011), PHB synthesis is supported by the PHA synthase expressed by phaC1, since its deletion abolishes PHB accumulation (Tirapelle et al., 2013). Furthermore, phbF (hereafter phaR) which encodes a transcriptional repressor protein was identified and characterized (Kadowaki et al., 2011). Probably, PHB plays important roles in nitrogen fixation and plant–bacteria interactions (Mandon et al., 1998; Kadouri et al., 2003; Wang et al., 2007; Quelas et al., 2013), but its action in H. seropedicae was not been fully characterized.
Poly-3-hydroxybutyrate is an aliphatic polyester member of the polyhydroxyalkanoates (PHA) family that some bacteria synthesize to store carbon and reducing equivalents (Anderson and Dawes, 1990; Madison and Huisman, 1999). In addition, the production of PHB is a hot topic in biotechnology due to its physicochemical properties very close to oil-based plastics, while PHB is readily degradable in the environment (Chen, 2009; Urtuvia et al., 2014). Therefore, PHB is a bio-sustainable alternative for synthetic plastic materials. PHB is usually produced under conditions of carbon excess and low levels of essential nutrients including nitrogen, phosphate, and oxygen (Hervas et al., 2008). At least three enzymes: 3-ketothiolase, acetoacetyl-CoA reductase and PHA synthase encoded by phaA, phaB, and phaC, respectively, are involved in its synthesis (Babel et al., 2001), which occurs by condensation of acetyl-CoA forming acetoacetyl-CoA, then reduction of acetoacetyl-CoA to 3-hydroxybutyryl-CoA (3HB-CoA) and finally polymerization of 3HB-CoA to yield PHB (Steinbuchel and Hein, 2001). When carbon/energy is required, the polymer is mobilized by PHA depolymerases encoded by phaZ genes (Babel et al., 2001). Polymeric PHB is stored as insoluble, intracellular granules that are coated with proteins (totaling 0.5–2% of the granule weight; Grage et al., 2009; Jendrossek, 2009). Phasins are small amphiphilic proteins attached on the surface of polyhydroxyalkanoate inclusions in Bacteria and Archaea (Neumann et al., 2008; Jendrossek, 2009; Cai et al., 2012). These proteins control the size and number of PHB granules (Wieczorek et al., 1995; Jurasek and Marchessault, 2002, 2004; Potter et al., 2004; Cho et al., 2012) and are present in all PHA producing bacteria. Although not highly conserved in terms of amino acid sequence, phasins perform similar functions in promoting granule formation and stabilization of PHA in different microbes (York et al., 2001a,b; Jurasek and Marchessault, 2002). Ralstonia eutropha H16, a well-studied model of PHB metabolism, contains seven phasin genes (Potter et al., 2005; Kuchta et al., 2007; Pfeiffer and Jendrossek, 2011, 2012) but it seems that PhaP1 is the major phasin affecting PHB accumulation (Potter et al., 2005). In plant-associated bacterium Sinorhizobium meliloti Rm1021, the genes SMc00777 and SMc02111 encode the phasins PhaP1 and PhaP2, respectively (Wang et al., 2007). The deletion of both genes resulted in a mutant defective in PHB production and plants of Medicago truncatula inoculated with this mutant exhibited reduced shoot dry weight. The occurrence of phasin-expressing genes in the genome of other plant-associated bacteria as Azospirillum brasilense Sp245, A. lipoferum 4B, Azoarcus sp. BH72 and Pseudomonas stutzeri A1501 (Krause et al., 2006; Yan et al., 2008; Wisniewski-Dye et al., 2011) indicates that PHB metabolism is important to bacteria during plant colonization, as previously suggested (Trainer and Charles, 2006). The genome sequencing of H. seropedicae SmR1 revealed two paralogous phasin genes (75% similarity, 59% identity), the products of which had been shown by proteomic analyses to be the main phasins associated to PHB granules (Pedrosa et al., 2011; Tirapelle et al., 2013). Nevertheless, in the ΔphaP1 mutant, PhaP2 replaced PhaP1 on the surface of the granules (Tirapelle et al., 2013). To see if this functional complementarity was also reflected in conservation of function, we deleted phaP1 or phaP2 or both genes to analyze the effect of phasin absence on PHB accumulation in the mutants. At the same time, we determined the expression patterns of the genes in transcriptional fusions to a lacZ reporter-gene housed in different genetic backgrounds, to address if phaP2 expression enhances upon deletion of phaP1, as related previously for other examples of redundant genes (Bratlie et al., 2010; Kondrashov, 2012).
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Strains and plasmids used in this work are listed in Table 1. Escherichia coli strain Top10 (Thermo Fisher Scientific Inc., Waltham, MA, USA) and S17.1 (Simon et al., 1983) were used for cloning and conjugation procedures, respectively, while strain ET8000 (MacNeil et al., 1982) was used in expression assays with lacZ fusions. E. coli strains were grown at 37°C in LB medium and shaken at 160 rpm. H. seropedicae parental strain SmR1 (Souza et al., 2000) and mutant strains were grown in NFbHP media with 37 mM DL-malate or 25 mM glucose and 20 mM NH4Cl at 30°C and shaken at 120 rpm (Pedrosa and Yates, 1984).
Table 1.
Bacterial strains and plasmids used in this work.
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Escherichia coli strains | ||
| Top10 | Cloning strain | Invitrogen |
| S17-1 | Conjugation strain | Simon et al., 1983 |
| ET8000 | Wild-type strain | MacNeil et al., 1982 |
| Herbaspirillum seropedicae strains | ||
| SmR1 | Parental strain, Nif+, SmR | Souza et al., 2000 |
| ΔphaP1 | Chromosomal deletion of phaP1 | Tirapelle et al., 2013 |
| ΔphaP2 | Chromosomal deletion of phaP2 | This work |
| ΔphaP1.2 | Chromosomal deletion of phaP1 and phaP2 | Tirapelle et al., 2013 |
| ΔphaC1 | Chromosomal deletion of phaC1 | Tirapelle et al., 2013 |
| Plasmids | ||
| pTZ18R | Cloning plasmid | Mead et al., 1986 |
| pTZ57R/T | T/A cloning plasmid | Fermentas |
| pDK6 | Expression vector tac promoter lacIq, KmR | Kleiner et al., 1988 |
| pMMS31 | Derivative of pDK6 encoding PhbF from H. seropedicae SmR1 | Kadowaki et al., 2011 |
| pMP220 | RK2 derivative, low-copy number, promoterless lacZ containing vector used to construct transcriptional fusions; TcR | Spaink et al., 1987 |
| pEFT11 | pMP220 harboring the 5′-flanking region of phaP1 cloned upstream of lacZ | This work |
| pEFT12 | pMP220 harboring the 5′-flanking region of phaP2 cloned upstream of lacZ | This work |
| pK18mobsacB | Suicide vector; KmR, sacB, mobilizable plasmid | Schafer et al., 1994 |
| pEFT13 | Deletion product ΔphaP2 cloned into the pK18mobsacB | This work |
| pBBR1MCS3 | Broad-host-range vector | Kovach et al., 1995 |
| pLPA01 | pBBR1MCS3 harboring phaP1 of H. seropedicae. Over-expression of PhaP1 | This work |
| pLPA02 | pBBR1MCS3 harboring the phaP2 of H. seropedicae. Over-expression of PhaP2 | This work |
Quantification of PHB
PHB was quantified by methanolysis and GC-FID (gas chromatography coupled to flame-ionization detector) analyses as described previously (Braunegg et al., 1978) on 5 to 10 mg of lyophilized bacteria. Amounts of PHB in each sample were normalized to the cell dry weight (cdw; weight of the lyophilized bacterial pellet) and expressed as % of PHB cell dry weight-1.
Construction of Mutants of H. seropedicae SmR1
The ΔphaP1 and ΔphaC1 mutants were constructed by in-frame deletion of the phaP1 (Hsero_1639, GenBank: 9402240) and phaC1 (Hsero_2999, GenBank: 9403600) in H. seropedicae SmR1, as previously described (Tirapelle et al., 2013). The in-frame marker-less deletion of phaP2 (Hsero_4759, GenBank: 9405360) was obtained by cloning upstream and downstream fragments of the gene into the non-replicating plasmid pK18mobsacB, which carries a kanamycin resistance cassette along with sacB that confers sucrose sensitivity (Schafer et al., 1994). Briefly, 500 bp fragments to either flank of phaP2 were amplified by PCR with primers Fw_phaP2_UP and Rev_phaP2_UP (sequences showed in Supplementary Table S1) for the upstream region whereas Fw_phaP2_DOWN and Rev_phaP2_DOWN were used for the downstream region. The PCR products were cloned into pTZ57R/T and sequenced using universal and reverse M13 primers. The downstream fragment was ligated to the upstream fragment using the KpnI site, resulting in the deletion product ΔphaP2. The entire construction was digested with BamHI and SalI, in order to ligate the ΔphaP2 fragment into pK18mobsacB digested with the same enzymes, yielding pEFT13. E. coli S17-1 was transformed with pEFT13 and the plasmid conjugated to H. seropedicae SmR1 using bi-parental mating. Single-recombinants were selected on NFb-malate agar containing streptomycin 80 μg mL-1, nalidixic acid 5 μg mL-1 and kanamycin 500 μg mL-1. A single-recombinant colony was collected in 3 mL NFb-malate and cultivated overnight without antibiotics. The culture was serially diluted and plated on NFb-malate agar containing 10% (w/v) sucrose. Colonies that grew on sucrose were screened for deletion by PCR using the primers Fw_phaP2_UP and Rev_phaP2_DOWN. To obtain the double mutant ΔphaP1.2, the plasmid pEFT13 was conjugated in the ΔphaP1 mutant of H. seropedicae and double-recombinants were selected as described above.
Construction of Transcriptional Fusions
The intergenic regions of phaP1 (333 bp including 28 bp of the phaP1 coding sequence) and phaP2 (224 bp including 54 bp of the phaP2 coding sequence) were amplified from H. seropedicae SmR1 genomic DNA by PCR (primer sequences showed in Supplementary Table S1) and cloned into pMP220 (Spaink et al., 1987) upstream of a promoterless, rbs-containing lacZ to yield plasmids pEFT11 and pEFT12, respectively.
RNA Extraction and RT-PCR
Strains were grown on NFb-malate medium with 20 mM of ammonium chloride at 30°C and shaken at 120 rpm. Cells from 1.5 mL of culture at OD600 of 1.0 were collected by centrifugation (10,000 × g, 4°C, 5 min) and re-suspended in 1 mL of TRIzol® Reagent (Life Technologies, USA). The homogenized sample was incubated for 5 min at room temperature and then, extracted with 0.25 mL of chloroform. After centrifugation (10,000 × g, 4°C, 5 min), the aqueous phase was precipitated with 0.5 mL of isopropanol. The pellet was collected by centrifugation (10,000 × g, 4°C, 5 min) and washed with 1 mL of ethanol 80%. The air dried RNA pellet was resuspended in 30 μL of RNAse-free water and the quality of RNA preparation was determined by A260/A280 ratio and checked by electrophoresis in 1.0% agarose gel. For cDNA preparation, 100 ng of total RNA was used in 20 μL reactions applying the High Capacity RNA-to-cDNA kit (Applied Biosystems, USA) according to the manufacturer’s instructions. From the cDNA preparations, 1 μL was used as template in a 20 μL PCR reaction with PfuX7 as described previously (Norholm, 2010), applying the specific primers described in Supplementary Table S1, 60°C as annealing temperature and 30 cycles of reaction. From each reaction, 2 μL were applied on a 1.5% agarose gel to visualize the intensity of the bands after ethidium bromide staining. The rrsA gene encoding the 16S rRNA was used as endogenous control. Negative controls were performed using as template samples of RNA untreated with reverse transcriptase.
Complementation of phaP Mutants
phaP1 (Hsero_1639) plus 333 bp of the intergenic region upstream of its start codon was amplified by PCR with Fw_Pro_phaP1 and Rev_Gen_phaP1 primers (Supplementary Table S1) from H. seropedicae SmR1 genomic DNA and cloned into the XhoI and XbaI sites of pBBR1MCS-3 (Kovach et al., 1995), generating pLPA01. Similarly, phaP2 (Hsero_4759) plus 224 bp of the intergenic region upstream of its start codon was amplified with Fw_Pro_phaP2 and Rev_Gen_phaP2 primers (Supplementary Table S1) by PCR and cloned into the XhoI and XbaI sites of the pBBR1MCS-3, generating pLPA02. Conjugation was performed by bi-parental mating between H. seropedicae SmR1 and E. coli S17-1.
β-Galactosidase Activity Assay
β-galactosidase activity was determined in E. coli ET8000 (grown in LB media) carrying the transcriptional fusion plasmids pEFT11 or pEFT12 in the presence of pMMS31 (that expresses the PhaR from H. seropedicae) or pDK6 (negative control). To perform similar analyses in H. seropedicae parental strain SmR1 and mutant strains, pEFT11 or pEFT12 was introduced by bi-parental mating with E. coli S17-1. Transconjugants were selected on NFb-malate agar containing 20 mM of ammonium chloride and tetracycline (10 μg.mL-1). β-galactosidase activity was assayed following previous protocol (Miller, 1972).
Fluorescence and Transmission Electron Microscopy
To visualize PHB granules in SmR1 and its phaP mutants, fluorescence microscopy was performed after staining with the fluorescent probe Nile Red, which stains neutral lipids (Spiekermann et al., 1999). Bacterial cultures (1 mL) were harvested by centrifugation for 60 s at 10,000 × g. The pellets were resuspended in 30% (v/v) ethanol in PBS (Na2HPO4 10 mM, KH2PO4 1.8 mM, NaCl 130 mM) and 3 μL of 1.6 mM Nile Red (λex 586–579 nm; λem 637–597 nm) dissolved in DMSO was added and incubated in the dark for 5 min. Then, the samples were centrifuged again for 60 s at 10,000 × g, resuspended in PBS and viewed under a fluorescent microscope. The images were obtained using an Axio Imager Z2 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany), equipped with four Metafer automated capture software (Metasystems GmbH, Altlussheim, Germany) and a CoolCube 1 camera (with 100× magnification). To TEM analyses, cell pellets from H. seropedicae cultures were fixed with Karnovsky’s fixative (Karnovsky, 1965), post-fixed with 2% OsO4 in 0.1 M cacodylic acid buffer (pH 7.2) for 1 h and embedded in Epon 812 (Luft, 1961). After contrasting with 2% uranyl acetate (Watson, 1958) and lead citrate (Reynolds, 1963), samples were examined with a JEOL-JEM 1200 EX II transmission electron microscope.
Phylogenetic Analysis of Phasin Sequences
The amino acid sequences used in the analysis are listed in the Supplementary Figure S1. The alignment was performed by Muscle (Edgar, 2004) into the MEGA software package (Tamura et al., 2013), using default parameters. The resulting alignment was cured by Gblocks to remove poorly aligned positions and highly divergent regions (Castresana, 2000). The phylogeny reconstruction of the sequences in the cured alignment was obtained by the Neighbor-joining method and tested by bootstrap (10,000 replicates).
Statistical Analysis
Where appropriate, statistical analysis was carried out using independent two-sample t-test with the R package (R Development Core Team, 2015).
Results
H. seropedicae SmR1 Contains Two Paralogous Phasin and a Third Less Conserved Putative Phasin
Three genes coding putative phasins (phaP1, locus-tag Hsero_1639; phaP2, Hsero_4759 and phaP3, Hsero_2402; Kadowaki et al., 2011; Pedrosa et al., 2011) are present in H. seropedicae SmR1. The alignments and pair-wise distances between PhaP1 and PhaP2 showed a short evolutionary distance (0.412 using the p-distance method), but PhaP3 was further removed, indicating significant sequence divergence and possibly also function (Figures 1A–D). Indeed, PhaP3 was only detected on PHB granules when phaP1 was deleted and, it was clearly less abundant than PhaP2, the main phasin in the absence of PhaP1. The phaP1 and phaP2 genes could have been generated by gene duplication based on 59% identity between their encoded amino acid sequences, as well as a region of more than 150 amino acids that can be aligned (Figure 1A), according to the classification of duplicated genes proposed by Gevers et al. (2004). Nevertheless, the horizontal gene transfer (HGT) of an additional phasin gene cannot be totally ruled out, as intra-genome homologs are also acquired via HGT (Maerk et al., 2014). Accordingly, the co-occurrence of phaP1 and phaP2 homologs was found in other bacteria phylogenetically close to H. seropedicae SmR1 (Figure 1E). The phylogenetic analysis of PhaP1 and PhaP2 homolog sequences showed that the appearance of a second phasin occurred early in evolution of the Herbaspirillum genus and was conserved in subsequent speciation events (Figure 1E), indicating that the presence of both phasins should be important for PHB accumulation.
FIGURE 1.
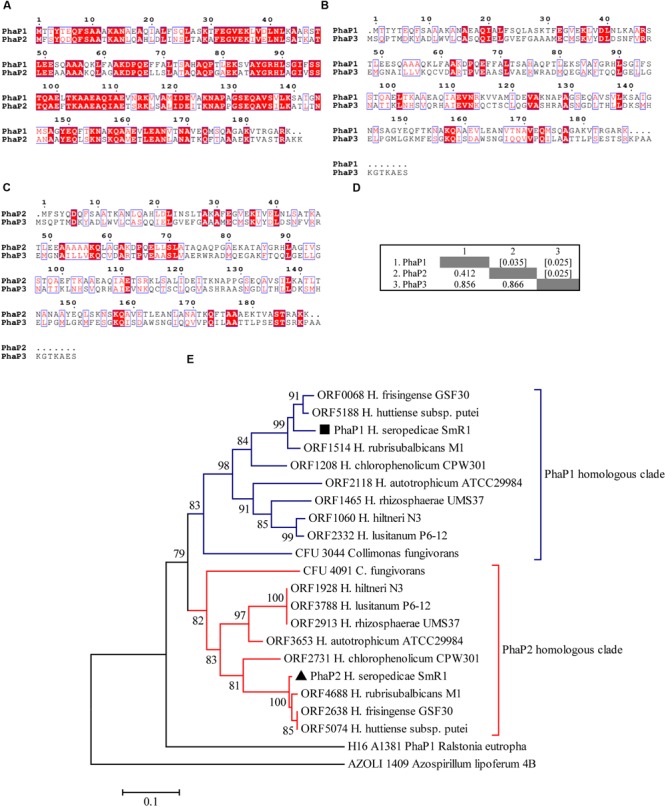
Protein sequence alignments of phasins from Herbaspirillum seropedicae SmR1 and phylogenetic analysis of homologous sequences. Multiple alignments of protein sequences of (A) PhaP1 (Hsero_1639) and PhaP2 (Hsero_4759), (B) PhaP1 and PhaP3 (Hsero_2402), and (C) PhaP2 and PhaP3 were carried out using Espript 3.0 and the default parameters (Robert and Gouet, 2014). The red shaded letters indicate identical residues, while red letters represent conserved residues. (D) The number of amino acid differences per site from between PhaP1, PhaP2, and PhaP3 sequences are shown. Standard error estimate(s) are shown above the diagonal (enclosed within brackets) and were obtained by a bootstrap procedure (10000 replicates). All ambiguous positions were removed for each sequence pair. There were a total of 199 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). (E) The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The putative phasin of Azospirillum lipoferum encoded by the AZOLI 1409 (Wisniewski-Dye et al., 2011) was applied as an outgroup. Only nodes with bootstrap test (10000 replicates) bigger than 70% are shown next to the branches (Efron et al., 1996). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of amino acid differences per site. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). The PhaP sequences used to construct the tree are shown in Supplementary Figure S1.
Deletion of phaP1 Reduces PHB Accumulation
To verify the roles of PhaP1 and PhaP2 in PHB accumulation, three isogenic deletion mutants – ΔphaP1, ΔphaP2, and ΔphaP1.2 – were constructed. The mutant strains grew equally well as the parental strain on NFb-malate medium (Figure 2A; Table 2). Since cells accumulating PHB are less translucent than those without PHB, which can affect optical density measurements, the growth of parental and mutant strains was also determined by counting the number of viable cells in culture (Supplementary Figure S2). Intracellular accumulation of PHB in the strains grown on malate was determined: (i) in early exponential growth (OD600 = 0.6); in mid-exponential growth (OD600 = 0.8); (iii) early stationary phase (OD600 = 1.2); and, (iv) in late stationary growth (OD600 = 1.4). The parental strain accumulated PHB up to 13% (w/w) of cell dry weight (cdw) after 12 h of growth (early stationary phase; Figure 2B), but decreased to 11% of PHB at late stationary phase. Similar behavior was observed with ΔphaP2 which reached 15% of cdw at early stationary phase. On the other hand, ΔphaP1 only accumulated approximately 50% of parental levels of PHB at all tested growth phases (Figure 2B). Deletion of both phasins (ΔphaP1.2) drastically reduced PHB accumulation to <1% of cdw (Figure 2B).
FIGURE 2.
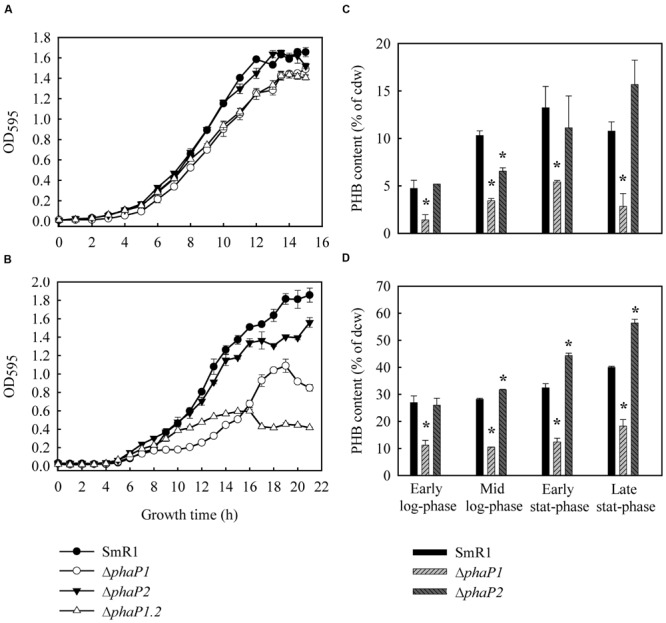
Growth and PHB accumulation profiles of H. seropedicae SmR1 (parental strain) and the mutants ΔphaP1, ΔphaP2, and ΔphaP1.2. Strains were grown in NFb medium with 20 mM of ammonium chloride and 37 mM DL-malate (A,B) or 25 mM (w/v) D-glucose (C,D) at 30°C (orbital agitation at 120 rpm). OD595 growth data were obtained from three independent cultures, while PHB contents were determined on four independent samples. PHB levels in ΔphaP1.2 were less than 1% at all growth phases. Where appropriate, statistical significance is shown (∗p-value ≤ 0.05, independent two-sample t-test). The data obtained to SmR1 was used for normalization and determination of statistical significance.
Table 2.
Growth rates of phaP mutants in NFb media containing malate or glucose as carbon source.
| Strain | Growth rate (ΔOD595/h)a |
|
|---|---|---|
| Malateb | Glucose | |
| SmR1 (wt) | 0.237 ± 0.007 | 0.184 ± 0.026c |
| ΔphaP1 | 0.182 ± 0.029 | 0.123 ± 0.022d |
| ΔphaP2 | 0.200 ± 0.018 | 0.151 ± 0.017c |
| ΔphaP1.2 | 0.164 ± 0.019 | 0.055 ± 0.009e∗ |
aThe values are averages ± standard errors for two independent experiments. bGrowth rate was calculated with the data of OD600 between 8 and 13 h of growth for all strains. cGrowth rate calculated between 10 and 16 h of growth. dGrowth rate calculated between 12 and 19 h of growth. eGrowth rate calculated between 7 and 16 h of growth. Where appropriate, statistical significance is shown (∗p-value ≤ 0.05). The data obtained to the SmR1 strain was used for normalization and determination of statistical significance.
To exclude the possibility that it was growth on malate rather than the mutations per se that affected PHB accumulation, similar experiments were performed using glucose as the carbon source. Under these conditions, SmR1 also accumulated PHB as previously reported for the strains Z67, Z69, and Z78 of H. seropedicae (Catalan et al., 2007) and the parental strain reached a maximum of 40% of cdw in late-stationary phase. The production of PHB by ΔphaP2 was similar to the parental strain at the early stat-phase and reached 56% of cdw at the late stat-phase. On the other hand, the ΔphaP1 accumulated only 18% of PHB as its maximum production, corresponding to 50% of the PHB content of the parental strain (Figure 2C). In ΔphaP1.2, PHB accumulation was not detected when cultivated on glucose. Noteworthy, the ΔphaP1.2 strain presented a growth penalty when cultivated in glucose (Table 2), suggesting that the absence of PHB affects the glucose metabolism in H. seropedicae. Despite the growth rate in glucose of the ΔphaP1 mutant did not present a statistical difference (Table 2), its growth pattern was atypical, since it stopped to grow early than parental strain (Figure 2A). This finding is also in agreement that reduction in PHB affects growth in glucose. In conclusion, regardless of the carbon source employed and the growth phase, the absence of PhaP1 or both phasins negatively affected PHB accumulation.
Microscopic Alterations in the Number of Granules Per Cell
After staining with Nile Red, 92% of native cells contained at least two PHB granules per cell (Figure 3) the rest only one (n = 100 counted cells), but in ΔphaP1 the situation was practically reversed – only 18% of cells contained two granules and 82% only one (n = 100 cells). ΔphaP2 had a similar granular distribution as the parental strain. No cells containing two PHB granules in ΔphaP1.2 were observed, but 9% of the counted cells (n = 100) contained one PHB granule. In the strict sense, it was not clear whether these PHB granules were coated by other proteins or a transient agglomeration of polymer within the cytoplasm, since at all times examined, the level of PHB accumulated in the double mutant was below 1% of cdw. The absence of granules in ΔphaP1.2 was also confirmed by TEM analysis (Figure 3B).
FIGURE 3.
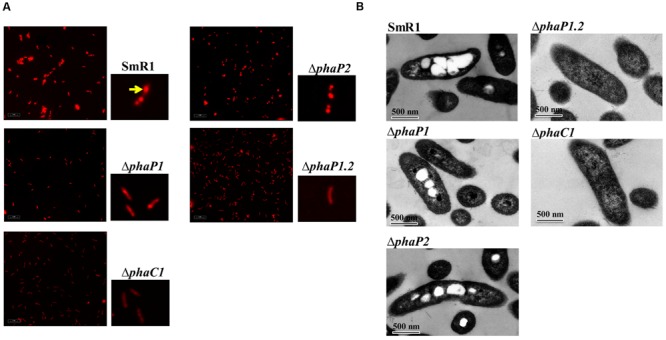
Fluorescence microscopy and transmission electron microscopy of H. seropedicae SmR1, phaP and phaC1 mutants. (A) Strains were grown in NFb medium with 37 mM DL-malate and 20 mM NH4Cl to an OD600 of 1.0. Cells were stained with Nile Red and visualized by excitation with 543 nm light. The yellow arrow in the SmR1 panel indicates a typical PHB granule stained with Nile Red. (B) The strains cultivated in NFb medium with 37 mM DL-malate and 20 mM NH4Cl to an OD600 of 1.0 were processed and visualized by TEM. The black bar represents 500 nm in scale.
Expression of phaP1 and phaP2 Is Repressed by PhaR
Previously, we showed that the negative regulator PhaR of H. seropedicae (a homolog of PhaR in R. eutropha H16) binds to the regulatory region of phaP1 thus repressing its expression (Maehara et al., 2001, 2002; Pötter et al., 2002; York et al., 2002; Kadowaki et al., 2011). To further examine the role of PhaR on expression of phaP1 and phaP2, we fused a promoterless rbs-containing lacZ downstream of the regulatory regions of both genes (hereafter denoted PphaP1-lacZ and PphaP2-lacZ). We decide to measure the activity of the fusions in E. coli as a heterologous non-PHB-producing model, to avoid any perturbation on PhaR activity caused by PHB production (Maehara et al., 2002). The fusions were transformed into E. coli ET8000 expressing or not H. seropedicae PhaR. Cells carrying PphaP1-lacZ and PphaP2-lacZ fusions showed similar, high β-galactosidase activities when PhaR was not expressed. However, cells expressing PhaR had a remarkable repression on expression levels of both PphaP1- and PphaP2-lacZ (Table 3).
Table 3.
Transcriptional analysis of PphaP1-lacZ and PphaP2-lacZ fusions in E. coli ET8000 expressing PhaR from H. seropedicae SmR1.
| Strain | β-galactosidase activity before induction (Miller units) | β-galactosidase activity 2 h after induction (Miller units) |
|---|---|---|
| ET8000/pDK6/PphaP1-lacZ | 8,516.5 ± 693.3 | 11,801.3 ± 642.1 |
| ET8000/pMMS31/PphaP1-lacZ∗ | 168.0 ± 6.4∗∗ | 89.7 ± 2.5∗∗ |
| ET8000/pDK6/PphaP2-lacZ | 11,180.8 ± 142.8 | 12,061.1 ± 55.5 |
| ET8000/pMMS31/PphaP2-lacZ∗ | 149.8 ± 3.2∗∗ | 85.5 ± 2.8∗∗ |
The cultures were grown in LB medium until OD600 of 0.6. At this point, the IPTG was added to final concentration of 1 mM. The activity was monitored for two more hours after inducer addition. ∗Strains expressing PhaR from H. seropedicae under control of the Ptac promoter. Where appropriate, statistical significance is shown (∗∗p-value ≤ 0.01, independent two-sample t-test). The data obtained to the strains harboring PphaP1- or PphaP2-lacZ not expressing PhaR were used for normalization and determination of statistical significance.
The phaP1 and phaP2 Genes Are Dissimilarly Expressed
To determine expression levels of phaP1 and phaP2, the PphaP1- and PphaP2-lacZ fusions were conjugated into the parental and mutant strains. Expression profiles were evaluated during growth in media containing DL-malate as the carbon source. Expression of PphaP1-lacZ in the parental strain was dependent on the growth stage, since expression increased after an OD600 of 0.5 was achieved and PHB began to accumulate (Figure 4A). Interestingly, expression of PphaP2-lacZ was 8-fold lower than that achieved by the PphaP1-lacZ fusion in the same strain (Figure 4A). However, expression of PphaP2-lacZ increased 6-fold in ΔphaP1, showing that upon deletion of the main phasin, PhaP2 can act as a backup phasin (Figure 4B). Since the ΔphaP1.2 did not accumulate PHB granules, one should anticipate that both fusions would be repressed by PhaR. Nevertheless, the PphaP1- and PphaP2-lacZ fusions were actives in ΔphaP1.2, indicating that not only PHB granules de-repress expression of phaP, but that newly synthesized chains of PHB have the same effect (Figure 4C). As expected, in ΔphaC1, which is unable to synthesize PHB, expression of both fusions was repressed (Figure 4D). Expression of phaP1 and phaP2 was also assayed in NFb media containing 25 mM glucose and 20 mM ammonium chloride, conditions which stimulated PHB accumulation. High β-galactosidase activities (4,000 Miller units) for PphaP1-lacZ were obtained in all strains analyzed (parental and the mutants), showing that even at low cell densities (OD600 ≥ 0.3, Figure 4E) enough PHB granules were present to almost completely de-repress phaP1 expression. Expression of phaP2 was not fully activated in the parental strain (Figure 4E), but in ΔphaP1 it was fivefold higher even at low cell densities (Figure 4F). The results of expression applying lacZ fusions were validated by RT-PCR. The same profile of transcription was observed when the intensities of the amplified bands from phaP1 and phaP2 cDNAs were compared in different genetic backgrounds (Figure 4G).
FIGURE 4.
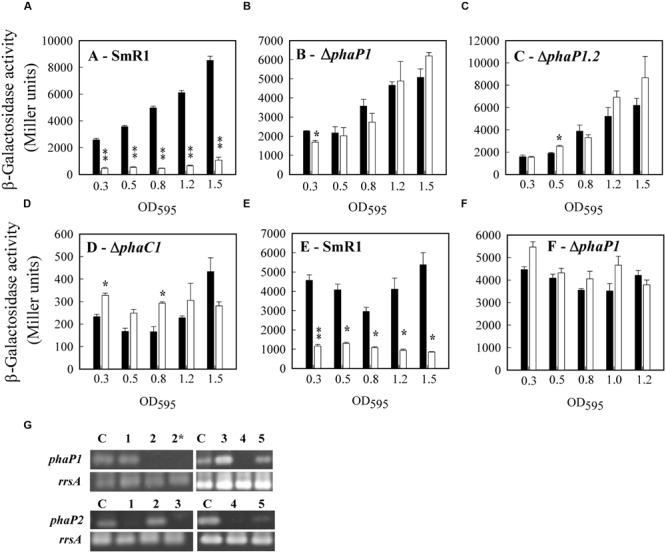
Transcriptional analyses of phaP1 and phaP2 expression in H. seropedicae. (A–D) Cells harboring PphaP1-lacZ (black bars) or PphaP2-lacZ (white bars) were grown in liquid NFb medium containing 37 mM DL-malate and 20 mM NH4Cl at 30°C. (E,F) Cells harboring PphaP1-lacZ (black bars) or PphaP2-lacZ (white bars) were grown in liquid cultures of NFb containing 25 mM D-glucose and 20 mM NH4Cl at 30°C. When the OD600 of the cultures reached the indicated values, 100 μL of culture was removed to determine β-galactosidase activity. H. seropedicae strains harboring pMP220 (promoterless lacZ plasmid) gave an average activity of 50 Miller units in all growth phases analyzed. Experiments were performed in biological triplicates. The means of β-galactosidase activity were tested in pairs for each sample point. Where appropriate, statistical significance is shown (∗p-value ≤ 0.05, ∗∗p-value ≤ 0.01, independent two-sample t-test). (G) RT-PCR analysis. The RNA from the strains SmR1 (lane 1), ΔphaP1 (lane 2 and 2∗ for phaP1, lane 2 for phaP2), ΔphaP2 (lane 3), ΔphaP1.2 (lane 4), and ΔphaC1 (lane 5) was purified and submitted to direct RT-PCR amplification of phaP1 and phaP2 gene as described in Methods. The lane C is a PCR product amplified from gDNA from H. seropedicae SmR1 used as positive control of the reaction. The 16S rRNA (rrsA) was used as an endogenous expression control. A representative gel from tree independent RNA extractions is showed.
Complementation with phaP1 Fully Restores PHB Accumulation, while phaP2 Expression Has a Partial Effect
To determine if both phasins were able to restore PHB accumulation, the phaP genes were cloned under control of their native promoter into a medium copy-number plasmid (pBBR1MCS-3) and expressed into the phaP mutants. The expression of phaP1 in the ΔphaP1, ΔphaP2, and ΔphaP1.2 mutants restored PHB accumulation to native levels (Figure 5A). The expression of a plasmid-borne phaP1 copy restored PHB granules formation in ΔphaP1.2 (Figure 5B). Complementation by expression of phaP2 significantly reduced PHB accumulation, regardless the strain tested. ΔphaP1 complemented with phaP2 presented 45% of reduction in PHB as compared to complementation with phaP1. Similarly, 51% less PHB was observed when ΔphaP1.2 was complemented with phaP2. Once again, these results show that PhaP1 is more effective in controlling PHB accumulation than its homolog PhaP2. As demonstrated before in this work, deletion of phaP1 or both phasin genes generated mutant strains with growth penalty in minimal medium containing glucose as sole carbon source (Figure 1C). Therefore, to verify if complementation would restore normal growth in NFb-glucose, the strains were complemented with phaP1 and their growth curves were determined (Figures 5C,D). The strains ΔphaP1 and ΔphaP1.2, which exhibited reduced growth on glucose, grew as the parental strain when complemented with phaP1. In conclusion, the expression of phaP1 recovered PHB accumulation in the deficient strains, consequently normalizing their metabolic status and turning them able to grow in glucose.
FIGURE 5.
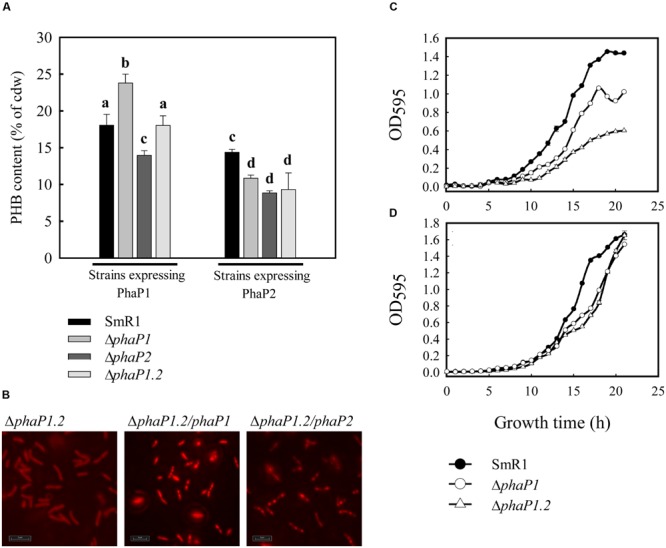
Poly-3-hydroxybutyrate contents of phaP mutants complemented with plasmids harboring phaP1 and phaP2. (A) H. seropedicae strains conjugated with pLPA01 were complemented with phaP1 in trans under control of its native promoter. The strains conjugated with pLPA02 were complemented with phaP2 in trans under control of its native promoter. Cells harboring pLPA01 or pLPA02 were grown in liquid NFb containing 37 mM DL-malate and 20 mM NH4Cl at 30°C. When the OD600 of the cultures reached 1.0, 10 mL was removed to determine the PHB content by gas chromatography as described in section “Materials and Methods.” The bars marked with different letters indicate means significantly different (independent two-sample t-test, p < 0.05). Experiments were performed in quadruplicate. (B) Cells of ΔphaP1.2 mutant complemented with pLPA01 were stained with Nile Red and visualized by excitation with 543 nm light. Strains harboring pBBR1MCS-3 (C) or pLPA01 (D) were grown in NFb medium with 20 mM of ammonium chloride and 25 mM (w/v) D-glucose at 30°C (orbital agitation at 120 rpm). OD595 growth data were obtained from three independent cultures.
Discussion
To analyze the role of phasins in H. seropedicae, we correlated their presence with PHB production and expression of phaP1 and phaP2. Deletion of phaP1 reduced PHB accumulation on 50%, showing that PhaP1 is the key phasin controlling synthesis and stability of PHB granules. Deletion of phaP2 had little effect, reinforcing the idea that when PhaP1 is expressed, PHB granules are well formed and stocked (Figures 2 and 3). The suggestion that H. seropedicae has other phasin-like proteins seems unlikely, since deletion of both genes completely suppressed PHB granules formation. A similar dependence of phasins on accumulation of PHB was also found in S. meliloti Rm1021 (Wang et al., 2007), in which deletion of phaP1, the main phasin in this bacterium, reduced PHB accumulation by 20% and increased the doubling time by 2.3 h (Wang et al., 2007). Similarly, deletion of phaP2 did not significantly affect growth and PHB production of S. meliloti, however, deletion of both phasins increased the doubling time by 3.6 h and fully abrogated PHB accumulation (Wang et al., 2007). In the insect gut symbiont Burkholderia sp. RPE75, the phaP deletion reduced PHB accumulation by 2.8-fold and the distribution of PHB granules was heterogeneous among the cells of the ΔphaP mutant (Kim et al., 2013). A total of six phaP genes were identified within the genome of Burkholderia sp. RPE75, therefore it was suggested that the redundancy among phasins could maintain PHB production in the ΔphaP mutant and render it with less impact on bacterial-insect symbiosis (Kim et al., 2013). In fact, from our results it is likely that the expression of a backup phasin ensures some level of PHB accumulation and, hence, reduces the impact on bacterial fitness. Possibly, the same mechanism might be occurring in the ΔphaP strain of Burkholderia sp. RPE75. Recently, Hauf et al. (2015) reported the deletion of the ssl2501 expressing a phasin in Synechocystis sp. PCC 6803. The Δssl2501 mutant presented a reduction in the number of PHB granules per cell and an increase in the mean PHB granule size, however, the PHB content was only slightly lower in the mutant. Once again, as the deletion of ssl2501 alone has not abolished PHB accumulation, it was also suggested that possibly in Synechocystis, other phasin-like proteins might be expressed (Hauf et al., 2015).
These data contrast with the situation in the well-studied model R. eutropha H16, in which deletion of the four phasin genes did not completely impair PHB accumulation (Kuchta et al., 2007). Besides these four phasins, three other proteins have been recently reported in R. eutropha H16, namely PhaP5, PhaP6, and PhaP7 (Pfeiffer and Jendrossek, 2011, 2012). The deletion of these additional phasin genes alone did not affect PHB granules formation and the content accumulated in R. eutropha (Pfeiffer and Jendrossek, 2011, 2012). To the best of our knowledge, a mutant of R. eutropha with all phasin genes deleted (phaP1 to phaP7) has not been constructed so far. This mutant could reveal whether all phasins are relevant to PHB granule biogenesis and accumulation in R. eutropha and, if the functional redundancy among phasins (including PhaP5-7) supports PHB accumulation in the multiple deletion mutants, as related to ΔphaP1234 strain (Kuchta et al., 2007).
A well accepted model for transcriptional regulation of phaP genes in R. eutropha H16 and Paracoccus denitrificans (Maehara et al., 2001, 2002; Pötter et al., 2002; York et al., 2002; Yamada et al., 2007, 2013) suggested that the transcriptional repressor PhaR binds to the regulatory region upstream of phaP genes, blocking gene transcription. At the onset of PHB synthesis, the PhaR repressor is sequestered from DNA by PHB and transcription is initiated. PhaR thus couples PHB synthesis with phasin expression (York et al., 2002). In H. seropedicae SmR1, we have shown that PhaR (previously named PhbF) also functions as a repressor of transcription (Kadowaki et al., 2011) and here we have extended these observations to include repression of phaP1 and phaP2 expression (in E. coli; Table 3). Furthermore, the pattern of phaP1 and phaP2 expression in different backgrounds of H. seropedicae demonstrated a backup regulation, whereas the genes are dissimilarly expressed (Figure 4). In other words, the simultaneously expression of both phasins seems to be unnecessary, however, upon mutation of phaP1, expression of phaP2 is reprogrammed to achieve a similar level compared to expression of phaP1 in the wild type. This mode of regulation is named responsive backup circuit (RBC) and has important consequences controlling expression of functional redundant proteins to increase the robustness of organisms when facing stressful conditions (Kafri et al., 2006). Clearly, our results demonstrated that the expression of phaP2 in the ΔphaP1 reduced the negative impact on PHB accumulation and growth in glucose, rendering a more fit phenotype than the ΔphaP1.2 strain.
The existence of two highly homologous genes encoding proteins with the same predicted function raises the question of whether the phasins of H. seropedicae are genuinely redundant. It is unlikely that phaP1 and phaP2 represent truly redundant genes, as deletion of phaP1 clearly resulted in a less fit phenotype. Therefore, it is possible that due to its greater efficiency, PhaP1 has been selected as the main phasin and, as consequence, the expression of phaP2 was attenuated, converting PhaP2 in a backup phasin. This assumption raises two important points: (i) it guarantees proper levels of phasin expression, avoiding perturbations on PHB synthesis and granule formation and (ii) it saves the cells of wasting unnecessary metabolic costs with superfluous gene expression. Other β-Proteobacteria including species of the Herbaspirillum genus, Collimonas fungivorans, Herminiimonas arsenicoxydans and Janthinobacterium sp. possess orthologous to phaP1 and phaP2 of H. seropedicae, indicating that the backup expression of phaP2 may be conserved among species phylogenetically related to H. seropedicae. In the genome of Azoarcus sp. BH72, a plant-associated and PHB-producing β-proteobacterium, four genes expressing putative phasins were found (Krause et al., 2006), suggesting that the backup expression of phasins may be an important mechanism to maintain PHB production also in other species less phylogenetically close to Herbaspirillum. Interestingly, in other well characterized plant-associated and PHB-producing bacteria as A. brasilense Sp245 and A. lipoferum 4B, the genome sequencings revealed only one probable gene encoding a phasin for each organism, AZOBR_p110146 and AZOLI_1409, respectively (Wisniewski-Dye et al., 2011). On the other hand, in A. brasilense FP2 (a mutant strain from A. brasilense Sp7 resistant to nalidixic acid and streptomycin) were found four phasin genes, which are expressed when the bacteria were epiphytically colonizing roots of Triticum aestivum (Camilios-Neto et al., 2014). To the best of our knowledge mutansts defective in phasin expression were not constructed to Azospirillum species so far. Therefore, these findings pave the way to investigate the impact of phasin gene deletions in other relevant plant-associated bacteria, as those from Azoarcus and Azospirillum genus.
Author Contributions
LA and CT designed and performed the most part of experiments, analyzed the data and wrote the manuscript. ET constructed mutants and some plasmids used in this work. LD performed the TEM analysis. MT-S prepared cDNA and performed the RT-PCR. MS, ES, and FO conceived and supervised the study. LC and MM-S conceived and supervised the study, analyzed the data and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very grateful to INCT of Biological Nitrogen Fixation/CNPq for financial support and to Valter de Baura, Roseli Prado, Marilza Lamour, and Alexsandro Albani for technical support. LA, CT, and ET are very grateful to CNPq for scholarships received during the performing of the experiments.
Footnotes
Funding. This research was funded by CNPq (Brazilian National Council for Scientific and Technological Development), project “Investigação do Papel das Proteínas Fasinas na Biossíntese de Polihidroxialcanoatos e na Resposta Anti-Estresse em Bactérias,” process 458417/2014-9.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00739
References
- Anderson A. J., Dawes E. A. (1990). Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54 450–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babel W., Ackermann J. U., Breuer U. (2001). Physiology, regulation, and limits of the synthesis of poly(3HB). Adv. Biochem. Eng. Biotechnol. 71 125–157. [DOI] [PubMed] [Google Scholar]
- Baldani J. I., Baldani V. L. D., Seldin L., Dobereiner J. (1986). Characterization of Herbaspirillum seropedicae gen-nov, sp-nov, a root-associated nitrogen-fixing bacterium. Int. J. Syst. Bacteriol. 36 86–93. 10.1099/00207713-36-1-86 [DOI] [Google Scholar]
- Bratlie M. S., Johansen J., Sherman B. T., Huang da W., Lempicki R. A., Drablos F. (2010). Gene duplications in prokaryotes can be associated with environmental adaptation. BMC Genomics 11:588 10.1186/1471-2164-11-588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunegg G., Sonnleitner B., Lafferty R. M. (1978). Rapid gas-chromatographic method for determination of poly-beta-hydroxybutyric acid in microbial biomass. Eur. J. Appl. Microbiol. Biotechnol. 6 29–37. 10.1007/Bf00500854 [DOI] [Google Scholar]
- Cai S., Cai L., Liu H., Liu X., Han J., Zhou J., et al. (2012). Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl. Environ. Microbiol. 78 1946–1952. 10.1128/AEM.07114-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilios-Neto D., Bonato P., Wassem R., Tadra-Sfeir M. Z., Brusamarello-Santos L. C., Valdameri G., et al. (2014). Dual RNA-seq transcriptional analysis of wheat roots colonized by Azospirillum brasilense reveals up-regulation of nutrient acquisition and cell cycle genes. BMC Genomics 15:378 10.1186/1471-2164-15-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Catalan A. I., Ferreira F., Gill P. R., Batista S. (2007). Production of polyhydroxyalkanoates by Herbaspirillum seropedicae grown with different sole carbon sources and on lactose when engineered to express the lacZlacY genes. Enzyme Microbial Technol. 40 1352–1357. 10.1016/j.enzmictec.2006.10.008 [DOI] [Google Scholar]
- Chen G. Q. (2009). A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem. Soc. Rev. 38 2434–2446. 10.1039/b812677c [DOI] [PubMed] [Google Scholar]
- Cho M., Brigham C. J., Sinskey A. J., Stubbe J. (2012). Purification of polyhydroxybutyrate synthase from its native organism, Ralstonia eutropha: implications for the initiation and elongation of polymer formation in vivo. Biochemistry 51 2276–2288. 10.1021/bi2013596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B., Halloran E., Holmes S. (1996). Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. U.S.A. 93 13429–13434. 10.1073/pnas.93.14.7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D., Vandepoele K., Simillon C., Van de Peer Y. (2004). Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends Microbiol. 12 148–154. 10.1016/j.tim.2004.02.007 [DOI] [PubMed] [Google Scholar]
- Grage K., Jahns A. C., Parlane N., Palanisamy R., Rasiah I. A., Atwood J. A., et al. (2009). Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules 10 660–669. 10.1021/bm801394s [DOI] [PubMed] [Google Scholar]
- Hauf W., Watzer B., Roos N., Klotz A., Forchhammer K. (2015). Photoautotrophic polyhydroxybutyrate granule formation is regulated by cyanobacterial phasin PhaP in Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 81 4411–4422. 10.1128/AEM.00604-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas A. B., Canosa I., Santero E. (2008). Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J. Bacteriol. 190 416–420. 10.1128/JB.01230-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D. (2009). Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J. Bacteriol. 191 3195–3202. 10.1128/JB.01723-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurasek L., Marchessault R. H. (2002). The role of phasins in the morphogenesis of poly(3-hydroxybutyrate) granules. Biomacromolecules 3 256–261. 10.1021/bm010145d [DOI] [PubMed] [Google Scholar]
- Jurasek L., Marchessault R. H. (2004). Polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha cells: a computer simulation. Appl. Microbiol. Biotechnol. 64 611–617. 10.1007/s00253-003-1551-9 [DOI] [PubMed] [Google Scholar]
- Kadouri D., Jurkevitch E., Okon Y. (2003). Involvement of the reserve material poly-beta-hydroxybutyrate in Azospirillum brasilense stress endurance and root colonization. Appl. Environ. Microbiol. 69 3244–3250. 10.1128/AEM.69.6.3244-3250.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki M. A., Muller-Santos M., Rego F. G., Souza E. M., Yates M. G., Monteiro R. A., et al. (2011). Identification and characterization of PhbF: a DNA binding protein with regulatory role in the PHB metabolism of Herbaspirillum seropedicae SmR1. BMC Microbiol. 11:230 10.1186/1471-2180-11-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R., Levy M., Pilpel Y. (2006). The regulatory utilization of genetic redundancy through responsive backup circuits. Proc. Natl. Acad. Sci. U.S.A. 103 11653–11658. 10.1073/pnas.0604883103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky M. J. (1965). A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27 A137. [Google Scholar]
- Kim J. K., Won Y. J., Nikoh N., Nakayama H., Han S. H., Kikuchi Y., et al. (2013). Polyester synthesis genes associated with stress resistance are involved in an insect-bacterium symbiosis. Proc. Natl. Acad. Sci. U.S.A. 110 E2381–E2389. 10.1073/pnas.1303228110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner D., Paul W., Merrick M. J. (1988). Construction of multicopy expression vectors for regulated over-production of proteins in Klebsiella pneumoniae and other enteric bacteria. J. Gen. Microbiol. 134 1779–1784. [DOI] [PubMed] [Google Scholar]
- Kondrashov F. A. (2012). Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 279 5048–5057. 10.1098/rspb.2012.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., II, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166 175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- Krause A., Ramakumar A., Bartels D., Battistoni F., Bekel T., Boch J., et al. (2006). Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 24 1385–1391. 10.1038/nbt1243 [DOI] [PubMed] [Google Scholar]
- Kuchta K., Chi L., Fuchs H., Potter M., Steinbuchel A. (2007). Studies on the influence of phasins on accumulation and degradation of PHB and nanostructure of PHB granules in ralstonia eutropha H16. Biomacromolecules 8 657–662. 10.1021/bm060912e [DOI] [PubMed] [Google Scholar]
- Luft J. H. (1961). Improvements in epoxy resin embedding methods. J. Biophys. Biochem. Cytol. 9 409–414. 10.1083/jcb.9.2.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil T., MacNeil D., Tyler B. (1982). Fine-structure deletion map and complementation analysis of the glnA-glnL-glnG region in Escherichia coli. J. Bacteriol. 150 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison L. L., Huisman G. W. (1999). Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63 21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara A., Doi Y., Nishiyama T., Takagi Y., Ueda S., Nakano H., et al. (2001). PhaR, a protein of unknown function conserved among short-chain-length polyhydroxyalkanoic acids producing bacteria, is a DNA-binding protein and represses Paracoccus denitrificans phaP expression in vitro. FEMS Microbiol. Lett. 200 9–15. 10.1111/j.1574-6968.2001.tb10685.x [DOI] [PubMed] [Google Scholar]
- Maehara A., Taguchi S., Nishiyama T., Yamane T., Doi Y. (2002). A repressor protein, PhaR, regulates polyhydroxyalkanoate (PHA) synthesis via its direct interaction with PHA. J. Bacteriol. 184 3992–4002. 10.1128/JB.184.14.3992-4002.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerk M., Johansen J., Ertesvag H., Drablos F., Valla S. (2014). Safety in numbers: multiple occurrences of highly similar homologs among Azotobacter vinelandii carbohydrate metabolism proteins probably confer adaptive benefits. BMC Genomics 15:192 10.1186/1471-2164-15-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon K., Michel-Reydellet N., Encarnacion S., Kaminski P. A., Leija A., Cevallos M. A., et al. (1998). Poly-beta-hydroxybutyrate turnover in Azorhizobium caulinodans is required for growth and affects nifA expression. J. Bacteriol. 180 5070–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. (1986). Single-stranded DNA ‘blue’ T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1 67–74. 10.1093/protein/1.1.67 [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Neumann L., Spinozzi F., Sinibaldi R., Rustichelli F., Potter M., Steinbuchel A. (2008). Binding of the major phasin, PhaP1 from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J. Bacteriol. 190 2911–2919. 10.1128/JB.01486-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norholm M. H. (2010). A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 10:21 10.1186/1472-6750-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa F. O., Monteiro R. A., Wassem R., Cruz L. M., Ayub R. A., Colauto N. B., et al. (2011). Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet. 7:e1002064 10.1371/journal.pgen.1002064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa F. O., Yates M. G. (1984). Regulation of nitrogen-fixation (Nif) genes of Azospirillum brasilense by Nifa and Ntr (Gln) type gene-products. FEMS Microbiol. Lett. 23 95–101. 10.1111/j.1574-6968.1984.tb01042.x [DOI] [Google Scholar]
- Pfeiffer D., Jendrossek D. (2011). Interaction between poly(3-hydroxybutyrate) granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology 157 2795–2807. 10.1099/mic.0.051508-0 [DOI] [PubMed] [Google Scholar]
- Pfeiffer D., Jendrossek D. (2012). Localization of poly(3-hydroxybutyrate) (PHB) granule-associated proteins during PHB granule formation and identification of two new phasins, PhaP6 and PhaP7, in Ralstonia eutropha H16. J. Bacteriol. 194 5909–5921. 10.1128/JB.00779-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pötter M., Madkour M. H., Mayer F., Steinbuchel A. (2002). Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148 2413–2426. 10.1099/00221287-148-8-2413 [DOI] [PubMed] [Google Scholar]
- Potter M., Muller H., Reinecke F., Wieczorek R., Fricke F., Bowien B., et al. (2004). The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150 2301–2311. 10.1099/mic.0.26970-0 [DOI] [PubMed] [Google Scholar]
- Potter M., Muller H., Steinbuchel A. (2005). Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology 151 825–833. 10.1099/mic.0.27613-0 [DOI] [PubMed] [Google Scholar]
- Quelas J. I., Mongiardini E. J., Perez-Gimenez J., Parisi G., Lodeiro A. R. (2013). Analysis of two polyhydroxyalkanoate synthases in Bradyrhizobium japonicum USDA 110. J. Bacteriol. 195 3145–3155. 10.1128/JB.02203-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2015). R: A Language and Environment for Statistical Computing. Vienna: The R Foundation for Statistical Computing. [Google Scholar]
- Reynolds E. S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17 208–212. 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42 W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schafer A., Tauch A., Jager W., Kalinowski J., Thierbach G., Puhler A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145 69–73. 10.1016/0378-1119(94)90324-7 [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer U., Puhler A. (1983). A broad host range mobilization system for invivo genetic-engineering - transposon mutagenesis in gram-negative bacteria. Bio-Technology 1 784–791. 10.1038/Nbt1183-784 [DOI] [Google Scholar]
- Souza E. M., Pedrosa F. O., Rigo L. U., Machado H. B., Yates M. G. (2000). Expression of the nifA gene of Herbaspirillum seropedicae: role of the NtrC and NifA binding sites and of the -24/-12 promoter element. Microbiology 146 1407–1418. 10.1099/00221287-146-6-1407 [DOI] [PubMed] [Google Scholar]
- Spaink H. P., Okker R. J. H., Wijffelman C. A., Pees E., Lugtenberg B. J. J. (1987). Promoters in the nodulation region of the Rhizobium leguminosarum sym plasmid Prl1ji. Plant Mol. Biol. 9 27–39. 10.1007/BF00017984 [DOI] [PubMed] [Google Scholar]
- Spiekermann P., Rehm B. H., Kalscheuer R., Baumeister D., Steinbuchel A. (1999). A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171 73–80. 10.1007/s002030050681 [DOI] [PubMed] [Google Scholar]
- Steinbuchel A., Hein S. (2001). Biochemical and molecular basis of microbial synthesis of polyhydroxyalkanoates in microorganisms. Adv. Biochem. Eng. Biotechnol. 71 81–123. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirapelle E. F., Muller-Santos M., Tadra-Sfeir M. Z., Kadowaki M. A., Steffens M. B., Monteiro R. A., et al. (2013). Identification of proteins associated with polyhydroxybutyrate granules from Herbaspirillum seropedicae SmR1 - old partners, new players. PLoS ONE 8:e75066 10.1371/journal.pone.0075066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainer M. A., Charles T. C. (2006). The role of PHB metabolism in the symbiosis of rhizobia with legumes. Appl. Microbiol. Biotechnol. 71 377–386. 10.1007/s00253-006-0354-1 [DOI] [PubMed] [Google Scholar]
- Urtuvia V., Villegas P., González M., Seeger M. (2014). Bacterial production of the biodegradable plastics polyhydroxyalkanoates. Int. J. Biol. Macromol. 70 208–213. 10.1016/j.ijbiomac.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Wang C., Sheng X., Equi R. C., Trainer M. A., Charles T. C., Sobral B. W. (2007). Influence of the poly-3-hydroxybutyrate (PHB) granule-associated proteins (PhaP1 and PhaP2) on PHB accumulation and symbiotic nitrogen fixation in Sinorhizobium meliloti Rm1021. J. Bacteriol. 189 9050–9056. 10.1128/JB.01190-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. L. (1958). Staining of tissue sections for electron microscopy with heavy metals. J. Biophys. Biochem. Cytol. 4 475–478. 10.1083/jcb.4.4.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek R., Pries A., Steinbuchel A., Mayer F. (1995). Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177 2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski-Dye F., Borziak K., Khalsa-Moyers G., Alexandre G., Sukharnikov L. O., Wuichet K., et al. (2011). Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments. PLoS Genet. 7:e1002430 10.1371/journal.pgen.1002430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Takahashi S., Okahata Y., Doi Y., Numata K. (2013). Monitoring and kinetic analysis of the molecular interactions by which a repressor protein, PhaR, binds to target DNAs and poly[(R)-3-hydroxybutyrate]. AMB Express 3 6 10.1186/2191-0855-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Yamashita K., Wakuda A., Ichimura K., Maehara A., Maeda M., et al. (2007). Autoregulator protein PhaR for biosynthesis of polyhydroxybutyrate [P(3HB)] possibly has two separate domains that bind to the target DNA and P(3HB): functional mapping of amino acid residues responsible for DNA binding. J. Bacteriol. 189 1118–1127. 10.1128/JB.01550-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Yang J., Dou Y., Chen M., Ping S., Peng J., et al. (2008). Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc. Natl. Acad. Sci. U.S.A. 105 7564–7569. 10.1073/pnas.0801093105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York G. M., Junker B. H., Stubbe J. A., Sinskey A. J. (2001a). Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 183 4217–4226. 10.1128/JB.183.14.4217-4226.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York G. M., Stubbe J., Sinskey A. J. (2001b). New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J. Bacteriol. 183 2394–2397. 10.1128/JB.183.7.2394-2397.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York G. M., Stubbe J., Sinskey A. J. (2002). The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184 59–66. 10.1128/JB.184.1.59-66.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


