Abstract
Introduction
Infectious diarrhoea particularly due to pathogenic bacteria is a major health problem in developing countries, including India. Despite significant reports of diarrhoeagenic Escherichia coli (DEC) pathotypes around the globe, studies which address genetic relatedness, antibiogram profile and their correlation with respect to their isolation from different sources are sparse. The present study determines isolation and identification of DEC pathotypes from different sources, their genetic characterisation, antibiogram profile and their correlation if any.
Materials and methods
A total of 336 samples comprising diarrhoeic stool samples from infants (n=103), young animal (n=106), foods (n=68) and associated environmental sources (n=59) were collected from Bareilly region of India. All the samples were screened by using standard microbiological methods for the detection of E. coli. The identified E. coli were then confirmed as DEC pathotypes using polymerase chain reaction–based assays. Those DEC pathotypes identified as Enteroaggregative E. coli (EAEC) were further confirmed using HEp-2 adherence assay. All the isolated DEC pathotypes were studied for their genetic diversity using pulsed-field gel electrophoresis (PFGE), and antimicrobial susceptibility testing was performed by using disc diffusion method as per Clinical Laboratory Standards Institute guidelines.
Results and discussion
Of the four DEC pathotypes investigated, EAEC was found to be the predominant pathogen with an isolation rate of 16.5% from infants, 17.9% from young animals, 16.2% from foods and 3.4% from the associated environmental sources. These EAEC isolates, on further characterisation, revealed predominance of ‘atypical’ EAEC, with an isolation rate of 10.7% from infants, 15.1% from young animals, 16.2% from foods, and 3.4% from the associated environmental sources. On PFGE analysis, discrimination was evident within DEC pathotypes as 52 unique pulsotypes were observed for 59 recovered DEC pathotypes. However, a few EAEC isolates were found to be clonal (clusters A, B, C, D, F, G, and H) irrespective of their source of isolation, suggests sharing and/or circulation among different sources. Further, a high antibiotic resistance pattern was observed among isolated DEC pathotypes as almost 86.4% of isolates were found to be resistant against ≥3 tested drugs.
Keywords: antibiotic resistance, ‘Atypical' enteroaggregative Escherichia coli, diarrhoeagenic Escherichia coli, genetic diversity, pulsed-field gel electrophoresis
Infectious diarrhoea remains a major global public health hurdle particularly in developing nations. Children under 5 years of age continue to be the victims of diarrhoea. Diarrhoea shares about 10.5% of the total under-5 mortality, which in turn has remained as one of the main blockades for achieving the millennium development goal(s) (1).
Worldwide, there are many reports on the isolation and identification of diarrhoeagenic Escherichia coli (DEC) pathotypes viz, enteroaggregative Escherichia coli (EAEC), shiga-toxin producing E. coli (STEC), enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC) and diffusely adherent E. coli (DAEC) from diarrhoeal cases of infants as well as young animals. Among the DEC pathotypes, EAEC along with the well-established ETEC and EPEC pathotypes causes a substantial health burden of infant diarrhoeal cases and they have also been isolated from a variety of animal species (2–4). Mostly, DEC outbreaks are often found to be associated with direct contact with infected animals or indirectly through consumption of vegetables, fruits and water contaminated with infected animal faeces (5).
In 2010, India accounted for 0.212 million deaths due to infant diarrhoea which constituted a huge share (12.6%) of global under-5 mortality burden (1). DEC pathotypes are widely distributed among humans, animals, foods, and environmental sources in different geographical regions of India (6). Despite significant reports of DEC pathotypes from across the globe including India, there is paucity of studies revealing the relatedness and/or diversity of the isolates recovered from various sources. Besides this, the problem of antimicrobial resistance in pathogenic and commensal bacteria is aggravated by the adoption of mobile genetic elements conferring resistance from their environmental counterparts (7). Moreover, in concordance with global reports, multiple antibiotic-resistant E. coli isolates were also reported from different parts of Indian subcontinent (8). The objectives of the present study were to isolate and identify DEC pathotypes from diarrhoeal cases of human infants, young animals, foods and associated environmental sources, with an aim to study their correlation with regard to genetic characterisation using pulsed-field gel electrophoresis (PFGE) and their antibiotic sensitivity profiling.
Methods
Sample collection and area of study
A total of 103 stool samples from children of age groups ≤5 years were randomly collected from a district government hospital and five other private paediatric hospitals in the Bareilly region; situated in the northern part of Uttar Pradesh, India. Most of the stool samples were collected within 24 h of onset of diarrhoea and also before the start of any antimicrobial treatment. The institutional review board of the Hospitals approved the study and consent was obtained from the parents wherever possible. Samples were collected only from those infants who fulfilled the World Health Organization criteria of acute diarrhoeal disease (9). Through the detailed case history reports of the hospitals and personal interactions with the patients, we found that a majority of the clinical cases were from semi-urban settings and were mostly associated with farming or companion–animal interface either through animal husbandry practices or by having pets in their home.
Diarrheal faecal samples (106) from young domestic animals (0–6 months of age) comprising canine (n=68), bovine (n=29) and caprine (n=9) were collected from private farms and veterinary polyclinics of Bareilly region. Subsequently, a total of 68 food samples (infant food formulae, vegetables, fish, milk, meat and their products) and 59 environmental samples (animal feed, drinking water, sewage waste and soil) adjacent to the animal and human dwellings were also collected from Bareilly region. The details of the samples are presented in Supplementary Table 1 and the areas of sampling are presented in Supplementary Fig. 1.
The diarrhoeic stool samples were collected aseptically using Cary-Blair transport medium swabs (HiMedia, Mumbai, India), whereas foods (infant food formulae, vegetables, fish, milk, meat and their products), animal feed, drinking water, sewage waste and surrounding soil of animal farms were collected in suitable aseptic containers. All the samples were transported to a laboratory under chilled conditions and were processed within 24 h of collection.
Reference strains
The DNA of DEC pathotypes (EAEC, ETEC, EPEC, and EHEC) was kindly provided by Dr. Chobi Debroy, National E. coli Referral Centre, Pennsylvania State University, USA.
Isolation of diarrhoeagenic E. coli
The diarrhoeal stool/faecal samples from human infants and young animals including foods and associated environmental samples were screened for E. coli using standard microbiological and molecular methods (10). The faeces/stool, foods (infant food formulae, vegetables, fish, meat and their products), animal feed, soil samples (approximately 5 g), and liquid samples such as milk, drinking water, and sewage waste (approx. 5 ml) were accordingly inoculated in nine parts of the E. coli enrichment (EE) broth, followed by incubation at 37°C for 18–24 h. A loopful of enriched inoculum was then plated onto eosin methylene blue (EMB) agar plates. Inoculated plates were then incubated at 37°C for 24 h. Colonies revealing the characteristic metallic sheen colonies on EMB agar (3–5 colonies for each sample) were subjected to biochemical tests for the identification of E. coli (11). Colonies exhibiting indole, methyl-red, and catalase tests as positive, Voges–Proskauer and citrate tests as negative, and giving fermentation of glucose and lactose sugars, were confirmed as E. coli isolates.
Identification of diarrhoeagenic E. coli pathotypes
All the confirmed E. coli isolates recovered from different sources were subjected to standardized polymerase chain reaction (PCR) assays for the detection of DEC pathotypes. The primers used are listed in Supplementary Table 2. The oligonucleotides used in present study were synthesised from Eurofins Genomics (Bengaluru, India). The eae gene was targeted for the identification of EPEC and/or EHEC. Further, bfp gene was targeted for the detection of EPEC pathotype and stx1 and/or stx2 genes for EHEC pathotype. The ETEC were detected by targeting LT and/or ST genes. The detection and characterisation of ‘typical’ and ‘atypical’ EAEC isolates were attempted by targeting plasmid borne (aggR, aatAA/cvd432) and chromosomal (aaiA, astA, irp2, pilS, pic, ecp) genes (12–17). E. coli DH5α DNA was taken as negative control. The optimized PCR protocol for 25 µL reaction mixture included 2.5 µL of 10× PCR buffer (100 mM Tris–HCl buffer, pH 8.3 containing 500 mM KCl, 15 mM MgCl2, and 0.01% gelatin), 400 µM of 10 mM deoxyribonucleotide phosphates (dNTPs), 2.0 µM of MgCl2, 0.4 µM of each primer set, 0.3 units of Taq DNA polymerase (3B Black Bio, Spain), and 3 µL of DNA template and sterilized MilliQ water to make up the reaction volume. The details of cycling conditions for targeted genes are listed in Supplementary Table 3. PCR was performed in the Eppendorf pro thermal cycler (Eppendorf, Germany).
The PCR products were separated by gel electrophoresis in 1.5% agarose (Sigma, USA) containing ethidium bromide in Tris–acetate–EDTA (TAE) buffer and visualized by UV transilluminator and digitally recorded by gel documentation system (UVP GelSeq Software). Materials contaminated with ethidium bromide were disposed as per the local guidelines.
Confirmation of EAEC pathotype using HEp-2 cell adherence assay
‘Typical’ and ‘atypical’ EAEC identified by PCR were also assayed by HEp-2 adherence assay for further confirmation (17). Thus, the isolates revealing ‘stacked brick appearance’ in HEp-2 cell culture assay were considered as confirmed EAEC pathotype (Supplementary Fig. 2).
Pulsed-field gel electrophoresis
PFGE was performed according to CDC PulseNet protocol (18). Briefly, agarose-embedded DNA was digested with 50 U of XbaI for 3 h in a water bath at 37°C. DNA fragments were separated by electrophoresis in 0.5× Tris–borate–EDTA buffer at 14°C for 18 h on a CHEF-II Mapper system (Bio-Rad Laboratories, USA), with a pulse time of 2.2–54.2s at a constant voltage of 6 V/cm. The gel was stained with ethidium bromide and visualized with the Alpha Innotech (AlphaImager HP). The dendrogram was prepared by using the default values of the Phoretix 1D pro software (v12.2) which gives Rf vector with a value 0.05 with minimum Rf 0 and maximum Rf 1. The similarity index of the isolates was calculated using the Dice correlation coefficient with a band position tolerance of 1% and an optimization of 1%. The unweighted-pair group method using average linkages (UPGMA) was used to construct a restriction profile dendrogram. The E. coli ATCC 25922 strain was used as standard-type culture on every gel run to compare both the images during dendrogram preparation. A standard molecular weight ladder (The Pulse Marker™ 50–1,000 kb; Sigma, cat. no. D2416) was used for comparison of the fingerprints over gels. In addition, reproducibility of the PFGE patterns was also verified by running same isolates twice.
Antimicrobial sensitivity test
Antimicrobial susceptibility testing was performed by using disc diffusion method as per the guidelines provided by Clinical Laboratory Standards Institute (CLSI) (19). The antimicrobials included in the present study were selected based on the information gathered from local veterinarians, paediatric clinicians, and available literature. The antimicrobials included were ampicillin (Amp, 10 µg), co-trimoxazole (Cot, 25 µg), cefotaxime (Ctx, 30 µg), imipenem (10 µg), ceftriaxone (Ctr, 30 µg), ciprofloxacin (Cip, 05 µg), and tetracycline (TE, 30 µg) (HiMedia Laboratories Ltd, Mumbai, India). E. coli ATCC 25922 was used as a quality control strain and the interpretations were carried out as per the CLSI standards (19).
Statistical analysis
The antimicrobial resistance data of the recovered isolates from human, animal, food, and environmental sources were statistically analysed by employing chi-squared test using SPSS software, 22.0 version.
Results
Isolation and identification of E. coli by cultural and biochemical methods
A total of 336 samples collected from various sources were screened for the DEC pathotypes. On microbiological and biochemical analyses, a total of 61 isolates from human diarrhoeal cases (n=103), 59 isolates from animal diarrhoeal cases (n=106) and 56 isolates from food and associated environmental sources (n=127) were confirmed as E. coli isolates.
Identification of DEC pathotypes
The results of the isolated and identified DEC pathotypes from various sources along with the respective amplification of target genes are presented in Table 1.
Table 1.
Details of the DEC pathotypes recovered from different sources along with their target gene amplification.
| Isolate ID | Source | Pathotype(s) | Virulence gene(s) |
|---|---|---|---|
| Human infant isolates | |||
| H005 | Human infant | ETEC | LT |
| H008 | Human infant | EAEC | irp2, ecp, irp2 |
| H010 | Human infant | EAEC | ecp, aaiA |
| H058 | Human infant | EPEC | eae, bfp |
| H115 | Human infant | EAEC | aaiA, irp2 |
| H121 | Human infant | EAEC | aggR, aatAA/cvd, ecp |
| H123 | Human infant | EAEC | aggR, aatAA/cvd, aaiA, irp2, ecp |
| H124 | Human infant | EAEC | aggR, aatAA/cvd, aaiA, ecp |
| H138 | Human infant | EAEC | aggR, aatAA/cvd, pilS, irp2, ecp |
| H140 | Human infant | EAEC | aggR, aatAA/cvd, irp2 |
| H197 | Human infant | EAEC | astA, ecp, irp2 |
| H198 | Human infant | EAEC | astA, ecp, irp2 |
| H313 | Human infant | EAEC | astA, irp2, ecp |
| H314 | Human infant | EAEC | aggR, aatAA/cvd, irp2, ecp |
| H316 | Human infant | EAEC | aaiA, astA, irp2, ecp |
| H319 | Human infant | EAEC | ecp, irp2, astA |
| H322 | Human infant | EAEC | irp2, ecp |
| H324 | Human infant | EAEC | aaiA, irp2 |
| H325 | Human infant | EAEC | irp2 |
| Young animal isolates | |||
| A099 | Canine | EHEC | eae, stx1 |
| A103 | Canine | EHEC | eae, stx1 |
| A105 | Canine | EHEC | eae, stx2 |
| A109 | Canine | EAEC | aggR, aatAA/cvd, aaiA, astA, ecp |
| A187 | Canine | EAEC | astA, irp2, ecp |
| A206 | Canine | EAEC | irp2 |
| A207 | Canine | EAEC | aaiA, irp2 |
| A209 | Canine | EPEC | eae, bfp |
| A210 | Canine | EAEC | aggR, aatAA/cvd, aaiA, astA, ecp |
| A216 | Canine | EAEC | irp2, ecp |
| A219 | Canine | EAEC | irp2, ecp |
| A258 | Canine | EAEC | ecp, irp2 |
| A262 | Canine | EAEC | ecp, aaiA |
| A265 | Canine | EAEC | ecp, aaiA |
| A266 | Canine | EAEC | astA, ecp |
| A278 | Canine | EPEC | eae, bfp |
| A279 | Canine | EAEC | aggR, aatAA/cvd, aaiA, astA, irp2, pic |
| A082 | Bovine | EAEC | astA, ecp |
| A084 | Bovine | EAEC | irp2, ecp |
| A183 | Bovine | EAEC | ecp, irp2 |
| A189 | Bovine | EAEC | astA, irp2, ecp |
| A185 | Bovine | EAEC | aaiA, irp2 |
| A217 | Bovine | EAEC | astA, ecp |
| A223 | Bovine | EAEC | astA, irp2, ecp |
| A101 | Caprine | EHEC | eae, stx1 |
| Foods and environmental isolates | |||
| E015 | Poultry meat | EAEC | aaiA, irp2 |
| E017 | Fish | EAEC | ecp, irp2 |
| E019 | Fish | EAEC | aaiA, irp2 |
| E022 | Mutton | EAEC | PilS |
| E023 | Chevon | EAEC | ecp, aaiA |
| E065 | Milk | EAEC | aaiA, irp2 |
| E066 | Milk | EAEC | aaiA, irp2 |
| E087 | Milk handler | EAEC | aaiA, irp2, ecp |
| E089 | Milk handler | EAEC | aaiA, astA, irp2 |
| E029 | Vegetable | EAEC | irp2 |
| E166 | Vegetable | EAEC | ecp, irp2 |
| E074 | Animal feed | EPEC | eae, bfp |
| E075 | Animal feed | EAEC | AaiA |
| E280 | Sewage | EAEC | aaiA, irp2 |
| E283 | Sewage | EPEC | eae, bfp |
From human infants, EAEC was found to be the predominant pathotype with an overall isolation rate of 16.5%, of which ‘atypical’ EAEC were 10.7% and ‘typical’ EAEC were 5.8% (Table 1). The other DEC pathotypes isolated and identified were 1.0% each of EPEC and ETEC (Table 1). Also in animals, EAEC was found to be the predominant pathotype. In bovine and canines, an overall isolation rate of 24.1 and 13.2% was observed for ‘atypical’ EAEC, whereas ‘typical’ EAEC were isolated only from canines with an isolation rate of 4.4% (Table 1). The other isolated and identified DEC pathotypes from animals were EHEC (3.8%) and EPEC (1.9%) (Table 1). In foods and associated environmental sources, ‘atypical’ EAEC pathotype revealed the predominance, with an isolation rate of 10.2%. Besides this, two environmental associated source samples, animal feed, and sewage waste yielded EPEC pathotype (Table 1).
PFGE analysis
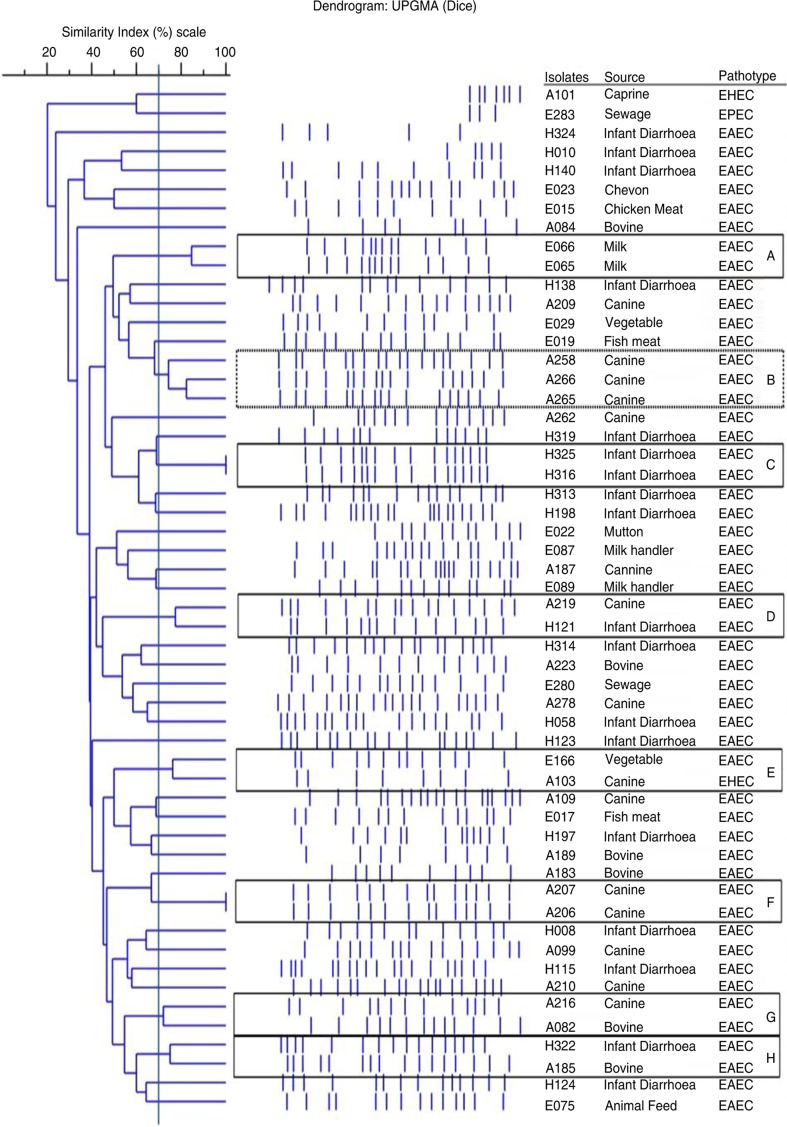
PFGE patterns revealed diverse genetic profiles among and between the EAEC, EPEC, and EHEC pathotype isolates recovered from humans, animals, foods, and associated environmental sources (Fig. 1). Of the 59 DEC pathotypes analysed, 52 unique pulsotypes for 54 DEC pathotypes were observed, whereas five DEC pathotypes were untypeable. At a genetic similarity of ≥70%, eight clusters were formed, designated as A–H with two to three isolates per cluster. Of these eight clusters, seven clusters were represented by various EAEC isolates. EAEC isolates of cluster C (H316 and H325) and cluster F (A206 and A207) were exhibiting 100% identical PFGE profiles, and the isolates of both clusters were recovered from similar sources. Similarly, at 70% genetic similarity, EAEC isolates from cluster A (E065 and E066) and cluster B (A258, A265, and A266) were also isolated from similar sources. On the contrary, EAEC isolates from cluster D (A219 and H121), cluster G (A216 and A082), and cluster H (H322 and A185) were exhibiting 70% genetic similarity in their respective cluster although their sources of isolation were different (Fig. 1). Interestingly, EAEC and EHEC isolates of cluster E (E166 and A103) were also exhibiting 70% genetic similarity (Fig. 1).
Fig. 1.
PFGE profile of DEC pathotypes isolated from different sources. The genotypic patterns generated by PFGE were analysed using the Phoretix 1D pro software (Total Lab, UK). The clustering was performed by UPGMA and the dice correlation coefficient.
Antibiogram profile
The present study results of antibiotic sensitivity profile of DEC pathotypes recovered from various sources are presented in Table 2. Overall, the recovered DEC pathotypes revealed a high degree of antimicrobial resistance against ampicillin (91.5%), tetracycline (88.1%), cefotaxime (86.4%), co-trimoxazole (74.6%), ceftriaxone (69.5%), and ciprofloxacin (45.8%). However, all the DEC pathotypes isolates were found sensitive for imipenem.
Table 2.
The antibiogram profile of the DEC pathotypes
| DEC pathotypes percentage (%) for tested antibiotics | ||||||||
|---|---|---|---|---|---|---|---|---|
| Target groups | Antibiotic resistance phenotype | AMP | CIP | COT | CTR | IPM | TE | CTX |
| Human cases (n=19) | R | 94.7% (18/19) | 68.4% (13/19) | 73.7% (14/19) | 78.9% (15/19) | 0 | 84.2% (16/19) | 94.7% (18/19) |
| S | 5.3% (1/19) | 31.6% (6/19) | 26.3% (5/19) | 21.0% (4/19) | 100% (19/19) | 15.8% (3/19) | 5.3% (1/19) | |
| Animal cases (n=25) | R | 96.0% (24/25) | 52.0% (13/25) | 80.0% (20/25) | 64.0% (16/25) | 0 | 88.0% (22/25) | 84.0% (21/25) |
| S | 4.0% (1/25) | 48.0% (12/25) | 20.0% (5/25) | 36.0% (9/25) | 100% (25/25) | 12.0% (3/25) | 16.0% (4/25) | |
| Food and environmental | R | 80.0% (12/15) | 6.7% (1/15) | 66.7% (10/15) | 66.7% (10/15) | 0 | 93.3% (14/15) | 80.0% (12/15) |
| sources (n=15) | S | 20.0% (3/25) | 93.3% (14/15) | 33.3% (5/15) | 33.3% (5/15) | 100% (15/15) | 6.7% (1/15) | 20.0% (3/15) |
| Overall pattern (n=59) | R | 91.5% (54/59) | 45.8% (27/59) | 74.6% (44/59) | 69.5% (41/59) | 0 | 88.1% (52/59) | 86.4% (51/59) |
| S | 8.5% (5/59) | 54.2% (32/59) | 25.4% (15/59) | 30.5% (18/59) | 100% (59/59) | 11.9% (7/59) | 13.6% (8/59) | |
R, resistant; S, sensitive; AMP, ampicillin; CIP, ciprofloxacin; COT, co-trimoxazole; CTR, ceftriaxone; IPM, imipenem; TE, tetracycline; CTX, cefotaxim.
DEC isolates from human infants (n=19) showed the highest resistance against ampicillin and cefotaxime each (94.7%), followed by tetracycline (84.2%), ceftriaxone (78.9%), co-trimoxazole (73.7%), and ciprofloxacin (68.4%). Also, the DEC isolates from animals (n=25) exhibited a high resistance pattern, wherein the highest resistance was observed against ampicillin (96.0%), followed by tetracycline (88.0%), cefotaxime (84.0%), co-trimoxazole (80.0%), ceftriaxone (64.0%), and ciprofloxacin (52.0%). Whereas DEC isolates recovered from foods and associated environmental sources (n=15) revealed highest resistance against tetracycline (93.3%), followed by ampicillin and cefotaxime each (80.0%), co-trimoxazole and ceftriaxone each (66.7%); however, these isolates were comparatively less resistant to ciprofloxacin (6.7%). Further, the observed antimicrobial resistance pattern among the recovered isolates from different sources was statistically evaluated using chi-squared test (Table 3). A significant relationship was not observed for most of the antibiotics to which the recovered isolates were found resistant, except for ciprofloxacin antibiotic, wherein EAEC isolates recovered from human versus environmental and animal versus environmental sources were found statistically significant.
Table 3.
Statistical analysis for resistance pattern among human, animal, and environmental isolates using chi-squared test
| p-Value for different combinations | ||||
|---|---|---|---|---|
| Antibiotics | (a) | (b) | (c) | Inference |
| Ampicillin | 0.842 | 0.185 | 0.102 | All non-significant |
| Ciprofloxacin | 0.272 | 0.000 | 0.004 | Set (a) – non-significant Set (b) and (c) – significant |
| Co-trimoxazole | 0.620 | 0.656 | 0.346 | All non-significant |
| Ceftriaxone | 0.282 | 0.420 | 0.864 | All non-significant |
| Tetracycline | 0.717 | 0.412 | 0.586 | All non-significant |
| Cefotaxim | 0.266 | 0.185 | 0.747 | All non-significant |
(a) Human versus animal isolates; (b) human versus environmental isolates; (c) animal versus environmental isolates.
Correlation of PFGE and antibiotic sensitivity profiles of EAEC isolates
On comparison of antibiogram and PFGE profiles, all the EAEC pathotype isolates that clustered together in their respective clusters (A–H) on PFGE analysis were exhibiting almost similar antibiotic resistance profile for three or more tested antimicrobials agents and were showing more than 70% resistance similarity (Table 4).
Table 4.
Correlation of PFGE and antibiogram profile of DEC pathotypes isolates
| Antibiogram of the DEC pathotypes in clusters | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| PFGE clusters | DEC pathotypes | AMP | CIP | COT | CTR | IPM | TE | CTX | Resistant % similarity |
| A | E065 | S | S | R | S | S | R | S | 100% (7/7) |
| E066 | S | S | R | S | S | R | S | ||
| B | A258 | R | S | R | R | S | R | R | 1 and 2: 71.4% (5/7) |
| A265 | R | R | R | R | S | R | S | 1 and 3: 85.7% (6/7) | |
| A266 | R | R | R | R | S | R | R | 2 and 3: 85.7% (6/7) | |
| C | H316 | R | R | R | R | S | R | R | 100% (7/7) |
| H325 | R | R | R | R | S | R | R | ||
| D | A219 | R | R | R | R | S | R | R | 100% (7/7) |
| H121 | R | R | R | R | S | R | R | ||
| E | E166 | R | R | R | S | S | R | R | 85.7% (6/7) |
| A103 | R | R | R | R | S | R | R | ||
| F | A206 | R | R | R | R | S | R | R | 100% (7/7) |
| A207 | R | R | R | R | S | R | R | ||
| G | A082 | R | R | R | S | S | R | R | 100% (7/7) |
| A216 | R | R | R | S | S | R | R | ||
| H | H322 | R | R | R | R | S | R | R | 85.7% (6/7) |
| A185 | R | S | R | R | S | R | R | ||
R, resistant; S, sensitive; AMP, ampicillin; CIP, ciprofloxacin; COT, co-trimoxazole; CTR, ceftriaxone; IPM, imipenem; TE, tetracycline; CTX, cefotaxim.
Discussion
Escherichia coli, the common ‘laboratory workhorse’, diverges from commensal flora of warm-blooded animals to highly adaptive pathotypes. To the best of our knowledge, this appears to be the first study in Indian context highlighting the genetic diversity and antimicrobial sensitivity profile of DEC pathotypes recovered from various sources such as diarrhoeal cases of human infants and young animals, including foods and associated environmental sources.
In this study, the isolation of DEC pathotypes from diarrhoeic infants and young animals is in concordance with the earlier reports from Indian subcontinent and also across the globe (16). The majority of isolates recovered in the present study from human infants (10.7%), young animals (15.1%), and from foods and environmental sources (10.2%) were ‘atypical’ EAEC. However, the detection rate of EAEC was higher compared to earlier studies from the Indian subcontinent (20). The highest isolation rate of EAEC pathotype isolates in the present study might be attributed to the study area and/or to its inherent biofilm-forming tendency both, in vivo and in vitro (21). The bio?lm formation promotes persistent colonization of EAEC strains, probably by presenting a barrier to the penetration of antibiotics and host antibacterial factors (15). EAEC pathotype in particular is responsible for causing acute as well as persistent diarrhoea among children ≤5 years in developing countries. Moreover, it is an emerging cause of diarrhoea in animals (22).
In the present study, the predominance of ‘atypical’ EAEC strains from different sources might be because, most of the researchers have targeted ‘typical’ EAEC (AggR regulon-positive) strains in epidemiological studies while ‘atypical’ EAEC (AggR regulon-negative) strains might have been excluded (23). However, even though the ‘atypical’ EAEC isolates do not carry the virulence plasmid associated aggR regulon, still were found to cause diarrhoea and also have been reported as an enteric pathogen of food-borne outbreaks (24). Also, with regard to their biofilm formation, significant differences were not observed between the ‘typical’ and ‘atypical’ EAEC strains (15). Thus, the recovery of ‘atypical’ EAEC strains from diarrhoeal cases of human infants and young animals in the present study highlights the need for further understanding of the pathogenicity of ‘atypical’ EAEC which yet has not been clearly defined.
There are a number of reports on DEC pathotypes from India and across the globe; however, there is a paucity of studies which reveals their relatedness with respect to their sources of isolation. Hence, to explore the likelihood of exposure and magnitude of genetic diversity among DEC pathotypes, all the recovered DEC pathotypes were subjected to PFGE typing, which still remains as the gold standard test for the detection of genotypic diversity (25, 26).
Overall, a diverse genetic profile was obtained among and between EAEC, EPEC and EHEC pathotypes recovered from various sources. Similar type of genetic diversity within and among DEC pathotypes was observed for E. coli pathotypes (27). In general, the highly adaptive nature of E. coli with a short generation time interval including easy acquisition of mobile genetic elements under selection pressure furnishes greater degree of genetic diversity among E. coli strains. To analyse the diverse PFGE data, Tenover and his colleagues proposed guidelines, wherein minor band differences should not be regarded as major genetic shifts and that only isolates differing by seven or more bands (approximately <50% similarity) should be regarded as unrelated, while those differing by four to six bands (approximately >70% similarity) should be regarded as possibly related (25). Thus, based on these guidelines, at a genetic similarity of 70% and/or more, we observed eight clusters designated as A–H, and of these eight clusters, seven clusters were represented by EAEC isolates recovered from either similar or different sources (Fig. 1). Of all the clusters, EAEC isolates of cluster C and F revealed 100% identical PFGE profile and, moreover, the isolates in the respective cluster were recovered from similar sources. It infers that these isolates have either been maintained or circulated within similar source of origin. Similar findings have also been reported by few researchers, wherein isolates with identical PFGE patterns were considered to be of the same clonal origin (25). On the other hand, while characterising EAEC isolates from different parts of the world, a highly heterogeneous DNA pattern among the EAEC strains has been observed with PFGE (28). Moreover, while characterising 58 E. coli O104:H4 strains, 40 PFGE patterns were observed which divided (EAEC) into two main clusters with a similarity index of 64% (29). Despite the vast genetic diversity among E. coli, we cannot exclude the hypothesis that animals, their foods and surrounding environmental source remain a major route of transmission in case of E. coli infection in humans (21). Thus, based on these reported facts, EAEC isolates recovered in the present study were either shared or circulated within similar sources (EAEC isolates of cluster A, B, C, and F), however, their probability of sharing between different sources (EAEC isolates of clusters D, G, H) cannot be ruled out.
Escherichia coli is used as a sentinel for monitoring antimicrobial drug resistance in faecal bacteria since it is found more frequently in a wide range of hosts and also acquires resistance easily (30). Surveillance data indicate that resistance in E. coli is consistently high for antimicrobial agents that are routinely used in human and veterinary medicine (8). In the present study, the antibiotic sensitivity profile revealed a high rate of antimicrobial resistance amongst majority of DEC pathotype isolates. Of the total 59 DEC isolates, 51 isolates (86.4%) were found to be resistant to ≥3 tested drug classes. Irrespective of the source of isolation, all the DEC pathotypes were resistant against all the tested drugs except imipenem. Similar multidrug resistant (MDR) strains of E. coli have been reported worldwide from diarrheal stool samples of children, animals, foods, and other sources (8). In developing countries, the incidence of MDR E. coli has increased in the recent years and up to 75% incidence rate have been reported from India (31, 32). Generally, the overuse or misuse of antibiotics in human and veterinary medicine could be a major factor for this increased antimicrobial resistance (7). Moreover, the exchange of transferable plasmids encoding MDR genes was observed among DEC isolates from diarrhoeal stool and surface water in developing countries, which strengthens the probability of widespread circulation of MDR DEC in the environment sources (33).
Further, a significant relationship was not observed for most of the antibiotics to which the recovered isolates were found resistant, except for ciprofloxacin antibiotic, wherein EAEC isolates recovered from human versus environmental and animal versus environmental sources were found statistically significant. The fluoroquinolones are considered as the first-line drugs for the treatment of diarrhoea (32, 34). Thus, in view of above observations, it seems that ciprofloxacin-resistant EAEC isolates are been either directly or indirectly contributed by humans and animals to the environmental sources. In the present study, on comparison of antibiogram and PFGE profile, all the clustered MDR EAEC isolates (clusters A, B, C, D, F, G, and H) exhibited almost similar antibiotic resistance profile for more than three tested antimicrobials agents. This finding also supports that the clustered EAEC isolates are closely associated; however, further studies with more numbers of isolates, including relevant studies such as monitoring plasmid profile, or investigating frequently associated antimicrobial resistance genes among EAEC strains may provide a useful marker for future epidemiological and interventions studies.
In conclusion, among all the sources investigated, EAEC pathotype in particular ‘atypical’ strains was found to be the predominant pathogen. On PFGE analysis, the diversity was evident within isolated DEC pathotypes; however, a few EAEC isolates were found to be clonal, suggesting that these EAEC isolates might have been shared or circulated between human and animals, including foods and associated environmental sources. Besides this, a high degree of antimicrobial resistance profile was observed for majority of the recovered DEC pathotype isolates. Thus, in the light of the present study and public health perspective, addressing the problem of antibiotic resistance among EAEC strains with due emphasis to the epidemiological triad is a potential area for future research work.
Supplementary Material
Acknowledgements
The authors thank The Director, Indian Veterinary Research Institute, Izatnagar, for providing necessary facilities for undertaking the research. The research was supported by grants from the Department of Biotechnology, Government of India (BT/PR15148/GBD/27/339/2011) to DBR and Junior Research Fellowship to PD by Indian Council of Agricultural Research, New Delhi. We are grateful to Dr. Chobi Debroy and Dr. Bhushan Jayarao, Pennsylvania State University, USA, for providing us with DEC pathotype DNA. We also thank Mr K.K. Bhatt for his excellent technical assistance.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Conflict of interest and funding
The authors declare that they have no conflict of interests.
References
- 1.Liu L, Hope LJ, Simon C, Jamie P, Susana S, Joy EL, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2010;379:2151–61. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 2.Puño-Sarmiento J, Medeiros L, Chiconi C, Martins F, Pelayo J, Rocha S, et al. Detection of diarrheagenic Escherichia coli strains isolated from dogs and cats in Brazil. Vet Microbiol. 2013;166:676–80. doi: 10.1016/j.vetmic.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Shabana II, Zaraket H, Suzuki H. Molecular studies on diarrhoea-associated Escherichia coli isolated from humans and animals in Egypt. Vet Microbiol. 2013;167:532–9. doi: 10.1016/j.vetmic.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Chen X, Zheng S, Yu F, Kong H, Yang Q, et al. Serotypes, genotypes and antimicrobial resistance patterns of human diarrhoeagenic Escherichia coli isolates circulating in southeastern China. Clin Microbiol Infect. 2014;20:52–8. doi: 10.1111/1469-0691.12188. [DOI] [PubMed] [Google Scholar]
- 5.Bautista-De León H, Gómez-Aldapa CA, Rangel-Vargas E, Vázquez-Barrios E, Castro-Rosas J. Frequency of indicator bacteria, Salmonella and diarrhoeagenic Escherichia coli pathotypes on ready-to-eat cooked vegetable salads from Mexican restaurants. Lett Appl Microbiol. 2013;56:414–20. doi: 10.1111/lam.12063. [DOI] [PubMed] [Google Scholar]
- 6.Farooq S, Hussain I, Mir MA, Bhat MA, Wani SA. Isolation of atypical enteropathogenic Escherichia coli and Shiga toxin 1 and m2f producing Escherichia coli from avian species in India. Lett Appl Microbiol. 2009;48:692–7. doi: 10.1111/j.1472-765X.2009.02594.x. [DOI] [PubMed] [Google Scholar]
- 7.Wellington EMH, Boxall ABA, Cross P, Feil EJ, Gaze WH, Hawkey PM, et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis. 2013;13:155–65. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 8.Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, et al. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg Infect Dis. 2012;18:741–9. doi: 10.3201/eid1805.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.UNICEF/World Health Organization. Diarrhoea: why children are still dying and what can be done. 2009. Available from: http://www.who.int/ topics/diarrhoea/en/ [cited 7 February 2014]
- 10.Brenner DJ, Krieg NR, Staley JR. The Gammaproteobacteria. In: GM Garrity., editor. Bergey's manual of systematic bacteriology. 2nd vol. New York: Springer; 2005. p. 1106. [Google Scholar]
- 11.Cheesbrough M. Medical laboratory manual for tropical countries. Microbiology. 1985;II:400–80. [Google Scholar]
- 12.Kudva IT, Evans PS, Perna NT, Barrett TJ, DeCastro GJ, Ausubel FM, et al. Polymorphic amplified typing sequences provide a novel approach to Escherichia coli O157:H7 strain typing. J Clin Microbiol. 2002;40:1152–59. doi: 10.1128/JCM.40.4.1152-1159.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishikawa Y, Zhou Z, Hase A, Ogasawara J, Kitase T, Abe N, et al. Diarrhoeagenic Escherichia coli isolated from stools of sporadic cases of diarrhoeal illness in Osaka city, Japan between 1997 and 2000: prevalence of enteroaggregative E. coli heat-stable enterotoxin 1 gene-possessing E. coli . J Infect Dis. 2002;55:182–90. [PubMed] [Google Scholar]
- 14.Jenkins C, Chart H, Willshaw GA. Genotyping of enteroaggregative Escherichia coli and identification of target genes for the detection of both typical and atypical strains. Diagn Microbiol Infect Dis. 2006;55:13–19. doi: 10.1016/j.diagmicrobio.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Tokuda K, Nishi J, Imuta N, Fujiyama R, Kamenosono A, Manago K, et al. Characterization of typical and atypical enteroaggregative Escherichia coli in Kagoshima, Japan: biofilm formation and acid resistance. Microbiol Immunol. 2010;54:320–9. doi: 10.1111/j.1348-0421.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 16.Vergis J. Izatnagar, India: Indian Veterinary Research Institute; 2013. Prevalence of diarrhoeagenic Escherichia coli pathotypes in human infants and young animals having diarrhoea (dissertation) p. 99. [Google Scholar]
- 17.Vijay D, Dhaka P, Vergis J, Negi M, Mohan V, Kumar M, et al. Characterization and biofilm forming ability of diarrhoeagenic enteroaggregative Escherichia coli isolates recovered from human infants and young animals. Comp Immunol Microbiol Infect Dis. 2015;38:21–31. doi: 10.1016/j.cimid.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 18.CDC. Standard operating procedure for PulseNet PFGE of E. coli O157:H7, E. coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei, and Shigella flexneri. 2013. Available from: http://www. cdc. gov/pulsenet/ PDF/ e.coli-shigella-salmonella-pfge-protocol-508c.pdf [cited 21 March 2014]
- 19.CLSI. Performance standards for antimicrobial susceptibility testing: seventeenth informational supplement, M100–S23. Pennsylvania, PA: Clinical and Laboratory Standards Institute; 2013. p. 200. [Google Scholar]
- 20.Dutta S, Guin S, Ghosh S, Pazhani GP, Rajendran K, Bhattacharya MK, et al. Trends in the prevalence of diarrheagenic Escherichia coli among hospitalized diarrheal patients in Kolkata, India. PLoS One. 2013;8:e56068. doi: 10.1371/journal.pone.0056068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Advances in understanding enteric pathogenic Escherichia coli . Clin Microbiol Rev. 2013;26:822–80. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estrada-Garcia T, Navarro-Garcia F. Enteroaggregative Escherichia coli pathotype: a genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunol Med Microbiol. 2012;66:1–18. doi: 10.1111/j.1574-695X.2012.01008.x. [DOI] [PubMed] [Google Scholar]
- 23.Huang DB, Mohanty A, DuPont HL, Okhuysen PC, Chiang T. A review of an emerging enteric pathogen: enteroaggregative Escherichia coli . J Med Microbiol. 2006;55:1303–11. doi: 10.1099/jmm.0.46674-0. [DOI] [PubMed] [Google Scholar]
- 24.Bouzari S, Jafari A, Zarepour M. Distribution of virulence related genes among enteroaggregative Escherichia coli isolates: using multiplex PCR and hybridization. Infect Genet Evol. 2005;5:79–83. doi: 10.1016/j.meegid.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goering RV, Fey PD. Pulsed field gel electrophoresis of Staphylococcus epidermidis . Methods Mol Biol. 2014;1106:55–60. doi: 10.1007/978-1-62703-736-5_4. [DOI] [PubMed] [Google Scholar]
- 27.Weiner M, Dacko J, Osek J. Molecular analysis of enterotoxigenic, Shiga toxigenic and enteroaggregative Escherichia coli strains isolated from suckling piglets with diarrhoea by the use of pulsed-field electrophoresis. Bull Vet Inst Pulawy. 2004;48:225–32. [Google Scholar]
- 28.Adachi JA, Jiang ZD, Mathewson JJ, Verenkar MP, Thompson S, Martinez-Sandoval F. Enteroaggregative Escherichia coli as a major etiologic agent in traveler's diarrhoea in 3 regions of the world. Clin Infect Dis. 2001;32:1706–9. doi: 10.1086/320756. [DOI] [PubMed] [Google Scholar]
- 29.Rump LV, Bodeis-Jones S, Abbott JS, Kase J, Lorenz S, Fischer M, et al. Genetic characterization of Escherichia coli O104 isolates from different sources in the United States. Appl Environ Microbiol. 2011;78:1615–18. doi: 10.1128/AEM.07533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erb A, Stürmer T, Marre R, Brenner H. Prevalence of antibiotic resistance in Escherichia coli: overview of geographical, temporal, and methodological variations. Eur J Clin Microbiol Infect Dis. 2007;26:83–90. doi: 10.1007/s10096-006-0248-2. [DOI] [PubMed] [Google Scholar]
- 31.Raju B, Ballal M. Multidrug resistant enteroaggregative Escherichia coli diarrhoea in rural southern Indian population. Scand J Infect Dis. 2009;41:105–8. doi: 10.1080/00365540802641856. [DOI] [PubMed] [Google Scholar]
- 32.Vaishnavi C, Kaur S. The epidemiological and resistogram patterns of enteropathogenic and enterotoxigenic Escherichia coli isolated from diarrhoeal stools in a north Indian hospital. Trop Gastroenterol. 2003;24:70–2. [PubMed] [Google Scholar]
- 33.Chigor VN, Umoh VJ, Smith SI, Igbinosa EO, Okoh AI. Multidrug resistance and plasmid patterns of Escherichia coli O157 and other E. coli isolated from diarrhoeal stools and surface waters from some selected sources in Zaria, Nigeria. Int J Environ Res Public Health. 2010;7:3831–41. doi: 10.3390/ijerph7103831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samal SK, Khuntia HK, Nanda PK, Satapathy CS, Nayak SR, Sarangi AK, et al. Incidence of bacterial enteropathogens among hospitalized diarrhea patients from Orissa, India. Jpn J Infect Dis. 2008;61:350–5. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.