Abstract
Background
Multiple genetic loci are associated with clinical cardiovascular (CV) disease and individual CV risk factors. Individuals with ideal levels of all major CV risk factors have very low risk for CVD morbidity or mortality. Ideal levels of risk factors can be attained by lifestyle modifications; however, little is known about gene variants associated with ideal CV health. Our objective was to carry out a genome-wide association study (GWAS) on the trait.
Methods and Results
We examined two dichotomous phenotypes of ideal CV health - Clinical (untreated cholesterol < 200 mg/dl; untreated blood pressure (BP) <120/<80; not diabetic) and Clinical+Behavioral (Clinical plus: not a current smoker; BMI <25 kg/m2) -among white participants aged 50 ± 5 years. We performed a meta-analysis of four GWAS (total n=11,708) from the MESA, CARDIA, ARIC and Framingham Heart Study cohorts. We identified a SNP (rs445925) in the APOC1/APOE region that was associated with Clinical ideal CV health at genome-wide level of significance (p<2.0×10−9). The significance of this region was validated using exome chip genotyping. The association with ideal CV health was attenuated after adjusting for LDL cholesterol.
Conclusion
A common SNP in the APOC1/APOE region, previously found to be associated with protective levels of cholesterol and lower cardiovascular risk, may be associated with ideal health. In future replication studies, larger sample sizes may be needed to detect loci with more modest effects on ideal CV health. In addition to the important impact of lifestyle modifications, we have identified evidence for gene variation that plays a role in ideal CV health.
Keywords: cardiovascular diseases, genetics, epidemiology
INTRODUCTION
The current American Heart Association 2020 Strategic Impact Goals introduce a concept – ideal cardiovascular (CV) health – defined by ideal levels of modifiable major risk factors, with the goal of improving the health of the nation in part by increasing the proportion of Americans who maintain ideal CV health.1 Individuals having ideal CV health during middle age have been found to have markedly lower risks for cardiovascular and other chronic diseases, lower mortality rates, lower healthcare costs, and improved quality of life in older age.2–7 Overall, among participants of Framingham, the lifetime risk for cardiovascular disease after age 50 has been found to be as high as 51% among men and 39% among women; but for individuals with ideal levels of cardiovascular risk factors, the remaining lifetime risk for cardiovascular disease (CVD) is only 5–8%.8 The relative contribution of genetic, behavioral and/or environmental factors to achieving or maintaining ideal CV health in middle age remains unclear. Although only about 5% of the U.S. population is in ideal CV health,9 understanding how these individuals achieve and maintain ideal cardiovascular health represents an important public health goal that could help reduce the future burden of cardiovascular disease.
Numerous genome-wide association studies (GWAS) have been completed to identify genetic variation underlying clinically apparent CV disease or levels of CVD risk factors.10–13 Two recent GWAS for the clustering of risk factors that comprise the metabolic syndrome have identified multiple loci, mostly near previously known lipid genes, underlying one or two metabolic syndrome components.14, 15 However, to date most studies have focused on individual phenotypes of risk and disease; examination of a healthy phenotype defined by the presence of several well defined health factors has not typically been studied. Accordingly, we conducted a meta-analysis of GWAS studies from community-based cohorts to identify common genetic variants associated with ideal CV health. We hypothesized that specific SNPs may be associated with the presence of ideal CV health in middle age.
METHODS
Discovery GWAS Studies
To identify common genetic variants associated with ideal CV health, we conducted a GWAS meta-analysis of a total of 11,708 white participants aged 50 ± 5 years at the time of an examination from four prospective cardiovascular cohort studies participating in the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) consortium.16 The four cohorts included the Multi-Ethnic Study of Atherosclerosis (MESA) (with examination cycles between 2000–2011), Coronary Artery Risk in Young Adults (CARDIA) Study (1985–2011), FHS Original Cohort and Offspring Cohorts (1948–2010) and Atherosclerosis Risk in Communities (ARIC) Study (1987–1998). Details about genome-wide genotyping in each cohort are found in Table 1. If participants were within the age range of 50 ± 5 years during multiple exams, the first/earliest exam within the age window was chosen and included in the analyses.
Table 1.
Study Characteristics
| COHORT | ARIC | CARDIA | FHS | MESA | |
| Name |
Atherosclerosis Risk in Communities Study (ARIC) |
Coronary Artery Risk Development in Young Adults (CARDIA) |
Framingham Heart Study (FHS) |
Multi-Ethnic Study of Atherosclerosis (MESA) |
|
| Ethnicity | European descent | European descent | European descent |
European- American |
|
| Country | United States | United States | United States | United States | |
| Collection type | Population-based | Cohort | Population- based |
Population-based (6 recruitment centers) |
|
|
EXOME GENOTYPING |
Genotyping platform | Illumina Exome Chip v1.0 |
Illumina Exome Chip v1.0 |
Illumina Exome Chip v1.0 |
Illumina Exome Chip v1.0 |
| Genotyping centre | UT Houston, Human Genetics Center |
UT Houston, Human Genetics Center |
Illumina FastTrack Genotyping facility |
Cedars Sinai | |
|
Genotyping calling algorithm |
Illumina GenomeStudio20 11.1 |
Illumina GenomeStudio20 11.1 |
Illumina GenomeStudio20 11.1 |
Illumina GenomeStudio20 11.1 |
|
|
Joint calling performed? |
Yes | Yes | Yes | Yes | |
| Call rate by variant | mean 99.7%, range 0–100% |
> 95% | ≥ 97% | mean 99.7%, range 0–100% |
|
|
HWE for MAF >5% & p<1×10−6? |
Yes | Yes | Yes | No | |
| Call rate by sample | Cutoff >95%, mean 99.77%, range 95–99.84% |
> 95% | - | - | |
|
AFFYMETRIX GENOTYPING |
Genotyping platform | Affymetrix Genome- Wide Human SNP Array 6.0 |
Affymetrix Genome- Wide Human SNP Array 6.0 |
Affymetrix 500K (250K Nsp & 250K Sty, MIPS 50K |
Affymetrix Genome- Wide Human SNP Array 6.0 |
| Genotyping centre | Broad Institute | Broad Institute | Affymetrix | Affymetrix | |
|
Genotyping calling algorithm |
BRLMM | Beaglecall | BRLMM | Birdseed v2 | |
| Call rate by variant | >95% | ≥95% | >97% | ≥95% | |
| IMPUTATION | Imputation Software | Mach (version 1.0.16) |
Mach (version 1.0.16) |
Mach (version 1.0.1.5) |
IMPUTE (version 2) |
| Reference Panel | BRLMM to Hapmap - V1 CEU backbone (build 35 |
HapMap Phase II, Build 36, Release 22 |
HapMap CEU, Build 36, Release 22 |
HapMap Phase I and II, Build 36, Release 24 |
|
|
Pre-Imputation MAF Filter |
<0.01 | <0.02 | <0.01 | <0.01 | |
|
Pre-Imputation HWE Filter |
<1×10−6 | <1×10−4 | <1×10−6 | <1×10−5 | |
|
Pre-Imputation Call Rate by Sample |
>95% | ≥95% | >97% | ≥95% | |
|
ANALYSIS AND REFERENCES |
Adjustments | Principal Components, Age, Sex, Center |
Principal Components, Age, Sex, Center |
Principal Components, Age, Sex, Center |
Principal Components, Age, Sex, Center |
| Software for analysis | PLINK, ProbABEL, SAS |
PLINK, ProbABEL, SAS |
PLINK, ProbABEL, SAS |
PLINK, SNPTest, SAS |
|
|
Reference GWAS (PMID) |
20031568 | 20400780, 20091798 |
- | 23966861 | |
| Website | http://www2.cscc.unc.edu/aric/ | http://www.cardia.dopm.uab.edu/ | http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v2.p1 | http://www.mesa-nhlbi.org/ |
Assessment of Cardiovascular Risk Factors
Methods for the measurement of CV health factor levels for each study have been published.17–20 In each of the studies, blood pressure was determined as the average of two measurements performed by trained study personnel with the participant seated. Fasting blood draws were used when possible to obtain total cholesterol and fasting glucose levels; however, casual glucose was used if no fasting sample was available. Medication use and current smoking status were assessed by self-report. Physical exams were conducted by trained study personnel in which height and weight were determined and used to calculate body mass index (BMI), defined as the weight in kg divided by the square of the height in meters.
Ideal Cardiovascular Health
Ideal CV health definitions were created based on the AHA 2020 goals.1 In this study, we examined the presence of two dichotomous phenotypes of ideal CV health at ages 50 ± 5 years: (1) Clinical, defined by the simultaneous presence of untreated serum cholesterol levels < 200 mg/dl (<5.16 mmol/l); untreated blood pressure of <120/<80 mm Hg; and not diabetic (fasting glucose < 126 mg/dL or casual glucose <201 mg/dL and no reported use of any anti-diabetic medications). (2) Clinical+Behavioral, defined using the same criteria as the Clinical definition plus: not being a current smoker and having BMI < 25 kg/m2.
Genome-Wide Association Analyses
We performed meta-analysis of four existing GWAS (n=11,708). For each of the four GWAS, genotypes were imputed using HapMap 2 data. Details about HapMap2 imputation in each cohort are found in Table 1. We filtered SNPs with MAF<0.05. Each study conducted logistic regression analyses for the approximately 2.5 million imputed SNPs using an additive SNP model for each of the two ideal CV health phenotypes. Models were adjusted for age, sex, site and study-specific principal components of ancestry. Study-specific results were combined using inverse-variance weighted meta-analysis implemented in METAL21 and corrected for residual inflation using genomic control. The genome-wide level of significance threshold was set at P< 5 × 10−8. There was no evidence of p-value inflation upon examination of the quantile-quantile plots. The genomic control inflation factor (λGC) was 1.019 for the Clinical ideal health phenotype and 1.017 for the Clinical+Behavioral phenotype.
Genetic determinants of ideal CV health, if observed, could be related to one or more of the individual cardiovascular health factors which together constitute the ideal CV health phenotype. In order to investigate this question, we examined the association of any SNPs that reached genome-wide significance for ideal CV health with each of the five individual dichotomous components included within either definition of ideal CV health: (1) ideal cholesterol, (2) ideal blood pressure, (3) not diabetic, (4) ideal BMI, and (5) non-smoker.
In order to determine whether the effect of the identified SNPs which reached genome-wide significance on ideal CV health were due to their association with LDL levels, we conducted logistic regression models in each of the cohorts after conditioning on continuous LDL in addition to the original covariates of age, sex, site and principal components of ancestry. Study-specific results of the conditional analyses were meta-analyzed in METAL using the same methods as the GWAS described above.
Validation Analysis
After completion of the GWAS, we conducted a validation analysis. Because the single SNP rs445925 that reached genome-wide significance in the GWAS meta-analysis was a low frequency variant with low imputation quality in some cohorts, we replicated the association of this SNP and of SNPs in the same region using directly genotyped SNPs from exome chip data from each of the same four cohorts. Exome chip data for participants of CARDIA, MESA, ARIC and FHS was obtained from the Illumina HumanExome Beadchip, after common genotype calling in these four cohorts and other cohorts in the CHARGE Consortium, as previously described.22 Details about genomewide exome chip genotyping in each cohort are found in Table 1.
Gene Expression Analysis
We queried over 90 datasets from multiple tissues for evidence of an expression quantitative trait locus (QTL) as well as 11 methylation QTL datasets to search for associations with our top SNP, rs445925, and 4 European ancestry LD proxies with r2>0.5 identified using SNAP.23
Multi-SNP Genetic Risk Scores
To further explore the role of genetic variation previously known to underlie lipids and CV disease with ideal CV health, including the contributory role of our top SNP, rs445925, we created 4 separate genetic risk scores (GRS) that were associated with coronary artery disease (CAD), LDL, HDL, and triglycerides (TG). The SNPs included in each of the Genetic Risk Scores can be found in Supplemental Table 1. Each GRS was calculated using single SNP meta-analysis association statistics from the meta-analysis described above and implemented using the gtx package in R as developed by Toby Johnson.24 The CAD GRS included 45 SNPs identified by DeLoukas et al.10 The lipid GRS included 144 lipid GWAS SNPs (n=57 for LDL, n=71 for HDL, n=39 for triglycerides).25 We examined the association of each GRS with ideal CV health with and without inclusion of our top SNP in the GRS, rs445925. Additionally, as a sensitivity analysis, we recalculated the CAD GRS, excluding SNPs known to be associated with LDL and TG (these eight loci are APOB, ABCG5-ABCG8, PCSK9, SORT1, ABO, LDLR, APOE and LPA),10 and examined its association with ideal CV health.
All studies included in this research have been supported by the National Institutes of Health. The authors are solely responsible for the design and conduct of this study, all analyses, the drafting and editing of the paper and its final contents.
RESULTS
Characteristics of Discovery GWAS Studies
Table 2 lists the characteristics of the four cohorts included in the GWAS meta-analysis for participants aged 50 years. Among our sample of 11,708 participants, the prevalence of Clinical ideal CV health was 19.2% (range: 10% to 29% among cohorts) and the prevalence of Clinical+Behavioral ideal CV health was 7.6% (range: 5% to 13% among cohorts). The mean age was just under 50 years and slightly more than half of the participants were female.
Table 2.
GWAS Cohort Characteristics
| ARIC | CARDIA | MESA | Framingham | |
|---|---|---|---|---|
| N | 4,751 | 1,311 | 757 | 4,889 |
| Mean Age (yr range) | 50.0 (45–55) | 48.3 (45–55) | 50.4 (45–55) | 48.2 (45–55) |
| % Female | 56.1% | 53.3% | 53.7% | 53.2% |
| BMI, mean (SD) | 26.7 (4.9) | 28.0 (6.1) | 27.9 (5.7) | 26.6 (4.9) |
| % Clinical Ideal CV Health | 24.3% | 28.4% | 29.3% | 10.1% |
| % Clinical+Behavioral Ideal CV Health | 9.2% | 12.9% | 6.9% | 4.6% |
Meta-Analysis of GWAS
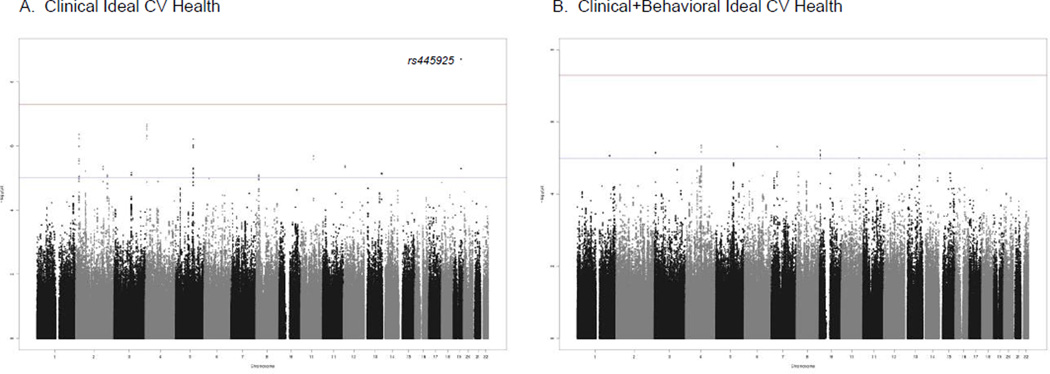
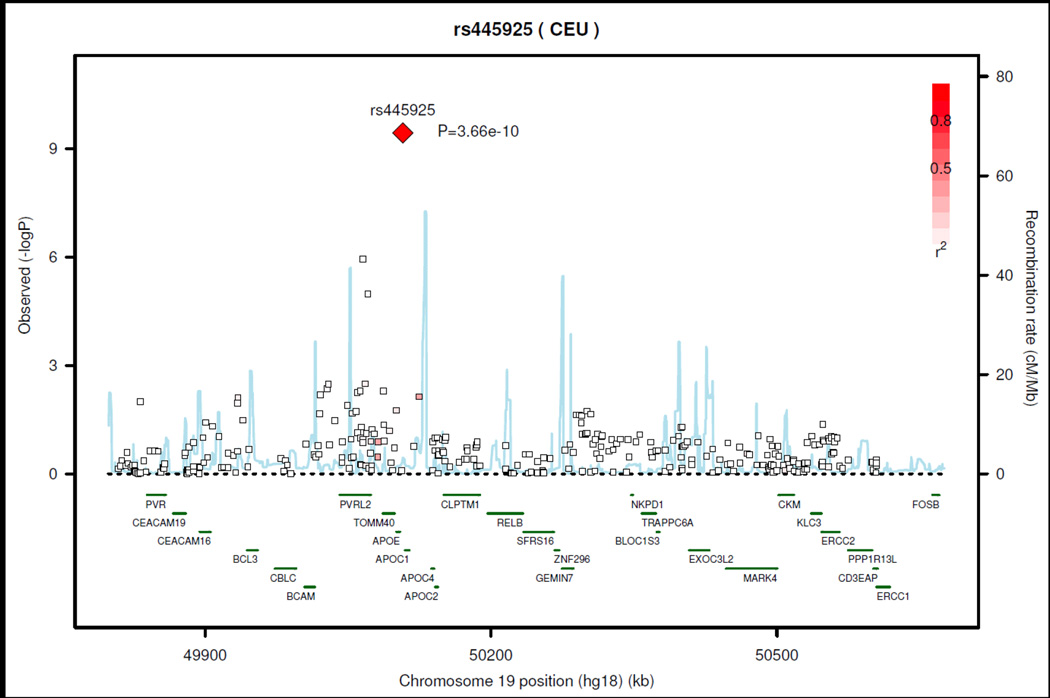
The Manhattan plots from the GWAS meta-analysis are shown in Figures 1A and 1B for the Clinical ideal CV health and Clinical+Behavioral ideal CV health, respectively. QQ plots for both analyses are shown in Supplemental Figures 1A and 1B. We identified one SNP (rs445925, range of MAF 0.13– 0.14) on chromosome 19 located between APOC1 and APOE for which the A (minor) allele was associated with Clinical ideal CV health at a genome-wide level of significance (p<1.97×10−9, OR= 1.72). These results were consistent across cohorts (Supplemental Table 2); however, the imputation quality was low (~ 0.3) across the cohorts and there were no other significant associations in this region (regional association plot in Figure 2). There was evidence for a nominally significant (p=0.0042) association for rs445925 with the Clinical+Behavioral ideal CV health phenotype. There were no SNPs associated at a genome-wide significance level with Clinical+Behavioral ideal CV health.
Figure 1.
Manhattan Plots
Figure 2.
Chrom 19 Regional Association Plot: Clinical Ideal CV Health
When the association between rs445925 and the individual dichotmous components of ideal CV health was examined, the only significant association for rs445925 was with ideal cholesterol levels (p=8.5 ×10−23) and not with the other clinical or behavioral components of ideal CV health (Table 3).
Table 3.
P-values for rs445925 associations with components of Ideal Health
| Component of Ideal CV Health | ARIC | CARDIA | MESA | Framingham | Meta-Analysis |
|---|---|---|---|---|---|
| Cholesterol | 4.75E-11 | 0.08 | 0.004 | 2.26E-11 | 8.50E-23 |
| Blood Pressure | 0.73 | 0.45 | 0.28 | 0.004 | 0.09 |
| Glucose | 0.76 | 0.28 | 0.54 | 0.22 | 0.13 |
| Smoking | 0.47 | 0.75 | 0.87 | 0.93 | 0.71 |
| BMI | 0.40 | 0.25 | 0.52 | 0.28 | 0.96 |
When adjusted for continuous LDL, the association of SNP rs445925 was attenuated and no longer significantly associated with either Clinical ideal CV health or with Clinical+Behavioral ideal CV health (in the meta-analysis, p=0.15 for Clinical ideal CV health and p=0.91 for Clinical+Behavioral ideal CV health, respectively). These results were consistent across cohorts.
Validation Analysis
In our GWAS meta-analysis we identified a significant association of rs445925 with Clinical ideal CV health. This SNP is in high LD (r2=0.588) with rs7412, known to be one of two SNPs defining the APOE2 allele. In order to determine whether rs7412 was driving the association between rs445925 and ideal CVD health, we used exome chip data from each of the cohorts to examine the associations between rs7412 and ideal CV health while adjusting for rs445925. Among cohort participants with exome chip data (n=12,230), rs445925 and rs7412 were both significantly associated with Clinical ideal CV health (p=8.6×10−10 and 0.1.45×10−8, respectively and Clinical+Behavioral ideal health (p=0.00019,) and (p=9.2×10−16) respectively. The association of rs445925 with ideal CV health was attenuated after adjustment for rs7412, such that the p-values for Clinical ideal CV health and Clinical+Behavioral ideal CV health were 0.017 and 0.078, respectively.
eQTL Gene Expression Analysis
We identified an eQTL of modest strength for this SNP in subcutaneous adipose tissue from the MuTHER consortium in which rs445925 is associated with increased APOE expression (p<3.85E-6, beta 0.28, SE 0.06 which exceeds a 1% FDR threshold in the dataset).26 The strongest eQTL SNP for APOE in this tissue is rs439401(P<2.38E-9), but the correlation is low between rs439401 and rs445925 (r^2 0.02, D’=0.51).
Multi-SNP Genetic Risk Scores
When we examined associations with genetic risk scores for lipid subfractions and CHD, the LDL GRS had the strongest association with ideal CV health (p-values were 5.04×10−25 and <0.001, for the Clinical and Clinical+Behavioral ideal CV health outcomes, respectively); these associations were only slightly attenuated with addition of rs445925. The TG GRS was also significantly associated with both ideal CV health outcomes (p=6.6×10−6 and 0.002, respectively), although these associations were not altered by the addition of rs445925. The CAD GRS was also significantly associated with ideal CV health. However, in sensitivity analysis after removal of eight SNPs in lipid loci known to be associated with LDL or triglycerides, the remaining CAD GRS was no longer significantly associated with ideal CV health (p=0.8). The HDL GRS was not associated with ideal CV health without or with inclusion of rs445925.
Funding for each of the cohort studies involved in the study is provided by the National Institutes of Health. The authors are solely responsible for the design and conduct of this study, all study analyses, the drafting and editing of the paper and its final contents.
DISCUSSION
To our knowledge, this is the first study to examine genetic determinants of ideal CV health and one of the few to examine a state of optimal health as opposed to a diseased state or shortened lifespan. In our results, we found that a common SNP in the APOC1/APOE region (rs445925) is associated with Clinical ideal CV health although this association appears to be driven largely by lipid levels. This specific SNP has been identified in previous GWAS to be associated with higher risk of coronary heart disease,10, 27 greater common carotid IMT and carotid plaque,28 and long-term higher LDL levels in older adults29 as well as in childhood through young adulthood.30 Additionally, SNPs in the APOC1/APOE region have been associated with Alzheimer’s disease,31, 32 longevity,33 lipids34 and metabolic syndrome.14 In our study we found that the A (minor) allele was associated with an increase in the odds of being in Ideal CV Health; this is consistent with the prior studies of CHD, carotid IMT and LDL cholesterol, in which the A (minor) allele was associated with a negative beta or decreased odds of disease. Our findings suggest that the effect of rs445925 on ideal CV health is likely mediated by its impact on LDL cholesterol levels. The SNP rs445925 remained significantly associated with ideal CV health in exome chip analyses even after adjustment for SNP rs7412 in the APOE region. Although adjustment for rs7412 attenuated the relationship, rs445925 remained significantly related to Clinical ideal CV health suggesting that this SNP (or a related variant) may have some independent effect on the trait above and beyond its direct effect on the APOE E2 allele. In analyses conditioning on LDL, rs445925 was no longer associated with ideal CV health. Beyond the finding that a lower prevalence of LDL raising risk loci are strongly and significantly associated with ideal health, we did not find strong evidence to support the association of other major gene loci.
There is substantial evidence demonstrating the benefits of the ideal cardiovascular health phenotype during middle age, and these benefits extend beyond the avoidance of incident cardiovascular disease. Prior research has shown that individuals who are able to achieve or maintain ideal CV health into middle age have greater longevity, lower morbidity from multiple chronic diseases, greater health-related quality of life, and lower healthcare costs in older age.2–7, 35 Similar to the findings in this study, the prevalence of Clinical (and Clinical+ Behavioral) ideal CV health is low across the United States.7, 9 Given the demonstrated benefits, the American Heart Association recently published their goal to improve the CV health of all Americans by 2020.1
We have identified a GWAS finding of a common SNP with potential biologic relevance and a consistent clinical impact. rs445925 is in the ApoC1/ApoE region. Apolipoprotein E (encoded by APOE) is primarily found in chylomicron and intermediate-density lipoproteins. In peripheral tissues, APOE mediates cholesterol metabolism while in the central nervous system it is involved in cholesterol transport. The APOE E2 allele (characterized by two SNPs including rs7412 in high LD with our top SNP rs445925) binds poorly to the cell surface receptors and has been shown to be associated with lower LDL-C levels, type III hyperlipoproteinemia and Parkinson’s disease.36 In more recent GWAS studies, SNPs in the APOE locus appear to have pleiotropic effects beyond simply lipid metabolism.36,37 Among African Americans of the PAGE study, variants in the APOE region were found to be associated with multiple components of metabolic syndrome.38 In addition to its effect on the functional form of APOE, rs445925 may also play a role in APOE expression. There is an eQTL of modest strength for this SNP in subcutaneous adipose tissue in which rs445925 is associated with increased APOE expression (p<3.85E-6, beta 0.28, SE 0.06 which exceeds a 1% FDR threshold in the dataset). This same SNP, rs445925, is also associated with levels of subclinical and clinical atherosclerosis with results consistently pointing to the A allele as being associated with greater health. In the CHARGE Consortium, the A (minor) allele of rs445925 is associated with lower common carotid cIMT (discovery p= 5.2 × 10−8) as well as with decreased risk of carotid plaque.28 Similarly, using data from the CHARGE Consortium, the A (minor) allele was associated with lower levels of coronary artery calcification (CAC) (p=6.99×10−5).39 Using data from CardioGram, rs445925 was found to be associated with coronary artery disease (p=0.019), such that the G (major) allele was associated with increased risk of coronary artery disease. In a more recent meta-analysis of 41,513 cases and 65,919 controls in the second stage of the CardioGramPlusC4D Consortium, the G (major) allele of rs445925 was associated with increased risk of coronary artery disease (OR 1.13, p=8.76 × 10−9).10 Thus, there is consistent evidence that this particular SNP provides strong protective effects on a range of traits related to cardiovascular health.
In this study, we found that rs445925 was primarily associated with Ideal Cardiovascular Health through an association with lipid levels; however, its impact may extend beyond simply lipid levels. There are data to suggest that lipid levels are prospectively associated with the development of other cardiovascular risk factors. For example, dyslipidemia is an independent predictor for the incidence of hypertension and diabetes.40–42 In addition, hypercholesterolemia tends to precede the development of other risk factors more often than would be expected by chance.43 These findings generate the hypothesis that persons with low lipid levels may be less likely to manifest other risk factors, and thus that individuals with the affected allele who are more likely to have ideal lipid levels may thus maintain ideal levels of the other risk factors as well (a.k.a. Ideal CV Health). Although lifestyle factors likely play an important role in ideal CV health, our GRS analyses reinforce the role that genetic predisposition to low LDL and possibly low triglycerides is an important contributor to ideal CV health and suggest that certain individuals with a beneficial genetic background (e.g. to lower LDL) are more likely to achieve ideal CV health. Given the association with ideal CV health of SNPs associated with both lower levels of lipids and lower CAD risk, future studies are warranted in larger populations to identify the protective genetic variants underlying ideal health.
This study took advantage of a several large, prospective cohorts with detailed risk factor information to conduct a collaborative meta-analysis of GWAS data. The findings in this study were generated in studies of Caucasians and are not necessarily applicable to racial/ethnic minorities. We examined genetic determinants of being in ideal health in middle-age (50 years of age); however, relatively few cardiovascular cohorts included participants within this age range. In addition, ideal CV health is a relatively low prevalence phenotype. In future replication studies, larger sample sizes may be needed to replicate and extend these findings as we are likely to have been underpowered to detect a more complete repertoire of loci with more modest effects on ideal CV health. In addition to the known important impact of lifestyle modifications, we here have identified evidence for lipid gene variation that underlies ideal CV health.
Supplementary Material
Acknowledgments
We acknowledge the important role of the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) consortium and the collaboration of the CHARGE Subclinical Atherosclerosis/Coronary Heart Disease Working Group in the development and support of this manuscript.
Funding Sources:
The Coronary Artery Risk Development in Young Adults Study (CARDIA) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with the University of Alabama at Birmingham (HHSN268201300025C & HHSN268201300026C), Northwestern University (HHSN268201300027C), University of Minnesota (HHSN268201300028C), Kaiser Foundation Research Institute (HHSN268201300029C), and Johns Hopkins University School of Medicine (HHSN268200900041C). CARDIA is also partially supported by the Intramural Research Program of the National Institute on Aging (NIA) and an intra-agency agreement between NIA and NHLBI (AG0005). This manuscript has been reviewed by CARDIA for scientific content.
The Multi-Ethnic Study of Atherosclerosis (MESA) is supported by contracts N01-HC-95159 through N01-HC-95169 from the National Heart, Lung, and Blood Institute and by grants UL1-RR-024156 and UL1-RR-025005 from National Center for Research Resources.
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
This work was supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No.N01-HC-25195)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
None
REFERENCES
- 1.Lloyd-Jones DM, Hong Y, Labarthe D, Mozaffarian D, Appel LJ, Van Horn L, Greenlund K, Daniels S, Nichol G, Tomaselli GF, Arnett DK, Fonarow GC, Ho PM, Lauer MS, Masoudi FA, Robertson RM, Roger V, Schwamm LH, Sorlie P, Yancy CW, Rosamond WD. Defining and setting national goals for cardiovascular health promotion and disease reduction: The american heart association's strategic impact goal through 2020 and beyond. Circulation. 2010;121:586–613. doi: 10.1161/CIRCULATIONAHA.109.192703. [DOI] [PubMed] [Google Scholar]
- 2.Stamler J, Stamler R, Neaton JD, Wentworth D, Daviglus ML, Garside D, Dyer AR, Liu K, Greenland P. Low risk-factor profile and long-term cardiovascular and noncardiovascular mortality and life expectancy: Findings for 5 large cohorts of young adult and middle-aged men and women. JAMA. 1999;282:2012–2018. doi: 10.1001/jama.282.21.2012. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Jones DM, Dyer AR, Wang R, Daviglus ML, Greenland P. Risk factor burden in middle age and lifetime risks for cardiovascular and non-cardiovascular death (chicago heart association detection project in industry) Am J Cardiol. 2007;99:535–540. doi: 10.1016/j.amjcard.2006.09.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daviglus ML, Liu K, Pirzada A, Yan LL, Garside DB, Greenland P, Manheim LM, Dyer AR, Wang R, Lubitz J, Manning WG, Fries JF, Stamler J. Cardiovascular risk profile earlier in life and medicare costs in the last year of life. Arch Intern Med. 2005;165:1028–1034. doi: 10.1001/archinte.165.9.1028. [DOI] [PubMed] [Google Scholar]
- 5.Daviglus ML, Liu K, Pirzada A, Yan LL, Garside DB, Feinglass J, Guralnik JM, Greenland P, Stamler J. Favorable cardiovascular risk profile in middle age and health-related quality of life in older age. Arch Intern Med. 2003;163:2460–2468. doi: 10.1001/archinte.163.20.2460. [DOI] [PubMed] [Google Scholar]
- 6.Daviglus ML, Liu K, Greenland P, Dyer AR, Garside DB, Manheim L, Lowe LP, Rodin M, Lubitz J, Stamler J. Benefit of a favorable cardiovascular risk-factor profile in middle age with respect to medicare costs. N Engl J Med. 1998;339:1122–1129. doi: 10.1056/NEJM199810153391606. [DOI] [PubMed] [Google Scholar]
- 7.Folsom AR, Yatsuya H, Nettleton JA, Lutsey PL, Cushman M, Rosamond WD. Community prevalence of ideal cardiovascular health, by the american heart association definition, and relationship with cardiovascular disease incidence. J Am Coll Cardiol. 2011;57:1690–1696. doi: 10.1016/j.jacc.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lloyd-Jones DM, Leip EP, Larson MG, D'Agostino RB, Beiser A, Wilson PW, Wolf PA, Levy D. Prediction of lifetime risk for cardiovascular disease by risk factor burden at 50 years of age. Circulation. 2006;113:791–798. doi: 10.1161/CIRCULATIONAHA.105.548206. [DOI] [PubMed] [Google Scholar]
- 9.Ford ES, Li C, Zhao G, Pearson WS, Capewell S. Trends in the prevalence of low risk factor burden for cardiovascular disease among united states adults. Circulation. 2009;120:1181–1188. doi: 10.1161/CIRCULATIONAHA.108.835728. [DOI] [PubMed] [Google Scholar]
- 10.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, Konig IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikainen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Muller-Nurasyid M, Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schafer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrieres J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kahonen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Tregouet DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvanen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimaki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O'Donnell C, Reilly MP, Marz W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparso T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proenca C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O'Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jorgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martinez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orru M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tanaka T, Thorand B, Tichet J, Tonjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Rios M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O'Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O'Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O'Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT, Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-wide association scan shows genetic variants in the fto gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristiansson K, Perola M, Tikkanen E, Kettunen J, Surakka I, Havulinna AS, Stancakova A, Barnes C, Widen E, Kajantie E, Eriksson JG, Viikari J, Kahonen M, Lehtimaki T, Raitakari OT, Hartikainen AL, Ruokonen A, Pouta A, Jula A, Kangas AJ, Soininen P, Ala-Korpela M, Mannisto S, Jousilahti P, Bonnycastle LL, Jarvelin MR, Kuusisto J, Collins FS, Laakso M, Hurles ME, Palotie A, Peltonen L, Ripatti S, Salomaa V. Genome-wide screen for metabolic syndrome susceptibility loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet. 2012;5:242–249. doi: 10.1161/CIRCGENETICS.111.961482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraja AT, Vaidya D, Pankow JS, Goodarzi MO, Assimes TL, Kullo IJ, Sovio U, Mathias RA, Sun YV, Franceschini N, Absher D, Li G, Zhang Q, Feitosa MF, Glazer NL, Haritunians T, Hartikainen AL, Knowles JW, North KE, Iribarren C, Kral B, Yanek L, O'Reilly PF, McCarthy MI, Jaquish C, Couper DJ, Chakravarti A, Psaty BM, Becker LC, Province MA, Boerwinkle E, Quertermous T, Palotie L, Jarvelin MR, Becker DM, Kardia SL, Rotter JI, Chen YD, Borecki IB. A bivariate genome-wide approach to metabolic syndrome: Stampeed consortium. Diabetes. 2011;60:1329–1339. doi: 10.2337/db10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JC, Boerwinkle E, Consortium C. Cohorts for heart and aging research in genomic epidemiology (charge) consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, Nelson JC, O'Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: Objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 18.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The framingham offspring study. Design and preliminary data. Prev Med. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 19.Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR, Jr, Liu K, Savage PJ. Cardia: Study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41:1105–1116. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 20.The atherosclerosis risk in communities (aric) study: Design and objectives. The aric investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 21.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, Gudnason V, Harris TB, Kathiresan S, Kraaij R, Launer LJ, Levy D, Liu Y, Mosley T, Peloso GM, Psaty BM, Rich SS, Rivadeneira F, Siscovick DS, Smith AV, Uitterlinden A, van Duijn CM, Wilson JG, O'Donnell CJ, Rotter JI, Boerwinkle E. Best practices and joint calling of the humanexome beadchip: The charge consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. Snap: A web-based tool for identification and annotation of proxy snps using hapmap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson T. Efficient calculation for multi-snp genetic risk scores. American Society of Human Genetics Annual Meeting. 2012 [Google Scholar]
- 25.Global Lipids Genetics C, Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O'Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Doring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, Bell JT, Yang TP, Meduri E, Barrett A, Nisbett J, Sekowska M, Wilk A, Shin SY, Glass D, Travers M, Min JL, Ring S, Ho K, Thorleifsson G, Kong A, Thorsteindottir U, Ainali C, Dimas AS, Hassanali N, Ingle C, Knowles D, Krestyaninova M, Lowe CE, Di Meglio P, Montgomery SB, Parts L, Potter S, Surdulescu G, Tsaprouni L, Tsoka S, Bataille V, Durbin R, Nestle FO, O'Rahilly S, Soranzo N, Lindgren CM, Zondervan KT, Ahmadi KR, Schadt EE, Stefansson K, Smith GD, McCarthy MI, Deloukas P, Dermitzakis ET, Spector TD. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ken-Dror G, Talmud PJ, Humphries SE, Drenos F. Apoe/c1/c4/c2 gene cluster genotypes, haplotypes and lipid levels in prospective coronary heart disease risk among uk healthy men. Mol Med. 2010;16:389–399. doi: 10.2119/molmed.2010.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bis JC, Kavousi M, Franceschini N, Isaacs A, Abecasis GR, Schminke U, Post WS, Smith AV, Cupples LA, Markus HS, Schmidt R, Huffman JE, Lehtimaki T, Baumert J, Munzel T, Heckbert SR, Dehghan A, North K, Oostra B, Bevan S, Stoegerer EM, Hayward C, Raitakari O, Meisinger C, Schillert A, Sanna S, Volzke H, Cheng YC, Thorsson B, Fox CS, Rice K, Rivadeneira F, Nambi V, Halperin E, Petrovic KE, Peltonen L, Wichmann HE, Schnabel RB, Dorr M, Parsa A, Aspelund T, Demissie S, Kathiresan S, Reilly MP, Taylor K, Uitterlinden A, Couper DJ, Sitzer M, Kahonen M, Illig T, Wild PS, Orru M, Ludemann J, Shuldiner AR, Eiriksdottir G, White CC, Rotter JI, Hofman A, Seissler J, Zeller T, Usala G, Ernst F, Launer LJ, D'Agostino RB, Sr, O'Leary DH, Ballantyne C, Thiery J, Ziegler A, Lakatta EG, Chilukoti RK, Harris TB, Wolf PA, Psaty BM, Polak JF, Li X, Rathmann W, Uda M, Boerwinkle E, Klopp N, Schmidt H, Wilson JF, Viikari J, Koenig W, Blankenberg S, Newman AB, Witteman J, Heiss G, Duijn C, Scuteri A, Homuth G, Mitchell BD, Gudnason V, O'Donnell CJ. Meta-analysis of genome-wide association studies from the charge consortium identifies common variants associated with carotid intima media thickness and plaque. Nat Genet. 2011;43:940–947. doi: 10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith EN, Chen W, Kahonen M, Kettunen J, Lehtimaki T, Peltonen L, Raitakari OT, Salem RM, Schork NJ, Shaw M, Srinivasan SR, Topol EJ, Viikari JS, Berenson GS, Murray SS. Longitudinal genome-wide association of cardiovascular disease risk factors in the bogalusa heart study. PLoS Genet. 2010;6:e1001094. doi: 10.1371/journal.pgen.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, Pankratz VS, Younkin SG, Saykin AJ, Jun G, Baldwin C, Logue MW, Buros J, Farrer L, Pericak-Vance MA, Haines JL, Sweet RA, Ganguli M, Feingold E, Dekosky ST, Lopez OL, Barmada MM. Genome-wide association study of alzheimer's disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, Swaminathan S, Shen L, Risacher SL, Nho K, Foroud T, Shaw LM, Trojanowski JQ, Potkin SG, Huentelman MJ, Craig DW, DeChairo BM, Aisen PS, Petersen RC, Weiner MW, Saykin AJ. Genome-wide association study of csf biomarkers abeta1-42, t-tau, and p-tau181p in the adni cohort. Neurology. 2011;76:69–79. doi: 10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanche H, Junge O, Wittig M, Ellinghaus D, Flachsbart F, Wichmann HE, Meitinger T, Nikolaus S, Franke A, Krawczak M, Lathrop M, Schreiber S. A genome-wide association study confirms apoe as the major gene influencing survival in long-lived individuals. Mech Ageing Dev. 2011;132:324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Lyytikainen LP, Kangas AJ, Soininen P, Wurtz P, Silander K, Dick DM, Rose RJ, Savolainen MJ, Viikari J, Kahonen M, Lehtimaki T, Pietilainen KH, Inouye M, McCarthy MI, Jula A, Eriksson J, Raitakari OT, Salomaa V, Kaprio J, Jarvelin MR, Peltonen L, Perola M, Freimer NB, Ala-Korpela M, Palotie A, Ripatti S. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkins JT, Ning H, Berry J, Zhao L, Dyer AR, Lloyd-Jones DM. Lifetime risk and years lived free of total cardiovascular disease. JAMA. 2012;308:1795–1801. doi: 10.1001/jama.2012.14312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X, Chen PC, Poole C. Apoe-[epsilon]2 allele associated with higher prevalence of sporadic parkinson disease. Neurology. 2004;62:2198–2202. doi: 10.1212/01.wnl.0000130159.28215.6a. [DOI] [PubMed] [Google Scholar]
- 37.Johnson AD, O'Donnell CJ. An open access database of genome-wide association results. BMC Med Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carty CL, Bhattacharjee S, Haessler J, Cheng I, Hindorff LA, Aroda V, Carlson CS, Hsu CN, Wilkens L, Liu S, Selvin E, Jackson R, North KE, Peters U, Pankow JS, Chatterjee N, Kooperberg C. Analysis of metabolic syndrome components in >15 000 african americans identifies pleiotropic variants: Results from the population architecture using genomics and epidemiology study. Circ Cardiovasc Genet. 2014;7:505–513. doi: 10.1161/CIRCGENETICS.113.000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Donnell CJ, Kavousi M, Smith AV, Kardia SL, Feitosa MF, Hwang SJ, Sun YV, Province MA, Aspelund T, Dehghan A, Hoffmann U, Bielak LF, Zhang Q, Eiriksdottir G, van Duijn CM, Fox CS, de Andrade M, Kraja AT, Sigurdsson S, Elias-Smale SE, Murabito JM, Launer LJ, van der Lugt A, Kathiresan S, Consortium CA, Krestin GP, Herrington DM, Howard TD, Liu Y, Post W, Mitchell BD, O'Connell JR, Shen H, Shuldiner AR, Altshuler D, Elosua R, Salomaa V, Schwartz SM, Siscovick DS, Voight BF, Bis JC, Glazer NL, Psaty BM, Boerwinkle E, Heiss G, Blankenberg S, Zeller T, Wild PS, Schnabel RB, Schillert A, Ziegler A, Munzel TF, White CC, Rotter JI, Nalls M, Oudkerk M, Johnson AD, Newman AB, Uitterlinden AG, Massaro JM, Cunningham J, Harris TB, Hofman A, Peyser PA, Borecki IB, Cupples LA, Gudnason V, Witteman JC. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011;124:2855–2864. doi: 10.1161/CIRCULATIONAHA.110.974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halperin RO, Sesso HD, Ma J, Buring JE, Stampfer MJ, Gaziano JM. Dyslipidemia and the risk of incident hypertension in men. Hypertension. 2006;47:45–50. doi: 10.1161/01.HYP.0000196306.42418.0e. [DOI] [PubMed] [Google Scholar]
- 41.Sesso HD, Buring JE, Chown MJ, Ridker PM, Gaziano JM. A prospective study of plasma lipid levels and hypertension in women. Arch Intern Med. 2005;165:2420–2427. doi: 10.1001/archinte.165.20.2420. [DOI] [PubMed] [Google Scholar]
- 42.Hanson RL, Imperatore G, Bennett PH, Knowler WC. Components of the "metabolic syndrome" and incidence of type 2 diabetes. Diabetes. 2002;51:3120–3127. doi: 10.2337/diabetes.51.10.3120. [DOI] [PubMed] [Google Scholar]
- 43.Tershakovec AM, Jawad AF, Stouffer NO, Elkasabany A, Srinivasan SR, Berenson GS. Persistent hypercholesterolemia is associated with the development of obesity among girls: The bogalusa heart study. Am J Clin Nutr. 2002;76:730–735. doi: 10.1093/ajcn/76.4.730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.