Summary
The mitochondrial inner membrane harbors three protein translocases. Presequence translocase and carrier translocase are essential for importing nuclear-encoded proteins. The oxidase assembly (OXA) translocase is required for exporting mitochondrial-encoded proteins; however, different views exist about its relevance for nuclear-encoded proteins. We report that OXA plays a dual role in the biogenesis of nuclear-encoded mitochondrial proteins. First, a systematic analysis of OXA-deficient mitochondria led to an unexpected expansion of the spectrum of OXA substrates imported via the presequence pathway. Second, biogenesis of numerous metabolite carriers depends on OXA, although they are not imported by the presequence pathway. We show that OXA is crucial for the biogenesis of the Tim18-Sdh3 module of the carrier translocase. The export translocase OXA is thus required for the import of metabolite carriers by promoting assembly of the carrier translocase. We conclude that OXA is of central importance for the biogenesis of the mitochondrial inner membrane.
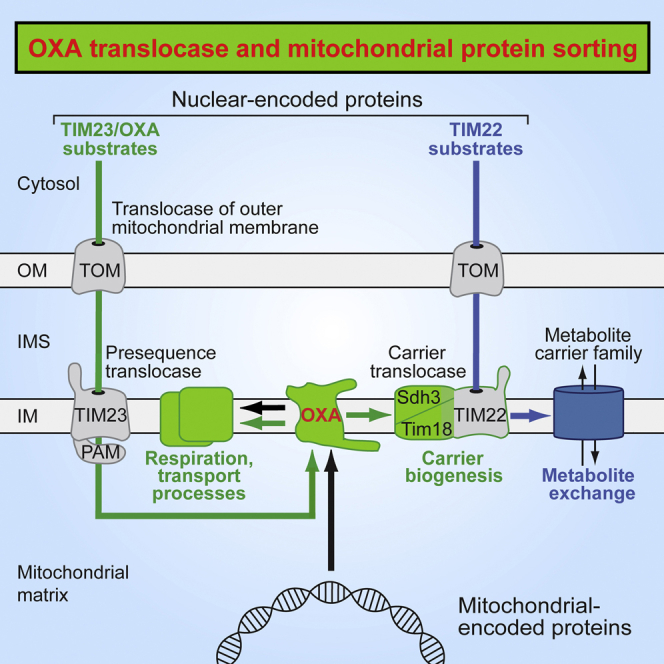
Graphical Abstract
Highlights
-
•
Systematic search for substrates of mitochondrial inner-membrane OXA translocase
-
•
OXA translocase has different functions in presequence and carrier pathways
-
•
Many cleavable preproteins use the presequence translocase-OXA import-export pathway
-
•
OXA promotes carrier import via assembly of Tim18-Sdh3 module of carrier translocase
Stiller et al. report that the sorting of many nuclear-encoded mitochondrial proteins depends on the OXA translocase, a homolog of bacterial YidC. The conservative sorting of transmembrane segments into the mitochondrial inner membrane is required for assembly of the carrier translocase and thus for the biogenesis of numerous metabolite carriers.
Introduction
The mitochondrial inner membrane is a protein-rich membrane harboring the respiratory chain complexes and numerous metabolite carriers. Two genetic systems are required for formation of the inner membrane (Neupert and Herrmann, 2007, Chacinska et al., 2009, Endo and Yamano, 2009). The mitochondrial genome encodes a small number of proteins. These typically hydrophobic proteins are synthesized in the mitochondrial matrix and are exported into the inner membrane by the cytochrome oxidase assembly (OXA) translocase (Hell et al., 2001, Funes et al., 2011). Oxa1, the major component of the OXA translocase, has been conserved from the prokaryotic ancestor of mitochondria and is thought to function as a membrane protein insertase like prokaryotic YidC (Hell et al., 2001, Funes et al., 2011, Kumazaki et al., 2014). Cox18 (Oxa2), an Oxa1 paralog, plays a minor role in the export of mitochondrial-encoded proteins (Funes et al., 2004, Bonnefoy et al., 2009).
Most inner-membrane proteins, however, are encoded by nuclear genes and are synthesized as precursors on cytosolic ribosomes. Targeting signals direct these precursor proteins to mitochondria and into the inner membrane. Two major classes of inner-membrane precursor proteins can be distinguished (Neupert and Herrmann, 2007, Chacinska et al., 2009). Preproteins transported by the presequence translocase of the inner membrane (TIM23 complex) typically carry cleavable amino-terminal targeting signals. These positively charged presequences are removed by the mitochondrial processing peptidase (MPP) in the matrix. Precursor proteins transported by the carrier translocase of the inner membrane (TIM22 complex) do not contain cleavable presequences, but internal targeting signals distributed over the mature part of the protein. Presequence translocase and carrier translocase are activated by the membrane potential (Δψ) across the inner membrane to drive translocation of precursor proteins.
Whereas all precursors known to be imported by the carrier translocase are laterally released into the inner membrane, two distinct pathways are possible with the presequence translocase. (1) Cleavable preproteins with hydrophobic stop transfer signals are arrested during translocation by the TIM23 complex and are laterally released into the lipid phase of the inner membrane (Glick et al., 1992, Botelho et al., 2011, Park et al., 2013, Ieva et al., 2014). (2) The presequence translocase-associated motor (PAM) cooperates with the TIM23 complex in the import of preproteins into the matrix. The mitochondrial heat shock protein 70 (mtHsp70, Ssc1) is the core component of PAM and drives matrix translocation of proteins in an ATP-dependent manner (Neupert and Herrmann, 2007, Chacinska et al., 2009). The TIM23-PAM pathway is obligatory for the import of all matrix proteins; however, it is also used by a few inner-membrane proteins. Upon import into the matrix, these inner-membrane precursors are exported into the membrane by the OXA translocase. This import-export pathway was termed the conservative sorting pathway, since the OXA-dependent protein export has been conserved from the prokaryotic ancestor of mitochondria (Neupert and Herrmann, 2007). To date, however, only three authentic nuclear-encoded proteins were shown to be exported by the OXA translocase in yeast: the precursors of Oxa1 and Cox18 themselves and the ABC transporter Mdl1 (Herrmann et al., 1997, Hell et al., 1998, Funes et al., 2004, Bohnert et al., 2010). Additionally, the Rieske Fe/S protein follows an import-export pathway, yet uses the AAA-ATPase Bcs1 as a precursor-specific export and assembly machine instead of the OXA translocase (Hartl et al., 1986, Wagener et al., 2011). The available data thus support the view that the vast majority of cleavable inner-membrane proteins are not sorted via OXA, but are laterally released from the TIM23 complex by a stop transfer mechanism (Glick et al., 1992, Neupert and Herrmann, 2007, Bohnert et al., 2010, Ieva et al., 2014).
Hildenbeutel et al. (2012) reported that Oxa1 is involved in the biogenesis of several metabolite carriers that are imported by the TIM22 complex. Since carrier precursors are inserted into the inner membrane from the intermembrane space side and are not translocated into the matrix (Neupert and Herrmann, 2007, Chacinska et al., 2009), the role of OXA cannot be explained by an export of the precursors from the matrix. It was discussed that OXA may promote the folding of imported carrier proteins or influence the lipid composition of the inner membrane (Hildenbeutel et al., 2012).
Here we identified numerous nuclear-encoded mitochondrial proteins that depend on OXA and establish the OXA translocase as a major machinery for the biogenesis of the inner membrane. OXA is also required for the biogenesis of the Tim18-Sdh3 module of the TIM22 carrier translocase, explaining the disturbed carrier import in OXA-deficient mitochondria.
Results and Discussion
Mitochondrial Inner-Membrane Proteins Depleted in OXA-Deficient Mitochondria
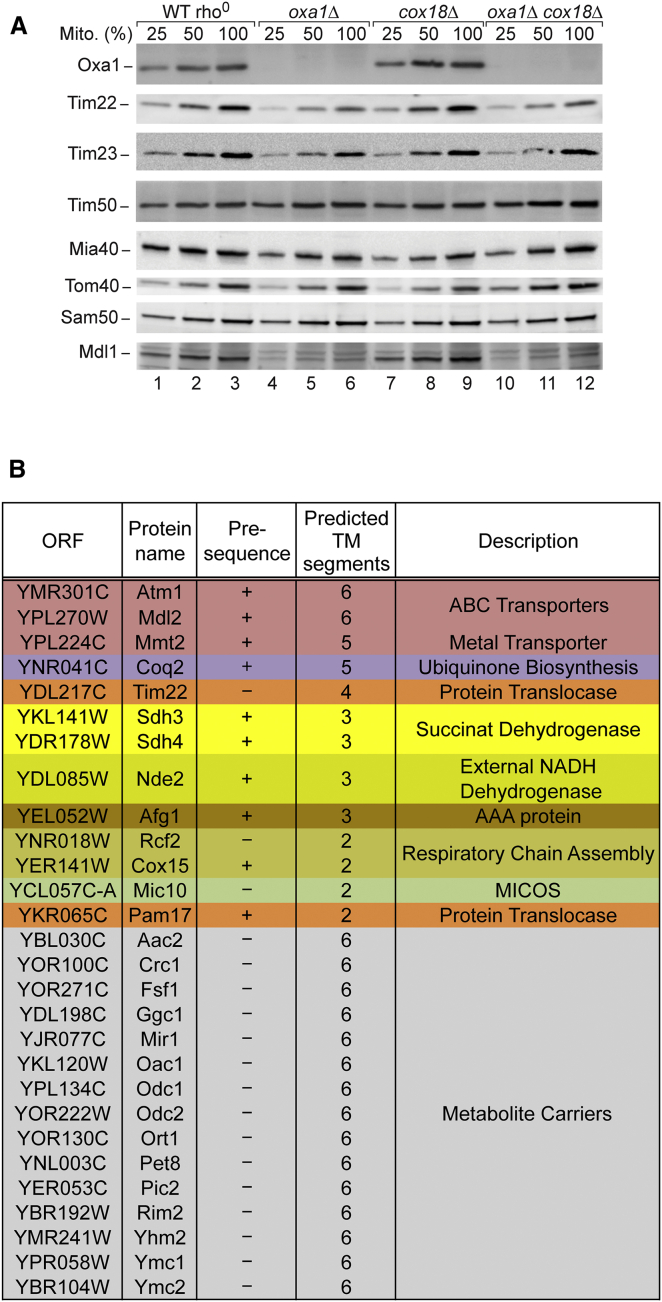
To study a possible influence of OXA on other preprotein translocases, we analyzed the protein levels in oxa1Δ mitochondria, cox18Δ mitochondria, and oxa1Δ cox18Δ mitochondria from budding yeast. The levels of Tim22 were reduced in oxa1Δ mitochondria and oxa1Δ cox18Δ mitochondria, whereas other protein import machineries, including presequence translocase (Tim23, Tim50), mitochondrial intermembrane space import and assembly machinery (Mia40), translocase of outer membrane (Tom40), and sorting and assembly machinery (Sam50), were not or were only mildly affected (Figure 1A). For comparison, the levels of the known Oxa1 substrate Mdl1 (Bohnert et al., 2010) were considerably reduced in oxa1Δ and oxa1Δ cox18Δ mitochondria.
Figure 1.
Mitochondrial Inner-Membrane Proteins Affected in OXA-Deficient Yeast Mutants
(A) The levels of translocase core subunits and of Mdl1 of wild-type (WT) rho0, oxa1Δ, cox18Δ, and oxa1Δ cox18Δ mitochondria were analyzed by SDS-PAGE and immunoblotting. Mito., total mitochondrial protein; 100% represents 40 μg protein (80 μg for Sam50).
(B) Differential analysis of the proteome of WT rho0 compared to oxa1Δ cox18Δ mitochondria using MS. WT rho0 and oxa1Δ cox18Δ cells were differentially labeled with stable isotopes, mixed and mitochondria were isolated and analyzed by quantitative MS (Table S1). Integral inner-membrane proteins found to be depleted in oxa1Δ cox18Δ mitochondria (heavy/light ratio >1.6) are listed with the yeast open reading frame (ORF).
A connection of OXA to the TIM22 complex could provide an explanation for its influence on the import of metabolite carriers (Hildenbeutel et al., 2012). So far, however, neither OXA nor any of the known OXA substrates have been linked to the TIM22 translocase. We performed a systematic approach to screen for mitochondrial inner-membrane proteins depleted in OXA-deficient mitochondria. We used a quantitative mass spectrometry (MS)-based approach to compare oxa1Δ cox18Δ mitochondria and control mitochondria. Since Oxa1-deficient mitochondria are strongly impaired in protein complexes containing mitochondrial-encoded subunits, including oxidative phosphorylation complexes III, IV, and V (see Figure S1 available online), the levels of many subunits of these complexes can be decreased independently if they are OXA substrates or not (Bonnefoy et al., 2009). Thus, wild-type mitochondria containing a functional mitochondrial genome (rho+) do not represent an appropriate control for oxa1Δ cox18Δ mitochondria. We found that wild-type rho0 mitochondria, which lack mtDNA but contain Oxa1 and Cox18, represented the most stringent control for screening for OXA-substrates. Wild-type rho0 cells and oxa1Δ cox18Δ cells were differentially labeled with stable isotopes and mixed, and isolated mitochondria were subjected to MS analysis (Table S1). Mitochondrial inner-membrane proteins depleted in oxa1Δ cox18Δ mitochondria are listed in Figure 1B. Tim22 was depleted in agreement with the analysis of its steady-state levels by immunodecoration (Figure 1A). We identified many members of the metabolite carrier family, and further noncleavable proteins (Mic10, Rcf2), as well as numerous cleavable proteins, including the ABC transporters Atm1 and Mdl2, the metal transporter Mmt2, and Sdh3 and Sdh4 of respiratory complex II (succinate dehydrogenase [SDH]) (Figure 1B). Thus, a broad spectrum of inner-membrane proteins were depleted in OXA-deficient mitochondria.
Oxa1 Is Required for Assembly of Both SDH Complex and TIM22 Complex
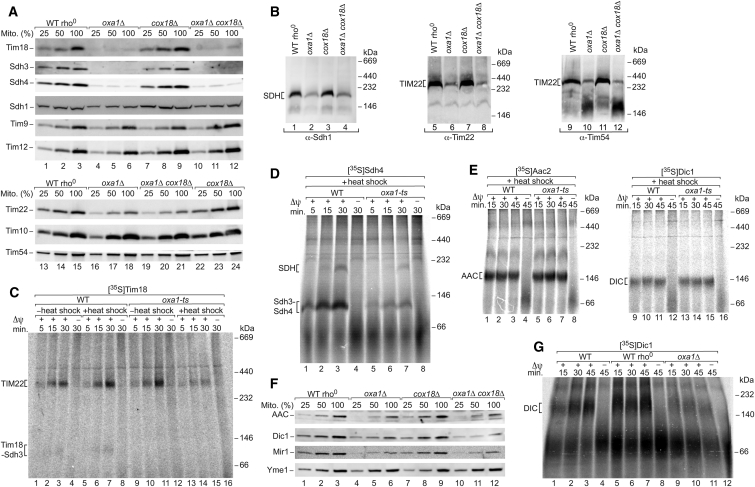
All subunits of the metazoan and fungal SDH complex are nuclear encoded. The membrane-integrated module Sdh3-Sdh4 is evolutionarily related to the Sdh3-Tim18 module of the TIM22 complex. Tim18 is a paralog of Sdh4, and Sdh3 is a subunit of both the SDH complex and the TIM22 complex (Gebert et al., 2011). Immunodecoration revealed that all three proteins were strongly depleted in oxa1Δ mitochondria and oxa1Δ cox18Δ mitochondria, but not in cox18Δ mitochondria (Figure 2A). The level of Sdh1 of the hydrophilic matrix-exposed portion of the SDH complex was only mildly affected in the mutant mitochondria. Similarly, the levels of the intermembrane space-exposed proteins of the TIM22 complex, Tim54 and Tim9-Tim10-Tim12, were close to that of control mitochondria (Figure 2A), whereas the levels of Tim22 were moderately reduced upon deletion of OXA1 (Figures 1A and 2A). The mature assembled SDH complex and TIM22 complex were analyzed by blue native electrophoresis of digitonin-lysed mitochondria. Consistent with the depletion of crucial subunits, the levels of the mature complexes were strongly decreased in Oxa1-deficient mitochondria (Figure 2B).
Figure 2.
Oxa1-Dependent Assembly of SDH Complex and TIM22 Complex
(A) Protein levels of TIM22 and SDH subunits were analyzed in the indicated mitochondria by SDS-PAGE and immunoblotting. Mito., total mitochondrial protein; 100% represents 160 μg protein (40 μg for Sdh3 and Sdh4; 80 μg for Tim9 and Tim12).
(B) SDH complex and TIM22 complex were analyzed by blue native electrophoresis of isolated mitochondria and immunoblotting.
(C) Mitochondria were either heat shocked for 12 min at 37°C or incubated at 25°C, followed by import of [35S]Tim18 precursor at 25°C. After Proteinase K treatment, assembly into the TIM22 complex was monitored by blue native electrophoresis and autoradiography.
(D) After an in vitro heat shock of mitochondria, [35S]Sdh4 precursor was imported at 30°C, followed by Proteinase K treatment. Assembly of SDH and the Sdh3-Sdh4 intermediate were analyzed by blue native electrophoresis.
(E) Mitochondria were subjected to a heat shock, followed by import of [35S]Aac2 and [35S]Dic1 at 25°C and Proteinase K treatment. Carrier assembly was monitored by blue native electrophoresis.
(F) Protein levels were analyzed by SDS-PAGE and immunoblotting. Mito., total mitochondrial protein; 100% represents 40 μg protein.
(G) Dic1 was imported into mitochondria. After Proteinase K treatment, carrier assembly was monitored by blue native electrophoresis.
See also Figures S2 and S3 and Table S2.
To discriminate between direct and indirect Oxa1 effects, we used a temperature-sensitive oxa1 yeast mutant. Upon growth of the mutant cells at permissive temperature, the levels of mitochondrial proteins were not or were only mildly affected in comparison to rho+ wild-type mitochondria (Figures S2A and S2B). The Oxa1 defect was induced by a short in vitro heat shock (12 min 37°C) of isolated oxa1-ts mitochondria (Hell et al., 2001, Preuss et al., 2001, Bohnert et al., 2010), leading to a degradation of the OXA complex (Figure S3A). The steady levels of other protein complexes including SDH and TIM22 were not affected by the in vitro heat shock (Figure S3A). To test the Oxa1 function in a kinetic range, we imported the 35S-labeled precursor of Tim18 into isolated energized mitochondria. Upon the in vitro heat shock, assembly of Tim18 into the TIM22 complex as well as formation of the Tim18-Sdh3 assembly intermediate (Gebert et al., 2011) was strongly impaired in oxa1-ts mitochondria (Figure 2C; Figures S3B and S3C). Similarly, the assembly of Sdh4 into the mature SDH complex and formation of the Sdh3-Sdh4 assembly intermediate (Gebert et al., 2011) were inhibited (Figure 2D; Figure S3D). Translocation to a protease-protected location and processing were not or were only mildly affected in heat-shocked oxa1-ts mitochondria, as shown for Tim18, Sdh3, and matrix-targeted preproteins (Figures S3E and S3F), indicating that functional Oxa1 is not crucial for the initial translocation into mitochondria. Upon import into mitochondria, the 35S-labeled precursor of Sdh4, but not fully assembled endogenous Sdh4, was copurified with tagged Oxa1 (Figure S3G). Taken together, we conclude that Oxa1 plays a specific role in the assembly pathways of Tim18, Sdh3, and Sdh4.
Δψ-dependent import and assembly of metabolite carriers were monitored by blue native electrophoresis (Gebert et al., 2011). Upon import of the 35S-labeled precursors of ADP/ATP carrier (Aac2) and dicarboxylate carrier (Dic1) into heat-shocked oxa1-ts mitochondria, their assembly was indistinguishable from that of wild-type mitochondria (Figure 2E). Hildenbeutel et al. (2012) showed, however, that the levels of Aac2 were diminished in oxa1Δ mitochondria and that the import of carrier precursors was reduced in Oxa1-depleted mitochondria (isolated from oxa1 mutant cells that had been heat shocked for 3 hr). Our analysis of oxa1Δ mitochondria agrees with the findings of Hildenbeutel et al. (2012). The levels of metabolite carriers analyzed by immunoblotting were reduced (Figure 2F), in agreement with the depletion of numerous metabolite carriers in the MS analysis (Figure 1B; Table S1). Assembly of 35S-labeled Dic1 was strongly inhibited in oxa1Δ mitochondria in comparison to wild-type mitochondria (rho+ as well as rho0 wild-type) (Figure 2G). Thus, the in vitro induction of the oxa1-ts mutant phenotype by a short heat shock of isolated mitochondria provides the basis for separating direct and indirect effects of Oxa1 on mitochondrial protein biogenesis. The assembly pathways of Tim18, Sdh3, and Sdh4, but not of metabolite carriers, directly depend on functional Oxa1. Since a defective assembly of the TIM22 complex impairs subsequent import of metabolite carriers (Gebert et al., 2011), depletion of Oxa1 will lead to defects in the biogenesis of carrier proteins.
Differential Roles of Import Motor and OXA in the Sorting of Sdh4/Sdh3/Tim18
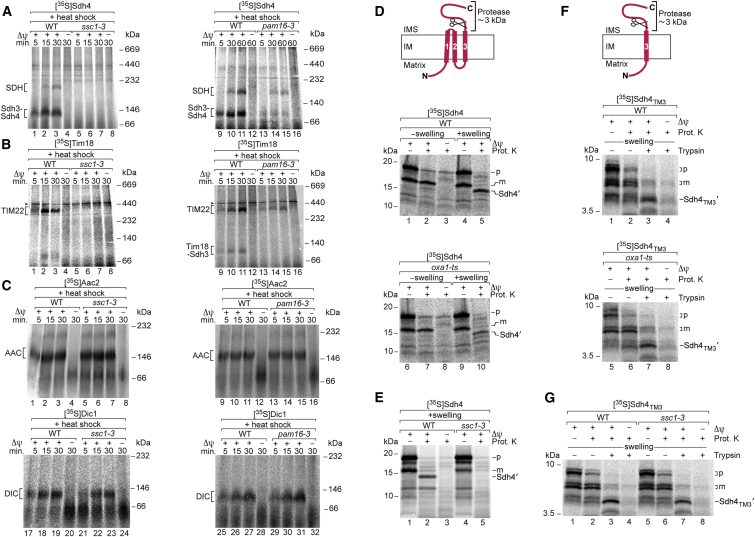
Since OXA exports polypeptides from the matrix, nuclear-encoded polypeptides that directly depend on OXA have to be translocated into the matrix. The mtHsp70 import motor PAM is essential for protein translocation into the matrix (Neupert and Herrmann, 2007, Chacinska et al., 2009). We asked if the dependence on PAM differed for the import pathways of Sdh4/Sdh3/Tim18 and metabolite carriers, similar to their differential dependence on functional Oxa1. We used temperature-sensitive yeast mutants of mtHsp70 (ssc1-3) and the cochaperone Pam16 (pam16-3). Isolated mitochondria were subjected to a short in vitro heat shock (12–15 min, 37°C) and incubated with the 35S-labeled preproteins. The biogenesis pathways of Sdh4/Sdh3/Tim18 were blocked in ssc1-3 mitochondria and diminished in pam16-3 mitochondria (Figures 3A and 3B; Figure S4A). In contrast, the biogenesis pathways of Aac2 and Dic1 were neither inhibited in ssc1-3 nor in pam16-3 mitochondria (Figure 3C), supporting the conclusion that Sdh4/Sdh3/Tim18 and metabolite carriers use different sorting routes.
Figure 3.
Mitochondrial Sorting of Sdh4 and Tim18 Depends on the mtHsp70 Import Motor
(A–C) Import of 35S-labeled Sdh4 (A), Tim18 (B), Aac2 and Dic1 (C) into in vitro heat-shocked (37°C for 12–15 min) wild-type (WT), ssc1-3, and pam16-3 mitochondria at 25°C (30°C for A, lanes 1–8), followed by Proteinase K treatment. Arrowhead, unspecific band.
(D–G) 35S-labeled Sdh4 (D and E) or TM3 fused to the Sdh4 presequence, [35S]Sdh4TM3 (F and G), were imported into WT, oxa1-ts, and ssc1-3 mutant mitochondria at 30°C. WT and mutant mitochondria were heat shocked prior to the import reaction. Nonimported [35S]Sdh4TM3 was removed by Proteinase K treatment. After washing, mitochondria were suspended in either isotonic buffer (−swelling) or hypotonic buffer (+swelling). Where indicated, Proteinase K or trypsin was added. The mitochondrial samples were analyzed by SDS-PAGE and autoradiography. p, precursor; m, mature form; Sdh4′ and Sdh4TM3′, protease-protected Sdh4 and Sdh4TM3 fragments. Cartoons in (D) and (F): the TMs of Sdh4 are numbered from 1 to 3; IM, inner membrane; IMS, intermembrane space.
Sdh4, Sdh3, and Tim18 each contain three transmembrane segments (TMs); the N termini are exposed to the matrix and the C termini to the intermembrane space (Sun et al., 2005, Huang et al., 2006, Gebert et al., 2011). We observed that the C-terminal segment of Sdh4 was accessible to protease from the intermembrane space side and used this assay to probe the insertion of [35S]Sdh4 into the inner membrane (Figure 3D; Figure S4B). The precursor of Sdh4 was imported in a Δψ-dependent manner and processed to the mature form, which was protected against Proteinase K added to the isolated mitochondria (Figure 3D, lane 2). Upon opening of the outer membrane by swelling of mitochondria, Proteinase K generated a truncated form Sdh4′ that was ∼3 kDa smaller (Figure 3D, lane 5), consistent with the removal of the ∼28-residue C-terminal segment that is located behind TM3 (TM1 and TM2 are connected by a short ∼5-residue loop that is not accessible to the protease; Sun et al., 2005). With heat-shocked ssc1-3 mitochondria, Sdh4 was processed to the mature-sized protein; however, formation of the truncated Sdh4′ form was blocked (Figure 3E), demonstrating that an active motor was required to drive efficient import of the mature portion of Sdh4. In contrast, with heat-shocked oxa1-ts mitochondria, truncated Sdh4′ was formed (Figure 3D, lane 10), indicating that a functional Oxa1 was not crucial for membrane insertion of the C-terminal TM3.
How can these differential effects of mtHsp70 and Oxa1 on the sorting of Sdh4 be explained? A comparison of TMs of mitochondrial inner-membrane proteins suggested that TMs arrested in the inner membrane by a stop transfer mechanism are more hydrophobic than TMs imported into the matrix; a proline residue in a TM also favors translocation into the matrix (Meier et al., 2005, Botelho et al., 2011, Park et al., 2013, Park et al., 2014). Park et al. (2013) analyzed membrane insertion of the TMs of Sdh4 in the context of fusion proteins and suggested that TM1 (containing a proline residue) and TM2 are translocated into the matrix, whereas TM3 is probably arrested in the inner membrane. Figure S4C presents a model that combines the analysis of TMs and our findings on the differential dependence on mtHsp70 and OXA: the motor PAM is required to drive import of the N-terminal portion of Sdh4 including TM1 and TM2 for subsequent export by OXA, whereas TM3 is arrested in the TIM23 complex by a stop-transfer mechanism and laterally released into the membrane in an OXA-independent manner.
To directly test the predicted independence of TM3 from mtHsp70 and OXA, we generated a shortened form of Sdh4 that lacked TM1 and TM2 but contained the presequence plus TM3 and the C-terminal segment (Figure S4B). 35S-labeled Sdh4TM3 was imported into mitochondria in a Δψ-dependent manner and processed to the mature-sized form. m-Sdh4TM3 was protected against protease added to mitochondria but cleaved to the truncated form Sdh4TM3′ upon opening of the outer membrane (Figure 3F, upper panel). Truncated Sdh4TM3′ was similarly generated with ssc1-3 mitochondria (Figure 3G) as well as oxa1-ts mitochondria (Figure 3F, lower panel), demonstrating that both functional mtHsp70 and Oxa1 were dispensable for import and inner-membrane sorting of TM3. Sdh4TM3 thus fulfills all requirements for sorting via a stop transfer mechanism (Figure S4D). Taken together with the study by Park et al. (2013), we conclude that the biogenesis of Sdh4 involves two sorting pathways: mtHsp70-dependent import into the matrix and OXA-mediated export for TM1 and TM2, as well as stop transfer and lateral release into the inner membrane for TM3 (Figure S4C).
Major Role of OXA in the Biogenesis of Inner-Membrane Proteins
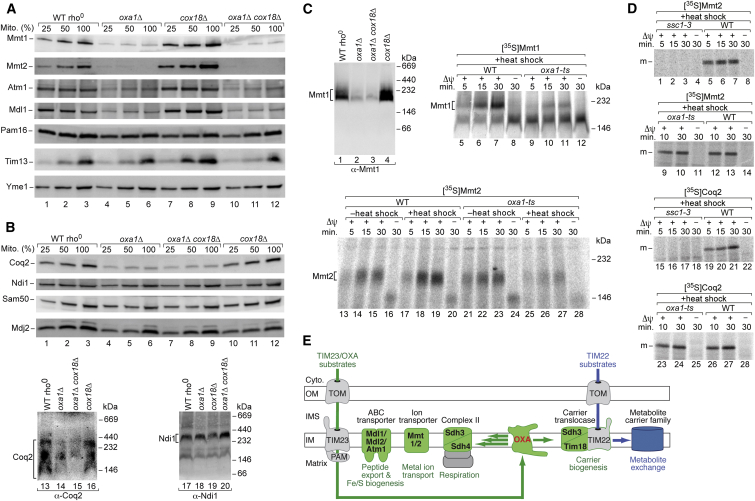
To further assess the importance of OXA for the biogenesis of nuclear-encoded inner-membrane proteins, we subjected additional candidate proteins derived from the MS analysis (Figure 1B) to a biochemical analysis. We tested proteins with lower MS intensities in oxa1Δ cox18Δ mitochondria as well as proteins belonging to the same protein family, including: the mitochondrial metal transporters Mmt1 and Mmt2, which are involved in mitochondrial iron accumulation; the ABC transporters Atm1 and Mdl1; and Coq2, an inner-membrane polyprenyl transferase functioning in the biosynthesis of ubiquinone. The levels of these proteins were considerably decreased in oxa1Δ mitochondria and oxa1Δ cox18Δ mitochondria (Figures 4A and 4B) in agreement with the MS findings and the reported OXA-dependence of Mdl1 (Bohnert et al., 2010; Figure 1A). Blue native analysis using oxa1-ts mitochondria showed that the assembly pathways of Mmt1 and Mmt2 depended on functional Oxa1 (Figure 4C; Figure S4E). To obtain further evidence that a conservative sorting pathway was used, we studied the initial import of Mmt2 and Coq2 to a protease-protected location. Both precursors contain proline within the predicted TMs, favoring translocation into the matrix (Meier et al., 2005). Indeed, the initial import strictly depended on functional mtHsp70, yet not Oxa1 (Figure 4D). The 35S-labeled precursor of Mmt2, but not mature endogenous Mmt2, was copurified with tagged Oxa1 (Figure S4F), strongly supporting an OXA-mediated sorting. Various inner-membrane control proteins did not reveal a dependence on OXA, including the AAA protease Yme1, the internal NADH dehydrogenase Ndi1 and the motor subunit Mdj2 (Figures 4A and 4B; Table S1).
Figure 4.
Role of OXA in the Biogenesis of Nuclear-Encoded Inner-Membrane Proteins
(A and B) Mitochondria isolated from the indicated yeast cells were either subjected to SDS-PAGE or lysed with 1% digitonin and analyzed by blue native electrophoresis, followed by immunoblotting. Mito., total mitochondrial protein; 100% represents 160 μg protein (40 μg for Tim13 and Yme1; 80 μg for Atm1 and Mdl1; 320 μg for Ndi1).
(C) Samples 1–4, the indicated mitochondria were lysed with digitonin and analyzed by blue native electrophoresis and immunoblotting. Samples 5–28, mitochondria were subjected to a heat shock where indicated; Mmt1 and Mmt2 were imported, followed by Proteinase K treatment. Mitochondria were analyzed by blue native electrophoresis and autoradiography.
(D) Heat-treated (15 min 37°C) mitochondria were incubated with Mmt2 or Coq2 precursors at 25°C, followed by Proteinase K treatment and SDS-PAGE.
(E) Model of Oxa1 function in the biogenesis of nuclear-encoded mitochondrial inner-membrane proteins. Cyto., cytosol; OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
See also Figure S4.
Thus, the MS-based depletion study represents an important source for finding mitochondrial inner-membrane proteins that depend on OXA, leading to the discovery of numerous nuclear-encoded substrates of the OXA export translocase. Since to date only Oxa1/Cox18 and Mdl1 have been known as authentic nuclear-encoded OXA substrates, this represents a major expansion of the role of OXA in inner-membrane biogenesis. However, two important points have to be considered. (1) The MS analysis reported here only represents a screening approach, and thus a biochemical analysis of inner-membrane sorting and assembly is required to define the molecular mechanisms of an OXA involvement. This is most evident in the case of the metabolite carriers of the inner membrane. The biogenesis pathway of carrier precursors is compromised in Oxa1-depleted mitochondria (Hildenbeutel et al., 2012; this study); however, not by a direct function of Oxa1 in carrier sorting and folding. We found that Oxa1 is crucial for the assembly pathway of the Tim18-Sdh3 module of the TIM22 carrier translocase, demonstrating that the export translocase is required for the proper assembly of this import translocase (Figure 4E). (2) To minimize false-positive identifications of OXA-dependent proteins, we used strict criteria for selecting the control mitochondria. It is thus conceivable that the number of OXA-dependent inner-membrane proteins is even larger. This is exemplified by the biochemical identification of OXA substrates such as Tim18, Mdl1, and Mmt1, which were not found by the MS analysis, but are related to proteins identified in the study.
The TIM23, PAM, and OXA machineries show a high versatility in the sorting of mitochondrial inner-membrane proteins. In the case of Sdh4, the first two TMs use the PAM/OXA-dependent import-export pathway, whereas TM3 is laterally released from the TIM23 complex into the inner membrane via the PAM/OXA-independent stop-transfer pathway (Figure S4C). In contrast, in the case of Mdl1, an opposite order of the two sorting pathways was observed (Bohnert et al., 2010). Thus, depending on the characteristics of sorted preproteins, lateral sorting via TIM23 and conservative sorting via OXA can be combined sequentially in order to properly assemble inner-membrane proteins.
In summary, the OXA export translocase plays a central role in the biogenesis of the mitochondrial inner membrane. In addition to its well-established role in the export of mitochondrial-encoded proteins (Hell et al., 2001, Bonnefoy et al., 2009), OXA promotes the proper sorting of numerous nuclear-encoded proteins with multiple TMs by two different mechanisms (Figure 4E). Inner-membrane precursors/domains, which are imported into the matrix by the presequence translocase/mtHsp70 motor machinery (TIM23-PAM), are subsequently exported by OXA. The OXA substrates characterized here and in previous studies have been conserved from bacteria to mitochondria in line with a conservative sorting mechanism (Herrmann et al., 1997, Hell et al., 1998, Funes et al., 2004, Bohnert et al., 2010), whereas the available evidence suggests that the metabolite carrier family has a eukaryotic origin (Palmieri, 2013). The large number of metabolite carriers do not use the TIM23-PAM-OXA pathway; however, two subunits of the carrier translocase TIM22 use this pathway. Thus, in OXA-depleted mitochondria, biogenesis and activity of the carrier translocase are strongly inhibited.
Experimental Procedures
Growth of Yeast and Isolation of Mitochondria
The Saccharomyces cerevisiae strains used in this study are listed in Table S2. Yeast cells were grown in YPS (1% [w/v] yeast extract, 2% [w/v] peptone, 2% [w/v] sucrose) or YPG (1% [w/v] yeast extract, 2% [w/v] peptone, 3% [v/v] glycerol) at 21°C or 30°C. Mitochondria were isolated by differential centrifugation and stored in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH [pH 7.2]) at −80°C. For MS analysis, cells were cultured in minimal medium (0.67% [w/v] yeast nitrogen base without amino acids, 0.69 g/l of CSM-ARG-LYS dropout medium [MP Biomedicals] and 2% [w/v] sucrose) containing either [13C6/15N2]lysine and [13C6/15N4]arginine (Eurisotop) (for YPH499 rho0) or natural lysine and arginine (for oxa1Δ cox18Δ; referred to as light amino acids). Cells were mixed in a ratio of 1:1 (based on their OD600) prior to isolation of mitochondria.
Import of Precursor Proteins into Mitochondria
Template DNA-encoding mitochondrial precursor proteins were amplified from genomic S. cerevisiae DNA using forward primers containing an SP6 promoter and reverse primers that bind at the 3′ end of the respective open reading frames. mRNA was obtained by in vitro transcription using the mMessage mMachine kit (Ambion). For purification of mRNA, the MEGAclear kit (Ambion) was employed. 35S-labeled proteins were synthesized in rabbit reticulocyte lysate (Promega) or wheat germ lysate (5 PRIME, for Mmt1). Import of radiolabeled precursor proteins into isolated yeast mitochondria was performed in the presence of 4 mM NADH, 4 mM ATP, and an ATP regenerating system (10 mM creatine phosphate and 0.2 mg/ml creatine kinase) in import buffer (3% [w/v] bovine serum albumin, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM KPi, 5 mM methionine and 10 mM MOPS-KOH [pH 7.2]) (Gebert et al., 2011, Ieva et al., 2014). Where indicated, mitochondria were subjected to an in vitro heat shock prior to the import reaction; they were resuspended in import buffer supplemented with 4 mM ATP and incubated for 12–15 min at 37°C. The import was performed at 25°C if not stated differently. For dissipation of Δψ, 1 μM valinomycin, 8 μM antimycin A, and 20 μM oligomycin (final concentrations) were added before the import reaction. Where indicated, a treatment with proteinase K (50 μg/ml for 15 min on ice) was performed after the import reaction, followed by the addition of PMSF (1 mM). Subsequently, mitochondria were washed and suspended in SEM buffer. Mitochondria were either solubilized in buffer containing 1% digitonin for blue native electrophoresis or were separated by SDS-PAGE, followed by digital autoradiography.
Protease Accessibility Assay for the Analysis of Membrane Insertion
Hypo-osmotic swelling was induced by 8-fold dilution of mitochondria (in SEM) with EM buffer (10 mM MOPS-KOH [pH 7.2], 1 mM EDTA); control mitochondria received SEM buffer. Where indicated, a treatment with proteinase K (7 μg/ml) or trypsin (8 μg/ml) was performed for 15 min on ice. Trypsin was inactivated by soybean trypsin inhibitor (30-fold excess). Mitochondria were pelleted, washed, and subjected to SDS-PAGE.
Data Analysis
Radiolabeled precursor proteins separated by gel electrophoresis were analyzed by digital autoradiography with the Storm systems 840 and 865, the ImageQuant software (GE Healthcare), the FLA 9000 image scanner and the Multi Gauge software (Fujifilm). Immunoblotting signals were detected using enhanced chemiluminescence and the LAS3000, LAS4000 Imager systems and the Multi Gauge software (Fujifilm). MS data were processed using MaxQuant version 1.3.0.5 and Andromeda. The results were generated and confirmed by using independent biological samples (different mitochondrial preparations, independent import and assembly experiments). Quantifications of independent assembly experiments (n = 3) are shown as mean ± SEM.
Author Contributions
S.B.S., J.H., S.O., C.S., S.G.S., J.V.-S., S.E.H., A.E.F., N.G., and M.B. performed experiments and analyzed data together with M.v.d.L., B.W., N.P., and N.W.; J.H.’s contributions include experiments on the sorting of Tim18 and Sdh4 via mtHsp70 and Oxa1 and assembly of the TIM22 complex; S.B.S.’s contributions include experiments on the specificity of oxa1-ts effects, membrane insertion of Sdh4, interaction of Sdh4 and Mmt2 with Oxa1, and the identification of different OXA substrates. N.W., N.P., B.W., M.B., and M.v.d.L. designed and supervised the project; S.B.S., J.H., S.O., C.S., and S.G.S. prepared the figures and tables; N.P. and N.W. wrote the manuscript; all authors discussed results from the experiments and commented on the manuscript.
Acknowledgments
We thank J.M. Herrmann, P. Rehling, and M. Andes for discussion, yeast strains, and basic import experiments and A. Schulze-Specking for expert technical assistance. Work included in this study has also been performed in partial fulfillment of the requirements for the doctoral theses of S.B.S., J.H., and S.G.S. at the University of Freiburg. This work was supported by the Deutsche Forschungsgemeinschaft (PF 202/8-1), the Sonderforschungsbereiche 746 and 1140, the Excellence Initiative of the German federal and state governments (EXC 294 BIOSS; GSC-4 Spemann Graduate School), the European Research Council (ERC) Consolidator Grant Number 648235, the German Academy of Sciences Leopoldina (LPDS 2013-08 to S.E.H.), and a Boehringer Ingelheim Fonds fellowship (to M.B.).
Published: May 10, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article at http://dx.doi.org/10.1016/j.cmet.2016.04.005.
Contributor Information
Nikolaus Pfanner, Email: nikolaus.pfanner@biochemie.uni-freiburg.de.
Nils Wiedemann, Email: nils.wiedemann@biochemie.uni-freiburg.de.
Supplemental Information
References
- Bohnert M., Rehling P., Guiard B., Herrmann J.M., Pfanner N., van der Laan M. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr. Biol. 2010;20:1227–1232. doi: 10.1016/j.cub.2010.05.058. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N., Fiumera H.L., Dujardin G., Fox T.D. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim. Biophys. Acta. 2009;1793:60–70. doi: 10.1016/j.bbamcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho S.C., Österberg M., Reichert A.S., Yamano K., Björkholm P., Endo T., von Heijne G., Kim H. TIM23-mediated insertion of transmembrane α-helices into the mitochondrial inner membrane. EMBO J. 2011;30:1003–1011. doi: 10.1038/emboj.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Yamano K. Multiple pathways for mitochondrial protein traffic. Biol. Chem. 2009;390:723–730. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- Funes S., Nargang F.E., Neupert W., Herrmann J.M. The Oxa2 protein of Neurospora crassa plays a critical role in the biogenesis of cytochrome oxidase and defines a ubiquitous subbranch of the Oxa1/YidC/Alb3 protein family. Mol. Biol. Cell. 2004;15:1853–1861. doi: 10.1091/mbc.E03-11-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funes S., Kauff F., van der Sluis E.O., Ott M., Herrmann J.M. Evolution of YidC/Oxa1/Alb3 insertases: three independent gene duplications followed by functional specialization in bacteria, mitochondria and chloroplasts. Biol. Chem. 2011;392:13–19. doi: 10.1515/BC.2011.013. [DOI] [PubMed] [Google Scholar]
- Gebert N., Gebert M., Oeljeklaus S., von der Malsburg K., Stroud D.A., Kulawiak B., Wirth C., Zahedi R.P., Dolezal P., Wiese S. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol. Cell. 2011;44:811–818. doi: 10.1016/j.molcel.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Glick B.S., Brandt A., Cunningham K., Müller S., Hallberg R.L., Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Schmidt B., Wachter E., Weiss H., Neupert W. Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell. 1986;47:939–951. doi: 10.1016/0092-8674(86)90809-3. [DOI] [PubMed] [Google Scholar]
- Hell K., Herrmann J.M., Pratje E., Neupert W., Stuart R.A. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl. Acad. Sci. USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Neupert W., Stuart R.A. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J.M., Neupert W., Stuart R.A. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 1997;16:2217–2226. doi: 10.1093/emboj/16.9.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildenbeutel M., Theis M., Geier M., Haferkamp I., Neuhaus H.E., Herrmann J.M., Ott M. The membrane insertase Oxa1 is required for efficient import of carrier proteins into mitochondria. J. Mol. Biol. 2012;423:590–599. doi: 10.1016/j.jmb.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Huang L.S., Shen J.T., Wang A.C., Berry E.A. Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the “oxaloacetate-inhibited” state. Biochim. Biophys. Acta. 2006;1757:1073–1083. doi: 10.1016/j.bbabio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R., Schrempp S.G., Opaliński L., Wollweber F., Höß P., Heißwolf A.K., Gebert M., Zhang Y., Guiard B., Rospert S. Mgr2 functions as lateral gatekeeper for preprotein sorting in the mitochondrial inner membrane. Mol. Cell. 2014;56:641–652. doi: 10.1016/j.molcel.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Kumazaki K., Chiba S., Takemoto M., Furukawa A., Nishiyama K., Sugano Y., Mori T., Dohmae N., Hirata K., Nakada-Nakura Y. Structural basis of Sec-independent membrane protein insertion by YidC. Nature. 2014;509:516–520. doi: 10.1038/nature13167. [DOI] [PubMed] [Google Scholar]
- Meier S., Neupert W., Herrmann J.M. Proline residues of transmembrane domains determine the sorting of inner membrane proteins in mitochondria. J. Cell Biol. 2005;170:881–888. doi: 10.1083/jcb.200505126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W., Herrmann J.M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Palmieri F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Aspects Med. 2013;34:465–484. doi: 10.1016/j.mam.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Park K., Botelho S.C., Hong J., Österberg M., Kim H. Dissecting stop transfer versus conservative sorting pathways for mitochondrial inner membrane proteins in vivo. J. Biol. Chem. 2013;288:1521–1532. doi: 10.1074/jbc.M112.409748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K., Jung S.J., Kim H., Kim H. Mode of membrane insertion of individual transmembrane segments in Mdl1 and Mdl2, multi-spanning mitochondrial ABC transporters. FEBS Lett. 2014;588:3445–3453. doi: 10.1016/j.febslet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Preuss M., Leonhard K., Hell K., Stuart R.A., Neupert W., Herrmann J.M. Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J. Cell Biol. 2001;153:1085–1096. doi: 10.1083/jcb.153.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Wagener N., Ackermann M., Funes S., Neupert W. A pathway of protein translocation in mitochondria mediated by the AAA-ATPase Bcs1. Mol. Cell. 2011;44:191–202. doi: 10.1016/j.molcel.2011.07.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.