Abstract
This study characterized EMRSA-15 isolates obtained from patients in Kuwait hospitals for their genotypic relatedness, antibiotic resistance and carriage of virulence genes using pulsed-field gel electrophoresis (PFGE), coagulase serotyping, SCCmec subtyping, spa typing, multilocus sequence typing and DNA microarray. The isolates were resistant to trimethoprim (75.6%), ciprofloxacin (29.7%), erythromycin and clindamycin (24.3%), tetracycline (19.0%), and gentamicin and kanamycin (21.6%). All 37 isolates belonged to sequence type (ST) 22, coagulase type XI, three PFGE types and eight subtypes, ten spa types including t223 (51.3%), t852 (13.5%), t032 (8.1%), t790 (8.1%), t3107 (5.4%) and one each of t309, t2251, t3935, t5708 and t5983. Twenty-six isolates (70.2%) carried SCCmec IVa, eight isolates carried SCCmec IV and three isolates carried SCCmec IVh. All isolates carried agr1, cap5 and egc gene cluster (seg, sei, selm, seln, selo, and selu). tst (toxic shock syndrome toxin) was detected in 23 isolates. Eight isolates (21.6%) were positive for Panton-Valentine leukocidin (PVL). Genotypic analysis revealed that 62.1% of the isolates comprising ST22-IVa-t223 (51.3%) and ST22-IVa-t309/t2251/t3935/t5708 (10.8%) were CC22-[tst1+] UK EMRSA-15/Middle Eastern variant, 21.6% were CC22-PVL+ EMRSA-15 variant and 16.2% were CC22-UK EMRSA-15/Barnim clone. These results show that the tst1 positive-ST22-IVa-t223 (Middle Eastern variant) and the CC22-PVL+ EMRSA-15 variant were the dominant EMRSA-15 variants in Kuwait hospitals.
Keywords: DNA microarray, EMRSA-15, MLST, MRSA, SCCmec typing, spa typing
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) strains continue to be isolated from both healthcare- and community-associated infections in different parts of the world. The increase in the number of MRSA infections reported worldwide has been accompanied by changes in the characteristics of MRSA strains emerging in different parts of the world [1]. Consequently, epidemiologic typing using a combination of phenotypic and genotypic typing methods has contributed to our understanding of changes in the clonal distribution of MRSA isolated in different geographical locations over time. Whereas MRSA isolates obtained in the 1960s (early or classic MRSA) were usually susceptible to majority of non-β-lactam antibiotics, those isolated in the late 1970s and beyond were multiply resistant to non-β-lactam antibiotics and were described as epidemic MRSA (EMRSA) because of their capacity to spread extensively and cause serious infections among hospitalized patients [1], [2]. Epidemiologic typing by Kerr et al. [3] and Aucken et al. [4] identified 17 different EMRSA clones, designated EMRSA-1 to EMRSA-17, in the United Kingdom.
The EMRSA-15 clone was first characterized in England on the basis of phage typing (a weak lysis pattern with phage 75), susceptibility to antimicrobial agents and failure to produce urease [5]. The urease-negative phenotype resulted from a single nucleotide deletion within the UreC gene, leading to a frameshift inactivation of the alpha subunit of the urease gene [6]. Subsequently, multilocus sequence typing of the EMRSA-15 strains classified them as multilocus sequence (ST) type 22 (ST22) [7].
EMRSA-15 strains are an important cause of nosocomial bacteraemia in many UK and Irish hospitals [8], and they are a major pathogen in other European healthcare facilities [9], [10]. EMRSA-15 strains have also been isolated in Australia [11], Singapore [12], India [13], [14], Malaysia [15], Kuwait [16], Saudi Arabia [17], United Arab Emirates [18], Qatar [19] and Oman [20]. The survival and widespread transmission of EMRSA-15 strains have been attributed to genetic changes within the bacterial genome due to the acquisition of antibiotic resistance and virulence genes and adaptation to the healthcare environment [6].
Molecular characterization of EMRSA-15 strains isolated in the Gulf Cooperative Council (GCC) countries have revealed different genetic backgrounds with different antibiotic susceptibility patterns. Whereas EMRSA-15 isolates from the United Arab Emirates belonged to spa types t032 or t005 [18], the majority of recent isolates from Qatar [19] and Oman [20] belong to t852.
In this study, we investigated EMRSA-15 isolates obtained in Kuwait public hospitals using a combination of molecular typing methods to establish their genetic relatedness to EMRSA-15 isolated in the United Kingdom and other GCC countries.
Materials and Methods
MRSA isolates
A total of 37 EMRSA-15 strains, isolated in the years 2005 (four isolates representing four pulsed-field gel electrophoresis (PFGE) patterns) and 2010 (33 isolates), were included in this study. These MRSA isolates were among clinical isolates submitted to the MRSA Reference Laboratory, Kuwait, for molecular typing. The MRSA isolates were obtained as part of routine bacteriology services in the individual hospital laboratories and were archived at the MRSA Reference Laboratory, Kuwait. The MRSA isolates were isolated at ten hospitals: Mubarak (nine isolates), Al Sabah (seven isolates), Ibn Sina (five isolates), maternity hospital (five isolates), Adan (three isolates), Al Razi (three isolates), Farwaniya (two isolates), Jahra (one isolates), Armed Forces (one isolate) and Chest Disease Hospital (one isolates). Isolates were identified by cultural characteristics, Gram staining, and positive tube coagulase and DNase tests. The isolates were preserved in glycerol 40% (v/v) in brain heart infusion broth (Oxoid, Basingstoke, UK) at −80°C. They were recovered by subculturing in brain–heart infusion broth at 37°C for 24 hours followed by two further subcultures on brain–heart infusion agar. Preliminary identification of MRSA isolates as EMRSA-15 was based on negative urease test [5] and carriage of SCCmec type IV genetic element.
Urease production
Urease production was detected on Christensen urea agar slope after 72 hours' incubation at 35°C as described previously [5], [16].
Antibacterial susceptibility testing
Antibacterial susceptibility testing was performed by the disk diffusion method [21] with the following antimicrobial disks (Oxoid): benzyl penicillin (10 U), cefoxitin (30 μg), kanamycin (30 μg), mupirocin (200 and 5 μg), gentamicin (10 μg), erythromycin (15 μg), clindamycin (2 μg), chloramphenicol (30 μg), tetracycline (10 μg), trimethoprim (2.5 μg), fusidic acid (10 μg), rifampicin (5 μg), ciprofloxacin (5 μg), teicoplanin (30 μg) and linezolid (30 μg). Minimum inhibitory concentration for cefoxitin, vancomycin and teicoplanin were determined with Etest strips (bioMérieux, Marcy l’Étoile, France) according to the manufacturer's instructions. S. aureus strain ATCC 25923 was used as a quality control strain for susceptibility testing. The Dtest was used to test for inducible resistance to clindamycin.
Molecular typing of isolates
Isolates were typed by PFGE, coagulase gene typing, SCCmec typing, spa typing and multilocus sequence typing (MLST). PFGE was performed on all 37 MRSA isolates as described previously [22]. DNA for PCR amplification was isolated and purified as described previously [20]. Coagulase typing was performed as described previously [23]. For the detection of coagulase type XI, primer pair (coaS-F) 5′-TGGGCAATTACATTTTG GAG-3′ and (coaS-XI-R) 5′-TCGTTTGGGTAAGTTGCTTT-3′ (395 bp) were designed and used in this study. PCR amplification was carried out on a Gene AMP PCR System 9700 instrument (Thermo Fisher Scientific Life Sciences, Waltham, MA, USA) in a total volume of 25 μL of reaction mixture containing 12.5 μL of AmpliTaq Gold master mix (Roche, Basel, Switzerland), 50 pmol of each primer and 2 μL of extracted DNA. The PCR reaction was 95°C for 15 minutes, followed by 30 cycles with 95°C for 30 seconds, 56°C for 40 seconds, 72°C for 30 seconds and a final extension step of 72°C for 5 minutes. PCR products were analysed by agarose gel electrophoresis using 2% (w/v) agarose in Tris-EDTA buffer.
SCCmec typing was performed by PCR assays as described previously [24], [25]. spa typing was performed as described by Harmsen et al. [26]. Clonal relatedness of the spa types was determined by the BURP (Based Upon Repeat Pattern) algorithm as described by Mellmann et al. [27]. MLST was performed on all 37 isolates as described by Enright et al. [7].
Detection of genes for antibiotic resistance and virulence
DNA microarray analysis using Identibac S. aureus Genotyping Kit 2 (Alere Technology, Jena, Germany) was used following protocols provided by the manufacturer. Data generated were analysed using the ArrayMate software and the ArrayMate Reader (Alere Technology), which assigned MRSA isolates to STs and clonal complexes (CCs) by comparing each isolate to STs and CCs of previously characterized isolates in the ArrayMate database [28], [29].
Results
The MRSA isolates were obtained from nasal swabs (n = 18), skin and soft tissues (n = 10), sputum (n = 3), groin (n = 2), ear (n = 2), and vaginal swab and axilla (n = 1 each). The patients consisted of 23 men and 13 women. The sex of one patient was not specified.
All 37 isolates were urease negative and were susceptible to vancomycin and teicoplanin (minimum inhibitory concentration ≤1.5 mg/L), linezolid, chloramphenicol, rifampicin, mupirocin and fusidic acid but resistant to trimethoprim (n = 28; 75.6%), ciprofloxacin (n = 11; 29.7%), erythromycin and clindamycin (n = 9; 24.3%), gentamicin and kanamycin (n = 8; 21.6%), and tetracycline (n = 7; 19.0%).
Molecular typing of MRSA isolates
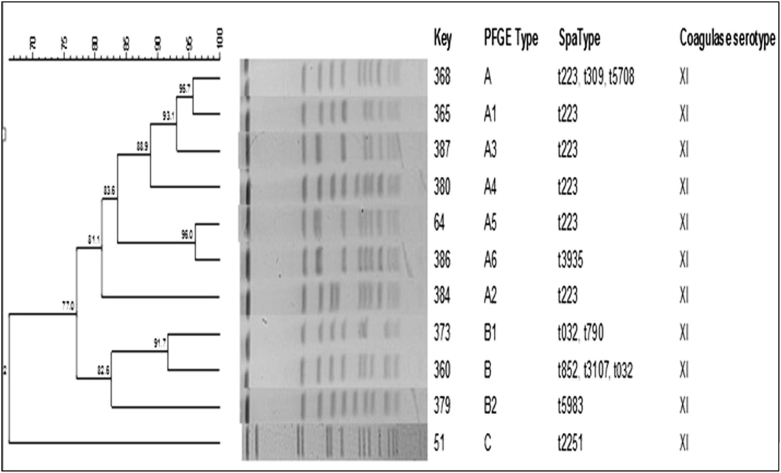
The isolates were grouped into three PFGE patterns, designated types A (22 isolates), B (14 isolates) and C (one isolate), as summarized in Table 1 and Fig. 1.
Table 1.
Source and characteristics of MRSA isolates in Kuwait hospitals
| Strain No. | Year | Hospital | Gender | Source | PFGE | SCCmec type | spa type |
|---|---|---|---|---|---|---|---|
| Kuwait_70 | 2005 | Armed Forces | M | Sputum | B1 | IVh | t032 |
| Kuwait_64 | 2005 | Mubarak | M | Swab | A5 | IVa | t223 |
| Kuwait_74 | 2005 | Mubarak | M | Wound | A3 | IVa | t223 |
| Kuwait_51 | 2005 | Mubarak | M | Nasal | C | IVa | t2251 |
| Kuwait_270 | 2010 | Farwaniya | M | Nasal | B | IVh | t032 |
| Kuwait_376 | 2010 | CDH | M | Wound | B1 | IVh | t032 |
| Kuwait_359 | 2010 | Sabah | F | Nasal | A | IVa | t223 |
| Kuwait_365 | 2010 | Maternity | F | Nasal | A1 | IVa | t223 |
| Kuwait_366 | 2010 | Al-Razi | M | Wound | A1 | IVa | t223 |
| Kuwait_368 | 2010 | Mubarak | M | Nasal | A | IVa | t223 |
| Kuwait_369 | 2010 | Sabah | M | Nasal | A | IVa | t223 |
| Kuwait_370 | 2010 | Ibn-Sina | M | Nasal | A | IVa | t223 |
| Kuwait_371 | 2010 | Al-Razi | F | Nasal | A | IVa | t223 |
| Kuwait_372 | 2010 | Jahra | F | Ear | A | IVa | t223 |
| Kuwait_378 | 2010 | Mubarak | M | Nasal | A | IVa | t223 |
| Kuwait_380 | 2010 | Maternity | — | Nasal | A4 | IVa | t223 |
| Kuwait_384 | 2010 | Ibn-Sina | M | Nasal | A2 | IVa | t223 |
| Kuwait_385 | 2010 | Ibn-Sina | M | Axilla | A2 | IVa | t223 |
| Kuwait_387 | 2010 | Mubarak | F | Nasal | A3 | IVa | t223 |
| Kuwait_388 | 2010 | Mubarak | M | Bed sore | A | IVa | t223 |
| Kuwait_391 | 2010 | Sabah | M | Nasal | A2 | IVa | t223 |
| Kuwait_398 | 2010 | Mubarak | M | Nasal | A1 | IVa | t223 |
| Kuwait_400 | 2010 | Sabah | F | Ear | A | IVa | t223 |
| Kuwait_229 | 2010 | Maternity | F | Nasal | A | IVa | t309 |
| Kuwait_361 | 2010 | Ibn-Sina | M | Swab | B | IV | t3107 |
| Kuwait_363 | 2010 | Sabah | M | Sputum | B | IV | t3107 |
| Kuwait_386 | 2010 | Al-Razi | M | Ulcer | A6 | IVa | t3935 |
| Kuwait_377 | 2010 | Sabah | M | Groin | A | IVa | t5708 |
| Kuwait_379 | 2010 | Adan | M | Nasal | B2 | IV | t5983 |
| Kuwait_373 | 2010 | Maternity | F | HVS | B1 | IVa | t790 |
| Kuwait_374 | 2010 | Sabah | M | Nasal | B1 | IVa | t790 |
| Kuwait_375 | 2010 | Ibn-Sina | F | Sputum | B1 | IVa | t790 |
| Kuwait_360 | 2010 | Adan | F | Abscess | B | IV | t852 |
| Kuwait_362 | 2010 | Farwaniya | F | Nasal | B | IV | t852 |
| Kuwait_364 | 2010 | Mubarak | M | Groin | B | IV | t852 |
| Kuwait_367 | 2010 | Adan | F | Pus | B | IV | t852 |
| Kuwait_399 | 2010 | Maternity | F | Pus | B | IV | t852 |
CDH, Chest Disease hospital; MRSA, methicillin-resistant Staphylococcus aureus; HVS, high vaginal swab; PFGE, pulsed-field gel electrophoresis.
Fig. 1.
Dendogram of pulsed-field gel electrophoresis patterns of ST22-IV-MRSA isolates. MRSA, methicillin-resistant Staphylococcus aureus.
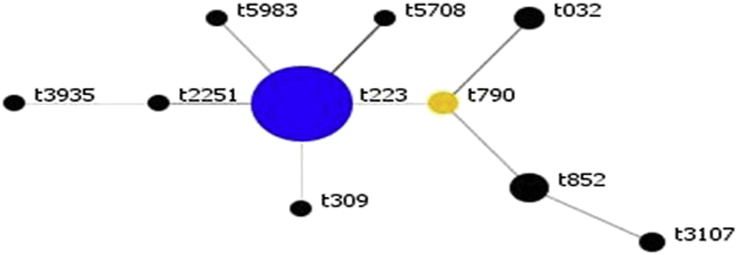
All 37 isolates belonged to coagulase type XI, ST22, and carried SCCmec IV genetic element. SCCmec subtyping distinguished the isolates into subtypes IVa (n = 26, 70.2%) and IVh (n = 3, 8.1%). Eight isolates (21.6%) had no subtypes. The isolates belonged to ten spa types with 19 isolates (51.3%) assigned to spa type t223, followed by t852 (n = 5; 13.5%), t790 (n = 3; 8.1%), t032 (n = 3; 8.1%) and t3107 (n = 2; 5.4%). spa types t5983, t309, t5708, t3935 and t2251 occurred in one isolate each. BURP analysis defined one spa CC (spa-CC) 223 with the founder as t223 (Fig. 2).
Fig. 2.
Population snapshot for MRSA BURP analysis. BURP grouping using default parameters resulted in one spa CC. Each dot represents unique spa type. Diameter of dot is proportional to quantity of corresponding spa type. Blue dots represent putative founder (i.e. spa type with highest score within CC); yellow dots, subfounder with second highest score. BURP, Based Upon Repeat Pattern); CC, clonal complex; MRSA, methicillin-resistant Staphylococcus aureus.
A combination of the molecular typing results revealed that the isolates were ST22-IVa-t223 (n = 19; 51.3%), ST22-IV-t852 (n = 5; 13.5%), ST22-IVa-t790 (n = 3; 8.1%), ST22-IVh-t032 (n = 3; 8.1%) and ST22-IV-t3107 (n = 2; 5.4%). ST22-IVa-t309, ST22-IVa-t2251, ST22-IVa-t3935, ST22-IVa-t5708 and ST22-IV-t5983 occurred in one isolate each (Table 2).
Table 2.
Molecular characteristics of ST22 MRSA isolates
| Year | No. of isolates | Strain definition | spa type | Antibiotic resistance (n) | Antibiotic resistance genes | Toxins encoding genes | Miscellaneous genes |
|---|---|---|---|---|---|---|---|
| UK EMRSA-15/Barnim | |||||||
| 2005 | 1 | ST22-MRSA-IVh | t032 | E, CC, CIP | ermC | seb, sec sel, egc | scn, hla, hlIII, hlgA |
| 2010 | 2 | ST22-MRSA-IVh | t032 | E (1), CC (1), CIP (2), W(1) | ermC | egc | scn, hla, hlgA |
| 2010 | 3 | ST22-MRSA-IVa | t790 | E (2), CC (2) | ermC | sec, egc | scn |
| UK EMRSA-15/Middle Eastern variant [tst+] | |||||||
| 2005 | 2 | ST22-MRSA-IVa | t223 | E (1), CC (1), TE (2), W (2) | ermC, tetK, dfrS1 | tst, egc | scn, hla, hlgA |
| 2010 | 17 | ST22-MRSA-IVa | t223 | TE (4), W(16) | tetK, dfrS1 | tst, egc | scn, hla, hlgA |
| 2010 | 1 | ST22-MRSA-IVa | t309 | W | dfrS1 | tst, egc | scn, hla, hlgA |
| 2005 | 1 | ST22-MRSA-IVa | t2251 | TE | tetK | tst, egc | scn, hla, hlgA |
| 2010 | 1 | ST22-MRSA-IVa | t3935 | W | dfrS1 | tst, sea egc | scn, hla |
| 2010 | 1 | ST22-MRSA-IVa | t5708 | W | tetK, dfrS1 | tst, egc | scn |
| CC22-MRSA-IV-PVL+ | |||||||
| 2010 | 5 | ST22-MRSA-IV | t852 | CN (5), K (5), E (1), CC (1), TOB (4), W (3), CIP (5) | aacA-aphD, aadD | PVL, egc | |
| 2010 | 2 | ST22-MRSA-IV | t3107 | CN (2), K (2), TOB (1), E (2), CC (2), W (2), CIP (2) | ermC, aacA-aphD, aadD, dfrS1 | PVL, egc | scn, hla, hlgA |
| 2010 | 1 | ST22-MRSA-IV | t5983 | E, CC, W, CIP, CN, K, TOB | ermC, aacA-aphD, aadD, dfrS1 | PV, egc | scn, hla |
All isolates carried hemolysin delta (hld) egc gene cluster (seg, sei, selm, seln, selo, selu).
CC, clindamycin; chp, chemotaxis-inhibiting protein; CIP, ciprofloxacin; CN, gentamicin; E, erythromycin; hl, putative membrane protein; hla, haemolysin alpha; hlb, haemolysin beta; hlgA, haemolysin gamma, component A; hlIII, putative membrane protein; K, kanamycin; MRSA, methicillin-resistant Staphylococcus aureus; PVL, Panton Valentine leukocidin; sak, staphylokinase; scn, staphylococcal complement inhibitor; TE, tetracycline; TOB, tobramycin; tst, toxic shock syndrome toxin; W, trimethoprim.
DNA microarray analysis of isolates
DNA microarray analysis revealed that the 37 isolates were positive for genes encoding accessory gene regulator type 1 (agrI), capsular polysaccharide type 5 (cap5), staphylococcal enterotoxin egc gene cluster (seg, sei, selm, seln, selo and selu), haemolysin beta (hlb), putative membrane protein (hl), staphylokinase (saK) and chemotaxis inhibition protein (chp) but differed in the carriage of sea, seb, sec, tst (toxic shock syndrome toxin), hlgA (haemolysin gamma A) and hla (haemolysin alpha). Eight isolates (21.6%) consisting of t852, t3107 and t5983 were positive for Panton-Valentine leukocidin (PVL) (Table 2). None of the isolates was positive for sed.
As shown in Table 2, DNA microarray analysis classified the isolates into three groups: CC22-UK EMRSA-15/Barnim-MRSA clone, CC22-[tst1+] UK EMRSA-15/Middle Eastern variant and CC22-PVL+ UK EMRSA-15. The CC22-UK EMRSA-15/Barnim-MRSA clone comprised six isolates, which belonged to SCCmec-IVh-t032 (three isolates) and SCCmec IVa-t790 (three isolates), that were resistant to erythromycin and clindamycin and carried ermC. The t790 isolates were positive for sec but lacked hla and hlgA, which were present in t032 isolates.
The CC22-[tst1+] UK EMRSA-15/Middle Eastern variant consisted of 23 isolates belonging to spa type t223 (n = 19) and one isolate each of t309, t2251, t3935 and t5708. These isolates harboured tst and were resistant to trimethoprim but differed in their resistance to tetracycline, erythromycin and clindamycin and carriage of virulence genes.
The CC22-PVL+ UK EMRSA-15 variant consisted of eight isolates that belonged to spa types t852 (n = 5), t3107 (n = 2) and t5983 (n = 1). These isolates were PVL positive and were resistant to gentamicin, kanamycin, tobramycin, erythromycin, clindamycin, trimethoprim and ciprofloxacin. The t852 isolates were negative for scn, hla and hlgA.
Discussion
The results of this study provide insight into the epidemiology of EMRSA-15 isolates in Kuwait hospitals. Similar to EMRSA-15 isolates reported elsewhere [6], [7], [29], the isolates investigated in this study belonged to ST22 and carried a type IV SCCmec genetic element. However, molecular subtyping and DNA microarray analysis revealed differences in their genetic backgrounds, suggesting multiple origins for EMRSA-15 in Kuwait hospitals.
The study revealed that isolates fitting the CC22-IV[tst1+] UK EMRSA-15/Middle Eastern variant, consisting of spa types t223 (51.3%), t309 (2.7%), t2251 (2.7%), t3935 (2.7%) and t5708 (2.7%), constituted the dominant EMRSA-15 variant in Kuwait hospitals in 2010. In addition, the t223 isolates widespread in the country evidenced by their isolation in six of the ten hospitals studied. The t223 isolates had antibiotic resistance and virulence profiles similar to PVL-negative, tst-positive ST22-IV-t223 isolates reported to have colonized children and parents in the Gaza Strip [30], [31] as well as PVL-negative, tst-positive ST22-IV-MRSA-t223 recovered from healthy individuals in Jordan [32], making it the dominant EMRSA-15 clade in the Middle East.
The multiresistant CC22-MRSA-IV-PVL+ variant, consisting of spa types t852 (13.5%), t3107 (5.4%) and t5983 (2.7%), was the second most common EMRSA-15 variant in this study. ST22-IV-MRSA-t852 isolates were reported among healthy carriers and patients in Indian hospitals as early as 2008 [13], [33], which may represent the origin of this EMRSA-15 variant. Since then, PVL-positive, multiresistant spa type t852 isolates have been reported among ST22-IV MRSA isolates in Saudi Arabia [17], Qatar [19] and Oman [20], suggesting an increasing transmission of this variant in the Gulf Cooperative Council countries. The t852 isolates were also reported among of ST22-IV MRSA isolated in Zurich, Switzerland, between 2012 and 2014 [34], pointing to their spread in European hospitals.
Surprisingly, the ST22-IVh-t032 isolates related to the UK EMRSA-15/Barnim MRSA clone [10] were less common in this study. Similarly, ST22-IV MRSA related to UK EMRSA-15/Barnim MRSA clone was detected only in 8.9% of MRSA in a hospital in Riyadh, Saudi Arabia [17]. Also, t032, which was the dominant spa type of ST22-IV-MRSA in a United Arab Emirates hospital in 2003, was replaced by t005 in 2008, with none of the 2008 isolates carrying t032 [18]. Furthermore, none of ST22-IV-MRSA reported recently in Qatar [19] and Oman [20] belonged to t032, suggesting a displacement of t032 isolates in Kuwait and other GCC hospitals, in contrast to its continued spread in Europe [8], [10], [29], Malaysia [15] and Singapore [12]. Similar to t032, t790 isolates, which were detected in small numbers in this study, constituted the dominant EMRSA-15 spa type in central Iran [35]. These studies highlight the genetic diversity of ST22-IV subtypes in different countries and the importance of molecular subtyping in understanding their epidemiology.
The UK EMRSA-15/Barnim clone has been associated with invasive infections and has been the dominant cause of bloodstream infections in European countries [3], [8]. In contrast, the ST22 isolates in this study were obtained mostly from skin and soft tissue infections and colonization sites. Similarly, the majority of ST22-IV isolates reported from Indian hospitals [33] and day care centres in the United States [36] were from carriers, probably reflecting the genetic diversity observed in this study and the ability of the clones to survive and proliferate under different environments which support their global spread.
All ST22-IV-MRSA isolates in this study possessed egc (seg, sei, selm, seln, selo and selu), as has been reported in other studies [29], [37], [38], implying that egc is a major virulence factor for ST22-IV-MRSA isolates. However, the PVL-positive t852 isolates lacked genes for staphylococcal complement inhibitor (scn), alpha haemolysin (hla) and the A component of haemolysins gamma (hlgA), highlighting genetic changes in the emerging variant.
The majority of the ST22-IV-MRSA isolates were resistant to trimethoprim, erythromycin and clindamycin, as has been reported in ST22-IV-MRSA isolates obtained in Ireland [39].
Conclusions
This study revealed that CC22-IV[tst1+] UK EMRSA-15/Middle Eastern variant is the dominant EMRSA-15 variant in Kuwait, followed by the PVL-positive, multiresistant t852 variant, with only few of the isolates related to the European EMRSA-15/Barnim variant of spa type t032. The presence of a mixed population of MRSA isolates poses unique problems for infection control. The study has enriched our understanding of the epidemiology of EMRSA-15 in Kuwait, emphasizing the need for continuous surveillance of MRSA in healthcare facilities to detect changes in their clonal composition and distribution.
Acknowledgements
Supported by grant YM 02/12 and Research Core Facility project SRUL02/13 from Kuwait University Research Sector. S. Boswihi received a graduate student fellowship from the College of Graduate Studies, Kuwait University, Kuwait. This study was presented as a poster at the 115th general meeting of the American Society for Microbiology, New Orleans, LA, USA, 30 May–2 June 2015.
Conflict of Interest
None declared.
References
- 1.Grundman H., Aanensen D.M., van den Wijngaard C.C., Spratt B.G., Harmsen D., Friedrich A.W. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe. A molecular epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grubb W.B. Genetics of MRSA. Rev Med Microbiol. 1988;9:153–162. [Google Scholar]
- 3.Kerr S., Kerr G.E., Mackintosh C.A., Marples R.R. A survey of methicillin-resistant Staphylococcus aureus affecting patients in England and Wales. J Hosp Infect. 1990;16:35–48. doi: 10.1016/0195-6701(90)90047-r. [DOI] [PubMed] [Google Scholar]
- 4.Aucken H.M., Ganner M., Murchan S., Cookson B.D., Johnson A.P. A new UK strain of epidemic methicillin-resistant Staphylococcus aureus (EMRSA-17) resistant to multiple antibiotics. J Antimicrob Chemother. 2002;50:171–175. doi: 10.1093/jac/dkf117. [DOI] [PubMed] [Google Scholar]
- 5.Richardson J.F., Reith S. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J Hosp Infect. 1993;25:45–52. doi: 10.1016/0195-6701(93)90007-m. [DOI] [PubMed] [Google Scholar]
- 6.Holden M.T., Hsu L.Y., Kurt K., Weinert L.A., Mather A.E., Harris S.R. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013;23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright M.C., Day N.P.J., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson A.P., Aucken H.M., Cavendish S., Ganner M., Wale M.C., Warner M. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS) J Antimicrob Chemother. 2001;48:143–144. doi: 10.1093/jac/48.1.143. [DOI] [PubMed] [Google Scholar]
- 9.Witte W., Enright M., Schmitz F.J., Cuny C., Braulke C., Heuck D. Characteristics of a new epidemic MRSA in Germany ancestral to United Kingdom EMRSA 15. Int J Med Microbiol. 2001;290:677–682. doi: 10.1016/S1438-4221(01)80006-0. [DOI] [PubMed] [Google Scholar]
- 10.Ghebremedhin B., Konig W., Witte W., Hardy K.J., Hawkey P.M., Konig B. Subtyping of ST22-MRSA-IV (Barnim epidemic MRSA strain) at a university clinic in Germany from 2002 to 2005. J Med Microbiol. 2007;56:365–375. doi: 10.1099/jmm.0.46883-0. [DOI] [PubMed] [Google Scholar]
- 11.Pearman J.W., Coomb G.W., Grubb W.B., O’Brien F. A British epidemic strain of methicillin-resistant Staphylococcus aureus (UK EMRSA-15) in Western Australia. Med J Aust. 2001;174:662. doi: 10.5694/j.1326-5377.2001.tb143485.x. [DOI] [PubMed] [Google Scholar]
- 12.Hsu L.Y., Koh T.H., Singh K., Kang M.L., Kurup A., Tan B.H. Dissemination of multisusceptible methicillin-resistant Staphylococcus aureus in Singapore. J Clin Microbiol. 2005;43:2923–2925. doi: 10.1128/JCM.43.6.2923-2925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadig S., Raju S.R., Arakere G. Epidemic meticillin-resistant Staphylococcus aureus (EMRSA-15) variants detected in healthy and diseased individuals in India. J Med Microbiol. 2010;59:815–821. doi: 10.1099/jmm.0.017632-0. [DOI] [PubMed] [Google Scholar]
- 14.Dhawan B., Rao C., Udo E.E., Gadepalli R., Vishnubhyatla S., Kapil A. Dissemination of methicillin-resistant Staphylococcus aureus SCCmec type IV and SCCmec type V epidemic clones in a tertiary hospital: challenge to infection control. Epidemiol Infect. 2015;143:343–353. doi: 10.1017/S095026881400065X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghaznavi-Rad E., Nor Shamsudin M., Sekawi Z., Khoon L.Y., Aziz M.N., Hamat R.A. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J Clin Microbiol. 2010;48:867–872. doi: 10.1128/JCM.01112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Udo E.E., Al-Sweih N., Noronha B. Characterization of non multiresistant methicillin-resistant Staphylococcus aureus (including EMRSA-15) in Kuwait Hospitals. Clin Microbiol Infect. 2006;12:262–269. doi: 10.1111/j.1469-0691.2005.01350.x. [DOI] [PubMed] [Google Scholar]
- 17.Monecke S., Skakni L., Hasan R., Ruppelt A., Ghazal S.S., Hakawi A. Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol. 2012;12:146. doi: 10.1186/1471-2180-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnevend A., Blair I., Alkaabi M., Jumaa P., al Haj M., Ghazawi A. Change in methicillin-resistant Staphylococcus aureus clones at a tertiary care hospital in the United Arab Emirates over a 5-year period. J Clin Pathol. 2012;65:178–182. doi: 10.1136/jclinpath-2011-200436. [DOI] [PubMed] [Google Scholar]
- 19.El-Mahdy T.S., El-Ahmady M., Goering R.V. Molecular characterization of MRSA isolated over a two year period in a Qatari hospital from multinational patients. Clin Microbiol Infect. 2014;20:169–173. doi: 10.1111/1469-0691.12240. [DOI] [PubMed] [Google Scholar]
- 20.Udo E.E., Al-Lawati B.A.H., Al-Muharmi Z., Thukral S.S. Genotyping of methicillin-resistant Staphylococcus aureus in the Sultan Qaboos University Hospital, Oman reveals the dominance of a Panton-Valentine leukocidin–negative ST6-IV/t304 clone. New Microbes New Infect. 2014;2:100–105. doi: 10.1002/nmi2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standard Institute . Vol. 32, no. 3. CLSI; Wayne, PA: 2012. (Performance standards for antimicrobial susceptibility testing). 22nd informational supplement M100-S22. [Google Scholar]
- 22.Udo E.E., Farook V.S., Mokadas E.M., Jacob L.E., Sanyal S.C. Molecular fingerprinting of mupirocin-resistant Staphylococcus aureus from a burn unit. Int J Infect Dis. 1999;3:82–87. doi: 10.1016/s1201-9712(99)90014-0. [DOI] [PubMed] [Google Scholar]
- 23.Hirose M., Kobayashi N., Ghosh S., Paul S.K., Shen T., Urushibara N. Identification of staphylocoagulase genotypes I–X and discrimination of type IV and V subtypes by multiplex PCR assay for clinical isolates of Staphylococcus aureus. Jpn J Infect Dis. 2010;63:257–263. [PubMed] [Google Scholar]
- 24.Oliveira D.C., de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang K., McClure J.A., Elsayed S., Louie T., Conly J.M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harmsen D., Claus H., Witte W., Rothanger J., Claus H., Turnwald D. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellmann A., Weniger T., Berssenbrügge C., Rothganger J., Sammeth M., Stoye J. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based upon spa polymorphisms. BMC Microbiol. 2007;7:98. doi: 10.1186/1471-2180-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monecke S., Slickers P., Ehricht R. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol Med Microbiol. 2008;53:237–251. doi: 10.1111/j.1574-695X.2008.00426.x. [DOI] [PubMed] [Google Scholar]
- 29.Monecke S., Coombs G., Shore A.C., Coleman D.C., Akpaka P., Borg M. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011;6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biber A., Abuelaish I., Rahav G., Raz M., Cohen L., Valinsky L. A typical hospital-acquired methicillin-resistant Staphylococcus aureus clone is widespread in the community in the Gaza strip. PLoS One. 2012;7:e42864. doi: 10.1371/journal.pone.0042864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al Laham N., Mediavilla J.R., Chen L., Abdelateef N., Elamreen F.A., Ginocchio C.C. MRSA clonal complex 22 strains harbouring toxic shock syndrome toxin (TSST-1) are endemic in the primary hospital in Gaza, Palestine. PLoS One. 2015;10:e0120008. doi: 10.1371/journal.pone.0120008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Bakri A.G., Al-Hadithi H., Kasabri V., Othman G., Kriegeskorte A., Becker K. The epidemiology and molecular characterization of methicillin-resistant staphylococci sampled from a healthy Jordanian population. Epidemiol Infect. 2013;141:2384–2391. doi: 10.1017/S0950268813000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shambat S., Nadig S., Prabhakara S., Bes M., Etienne J., Arakere G. Clonal complexes and virulence factors of Staphylococcus aureus from several cities in India. BMC Microbiol. 2012;12:64. doi: 10.1186/1471-2180-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seidl K., Leimer N., Marques M.P., Furrer A., Holzmann-Burgel A., Senn G. Clonality and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus at the University Hospital Zurich, Switzerland between 2012 and 2014. Ann Clin Microbiol Antimicrob. 2015;14:14. doi: 10.1186/s12941-015-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Japoni-Nejad A., Rezazadeh M., Kazemian H., Fardmousavi N., van Belkum A., Ghaznavi-Rad E. Molecular characterization of the first community-acquired methicicillin-resistant Staphylococcus aureus strains from central Iran. Int J Infect Dis. 2013;17:e949–e954. doi: 10.1016/j.ijid.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Moritz E.D., Hanson B.M., Kates A.E., Smith T.C. Molecular characteristics of Staphylococcus aureus isolated from employees, children, and environmental surfaces in Iowa child daycare facilities. Am J Infect Control. 2015;43:482–488. doi: 10.1016/j.ajic.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 37.Scicluna E.A., Shore A.C., Thurmer A., Ehricht R., Slickers P., Borg M.A. Characterisation of MRSA from Malta and the description of a Maltese epidemic MRSA strain. Eur J Clin Microbiol Infect Dis. 2010;29:163–170. doi: 10.1007/s10096-009-0834-1. [DOI] [PubMed] [Google Scholar]
- 38.Oksuz L., Dupieux C., Tristan A., Bes M., Etienne J., Gurler N. The high diversity of MRSA clones detected in a university hospital in Istanbul. Int J Med Sci. 2013;10:1740–1745. doi: 10.7150/ijms.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinnevey P.M., Shore A.C., Brennan G.I., Sullivan D.J., Ehricht R., Monecke S. Extensive genetic diversity identified among sporadic methicillin-resistant Staphylococcus aureus isolates recovered in Irish hospitals between 2000 and 2012. Antimicrob Agents Chemother. 2014;58:1907–1917. doi: 10.1128/AAC.02653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]