Abstract
Aims:
The aim of this study was to quantitatively investigate the hypermethylation of p16 gene in buccal cells and saliva of oral submucous fibrosis (OSMF) patients using real-time quantitative methylation-specific polymerase chain reaction (PCR) and to compare the values of two methods.
Subjects and Methods:
A total of 120 samples were taken from 60 subjects selected for this study, of which 30 were controls and 30 patients were clinically and histopathologically diagnosed with OSMF. In both groups, two sets of samples were collected, one directly from the buccal cells through cytobrush technique and the other through salivary rinse. We analyzed the samples for the presence of p16 hypermethylation using quantitative real-time PCR.
Results:
In OSMF, the hypermethylation status of p16 in buccal cells was very high (93.3%) and in salivary samples, it was partially methylated (50%). However, no hypermethylation was found in controls suggesting that significant quantity of p16 hypermethylation was present in buccal cells and saliva in OSMF.
Conclusions:
This study indicates that buccal cell sampling may be a better method for evaluation than the salivary samples. It signifies that hypermethylation of p16 is an important factor to be considered in epigenetic alterations of normal cells to oral precancer, i.e. OSMF.
Keywords: CpG islands, epigenetic, hypermethylation, oral submucous fibrosis, p16
Introduction
The fifth most common cancer in the world is oral cancer.[1] Approximately, 75,000 ± 80,000 new cases of oral cancer have been documented in India. In all nonrural cancer registries, oral cancer ranks highest among all types of cancers in the world.[2] Oral cancer and oral mucosal diseases such as oral submucous fibrosis (OSMF) and leukoplakia have a close association with the habit of smoking, tobacco, and betel nut chewing.[2] A chronic, progressive, disabling oral mucosal disease with a potential for malignant transformation is OSMF. It is seen predominantly in the South Asians, more prevalent in Indians. In India alone, the statistics for OSMF is about 5 million people (0.5%).[3]
Oral squamous cell carcinoma associated with betel quid causes specific genetic mutations, in the p53 gene. A part from gene mutation, epigenetic alterations such as DNA hypermethylation, histone acetylation, and phosphorylation are often observed in the tumor.[4] DNA methylation takes place in the mammalian DNA molecule predominantly at cytosine bases that are located 5΄ to a guanosine. It is basically a covalent biochemical modification.[5]
CpG islands mostly appear near the promoter regions, and it extends to the first exon of specific genes. Hypermethylation and unmethylation state of CpG islands are located in and around the promoter region, plays an important role in regulating gene expression. In normal cells, they are unmethylated and in human cancer they seem to be hypermethylated.[6] In fact, gene silencing occurs commonly due to promoter hypermethylation than genetic mutation. This event occurs earlier, preceding changes in protein expression level in oral carcinogenesis. This pathway makes promoter hypermethylation a very attractive diagnostic marker for the early detection of oral cancer.[7]
p16 is a tumor suppressor gene located at chromosome 9p21. In human cancers, one of the most frequently altered genomic regions is a 9p21 chromosomal band. This region contains cluster of three genes, p14ARF, p15INK4b, and p16INK4a within a short distance of 50 kb, all of which have putative tumor suppressor roles. Moreover, CpG islands are highly abundant in the promoter regions of all three genes, and they are more susceptible to hypermethylation.[8] In many human cancers including oral cancers, loss of p16 is frequently observed.[9,10] 83% of oral cancer and 60% of the premalignant lesion shows the loss of p16 expression, suggesting that p16 alteration is an early event in oral cancer.[11]
Only very few studies are available on hypermethylation of p16 in oral precancer even though hypermethylation of tumor suppressor genes in malignant tumors including oral cancer has been documented. The detection of p16 hypermethylation in precancer can predict the risk of malignant transformation[12] and may also be a used as a prognostic marker. Hence, this study was aimed to quantitatively investigate the promoter hypermethylation of p16 gene in buccal cells and saliva of OSMF patients using real-time quantitative methylation-specific polymerase chain reaction (PCR) and to compare the values of two methods.
Aim
To quantitatively evaluate p16 hypermethylation in buccal cells and saliva of OSMF patients and to compare the values in these two samples.
Objectives
To quantitatively determine the presence of p16 hypermethylation in buccal cells and salivary samples of OSMF patients
To quantitatively determine the presence of p16 hypermethylation in buccal cells and salivary samples of normal healthy individuals
To compare p16 hypermethylation in buccal cells and salivary samples of OSMF patients.
Subjects and Methods
Selection of patients
The study protocol was approved by Ethics Committee for student proposal, Sri Ramachandra University. A total of 120 samples were taken from 60 subjects selected for the study, of which 30 were controls (Group A) and 30 were patients with OSMF (Group B). In both groups two sets of samples were collected, one directly from the buccal cells through cytobrush technique and the other through salivary rinse. All 60 patients were reported to the department of oral medicine and radiology. Based on the clinical and histopathological evaluation these patients were confirmed as OSMF. The patient age group for the OSMF ranged from 21 to 67 years with mean age of 44. Out of 30 OSMF patients two of them were female, 28 of them were male. Samples from 30 study group (Group B) were taken from those who had not undergone any form of therapy for the presenting illness. Samples from 30 controls (Group A) who participated in the study were taken from the healthy volunteers of matched age and gender, who were free of oral and medical illness, without any habits of smoking, alcohol, chewing betel nut, and tobacco.[13] The age group for controls (Group A) ranged from 21 to 60 years with mean age of 40 years. The patients and controls were explained about the study, and written consent was taken.
Sampling of exfoliative cytology
Buccal samples collection was done with the help of cytobrush from 30 control group and 30 OSMF patients. All the participants were instructed to rinse the mouth with tap water for 10 s before collection. The buccal mucosa was scraped by simple counter pressure by twirling the brush while moving it downward and the counter pressure was applied with fingers against the external cheek for 30 s, and the brush was stored in a 15 ml centrifuge tube directly without additional processing in −80°C freezer. The Same method was followed for the control group.[14]
Sampling of saliva
Twenty milliliters of salivary rinses were collected by rinsing or gargling, for 60 s with 20 ml of sterile sodium chloride solution at 0.9%, in a 30 ml sterile centrifuge tube from 30 control group, and 30 OSMF patients. The salivary samples collected were transferred to the laboratory and was stored at −80°C.[14]
The samples were subjected to analyses for the presence of p16 hypermethylation using quantitative real-time PCR.
DNA extraction
Genomic DNA from samples of exfoliative cytology and salivary rinse was extracted with QIAamp DNA mini kit with 20 μl of QIAGEN Protease stock solution. DNA from salivary samples and exfoliative cytology of OSMF patients and from controls were modified with bisulfite and cleaned using EpiTect bisulfite conversion kit (Cat. No: 59104) purchased from QIAGEN. Briefly, 2 mg of genomic DNA was denatured in 0.2 mol/L of NaOH for 20 min at 50°C.
Real-time quantitative methylation-specific polymerase chain reaction
The modified DNA was used as a template for fluorescence-based real-time PCR. The quantitative real-time PCR was carried out on a fast real-time PCR 7900HT system. The primers were used to detect methylated p16 gene. ß-actin gene was used as a reference control. The ratio of methylated p16 promoter DNA to ß-actin DNA represented the relative p16 methylation level,[15] all reactions were performed in triplicate in 20 μl of total volume of PCR products. The component of PCR product is given in Table 1.
Table 1.
Component of PCR products

All reactions were performed with methylated and unmethylated primers and ß-actin endogenous control (positive control) and the nontemplate control (negative control) contained no template DNA.
During the annealing and the extension step of each cycle of real-time PCR, the Ct value of the amount of product amplified by the fluorescence of SYBR Green dye was plotted on an amplification curve. The cycle threshold value obtained was calculated for each amplification in each experimental sample by use of Applied Biosystems 7900HT fast real time pcr system soft ware SDS2.1.[15] The results derived for evaluation of the presence of p16 hypermethylation by real-time PCR.
The comparative Ct method (also called ΔCt) is calculated by formula:
ΔCt = avgCtGOI − avgCtref[16]
GOI - gene of interest, ref - the reference gene.
Subsequent to the derived Ct values, the results of p16 hypermethylation was evaluated.
Statistical analysis
Since Group A (controls) showed no methylation (0%), statistically odds ratio cannot be calculated to give a significant P value.
Results
The sample collected were analyzed to study the methylation status of p16 gene by quantitative methylation specific PCR and the values are tabulated in Tables 2–5, respectively.
Table 2.
Methylation status of p16 in the buccal cells from exfoliative cytology samples of Group A (controls)
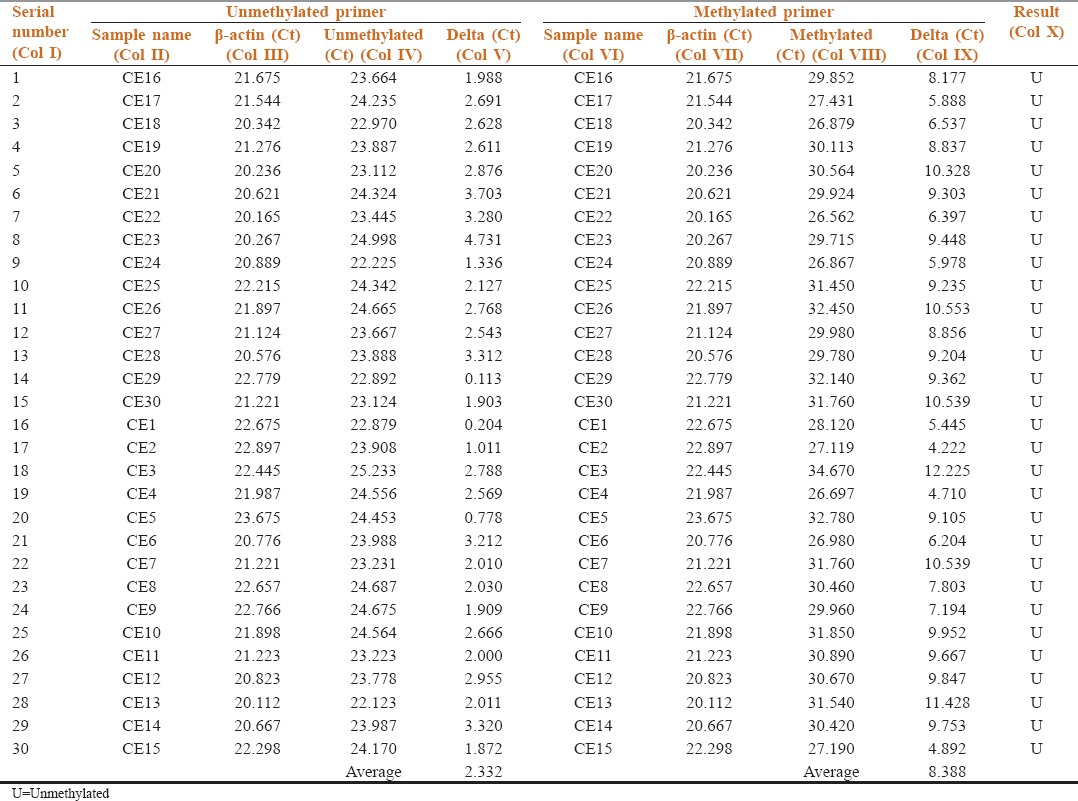
Table 5.
Methylation status of p16 in the salivary samples of Group B (OSMF patients)
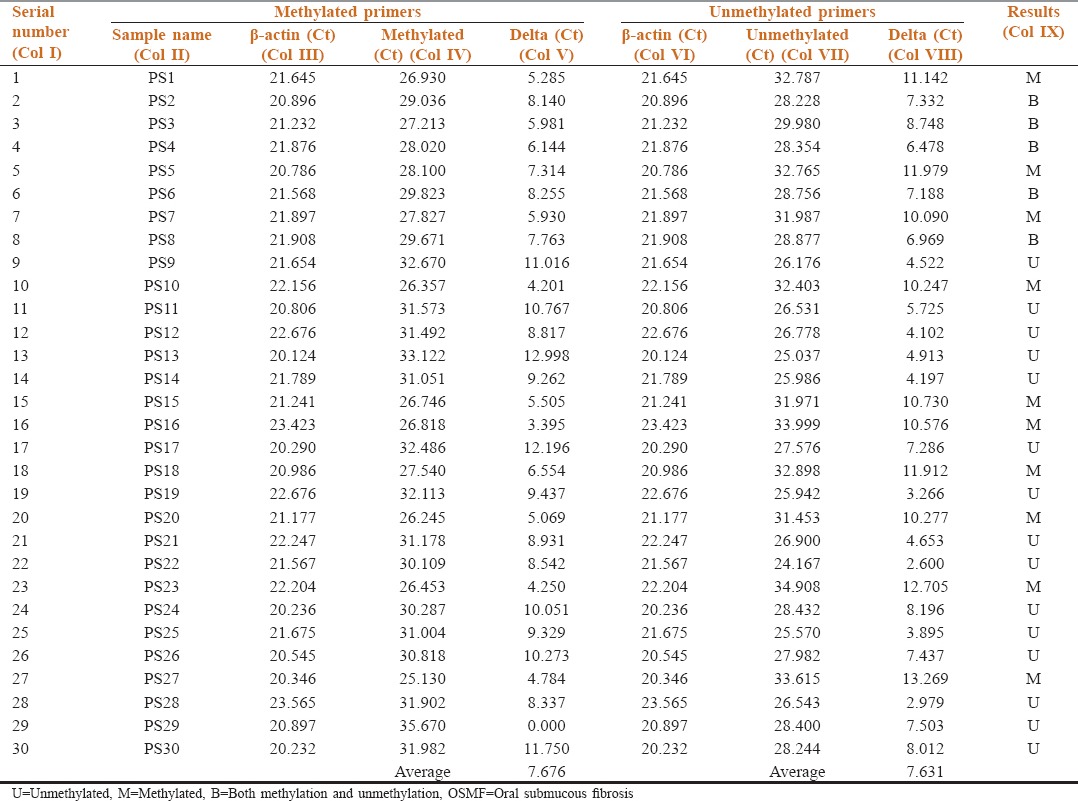
Table 2 shows the methylation status of p16 in buccal cells of Group A by comparing the target gene p16 (Col IV and VIII) with reference gene β-actin (Col III and VII) in unmethylated and methylated primers respectively. Delta Ct values, the difference in threshold cycles for the target gene and reference gene were calculated. The average Delta Ct value of unmethylated p16 gene was 2.332 and methylated 8.388. No methylation was detected in all 30 samples, and the results suggest that Group A (controls) is unmethylated (Col X).
Table 3 shows the methylation status of p16 in buccal cells of Group B by comparing the target gene p16 (Col IV and VIII) with reference gene β-actin (Col III and VII) in the unmethylated and methylated primers respectively. Of 30 samples tested 22 showed methylation, 2 showed unmethylation, and 6 samples showed both methylation and unmethylation (Col X).
Table 3.
Methylation status of p16 in the buccal cells from exfoliative cytology samples of Group B (OSMF patients)
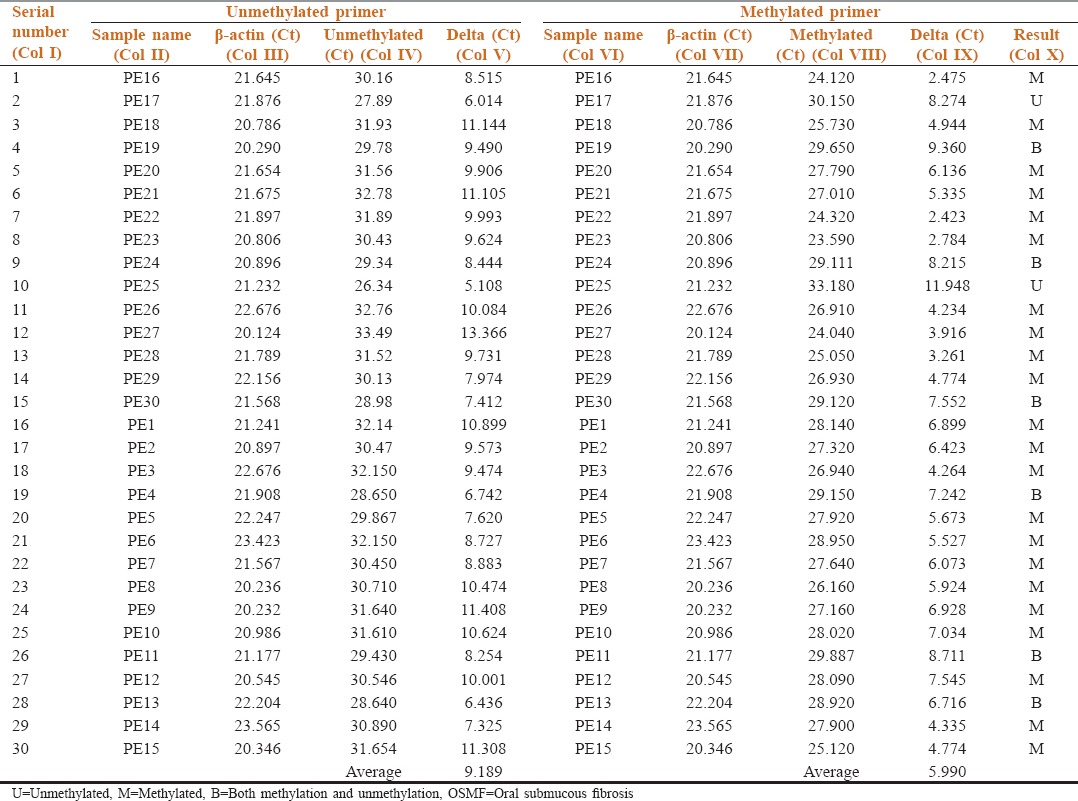
6 samples that showed both methylation and unmethylation were considered to be methylated as done in a similar study.[4] The methylated value (5.990) increased when compared with a unmethylated value (9.189), which suggests that majority in Group B (OSMF patients) is methylated.
Table 4 shows methylation status of p16 in salivary samples of Group A by comparing the target gene p16 (Col IV and VII) with reference gene β-actin (Col III and VI) in the unmethylated and methylated primers, respectively. The average Delta Ct value of unmethylated p16 gene was 6.214 and methylated 12.785. All 30 samples (100%) were unmethylated, and the results suggest that Group A (controls) is unmethylated (Col IX).
Table 4.
Methylation status of p16 in the salivary samples of Group A (controls)
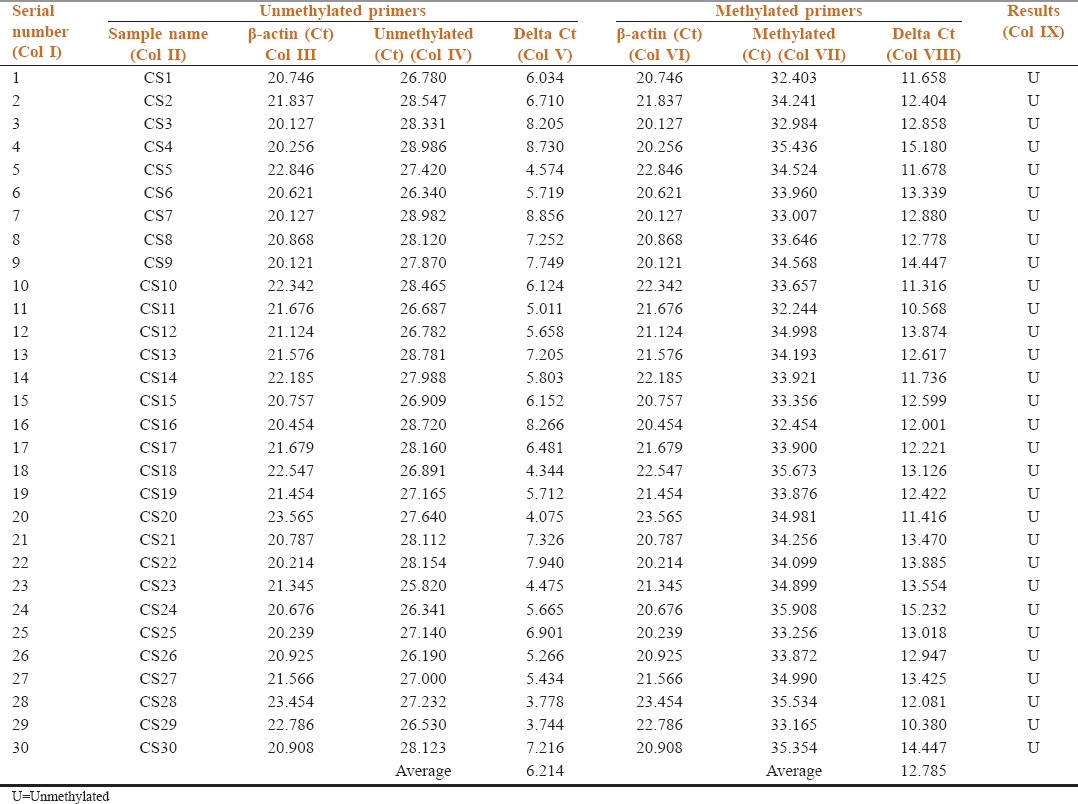
Table 5 shows methylation status of p16 in salivary samples of Group B by comparing the target gene p16 (Col IV and VII) with reference gene β-actin (Col III and VI) in the methylated and unmethylated primers, respectively. Out of 30 samples tested 10 were methylated, 15 unmethylated, and 5 showed equal methylation and unmethylation (Col IX). Five samples which showed both methylation and unmethylation were considered to be methylated as done in an earlier study.[4] Since, there was no significant difference between the unmethylated and methylated values the average delta Ct value of Group B (OSMF patients) is considered to be partially methylated.
Discussion
The current study was done to quantitatively evaluate the hypermethylation status of p16 gene in 30 patients with oral submucous fibrosis (OSMF) and to compare it with 30 healthy individuals in their buccal cells and saliva. As the incidence rate of this condition in India is about 5 million people (0.5%) of the total population[3] and as this precancerous condition possesses a risk of 7.6% for malignant transformation over a period of 17 years,[17] this study was designed to investigate an objective marker in early carcinogenesis.
Various studies have evaluated the genetic alteration where there is a progression of normal cells to precancer and cancer.[18,19] A change in gene expression is evident in cancer cells at the epigenetic level via transcriptional inactivation.[6] These epigenetic changes have been identified as an important component of carcinogenesis.[5] DNA methylation is the most important epigenetic alterations that lead to altered gene expression.[6] An increasingly recognized epigenetic mechanism of transcription activation of tumor suppressor genes or DNA repair genes is methylation of normally unmethylated CpG islands in gene promoter region.[20]
p16 is a tumor suppressor gene, normally block cellular proliferation by binding to complexes of cyclin dependent kinase CDK4 and CDK6. This binding prevents entry into the S phase of cell cycle. In many human cancers including oral cancers, frequent loss of p16 is observed. According to Reed et al.,[11] p16 expression is lost in 83% of oral cancers and 60% of premalignant lesions, suggesting that p16 alteration is an early event in oral cancer progression.[17] In a similar study by Takeshima et al.[4] has shown 70% hypermethylation of p16 gene in OSMF patients in Srilanka. Even though hypermethylation of tumor suppressor genes in malignant tumors including oral cancer has been well documented, very few studies are available on hypermethylation of p16 in oral precancerous conditions. Since, the detection of hypermethylation in precancer can predict the risk of malignant transformation, this study evaluated p16 hypermethylation in OSMF patients.
Sources of free DNA includes serum, plasma, saliva/oral rinse, urine,[21] and cell collection through scraping of the oral mucosa.[14] Compared to serum, saliva has a significant diagnostic advantage as a diagnostic fluid as its collection is noninvasive and simple. According to Righini et al.,[22] malignant cells collected in body cavity fluids at direct contact with the tumor shows gene methylation; therefore, salivary analysis has been used in this study for early detection of oral cancer. The advantage of swish methods is higher average DNA yields and longer DNA fragments. According to Mehrotra et al.,[23] buccal cell collection through cytobrush technique is a simple, relatively inexpensive, and risk-free method for obtaining cell samples. These advantages justify that salivary rinse and cytobrush technique can be used as an easily accessible method of DNA collection. In this study, salivary rinses were collected by modified method[14] as from the protocol described by Carvalho et al.[24] and the buccal samples through cytobrush technique.[14]
Takeshima et al.[4] evaluated the hypermethylation status of p14, p15, and p16 in various oral precancers by immunohistochemical methods and has inferred that all frequencies were high in OSMF patients. In this study, hypermethylation of p16 was quantified in salivary rinse and buccal cells of OSMF patients and compared with healthy individuals. Exfoliative cytology samples of Group A (controls) showed no methylation and all 30 samples were unmethylated (100%), whereas in Group B (OSMF patients), 93.3% were methylated, and 6.7% were unmethylated, which suggests that hypermethylation of p16 is very highly significant in buccal cell samples and is consistent with previous studies.[25,26,27] Although the unmethylated percent (6.7%) in the buccal cell samples are negligible, it can be attributed to sampling and storage/processing errors.
According to Lee et al.,[28] hypermethylation rate can be low in salivary rinse due to the dilution effect of normal-unmethylated genomes present from normal mucosa. However, in the present study, results from the salivary rinse shows that 50% of Group B are methylated, and 50% are unmethylated. No methylated samples were present in Group A (control), and all 30 samples (100%) were unmethylated. These results suggest that percentage of methylation and unmethylation is same, and Group B is considered to be partially methylated, which is consistent with previous studies.[4]
Comparison of the methylation status of p16 between buccal cells and salivary samples of Group B shows that 22 samples in buccal cells were methylated, 2 samples were unmethylated, and 6 samples were both unmethylated and methylated. Among the salivary samples, 10 were methylated, 15 were unmethylated, and 5 samples were both unmethylated and methylated, which infers that the hypermethylation of p16 was significantly higher in buccal cell samples than the salivary samples and consistent with earlier studies.[25,26,27,28,29,30] Statistical analysis to indicate the P value through odds ratio was not done in this t study as Group A was completely unmethylated (0%).
Conclusion
To conclude significant quantity of p16 hypermethylation were present in buccal cells and saliva. Buccal cell sampling may be a better sampling method for evaluation in this study than the salivary samples. However, the quantification of p16 hypermethylation in oral cancer group as a part of the study may lead to a definitive conclusion about the transformation of precancer to cancer and will serve as a very early indicator of carcinogenic activity and aid in the prognosis of the disease.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
My sincere thanks to Dr. V. Vetrichelvi Ph.D., Assistant Professor, Department of Human Genetics for her immense help, suggestion and expertise and MS.C.D. Mohanapriya Msc., human genetics, application specialist (genomics and molecular biology) for her unhesitating help and support.
My special thanks to Dr. Sankara Aravind Warrier M.D.S., Reader for enthusiastic suggestions, critical judgment and advices and Dr. T. Malarkodi M.D.S. Senior Lecturer, for giving untiring support and offering her guidance, ideas and suggestions through every step of my project.
References
- 1.Kao SY, Chu YW, Chen YW, Chang KW, Liu TY. Detection and screening of oral cancer and pre-cancerous lesions. J Chin Med Assoc. 2009;72:227–33. doi: 10.1016/s1726-4901(09)70062-0. [DOI] [PubMed] [Google Scholar]
- 2.Nair U, Bartsch H, Nair J. Alert for an epidemic of oral cancer due to use of the betel quid substitutes gutkha and pan masala: A review of agents and causative mechanisms. Mutagenesis. 2004;19:251–62. doi: 10.1093/mutage/geh036. [DOI] [PubMed] [Google Scholar]
- 3.Angadi PV, Rao SS. Areca nut in pathogenesis of oral submucous fibrosis: Revisited. Oral Maxillofac Surg. 2011;15:1–9. doi: 10.1007/s10006-010-0219-8. [DOI] [PubMed] [Google Scholar]
- 4.Takeshima M, Saitoh M, Kusano K, Nagayasu H, Kurashige Y, Malsantha M, et al. High frequency of hypermethylation of p14, p15 and p16 in oral pre-cancerous lesions associated with betel-quid chewing in Sri Lanka. J Oral Pathol Med. 2008;37:475–9. doi: 10.1111/j.1600-0714.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 5.Barros SP, Offenbacher S. Epigenetics: Connecting environment and genotype to phenotype and disease. J Dent Res. 2009;88:400–8. doi: 10.1177/0022034509335868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radhakrishnan R, Kabekkodu S, Satyamoorthy K. DNA hypermethylation as an epigenetic mark for oral cancer diagnosis. J Oral Pathol Med. 2011;40:665–76. doi: 10.1111/j.1600-0714.2011.01055.x. [DOI] [PubMed] [Google Scholar]
- 7.Viet CT, Jordan RC, Schmidt BL. DNA promoter hypermethylation in saliva for the early diagnosis of oral cancer. J Calif Dent Assoc. 2007;35:844–9. [PubMed] [Google Scholar]
- 8.Xing EP, Nie Y, Song Y, Yang GY, Cai YC, Wang LD, et al. Mechanisms of inactivation of p14ARF, p15INK4b, and p16INK4a genes in human esophageal squamous cell carcinoma. Clin Cancer Res. 1999;5:2704–13. [PubMed] [Google Scholar]
- 9.Timmermann S, Hinds PW, Münger K. Re-expression of endogenous p16ink4a in oral squamous cell carcinoma lines by 5-aza-2’- deoxycytidine treatment induces a senescence-like state. Oncogene. 1998;17:3445–53. doi: 10.1038/sj.onc.1202244. [DOI] [PubMed] [Google Scholar]
- 10.Sartor M, Steingrimsdottir H, Elamin F, Gäken J, Warnakulasuriya S, Partridge M, et al. Role of p16/MTS1, cyclin D1 and RB in primary oral cancer and oral cancer cell lines. Br J Cancer. 1999;80:79–86. doi: 10.1038/sj.bjc.6690505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed AL, Califano J, Cairns P, Westra WH, Jones RM, Koch W, et al. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56:3630–3. [PubMed] [Google Scholar]
- 12.Kresty LA, Mallery SR, Knobloch TJ, Song H, Lloyd M, Casto BC, et al. Alterations of p16(INK4a) and p14(ARF) in patients with severe oral epithelial dysplasia. Cancer Res. 2002;62:5295–300. [PubMed] [Google Scholar]
- 13.Bahar G, Feinmesser R, Shpitzer T, Popovtzer A, Nagler RM. Salivary analysis in oral cancer patients: DNA and protein oxidation, reactive nitrogen species, and antioxidant profile. Cancer. 2007;109:54–9. doi: 10.1002/cncr.22386. [DOI] [PubMed] [Google Scholar]
- 14.King IB, Satia-Abouta J, Thornquist MD, Bigler J, Patterson RE, Kristal AR, et al. Buccal cell DNA yield, quality, and collection costs: Comparison of methods for large-scale studies. Cancer Epidemiol Biomarkers Prev. 2002;11(10 Pt 1):1130–3. [PubMed] [Google Scholar]
- 15.Hong J, Resnick M, Behar J, Wang LJ, Wands J, DeLellis RA, et al. Acid-induced p16 hypermethylation contributes to development of esophageal adenocarcinoma via activation of NADPH oxidase NOX5-S. Am J Physiol Gastrointest Liver Physiol. 2010;299:G697–706. doi: 10.1152/ajpgi.00186.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal. 2003;1:e012. doi: 10.1621/nrs.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tilakaratne WM, Klinikowski MF, Saku T, Peters TJ, Warnakulasuriya S. Oral submucous fibrosis: Review on aetiology and pathogenesis. Oral Oncol. 2006;42:561–8. doi: 10.1016/j.oraloncology.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: Implications for therapy. J Dent Res. 2008;87:14–32. doi: 10.1177/154405910808700104. [DOI] [PubMed] [Google Scholar]
- 19.Campo-Trapero J, Cano-Sánchez J, Palacios-Sánchez B, Sánchez-Gutierrez JJ, González-Moles MA, Bascones-Martínez A. Update on molecular pathology in oral cancer and precancer. Anticancer Res. 2008;28:1197–205. [PubMed] [Google Scholar]
- 20.Liu M, Feng L, Tang X, Guo S. Gene promoter hyper methylation in leukoplakia of the oral mucosa. Pathol Lab Med Int. 2010;2:71–7. [Google Scholar]
- 21.Shaw R. The epigenetics of oral cancer. Int J Oral Maxillofac Surg. 2006;35:101–8. doi: 10.1016/j.ijom.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Righini CA, de Fraipont F, Timsit JF, Faure C, Brambilla E, Reyt E, et al. Tumor-specific methylation in saliva: A promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007;13:1179–85. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- 23.Mehrotra R, Gupta A, Singh M, Ibrahim R. Application of cytology and molecular biology in diagnosing premalignant or malignant oral lesions. Mol Cancer. 2006;5:11. doi: 10.1186/1476-4598-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Carvalho AL, Jeronimo C, Kim MM, Henrique R, Zhang Z, Hoque MO, et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 25.Kulkarni V, Saranath D. Concurrent hypermethylation of multiple regulatory genes in chewing tobacco associated oral squamous cell carcinomas and adjacent normal tissues. Oral Oncol. 2004;40:145–53. doi: 10.1016/s1368-8375(03)00143-x. [DOI] [PubMed] [Google Scholar]
- 26.Ruesga MT, Acha-Sagredo A, Rodríguez MJ, Aguirregaviria JI, Videgain J, Rodríguez C, et al. p16(INK4a) promoter hypermethylation in oral scrapings of oral squamous cell carcinoma risk patients. Cancer Lett. 2007;250:140–5. doi: 10.1016/j.canlet.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Hall GL, Shaw RJ, Field EA, Rogers SN, Sutton DN, Woolgar JA, et al. p16 Promoter methylation is a potential predictor of malignant transformation in oral epithelial dysplasia. Cancer Epidemiol Biomarkers Prev. 2008;17:2174–9. doi: 10.1158/1055-9965.EPI-07-2867. [DOI] [PubMed] [Google Scholar]
- 28.Lee KD, Lee HS, Jeon CH. Body fluid biomarkers for early detection of head and neck squamous cell carcinomas. Anticancer Res. 2011;31:1161–7. [PubMed] [Google Scholar]
- 29.Rosas SL, Koch W, da Costa Carvalho MG, Wu L, Califano J, Westra W, et al. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939–42. [PubMed] [Google Scholar]
- 30.Lopez M, Aguirre JM, Cuevas N, Anzola M, Videgain J, Aguirregaviria J, et al. Gene promoter hypermethylation in oral rinses of leukoplakia patients - A diagnostic and/or prognostic tool? Eur J Cancer. 2003;39:2306–9. doi: 10.1016/s0959-8049(03)00550-1. [DOI] [PubMed] [Google Scholar]


