Abstract
Hemocyanins induce a potent Th1-dominant immune response with beneficial clinical outcomes when used as a carrier/adjuvant in vaccines and nonspecific immunostimulant in cancer. However, the mechanisms by which hemocyanins trigger innate immune responses, leading to beneficial adaptive immune responses, are unknown. This response is triggered by a proinflammatory signal from various components, of which macrophages are an essential part. To understand how these proteins influence macrophage response, we investigated the effects of mollusks hemocyanins with varying structural and immunological properties, including hemocyanins from Concholepas concholepas, Fissurella latimarginata, and Megathura crenulata (keyhole limpet hemocyanin), on cultures of peritoneal macrophages. Hemocyanins were phagocytosed and slowly processed. Analysis of this process showed differential gene expression along with protein levels of proinflammatory markers, including IL-1β, IL-6, IL-12p40, and TNF-α. An extended expression analysis of 84 cytokines during a 24-h period showed a robust proinflammatory response for F. latimarginata hemocyanin in comparison with keyhole limpet hemocyanin and C. concholepas hemocyanin, which was characterized by an increase in the transcript levels of M1 cytokines involved in leukocyte recruitment. These cytokine genes included chemokines (Cxcl1, Cxcl3, Cxcl5, Ccl2, and Ccl3), ILs (Il1b and Ifng), growth factors (Csf2 and Csf3), and TNF family members (Cd40lg). The protein levels of certain cytokines were increased. However, every hemocyanin maintains downregulated key M2 cytokine genes, including Il4 and Il5. Collectively, our data demonstrate that hemocyanins are able to trigger the release of proinflammatory factors with different patterns of cytokine expression, suggesting differential signaling pathways and transcriptional network mechanisms that lead to the activation of M1-polarized macrophages.
Introduction
Hemocyanins from mollusks are enormous glycoproteins with multiple functions, where oxygen transport is the best known (1, 2). Hemocyanins alone are capable of potently activating the immune system of mammals. Therefore, hemocyanins have been used as a biotechnological tool in the development of Abs and vaccines, and also as immunomodulators such as nonspecific immunostimulants in superficial bladder cancer (3). The hemocyanin purified from the giant keyhole limpet gastropod Megathura crenulata, commonly known as keyhole limpet hemocyanin (KLH), has traditionally been used for those purposes (4–6). However, the biodiversity of hemocyanins has prompted interest in developing new candidates with better biochemical and immunological properties because the supply depends on natural resources. Therefore, hemocyanins from Concholepas concholepas (CCH) (7) and Fissurella latimarginata (FLH) (8), among others (6), have been studied. CCH is used as a carrier (9) and experimental Ag (10), and it has been preclinically evaluated in a murine model of superficial bladder cancer and showed antitumor effects similar to KLH (11). Additionally, CCH has been used as an adjuvant in dendritic cell (DC) immunotherapy of patients with prostate cancer, and it is considered a safe complement or alternative to KLH (12, 13). In contrast, FLH has shown intrinsically greater immunogenic capabilities, exhibiting a more potent antitumor activity than CCH and KLH against melanoma in the B16F10 mouse melanoma model (8).
Therefore, the immunostimulatory properties of these hemocyanins show differences in immunogenicity and immunomodulatory effects, indicating that different hemocyanins can activate diverse molecular and cellular pathways to promote Th1 immune responses. Studying the effects of these hemocyanins on innate immune cells might help us to understand their different properties; however, the effect of hemocyanins at this level and the mechanism by which the target cell types incorporate these proteins and promote their immunostimulatory effects have not been thoroughly studied.
We propose that when hemocyanins are incorporated by APCs, they induce a proinflammatory milieu that produces a bystander effect, which is a key event in their nonspecific immunostimulatory activity (14). Indeed, macrophages play an essential role in both innate and adaptive immunity as regulatory and effectors cells, and as one of the primary danger sensors in the host (15, 16). Macrophages display remarkable plasticity and flexibility; in response to Ags, they have the ability to polarize innate immune responses to produce various M1 or M2 patterns of cytokines that can be reversed in vitro and in vivo and that are stages that mirror the Th1/Th2 polarization of T cells (17–19). Pioneer investigations by Unanue and Cerottini (20) demonstrated that macrophages take up and catabolize hemocyanins; however, whether hemocyanins induce macrophage activation remains unclear.
Several authors have explained that the immunomodulatory effect of hemocyanins is a result of the extremely complex xenogeneic structures (4, 6, 21), which resemble an icosahedral virus capsid (22). In fact, mollusk hemocyanins are enormous glycoproteins of ∼4 MDa, composed by an intricate arrangement of non–covalently linked subunits. The basic quaternary structure is a cylindrical decamer of 35 nm in diameter and 18 nm in height. The decamers in gastropod hemocyanins may associate in pairs to form colossal molecules called didecamers, with molecular masses of ∼8 MDa (23). The hemocyanins used in this study present clear structural differences at this level, and experimental data have demonstrated that the KLH preparation is made up of two independent isoforms that coexist in variable proportions in the hemolymph of the animals, each composed of one type of subunit (24). In contrast, the two subunits of CCH are intermingled in the molecule, forming heterodidecamers (7). FLH is composed of a single type of subunit that forms homodidecamers (8). Another significant feature of the hemocyanin structure is the presence of carbohydrates, as heterogeneous N- and O-glycosylation ramifications, with mannose being the common and most abundant oligosaccharide (25–30). Although the three hemocyanins present N- and O-linked oligosaccharides, there are differences in their arrangements. For instance, KLH and FLH present xylose and sialic acid, but CCH is devoid of these sugars (8).
To better understand how these hemocyanins influence macrophage responses, we investigated the effects of CCH, FLH, and KLH on in vitro cultures of murine peritoneal macrophages. We studied their endocytosis and the temporal patterns of mRNA expression and protein levels of a panel of cytokines, using quantitative PCR (qPCR) and ELISA. Our main findings showed that hemocyanins are slowly processed by these cells. In response to structurally diverse hemocyanins, macrophages undergo activation and display different temporal patterns of cytokine gene expression and protein secretion, leading to an M1-polarized proinflammatory milieu. We propose that in vivo these effects activate bystander cell signaling that might increase tumor killing and other latent-specific responses.
Materials and Methods
Hemocyanin sources
The hemocyanins from C. concholepas and F. latimarginata that were isolated and purified under sterile and pyrogen-free conditions were provided by Biosonda (Santiago, Chile), and both proteins were suspended in PBS (0.1 M sodium phosphate, 0.15 M NaCl [pH 7.2]). Lyophilized KLH (from M. crenulata) in PBS was purchased from Thermo Scientific (Waltham, MA) and Calbiochem (San Diego, CA). Deglycosylated CCH and FLH were produced by sodium periodate treatment using the procedure and controls in Arancibia et al. (8). To ensure that the biological effects observed were due to the hemocyanin component, the endotoxin contents of the hemocyanin preparations were determined using a PyroGene recombinant factor C endotoxin detection assay kit (Lonza Group, Walkersville, MD). All of the chemicals were analytical-grade reagents, and the solutions were prepared using water for human irrigation (Baxter Healthcare, Charlotte, NC) and filtered through a 0.2-μm membrane filter (Millipore, Billerica, MA).
Mice
C57BL/6 mice were obtained from the Universidad de Chile (Santiago, Chile) and bred at Biosonda. The experimental mice were housed between 22 and 24°C with a light/dark cycle of 12/12 h. Sterile water and food were available ad libitum. The study was performed in strict accordance with the Guidelines for the Care and Use of Laboratory Animals (National Commission for Scientific and Technological Research of Chile). The National Commission for Scientific and Technological Research Committee on Animal Welfare approved all of the animal protocols used in this study.
Preparation and characterization of macrophages
Macrophages from C57BL/6 mice were prepared according to the method described by Zhang et al. (31) with modifications. Briefly, the mice were injected i.p. with 1.6 ml sterile solution of thioglycollate-enriched medium. After 4 d, the animals were euthanized and immediately injected with 5 ml sterile ice-cold PBS into the peritoneal cavity and the peritoneal fluid was collected in 15-ml conical Falcon tubes. The tubes were centrifuged at 1500 rpm for 10 min at 25°C, and the pellets were collected. The cells were resuspended for 5 min at 37°C in 2.5 ml erythrocyte lysis solution, and the lysis was halted by adding 2.5 ml DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated FBS (HyClone Laboratories, St. Louis, MO), 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 1 mM l-glutamine, and 1 mM nonessential amino acids (Invitrogen). After centrifugation for 10 min at 25°C, the pellets were resuspended in complete DMEM and quantified. The cells in complete DMEM were seeded at 1 × 106 cells/ml in a plastic petri dish and cultured for 1 h at 37°C in 10% CO2. The culture supernatant containing no adherent cells was discarded. Adherent cells were washed with the same culture conditions. The cultures were observed and imaged on the culture plates under a Zeiss inverted phase-contrast light microscope. The purity and homogeneity of the cultures were determined by flow cytometry using a FACScan flow cytometer (Flow Cytometry Facility, Faculty of Science, Universidad de Chile) and analyzed using Weasel software. In this assay, we used Escherichia coli LPS (0111.B4 type; Sigma-Aldrich, St. Louis, MO) as a positive control at 100 ng/ml. Consequently, the macrophages used in this study were of a larger cell size with a granular cytoplasm and were practically homogeneously stained to an anti-F4/80 mAb (BD Biosciences, San Diego, CA). mAbs to CD80, CD86, CD40, and MHC class II (MHC-II; I-Ab) were all from BD Biosciences.
Transmission electron microscopy
The in vitro–cultured macrophages that were adhered to the 24-well cultured plates that had been previously treated with hemocyanins for 10 and 60 min were washed with PBS and fixed with a solution of 2% glutaraldehyde (Polysciences, Warrington, PA) in 0.1 M sodium cacodylate solution (pH 7.4) for 3 min; then, the macrophages were carefully scraped with a Teflon spatula. The cells were centrifuged at 5000 rpm for 5 min and maintained in the fixative for 1 h. The cells were postfixed with OsO4, dehydrated, and embedded in Epon (Polysciences) according to the method of Luft (32). Then, the samples were stained with lead citrate according to the procedure of Reynolds (33). The preparations were examined and photographed at 80 kV with a Philips Tecnai 12 electron microscope (Electronic Microscopy Facility, Pontificia Universidad Católica de Chile).
SDS-PAGE and Western blotting
To determine the processing pattern of CCH, FLH, and KLH by macrophages, we used the general procedure previously described by Arancibia et al. (14). Briefly, the macrophages were incubated with 100 μg/ml each protein at 37°C for 4, 24, 72, 120, and 144 h. For the electrophoretic analysis of macrophage extracts, the cells were washed with PBS, harvested with trypsin/EDTA for 30 min at 37°C, and lysed with a low-detergent buffer (20 mM Tris/HCl [pH 7.5], 2 mM EDTA, 150 mM NaCl, and 0.5% Triton X-100). The macrophage extracts were mixed with a buffer containing SDS and 2-ME, heated for 5 min at 100°C, and analyzed by SDS-PAGE on a gradient (5–15%) polyacrylamide separating gel as described by Laemmli (34). For the Western blot analyses, polyacrylamide gels containing samples were transferred onto a 0.45-μm pore nitrocellulose membrane (Pierce/Endogen, Rockford, IL) at 100 V for 1.5 h as described by Towbin et al. (35) with minor modifications. The membranes were incubated overnight at 4°C with TBS containing 1% casein and 0.02% Tween 20, and the hemocyanin bands were then visualized with an anti-hemocyanin rabbit antiserum developed by our laboratory as previously described (36). An anti–β-actin Ab (Sigma-Aldrich) was used as a loading control.
Real-time quantitative RT-PCR analysis
The procedures of Barksby et al. (37) and Schmid et al. (38) were used with modifications. Total RNA was isolated from freshly harvested cultured macrophages using the RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Total RNA was quantified at 260 nm, and its integrity and absence of genomic DNA were examined by 1% agarose-formaldehyde gel electrophoresis. cDNA was synthesized using the Sensiscript RT kit (Qiagen) according to the manufacturer’s instructions. Then, single-stranded cDNA was subjected to PCR using the TopTaq polymerase kit (Qiagen). The oligonucleotide primers for well-characterized markers of inflammation, including IL-1β, IL-6, IL-12p40, and TNF-α, as well as those of the housekeeping genes, β-actin and GAPDH, were prepared by Eurofins MWG Operon (Huntsville, AL) and are provided in Supplemental Table I (39–41). The resulting PCR products were electrophoresed on a 2% agarose gel, stained with GelRed (Biotium, Hayward, CA), and photographed.
Real-time PCR was performed using RT2 SYBR Green qPCR master mixes (SABiosciences, Hilden, Germany) as a detection dye to determine the relative cDNA levels of IL-1β, IL-6, IL-12p40, and TNF-α genes in each sample on the Rotor-Gene Q 100 (Qiagen). β-Actin was chosen as the housekeeping gene for normalization. Threshold cycle (Ct) values were collected and used for ΔΔCt analysis. The relative amount of each target gene mRNA normalized to the housekeeping gene was calculated as 2−(ΔCt cytokine −ΔCt housekeeping gene). The fold change of each target gene’s mRNA was obtained relative to the control macrophages, that is, macrophages incubated with culture medium only and calculated as ΔΔCt = 2(ΔCt target gene in hemocyanins or LPS-cultured macrophages − ΔCt target gene in control macrophages) according to the RT2 Profiler PCR array system handbook (SABiosciences). Experiments were performed in triplicate and repeated at least three times.
PCR array
The RT2 Profiler PCR array (SABiosciences) was used for cytokine/chemokine expression analysis according to the manufacturer’s instructions. RNA (1 μg) was extracted and converted into cDNA with the RT2 first-strand kit (SABiosciences). This array contains primers for 84 different cytokine-related genes, including the following: 15 growth factors (Bmp2, Bmp4, Bmp6, Bmp7, Cntf, Csf1, Csf2, Csf3, Gpi1, Lif, Mstn, Nodal, Osm, Thpo, Vegfa); 25 chemokines (Ccl1, Ccl11, Ccl12, Ccl17, Ccl2, Ccl20, Ccl22, Ccl24, Ccl3, Ccl4, Ccl5, Ccl7, Cx3cl1, Cxcl1, Cxcl10, Cxcl11, Cxcl12, Cxcl13, Cxcl16, Cxcl3, Cxcl5, Cxcl9, Pf4, Ppbp, Xcl1); 13 ILs (Il15, Il16, Il17a, Il17f, Il1a, Il1b, Il1rn, Il21, Il23a, Il27, Il5, Il7, Il9); 2 IFNs (Ifna2, Ifng); 14 anti-inflammatory cytokines (Ccl19, Il10, Il11, Il12a, Il12b, Il13, Il18, Il2, Il22, Il23a, Il24, Il4, Il6, Tgfb2); 10 TNF superfamily members (Cd40lg, Cd70, Fasl, Lta, Ltb, Tnf, Tnfrsf11b, Tnfsf10, Tnfsf11, Tnfsf13b); and 5 cytokines (Adipoq, Ctf1, Hc, Mif, Spp1). Experiments were performed in triplicate and repeated at least three times. The PCR array data were analyzed using the RT2 Profiler PCR array data analysis Web portal from SABiosciences based on the ΔΔCt method with five different housekeeping genes (Hsp90ab1, Gapdh, Actb, Gusb, and Hprt).
Measurement of cytokine protein levels
The levels of the cytokines IL-1β, TNF-α, IL-6, and IL-12p40 were measured in the supernatants of macrophages cultured with hemocyanins or with LPS (positive control) at 100 ng/ml or in culture medium (negative control), using quantitative commercial ELISA kits according to the manufacturer’s instructions (DuoSet ELISA development systems, R&D Systems, Minneapolis, MN). Additionally, a panel of 32 cytokines, including chemokines (Eotaxin-1/Ccl11, MCP-1/Ccl2, MIP-1α/Ccl3, MIP-1β/Ccl4, MIP-2/Cxcl2, RANTES/Ccl5, KC/Cxcl1, IP-10/Cxcl10, LIX/Cxcl5, and MIG/Cxcl9), growth factors (M-CSF/Csf1, G-CSF/Csf3, GM-CSF/Csf2, LIF/Lif), IFNs (IFN-γ/Ifng), ILs (IL-10/Il10, IL-12p40/Il12b, IL12p70, IL-13/Il13, IL-15/Il15, IL-17A/Il17a, IL-1α/Il1a, IL-1β/Il1b, IL-3/Il3, IL-4/Il4, IL-5/Il5, IL-6/Il6, IL-7/Il7, Il9/Il9, IL-10/Il10), TNF superfamily member (TNF-α/Tnf), and others (VEGF/Vegfa) in the supernatants of macrophages cultured as above was analyzed by Eve Technologies (Calgary, AB, Canada) with a mouse cytokine array/chemokine array 32-plex panel using multiplexing laser bead technology. As a control, a sample of the culture medium alone was included in both types of ELISA analysis. The experiments were performed in triplicate and repeated at least three times.
Statistical analyses
For the cytokine determination experiments by ELISA and ELISArray, the data were subjected to a square root transformation (42). Comparisons between groups were performed using an unpaired two-tailed Student t test. The qPCR for normally distributed data used the same analysis; however, for data that were not normally distributed, the Kruskal–Wallis rank test was used. The analyses were performed using GraphPad Prism version 6.0 (GraphPad Software, San Diego, CA). To analyze the PCR array data, an integrated Web-based software, that is, the RT2 Profiler PCR array software package (SABiosciences), was used. An analysis of the relative quantification was performed using the ΔΔCt method (43). For each PCR reaction, two normalized average Ct values (control and test), an unpaired two-tailed t test (p value), and fold changes were calculated. Based on the Student t test, differences were considered significant when p < 0.05.
Results
Hemocyanins are phagocytosed by macrophages and slowly processed into smaller peptide fragments
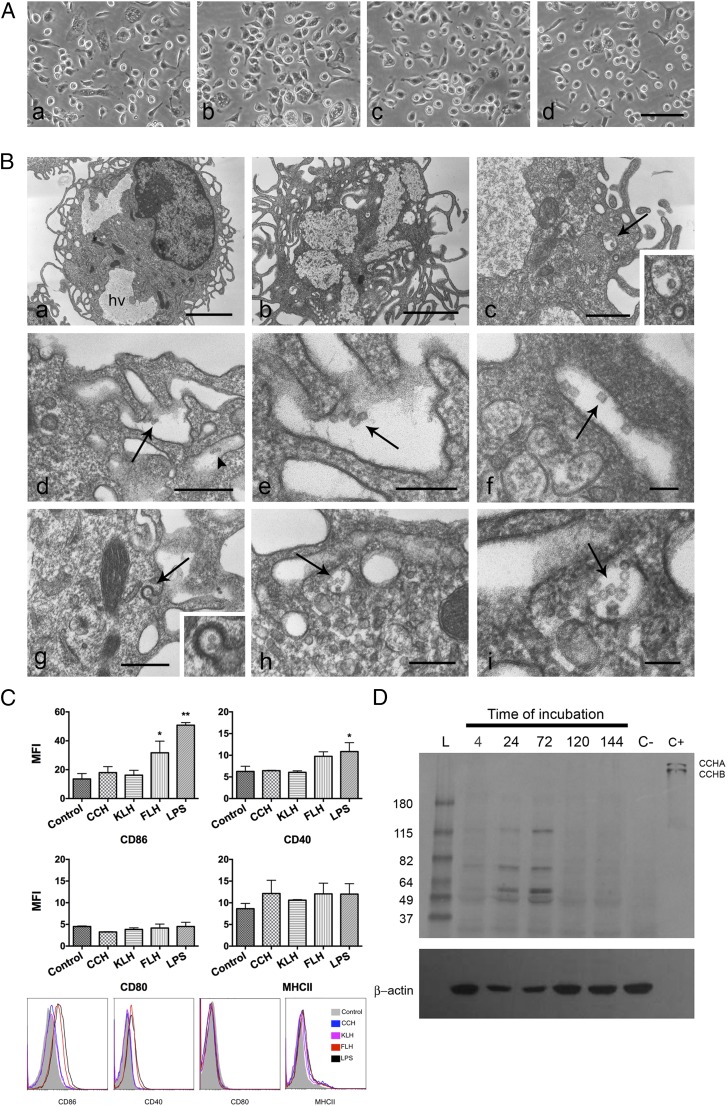
Peritoneal macrophages were cultured at different time points, up to 6 d, with CCH, KLH, or FLH at a concentration range from 100 μg/ml to 1 mg/ml. Culture media supplemented with or without LPS were used as the positive and negative controls, respectively. Direct visual analysis of the culture plates by phase-contrast light microscopy did not show any morphological changes characteristic of cell death induced by hemocyanins (Fig. 1A). Transmission electron microscopy analyses were performed at early time points to visualize the localization of hemocyanin molecules by taking advantage of their large size and hollow cylindrical features. These analyses showed that hemocyanin molecules, both native and deglycosylated, were quickly incorporated by macrophages, primarily through macropinocytosis (Fig. 1Bb–f) and clathrin-coated pits (Fig. 1Bg). Finally, the molecules were recognized inside endosome-like vesicles (Fig. 1Bh, 1Bi). Next, we used flow cytometry to analyze Ag processing and presentation by macrophages. We found that after 24 h of incubation with the different hemocyanins, the macrophages exhibited a statistically significant difference in the upregulation of the costimulatory molecule of CD86 only under FLH stimulation; neither KLH nor CCH was able to increase the levels of costimulatory molecules (Fig. 1C). The expressions of NO, a marker of fully activated macrophages (19), and Arginase-1, an M2 macrophage marker (44), were assessed to explore the functional effects of hemocyanins, and no changes in either marker occurred at 24 h of treatment (Supplemental Fig. 1).
FIGURE 1.
Hemocyanins are phagocytosed by macrophages and slowly processed. (A) Morphological characteristics of cultures at 24 h observed directly on tissue culture plates by phase-contrast microscopy. (Aa) Negative control: culture media. Cells show the characteristic spherical and elongated forms of macrophages (88). (Ab) Positive control: macrophages incubated with LPS. Cells show a predominant elongated form with a granular cytoplasm. (Ac and Ad) Cultures with CCH and FLH, respectively, showing spherical and elongated cells. Scale bars, 50 μm. (B) Transmission electron microscopy images of hemocyanin internalization by macrophages. The hollow cylinder structure of the hemocyanin molecules viewed from the top (circles) and side (rectangles) (shown with arrows). (Ba) Negative control. Macrophages show an irregular nucleus, a cytoplasm with heterophagic vacuoles (hv) and superficial undulant folds, and pseudopodia. Scale bar, 3 μm. (Bb) Macrophages cultured with FLH for 30 min show larger hv and superficial cytoplasm with signs of intense phagocytic activity. Scale bar, 2 μm. (Bc) Hemocyanin molecules inside of a clear vacuole at 1 h of coincubation. Inset is enlarged image of the area indicated with arrow. Scale bar, 0.5 μm. (Bd) Macrophages cultured for 10 min with deglycosylated CCH showing molecules being internalized by macropinocytosis. Scale bar, 0.5 μm. (Be) Higher magnification of the area indicated with arrows in (Bd). Scale bar, 0.2 μm. (Bf) Higher magnification of hemocyanin molecules in a pinocytic vacuole. Scale bar, 0.1 μm. (Bg) Macrophages cultured for 10 min with deglycosylated CCH showing molecules internalized by clathrin-coated vesicles showing their characteristic basket-like structure; inset, area indicated with arrows. Scale bar, 0.5 μm. (Bh) Hemocyanin molecules inside of an early endosome. (Bi) Higher magnification of the area indicated with arrows in (Bh). Scale bar, 0.3 μm. (C) Hemocyanins and macrophage maturation markers. Flow cytometry analysis of maturation markers. Macrophages were cultured with CCH, FLH, and KLH for 24 h and then analyzed CD80, CD86, CD40, and MHC-II. F4/80 was used to identify macrophages (not shown). Culture media alone and LPS were used as the negative and positive control, respectively. Results are presented as mean ± SD of the mean fluorescence intensity (MFI) of n = 2 experiments. *p < 0.05, **p < 0.01. (D) Kinetics of hemocyanin processing by macrophages analyzed by Western blotting. Macrophages were cultured for 4–144 h with CCH. The cell extracts were immunoblotted with rabbit anti-CCH Abs followed by enzymatic detection. As a negative control (C−), extracts were prepared with macrophages alone and cultured without hemocyanin. As a positive control (C+), CCH alone was used. The characteristic banding pattern of CCH subunits (CCHA and CCHB) is observed. β-Actin was used as a loading control using a specific mAb and chemiluminescent detection. The results are representative of at least 10 independent experiments performed with the different hemocyanins.
Complementary analyses by immunoblotting showed the presence of hemocyanin fragments as faint bands after 4 h of incubation (Fig. 1D), with a maximum intensity at 72 h and with a persistent faint band at 46 kDa. We think that this band corresponds to a functional unit, which is the minimal organizational structural/functional unit of hemocyanins (1). Similar results were observed for the three hemocyanins.
Collectively, these data indicate that the molecules were internalized and processed slowly by macrophages, independently of the structural differences between these three hemocyanins. Our early in vivo studies were consistent with these observations. When we compared the incorporation of CCH labeled with Alexa Fluor 488 (CCH-488) and OVA (OVA-488) as a control, we observed that both Ags were incorporated by a similar percentage of peritoneal macrophages (Supplemental Fig. 2A). However, OVA-488 endocytosis occurred faster than that of CCH-488, with peak incorporation at ∼14 and 24 h, respectively, and decay in fluorescence, which was attributed to protein processing, observed until 20 and 72 h, respectively (Supplemental Fig. 2B). Therefore, these results strongly suggest that the slow Ag processing in macrophages, which also occurs in DCs (8, 14), allows the sequestration of these proteins for presentation to T lymphocytes several days later, which is a key feature of Ags that enhance immunogenicity (45).
Different hemocyanins induce distinctive temporal patterns of proinflammatory cytokine expression in macrophages
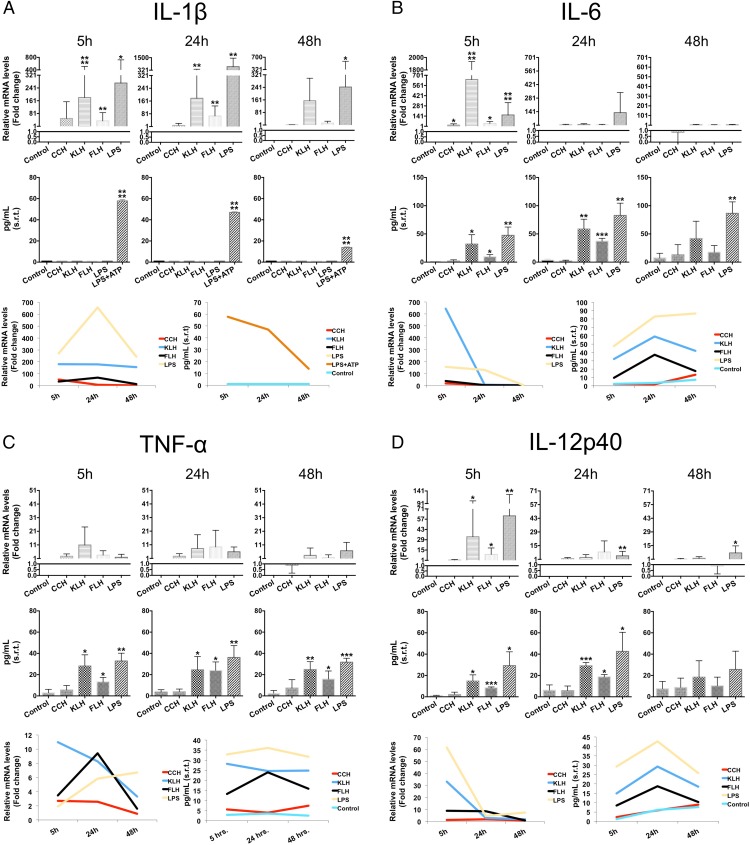
To address whether hemocyanins induce a proinflammatory state in macrophages, we evaluated temporal mRNA expression changes along with protein levels using qPCR and a specific capture ELISA analysis, respectively. We limited our analyses to four well-characterized markers of inflammation: IL-1β, IL-6, IL-12p40, and TNF-α. Macrophages were incubated at different time points (5, 24, and 48 h) with 1 mg/ml of each hemocyanin. We used untreated macrophages and macrophages cultured with LPS (100 ng/ml) as negative and positive controls, respectively. The results are shown in Fig. 2.
FIGURE 2.
Hemocyanins induce the expression of certain characteristic markers of inflammation at different times and intensities. In parallel with the reverse transcription quantitative PCR (RT-qPCR) results (upper panel), the conventional ELISA results (middle panel) were analyzed for IL-1β, IL-6, TNF-α, and IL-12p40. The lower panels correspond to a temporal profile of each cytokine using the average of the data presented in each graph. Macrophages were stimulated for different durations (5, 24, and 48 h) with CCH, KLH, FLH (1 mg/ml), or LPS (100 ng/ml; positive control). Culture medium was used as a negative control. For the qPCR data analysis, triplicate Ct values obtained from three different experimental mRNA samples were normalized to β-actin mRNA. Relative mRNA levels were calculated using the ΔΔCt method, and normalized Ct values were obtained from untreated macrophages (negative control). (A) RT-qPCR and ELISA for Il-1β. (B) RT-qPCR and ELISA for IL-6. (C) RT-qPCR and ELISA for TNF-α. (D) RT-qPCR and ELISA for IL-12p40. Results are shown as square root–transformed (s.r.t) data and are representative of three experiments performed in triplicate. A Student t test was used. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
We found that all hemocyanins induced Il1b gene expression; however, CCH upregulated mRNA levels early, at 5 h after stimulation, whereas KLH and FLH induced the upregulation at 24 h, similar to the results with LPS (Fig. 2A, upper panel). Interestingly, IL-1β protein was not found in the culture medium (or the protein levels were lower than the detection limit of the capture ELISA). The previous results can be explained based on the fact that IL-1β secretion is dependent on inflammasome activation (46, 47). To confirm this explanation, we incubated the cultures with ATP for 30 min before culture medium collection in the cells treated with LPS. Under these conditions, we observed a maximum level of IL-1β protein after 5 h, and this then decreased to baseline levels at 48 h (Fig. 2A, middle panel). Additionally, all three hemocyanins induced (to different degrees) a significant maximal upregulation of Il6 gene expression at 5 h after incubation, highlighting the fact that KLH had an even greater effect than LPS (Fig. 2B, upper panel). However, these increases in mRNA levels do not correlate with the protein secretion during the time course, where its maximum expression level was reached at 24 h in response to KLH and FLH. In contrast, CCH induced basal protein levels at all of the time points (Fig. 2B, middle panel). Tnf gene expression was essentially at baseline in every time point studied for all hemocyanins (Fig. 2C, upper panel). However, we detected a significant increase in TNF-α protein levels in the culture medium at 5 h after incubation with KLH and FLH relative to CCH (Fig. 2C, middle panel). Additionally, only KLH and FLH upregulated the Il12b maximally at 5 h after stimulation, whereas CCH resulted in basal gene expression and protein levels at all times analyzed (Fig. 2D, upper panel). However, KLH and FLH significantly increased the gene expression of IL-12p40 at 5 h after incubation and induced higher protein levels at 24 h incubation (Fig. 2D, middle panel), indicating that this cytokine is also under transcriptional control (48).
Collectively, these data show different temporal expression patterns of proinflammatory cytokines in macrophages following treatment with the different hemocyanins. This result suggests that these proteins activate different signaling pathways and transcriptional networks (Fig. 2, lower panel).
Hemocyanins induced an M1-dominant immune profile in macrophages and maintained the downregulation of key M2 cytokines
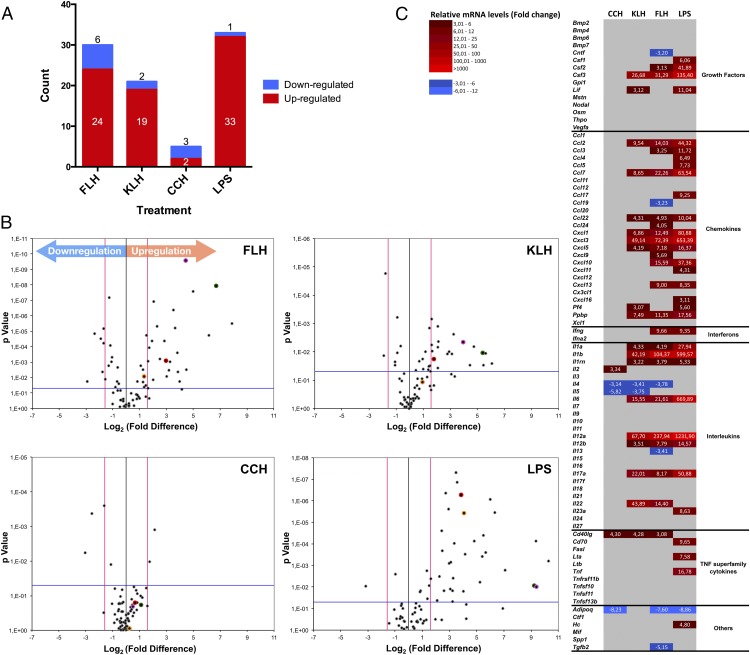
To further investigate the effect of hemocyanins on macrophage activation, we extended our analysis using a PCR array to evaluate differences in the expression of genes encoding 84 cytokines that mediate innate and adaptive immune responses. Thus, macrophages were cultured during a 24-h time period with 1 mg/ml of each hemocyanin. Of the 84 genes analyzed, only certain genes showed significant differences in expression (fold change ≥ 3 and p < 0.05) after incubation with hemocyanins, which is summarized in Fig. 3A and Table I. In macrophages treated with LPS (the positive control), only 34 genes showed significant differences with respect to culture medium used as negative control. Of these genes, 33 (39.3%) were upregulated. Only one (1.2%) gene was downregulated, that is, Adipoq, which encodes an anti-inflammatory cytokine that inhibits macrophage functions (49). In the presence of FLH, 30 genes (35.7%) showed significant differences, with 24 (28.6%) upregulated and 6 (7.1%) downregulated (Adipoq, Tgfb2, Il4, Il13, Ccl19, Cntf). In the presence of KLH, 21 genes (25%) showed significant differences, with 19 (22.6%) upregulated and 2 (2.4%) downregulated (Il5, Il4). In the presence of CCH, only five genes (6%) showed significant differences, with two (2.4%) upregulated (Il2, Cd40lg), and three (3.6%) downregulated (Adipoq, Il5, Il4).
FIGURE 3.
Hemocyanins induced the differential expression of cytokine genes in macrophages. Summary of the PCR array profile results for the expression of 84 cytokine genes analyzed using the RT2 Profiler PCR array is shown. This array includes chemokines, ILs, IFNs, growth factors, and TNF superfamily members. Macrophages were stimulated for 24 h with CCH, KLH, FLH (1 mg/ml), or LPS (100 ng/ml) as the positive control. Culture medium was used as the negative control. Reverse transcription quantitative PCR (RT-qPCR) data were analyzed using the ΔΔCt method. The criteria for a change in cytokine mRNA levels detected with the array were as follows: a Ct value of <33; a reaction efficiency of >55%; a melting curve profile corresponding to the specific amplicon; and a threshold fold change of >3-fold. (A) FLH induced a stronger proinflammatory response than did KLH or CCH. Graphic indicates the number of significantly upregulated (red) and downregulated (blue) genes for each treatment (FLH, KLH, CCH, and LPS). (B) Volcano plots. Differences in the expression of genes encoding the 84 inflammation-related factors are shown. Each dot in the plot represents one gene: the x-axis shows log2, indicating the biological impact (fold change in response to hemocyanin treatment [CCH, FLH, KLH] or LPS), whereas the y-axis shows −log10, indicating the statistical reliability of the fold change (p value). Gene markers of inflammation evaluated in the experiments of Fig. 2 are indicated by colored dots: IL-1β, green; IL-6, pink; TNF-α, yellow; and IL-12p40, red. (C) Heat maps. The figure displays mRNA expression levels in macrophages in response to hemocyanins. The map compares CCH, FLH, and KLH with the positive control LPS. Upregulated cytokines are represented in graded shades of red, and downregulated cytokines are shown in graded shades of blue. The data are representative of three independent experiments.
Table I. Summary of the PCR array analysis of 84 genes encoding cytokines expressed by murine macrophages after 24 h of coculture with hemocyanins.
| Cytokine Genes | LPS |
FLH |
KLH |
CCH |
||||
|---|---|---|---|---|---|---|---|---|
| No. of Genes | % p < 0.05 | No. of Genes | % p < 0.05 | No. of Genes | % p < 0.05 | No. of Genes | % p < 0.05 | |
| Total | 42 | 50 | 48 | 57.1 | 37 | 44 | 6 | 7.1 |
| Upregulated | 33 | 39.3 | 24 | 28.6 | 19 | 22.6 | 2 | 2.4 |
| Downregulated | 1 | 1.2 | 6 | 7.1 | 2 | 2.4 | 3 | 3.6 |
| Unaffected | 8 | 9.5 | 18 | 21.4 | 16 | 19 | 1 | 1.2 |
Only the genes that were statistically significant were considered.
The data are presented as volcano plots to show the differences in the expression of the 84 cytokine genes in the macrophages treated with the different hemocyanins or LPS (positive control) with respect to the control media (negative control; Fig. 3B). Gene markers of inflammation evaluated in the experiments in Fig. 2 are indicated by colored dots in Fig. 3B (IL-1β, green; IL-6, pink; TNF-α, yellow; and IL-12p40, red), showing concordance among methods. At first glance, differences in expression patterns are indicated for the three hemocyanins and LPS, which are shown in the heat map visualization of the results, where the cytokines are grouped into different categories based on their functions (Fig. 3C).
Macrophages that were incubated with FLH showed an elevated M1 transcript cytokine profile compared with macrophages that were incubated with CCH or KLH (Fig. 3B, 3C). A significant (p < 0.05) upregulation that produced a >3-fold change in the FLH treatment compared with the control was observed for the following cytokine genes: the chemotactic chemokines genes Ccl3, Ccl24, Ccl22, Cxcl9, Cxcl5, Cxcl13, Ppbp, Cxcl10, Ccl2, Cxcl3, Ccl7, and Cxcl1 (the last of which is the first chemokine synthesized during an acute inflammatory response by macrophages) (50); the IL genes Il1rn, Il1a, Il12b, 1l17a, Il6, Il1b, Il12a, and Il22; the growth factor genes Csf2 and Csf3; the IFN gene Ifng; and the TNF superfamily member gene Cd40lg (3.08-fold). In contrast, the anti-inflammatory cytokine genes that promote M2 immune responses, which were significantly downregulated in response to FLH, included Il4 and Il13 (−3.78- and −3.41-fold, respectively). Additionally, FLH was unique in inducing transcriptional suppression of Ccl19 (−3.23-fold) and Tgfb2 (−5.15-fold), the latter of which is an anti-inflammatory cytokine secreted by murine macrophages to counteract inflammation by promoting the termination of leukocyte trafficking and the clearance of inflammatory cells (51).
A slightly elevated Th1 transcript cytokine profile was obtained in response to FLH relative to KLH (Fig. 3B, 3C), and the significant upregulation of 8 of the 12 chemotactic chemokine genes (Ccl3, Ccl22, Cxcl5, Ppbp, Cxcl1, Ccl2, Cxcl3, Ccl7) was consistent with the results for FLH. Additionally, an upregulation of the growth factor genes Csf3 and the TNF superfamily member Cd40lg (4.28-fold) was found. With regard to anti-inflammatory cytokines, differences between the KLH treatment and the control were not detected for transcript levels of Il13, whereas the Il4 and Il5 transcript levels were significantly downregulated (−3.41- and −3.75-fold, respectively).
Different results were obtained for the cytokine transcript levels in macrophages incubated with CCH relative to the control (Fig. 3B, 3C), although the only significantly upregulated transcripts were Il2 (3.34-fold), as IL increases T cell proliferation and activates B cells (49), and a member of the TNF superfamily (Cd40lg; 4.30-fold). Macrophages treated with LPS exhibited increased M1 subset cytokine mRNA levels.
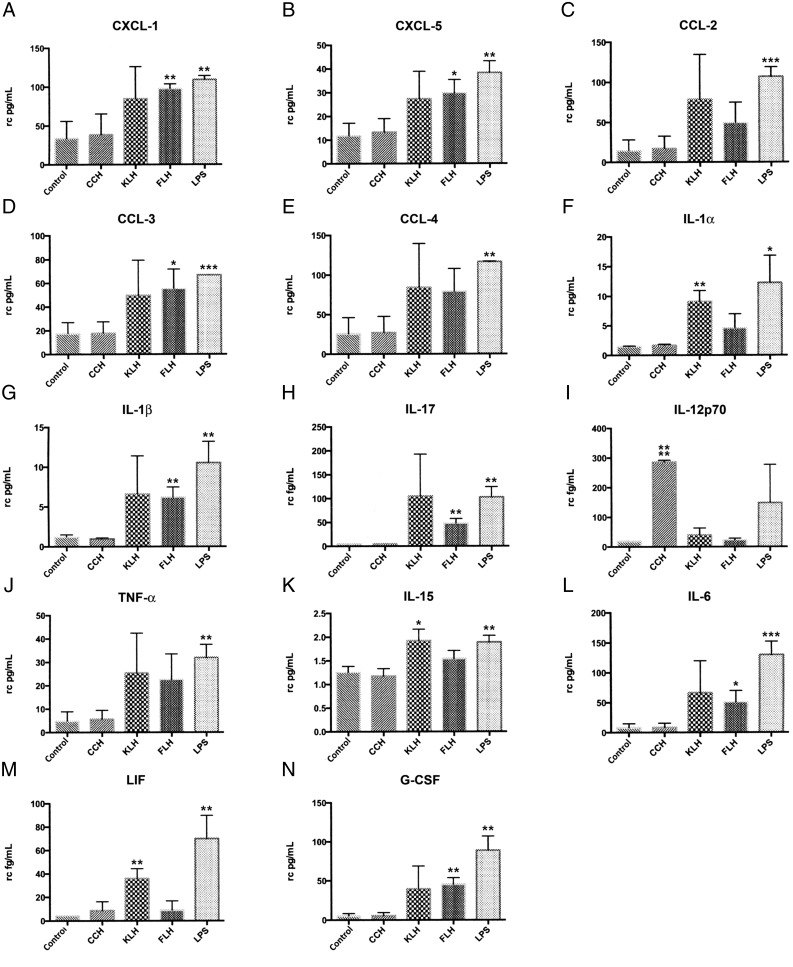
To verify the results of the PCR array, we evaluated the secretion of 32 cytokines (31 of which were included in the PCR array) using a mouse cytokine/chemokine/growth factor 32-plex array panel. Significant protein levels of 14 (43.8%) cytokines were found in the supernatants of macrophages treated with the different hemocyanins (Fig. 4). Along with inducing the expression of cytokines known to be potent attractors of various leukocytes, FLH and KLH considerably induced the protein expression of CXCL-1 (Fig. 4A), CXCL-5 (Fig. 4B), and CCL-3 (Fig. 4D), which is consistent with the PCR array results for FLH CXCL-1, CXCL-5, and CCL-3.
FIGURE 4.
In macrophages, hemocyanins induce differential protein expression levels of several cytokines, as measured by an ELISArray kit. The amount of 32 cytokines in culture supernatants from peritoneal macrophages exposed to the different treatments (CCH, FLH, and KLH; culture medium and LPS as the negative and positive controls, respectively) was determined for up to 24 h using the mouse cytokine array/chemokine array 32-plex. The data represent the following: chemokines, (A) CXCL-1, (B) CXCL-5, (C) CCL-2, (D) CCL-3, (E) CCL-4; cytokines, (F) IL-1α, (G) IL-1β, (H) IL-17, (I) IL-12p70, (J) TNF-α, (K) IL-15, (L) IL-6; and growth factors, (M) LIF and (N) G-CSF. The data are representative of three independent experiments and are presented as picograms cytokine level per milliliter cell culture (mean ± SD). *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001. rc, square root transformation.
With regard to proinflammatory cytokines, IL-1α protein and IL-1β protein were detected in response to KLH and FLH, respectively (Fig. 4F and 4G, respectively), which is consistent with the PCR array results and the secretion of IL-1β observed by ELISA (Fig. 4G). However, significant levels of IL-17 were detected only in response to FLH (Fig. 4H), and CCH induced a sharp increase in the protein levels of IL-12p70 to levels significantly higher than those induced by KLH or FLH (Fig. 4I), although the PCR array results did not show a significant rise of either the Il12a or Il12b gene transcripts (which correspond to the IL-12p35 and IL-12p40 subunits of this cytokine) (Fig. 3). According to ELISArray results, hemocyanins did not significantly induce the release of TNF-α (Fig. 4J); nonetheless, these findings showed more similar protein levels than did the results from ELISA, where this cytokine was found to be significantly induced by KLH and FLH (Fig. 2C). In contrast, KLH increased the protein level of IL-15 (Fig. 4K), a proinflammatory cytokine, and the corresponding gene was not in the PCR array panel. With regard to IL-6, it was detected in response to FLH and KLH; however, the differences were only significant in response to FLH (Fig. 4L). This result is consistent with the ELISA results (Fig. 2B).
Concerning the growth factors, although the results of the PCR array after treatment with KLH and FLH indicated increases in the mRNA levels of Csf3 (corresponding to G-CSF), the only significant increase in protein level was observed after treatment with FLH (Fig. 4N). However, the results of the PCR array for the mRNA of Lif (LIF) corresponded to a significant increase in protein levels after treatment with KLH (Fig. 4M). LPS induced a significantly higher secretion of all cytokines, which is consistent with the PCR array results, and the LPS results were greater than those with the medium alone or hemocyanins (Fig. 4A–M).
Overall, our data showed that the cytokine gene expression profile and the cytokine protein secretion induced by CCH, FLH, and KLH were significantly different. Additionally, our data confirmed that hemocyanins strongly promote M1-directed immune responses by downregulating key cytokines that drive M2 immune responses.
Discussion
The aim of this study was to determine how mollusk hemocyanins, in the absence of additional adjuvants, trigger an innate immune response that leads to the maturation of a Th1-specific adaptive immune response together with powerful nonspecific immunomodulatory effects, including an antitumor capacity in certain cancers (3, 5, 6). The immune response is triggered by a proinflammatory signal that arises from various components of the innate immune system that sense invading pathogens (52) or danger/alarm signals from injured cells (53), phenomena in which phagocytic cells and APCs are fundamental. In the present study, we focused our attention on macrophages, which are central regulators of immune responses and one of the most active secretory cell types in the body (54). Macrophages can internalize almost any form of Ag (whether cell associated or soluble) either nonspecifically or via specific receptors, which allows them to stimulate T cells (55). Consequently, we studied whether hemocyanins can drive the expression of proinflammatory cytokines by peritoneal macrophages to generate a proinflammatory milieu. Therefore, we compared three hemocyanins that have different quaternary structures, oligosaccharides, and immunomodulatory properties (6). We hypothesized that they would show differences at the innate immune level.
The main results show that hemocyanins were internalized by macrophages through pinocytic vesicles and by clathrin-dependent endocytosis, and these proteins were then slowly processed, an idea supported by their persistence for several days in vitro and in vivo. Surprisingly, the first observed differences between these proteins appear at 24 h. FLH and KLH (to a lesser extent) induced a significant upregulation of the M1 proinflammatory cytokine mRNA response in macrophages, which did not always correlate with protein levels. This response was less intense than that induced by LPS, which was used as a positive control. CCH showed a restrained but balanced action at this time. However, all of the hemocyanins maintained the downregulation of key M2 cytokine genes, such as Il4, Il5, Il13, and Tgfb2, with differences in the type of gene and/or intensity. Thus, although treatment with hemocyanins does not mirror the classical activation pattern (19, 56, 57), our results, in combination with no change in arginase-1 expression, strongly suggest a shift toward a proinflammatory state. There is a consensus that M1 macrophages activate a tumor-killing mechanism and antagonize the suppressive activity of M2 macrophages, which promote tumor growth and metastasis (16).
In contrast to murine macrophages, murine DCs did not mature in vitro after 72 h in the presence of CCH or KLH. On the contrary, FLH induced poor maturation, which was demonstrated by the partial upregulation of costimulatory molecules and MHC-II. However, the response was sufficient to induce the secretion of several proinflammatory cytokines, including IL-6, IL-12p40, IL-23, and TNF-α (58). It has been shown that the lysosomal activity in DCs is less efficient in macrophages (59), and this activity cannot significantly degrade large proteins (60), such as KLH or CCH (14). It is unlikely that these structures hydrolyze these proteins to smaller peptide fragments equivalent to those found in macrophages (Fig. 1D). Assuming that the proteins bind to similar receptors, a possible explanation of these differences could be different trafficking patterns of hemocyanin in both types of APCs. Although DCs and macrophages phagocytose Ags in a process that involves phagosomes, these structures can be modified over time by a range of cellular machinery before fusing with lysosomes to produce antigenic peptides (60).
Our data show that the increment of mRNA levels of certain cytokines did not correspond to the secreted protein levels, such as IL-6 and IL-12p40 (Fig. 2). Thus, increases in mRNA expression may not necessarily promote the immediate synthesis of the respective protein due to posttranscriptional or translational control mechanisms for cytokine protein production. Therefore, the differences between Il6 mRNA expression and IL-6 secretion can be attributed to posttranscriptional regulation of this cytokine (61, 62) and may be triggered by various factors, including other cytokines such as TNF-α (63). Regarding TNF-α, its mRNA level was not increased in response to hemocyanins, which was inconsistent with the protein level. We suggest that it is possible that hemocyanins activate control mechanisms because TNF-α is a major proinflammatory cytokine under transcriptional regulation, and these mechanisms may regulate TNF-α mRNA stability and TNF-α protein shedding from the cell surface by a TNF-α–converting enzyme (64). A similar phenomenon has been observed in human macrophages stimulated with LPS: macrophages have a posttranscriptional mechanism that potentiates TNF-α protein secretion, although the transcription of the mRNA is apparently slower because of the increased stability of the mRNA, and this leads to a significant increase in TNF-α protein secretion without notable differences in TNF-α mRNA induction (65). The results that we obtained for the induction of TNF-α by LPS differ from those of other authors in terms of the timing at which Tnf mRNA increased (66–68). Such discrepancies can be attributed to methodological differences (63). The gene expression of IL-2 in macrophages cultured with CCH, a cytokine produced mainly by CD4+ T cells following its activation, was an unexpected finding; however, its quantification by ELISArray showed traces (on the order of femtomolar) of the cytokine, in all experimental treatment conditions of macrophages, but found no significant differences between conditions (data not shown). One possible explanation is that the low amount of IL-2 produced by the macrophages could result from contamination of the peritoneal macrophage preparations with low numbers of T lymphocytes.
One of the immediate effects of an acute inflammatory response is the rapid recruitment of leukocytes from the blood to sites of inflammation; therefore, it is remarkable that FLH and KLH produce significantly elevated mRNA levels of CC chemokines (five for FLH and three for KLH, with Ccl2, Ccl7, and Ccl22 common to both), which induce the migration and activation of macrophages/monocytes and lymphocytes. Alternatively, CXC chemokines (six for FLH and three for KLH, with Cxcl1, Cxcl3, and Cxcl5 common to both) are primarily neutrophil chemoattractants and activators (54). These effects are represented in Fig. 5. Consistent with these results, the delayed-type hypersensitivity induced by KLH in mouse ear tissues corresponded to a significant increase in the mRNA and protein levels of CXCL-1, CCL-2, and CCL-3, among others (69). Additionally, it has been reported that human bladder cancer cell lines, after treatment with hemocyanin from the gastropod Helix lucorum and Rapana venosa, upregulated genes associated with apoptosis and immune responses, including genes involved in leukocyte activation and downregulated genes related to cell proliferation and wounding (70).
FIGURE 5.
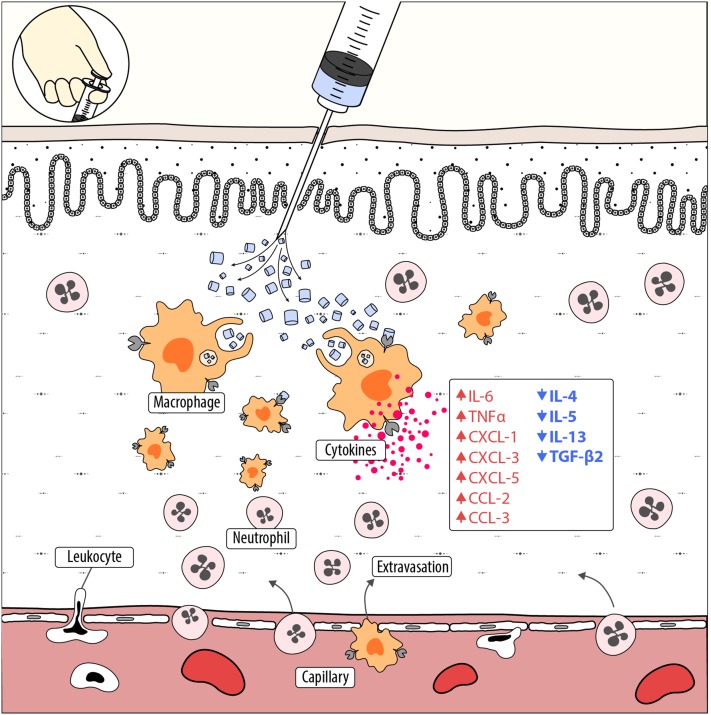
Hemocyanins stimulate innate immunity by inducing macrophage activation. A simplified scheme of the early immune response of mammals against mollusk hemocyanins, supported by the principal results presented in this study, is shown. When hemocyanin molecules are inoculated, macrophages localized in the tissue incorporate these huge proteins, both by macropinocytosis and clathrin-mediated endocytosis, and they become activated while processing these molecules slowly. As a consequence, hemocyanins drive gene expression toward the M1 macrophage phenotype, triggering the secretion of inflammatory mediators, including chemokines of the C-X-C and C-C family (red, upward arrows), which are important for leukocyte extravasation and recruitment, especially of neutrophils, at the site of inoculation. Thus is generated a proinflammatory milieu that produces a bystander effect. During this early phase of immune response, hemocyanins maintain the downregulation of several M2 cytokine genes (blue, downward arrows) in macrophages. Hemocyanins are colored blue when in their oxygenated form.
Interestingly, the three hemocyanins induced significant elevations in the mRNA levels of CD40L (Cd40lg) in macrophages, which might support the observed nonspecific antitumor activity of hemocyanins in several cancers (6, 71). CD40–CD40L interactions promote the activation of NK cells by murine macrophages (72). These cells play a fundamental role in innate immune surveillance via their direct cytolytic functions (e.g., destruction of nascent tumors) or regulatory capabilities (e.g., prevention of tumor metastasis dissemination via the expression of IFN-γ) (73, 74). We previously demonstrated that CCH and KLH increased NK cell activity in vivo in a superficial bladder cancer model (11), and in a melanoma model, we demonstrated that FLH augmented NK cell infiltration in the tumor. Moreover, in a recently reported murine model of sarcoma cells, KLH was found to increase the antitumor activity of NK cells in vitro (75).
Our data showed a significant upregulation of the IL-1α and IL-1β genes in response to FLH and KLH (Fig. 3) and indicated the presence of these cytokines in the culture medium of macrophages; the ELISArray detected small amounts (Fig. 4), whereas conventional ELISA did not detect their presence (Fig. 2). Supporting these data, it has been reported that patients with superficial bladder cancer who responded to intravesical instillation therapy with KLH had higher IL-1α levels in the urine than patients who failed to respond (76).
Importantly, note that our PCR array and ELISArray analyses had inherent limitations because it was necessary to select a single time point at which to perform the comparison between hemocyanins so that the upregulated or downregulated genes were limited up to 24 h of analyses. Therefore, additional time course experiments will be required, especially in the case of CCH, which produced a low proinflammatory profile at this time point. This phenomenon may have been partially caused by the significantly improved stability of CCH compared with FLH and KLH, a phenomenon that could be attributed to a lower hydrophobicity at the surface of the protein (77), differences in glycosylations (8), in addition to oligomerization and the copper/dioxygen system at the active site (78). Thus, the conformational and structural stability of a protein Ag may play crucial roles in its proteolysis, APC processing, T cell stimulation, and immunogenicity (79).
Our previous studies demonstrated that FLH, unlike CCH and KLH, has potent antitumor activity on murine melanomas even without previous priming, an effect that we suggested might have been caused by FLH inducing a more potent proinflammatory milieu than did CCH and KLH (8). Thus, the present data support this assumption and suggest potential mechanisms, but they do not provide evidence on the cause of these differences. We think that differences in the intrinsic structural features of these proteins are the key to activation of APCs, and several authors have proposed that sugar moieties may play a role during the immune response against hemocyanins (4, 8, 26, 29). Because mannose is the major sugar found in these proteins, we proposed that APCs might recognize mollusk hemocyanins as a “highly mannosylated infectious agent” (6). This assumption is supported by the promotion of human DC maturation by KLH in vitro through its engagement of the membrane C-type lectin mannose receptor (MR) (80), a phenomenon that is not observed in murine DCs cultured in vitro with KLH and CCH (14, 81). However, FLH promotes the secretion of several proinflammatory cytokines, although this ability ceases when DCs are treated with deglycosylated FLH, suggesting a crucial role for oligosaccharides in the immunomodulatory effects of FLH (8). In this study, we performed experiments to evaluate whether the MR is involved in the endocytosis of hemocyanins in murine peritoneal macrophages. First, the expression of the MR in macrophages was assessed using OVA as positive control, because it has been described as a classical MR ligand (82) (Supplemental Fig. 3A); then, a hemocyanin uptake inhibition assay was developed, showing that the endocytosis of hemocyanins was calcium-dependent and was partially inhibited by the presence of d-mannose, the natural ligand of MR (Supplemental Fig. 3B). These results suggest that other C-type lectin receptors participate in the endocytosis of hemocyanins by macrophages. Notably, MR does not have a “signaling motif,” and therefore cooperation with other receptors is required to trigger the signaling cascade leading to cytokine secretion (83), such as DC-SIGN (84), Dectin-2 (85), or TLRs (e.g., TLR4 or TLR2) (86, 87).
Thus, we reasoned that macrophages might bind hemocyanins through receptors with different carbohydrate recognition domains that occur in certain C-type lectin receptors depending on the oligosaccharide (number, type, and substitution patterns of outer sugar branches) on the protein. These receptors directly or indirectly trigger distinct signaling pathways, leading to the differential production of proinflammatory cytokines and promotion of the M1 immune response; thus, a bystander signaling pathway might be activated, increasing tumor cell killing or other latent-specific responses. Hence, because CCH lacks oligosaccharides that contain sialic acid, their endocytosis may be mediated by different receptors than those for KLH and FLH, which have these oligosaccharides (8).
In conclusion, our data demonstrate that CCH, KLH, and FLH have early differential effects at the molecular level in macrophages, reflected in quantitative differences in cytokine gene expression and protein secretion patterns, leading to an M1 macrophage polarization. Additionally, the duration and magnitude of the cytokine cascade after APC uptake of these proteins have been mostly unclear until now. Thus, the data reported in the present study provide an initial roadmap for further in-depth investigations into the immunological mechanism of action of these proteins, thus supporting their application, especially in the biomedical sciences.
Supplementary Material
Acknowledgments
We thank Fabián Salazar (Ph.D. student in immunology, University of Nottingham, Nottingham, U.K.), José Jiménez (Ph.D. student in pharmacology, Universidad de Chile, Santiago, Chile), Dr. Claudia d’Alençon (Fundación Ciencia y Tecnología para el Desarrollo), Dr. Pablo De Ioannes (Department of Structural Biology, Skirball Institute of Biomolecular Medicine, New York University School of Medicine, New York, NY), and Alfredo De Ioannes (Biosonda Corporation, Santiago, Chile) for valuable discussions and suggestions. We also thank Alejandro Munizaga (Servicio de Microscopía Electrónica, Pontificia Universidad Católica de Chile, Santiago, Chile) for outstanding technical assistance and Gabriel De Ioannes (Faculty of Engineering in Development of Video Games and Virtual Reality, Universidad de Talca, Talca, Chile) for the design of Fig. 5.
This work was partially supported by Fondo Nacional de Desarrollo Científico y Tecnológico Grants 1110651 and 1151337 (to M.I.B.).
The online version of this article contains supplemental material.
- CCH
- Concholepas concholepas hemocyanin
- CCH-488
- CCH labeled with Alexa Fluor 488
- Ct
- threshold cycle
- DC
- dendritic cell
- FLH
- Fissurella latimarginata hemocyanin
- KLH
- keyhole limpet hemocyanin
- MHC-II
- MHC class II
- MR
- mannose receptor
- OVA-488
- OVA labeled with Alexa Fluor 488
- qPCR
- quantitative PCR.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.van Holde K. E., Miller K. I. 1995. Hemocyanins. Adv. Protein Chem. 47: 1–81. [DOI] [PubMed] [Google Scholar]
- 2.Coates C. J., Nairn J. 2014. Diverse immune functions of hemocyanins. Dev. Comp. Immunol. 45: 43–55. [DOI] [PubMed] [Google Scholar]
- 3.Arancibia S., Salazar F., Becker M. I. 2012. Hemocyanins in the immunotherapy of superficial bladder cancer. In Bladder Cancer–From Basic to Robotic Surgery. Canda A., ed. InTech, Rijeka, Croatia, p. 221–242. [Google Scholar]
- 4.Harris J. R., Markl J. 1999. Keyhole limpet hemocyanin (KLH): a biomedical review. Micron 30: 597–623. [DOI] [PubMed] [Google Scholar]
- 5.Patil U. S., Jaydeokar A. V., Bandawane D. D. 2012. Immunomodulators: a pharmacological review. Int. J. Pharm. Pharm. Sci. 4(Suppl. 1): 30–36. [Google Scholar]
- 6.Becker M. I., Arancibia S., Salazar F., Del Campo M., De Ioannes A. E. 2014. Mollusk hemocyanins as natural immunostimulants in biomedical applications. In Immune Response Activation. Duc G. H. T., ed. InTech, Rijeka, Croatia, p. 45–72. [Google Scholar]
- 7.De Ioannes P., Moltedo B., Oliva H., Pacheco R., Faunes F., De Ioannes A. E., Becker M. I. 2004. Hemocyanin of the molluscan Concholepas concholepas exhibits an unusual heterodecameric array of subunits. J. Biol. Chem. 279: 26134–26142. [DOI] [PubMed] [Google Scholar]
- 8.Arancibia S., Espinoza C., Salazar F., Del Campo M., Tampe R., Zhong T. Y., De Ioannes P., Moltedo B., Ferreira J., Lavelle E. C., et al. 2014. A novel immunomodulatory hemocyanin from the limpet Fissurella latimarginata promotes potent anti-tumor activity in melanoma. PLoS One 9: e87240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Campo M., Arancibia S., Nova E., Salazar F., González A., Moltedo B., De Ioannes P., Ferreira J., Manubens A., Becker M. I. 2011. [Hemocyanins as immunostimulants]. Rev. Med. Chil. 139: 236–246. [PubMed] [Google Scholar]
- 10.Ebensperger L. A., León C., Ramírez-Estrada J., Abades S., Hayes L. D., Nova E., Salazar F., Bhattacharjee J., Becker M. I. 2015. Immunocompetence of breeding females is sensitive to cortisol levels but not to communal rearing in the degu (Octodon degus). Physiol. Behav. 140: 61–70. [DOI] [PubMed] [Google Scholar]
- 11.Moltedo B., Faunes F., Haussmann D., De Ioannes P., De Ioannes A. E., Puente J., Becker M. I. 2006. Immunotherapeutic effect of Concholepas hemocyanin in the murine bladder cancer model: evidence for conserved antitumor properties among hemocyanins. J. Urol. 176: 2690–2695. [DOI] [PubMed] [Google Scholar]
- 12.Reyes D., Salazar L., Espinoza E., Pereda C., Castellón E., Valdevenito R., Huidobro C., Inés Becker M., Lladser A., López M. N., Salazar-Onfray F. 2013. Tumour cell lysate-loaded dendritic cell vaccine induces biochemical and memory immune response in castration-resistant prostate cancer patients. Br. J. Cancer 109: 1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salazar-Onfray F., Pereda C., Reyes D., López M. N. 2013. TAPCells, the Chilean dendritic cell vaccine against melanoma and prostate cancer. Biol. Res. 46: 431–440. [DOI] [PubMed] [Google Scholar]
- 14.Arancibia S., Del Campo M., Nova E., Salazar F., Becker M. I. 2012. Enhanced structural stability of Concholepas hemocyanin increases its immunogenicity and maintains its non-specific immunostimulatory effects. Eur. J. Immunol. 42: 688–699. [DOI] [PubMed] [Google Scholar]
- 15.Adams D. O., Hamilton T. A. 1984. The cell biology of macrophage activation. Annu. Rev. Immunol. 2: 283–318. [DOI] [PubMed] [Google Scholar]
- 16.Murray P. J., Wynn T. A. 2011. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11: 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosser D. M., Edwards J. P. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8: 958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon S. 2003. Alternative activation of macrophages. Nat. Rev. Immunol. 3: 23–35. [DOI] [PubMed] [Google Scholar]
- 19.Sica A., Mantovani A. 2012. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122: 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unanue E. R., Cerottini J. C. 1970. The immunogenicity of antigen bound to the plasma membrane of macrophages. J. Exp. Med. 131: 711–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gesheva V., Idakieva K., Kerekov N., Nikolova K., Mihaylova N., Doumanova L., Tchorbanov A. 2011. Marine gastropod hemocyanins as adjuvants of non-conjugated bacterial and viral proteins. Fish Shellfish Immunol. 30: 135–142. [DOI] [PubMed] [Google Scholar]
- 22.Orlova E. V., Dube P., Harris J. R., Beckman E., Zemlin F., Markl J., van Heel M. 1997. Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 A resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol. 271: 417–437. [DOI] [PubMed] [Google Scholar]
- 23.Markl J. 2013. Evolution of molluscan hemocyanin structures. Biochim. Biophys. Acta 1834: 1840–1852. [DOI] [PubMed] [Google Scholar]
- 24.Swerdlow R. D., Ebert R. F., Lee P., Bonaventura C., Miller K. I. 1996. Keyhole limpet hemocyanin: structural and functional characterization of two different subunits and multimers. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 113: 537–548. [DOI] [PubMed] [Google Scholar]
- 25.Hall R. L., Wood E. J. 1976. The carbohydrate content of gastropod haemocyanins. Biochem. Soc. Trans. 4: 307–309. [DOI] [PubMed] [Google Scholar]
- 26.Wirguin I., Suturkova-Milosević L., Briani C., Latov N. 1995. Keyhole limpet hemocyanin contains Gal(β1–3)-GalNAc determinants that are cross-reactive with the T antigen. Cancer Immunol. Immunother. 40: 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geyer H., Wuhrer M., Resemann A., Geyer R. 2005. Identification and characterization of keyhole limpet hemocyanin N-glycans mediating cross-reactivity with Schistosoma mansoni. J. Biol. Chem. 280: 40731–40748. [DOI] [PubMed] [Google Scholar]
- 28.Gatsogiannis C., Markl J. 2009. Keyhole limpet hemocyanin: 9-A CryoEM structure and molecular model of the KLH1 didecamer reveal the interfaces and intricate topology of the 160 functional units. J. Mol. Biol. 385: 963–983. [DOI] [PubMed] [Google Scholar]
- 29.Dolashka P., Velkova L., Shishkov S., Kostova K., Dolashki A., Dimitrov I., Atanasov B., Devreese B., Voelter W., Van Beeumen J. 2010. Glycan structures and antiviral effect of the structural subunit RvH2 of Rapana hemocyanin. Carbohydr. Res. 345: 2361–2367. [DOI] [PubMed] [Google Scholar]
- 30.Velkova L., Dolashka P., Lieb B., Dolashki A., Voelter W., Van Beeumen J., Devreese B. 2011. Glycan structures of the structural subunit (HtH1) of Haliotis tuberculata hemocyanin. Glycoconj. J. 28: 385–395. [DOI] [PubMed] [Google Scholar]
- 31. Zhang, X., R. Goncalves, and D. M. Mosser. 2008. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. Chapter 14: Unit 14.1. doi:10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed]
- 32.Luft J. H. 1961. Improvements in epoxy resin embedding methods. J. Biophys. Biochem. Cytol. 9: 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17: 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 35.Towbin H., Staehelin T., Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliva H., Moltedo B., De Ioannes P., Faunes F., De Ioannes A. E., Becker M. I. 2002. Monoclonal antibodies to molluskan hemocyanin from Concholepas concholepas demonstrate common and specific epitopes among subunits. Hybrid. Hybridomics 21: 365–374. [DOI] [PubMed] [Google Scholar]
- 37.Barksby H. E., Nile C. J., Jaedicke K. M., Taylor J. J., Preshaw P. M. 2009. Differential expression of immunoregulatory genes in monocytes in response to Porphyromonas gingivalis and Escherichia coli lipopolysaccharide. Clin. Exp. Immunol. 156: 479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid C. D., Melchior B., Masek K., Puntambekar S. S., Danielson P. E., Lo D. D., Sutcliffe J. G., Carson M. J. 2009. Differential gene expression in LPS/IFNγ activated microglia and macrophages: in vitro versus in vivo. J. Neurochem. 109(Suppl. 1): 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giulietti A., Overbergh L., Valckx D., Decallonne B., Bouillon R., Mathieu C. 2001. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25: 386–401. [DOI] [PubMed] [Google Scholar]
- 40.Wang G., Petzke M. M., Iyer R., Wu H., Schwartz I. 2008. Pattern of proinflammatory cytokine induction in RAW264.7 mouse macrophages is identical for virulent and attenuated Borrelia burgdorferi. J. Immunol. 180: 8306–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan Y., Weifeng W., Yuluan Y., Qing K., Yu P., Yanlan H. 2011. Treatment with a neutralizing anti-murine interleukin-17 antibody after the onset of coxsackievirus b3-induced viral myocarditis reduces myocardium inflammation. Virol. J. 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suthanthiran M., Gerber L. M., Schwartz J. E., Sharma V. K., Medeiros M., Marion R., Pickering T. G., August P. 2009. Circulating transforming growth factor-β1 levels and the risk for kidney disease in African Americans. Kidney Int. 76: 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 44.Rőszer T. 2015. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm. 2015: 816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delamarre L., Pack M., Chang H., Mellman I., Trombetta E. S. 2005. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307: 1630–1634. [DOI] [PubMed] [Google Scholar]
- 46.Netea M. G., Simon A., van de Veerdonk F., Kullberg B. J., Van der Meer J. W., Joosten L. A. 2010. IL-1β processing in host defense: beyond the inflammasomes. PLoS Pathog. 6: e1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Veerdonk F. L., Netea M. G., Dinarello C. A., Joosten L. A. 2011. Inflammasome activation and IL-1β and IL-18 processing during infection. Trends Immunol. 32: 110–116. [DOI] [PubMed] [Google Scholar]
- 48.Saito S., Matsuura M., Hirai Y. 2006. Regulation of lipopolysaccharide-induced interleukin-12 production by activation of repressor element GA-12 through hyperactivation of the ERK pathway. Clin. Vaccine Immunol. 13: 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dinarello C. A. 2007. Historical insights into cytokines. Eur. J. Immunol. 37(Suppl. 1): S34–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Filippo K., Henderson R. B., Laschinger M., Hogg N. 2008. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J. Immunol. 180: 4308–4315. [DOI] [PubMed] [Google Scholar]
- 51.Stables M. J., Shah S., Camon E. B., Lovering R. C., Newson J., Bystrom J., Farrow S., Gilroy D. W. 2011. Transcriptomic analyses of murine resolution-phase macrophages. Blood 118: e192–e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mogensen T. H. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22: 240–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matzinger P. 2002. The danger model: a renewed sense of self. Science 296: 301–305. [DOI] [PubMed] [Google Scholar]
- 54.Laskin D. L., Sunil V. R., Gardner C. R., Laskin J. D. 2011. Macrophages and tissue injury: agents of defense or destruction? Annu. Rev. Pharmacol. Toxicol. 51: 267–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trombetta E. S., Mellman I. 2005. Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 23: 975–1028. [DOI] [PubMed] [Google Scholar]
- 56.Biswas S. K., Mantovani A. 2010. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11: 889–896. [DOI] [PubMed] [Google Scholar]
- 57.Murray P. J., Wynn T. A. 2011. Obstacles and opportunities for understanding macrophage polarization. J. Leukoc. Biol. 89: 557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Savina A., Amigorena S. 2007. Phagocytosis and antigen presentation in dendritic cells. Immunol. Rev. 219: 143–156. [DOI] [PubMed] [Google Scholar]
- 59.Chain B. M., Kay P. M., Feldmann M. 1986. The cellular pathway of antigen presentation: biochemical and functional analysis of antigen processing in dendritic cells and macrophages. Immunology 58: 271–276. [PMC free article] [PubMed] [Google Scholar]
- 60.Vyas J. M., Van der Veen A. G., Ploegh H. L. 2008. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 8: 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Snick J. 1990. Interleukin-6: an overview. Annu. Rev. Immunol. 8: 253–278. [DOI] [PubMed] [Google Scholar]
- 62.Paschoud S., Dogar A. M., Kuntz C., Grisoni-Neupert B., Richman L., Kühn L. C. 2006. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol. Cell. Biol. 26: 8228–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kontoyiannis D., Pasparakis M., Pizarro T. T., Cominelli F., Kollias G. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10: 387–398. [DOI] [PubMed] [Google Scholar]
- 64.Dumitru C. D., Ceci J. D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J. H., Patriotis C., Jenkins N. A., Copeland N. G., Kollias G., Tsichlis P. N. 2000. TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103: 1071–1083. [DOI] [PubMed] [Google Scholar]
- 65.MacKenzie S., Fernàndez-Troy N., Espel E. 2002. Post-transcriptional regulation of TNF-α during in vitro differentiation of human monocytes/macrophages in primary culture. J. Leukoc. Biol. 71: 1026–1032. [PubMed] [Google Scholar]
- 66.Kunkel S. L., Spengler M., Kwon G., May M. A., Remick D. G. 1988. Production and regulation of tumor necrosis factor α. A cellular and molecular analysis. Methods Achiev. Exp. Pathol. 13: 240–259. [PubMed] [Google Scholar]
- 67.Takasuka N., Matsuura K., Yamamoto S., Akagawa K. S. 1995. Suppression of TNF-α mRNA expression in LPS-primed macrophages occurs at the level of nuclear factor-κB activation, but not at the level of protein kinase C or CD14 expression. J. Immunol. 154: 4803–4812. [PubMed] [Google Scholar]
- 68.Simpson A. E., Tomkins P. T., Cooper K. L. 1997. An investigation of the temporal induction of cytokine mRNAs in LPS-challenged thioglycollate-elicited murine peritoneal macrophages using the reverse transcription polymerase chain reaction. Inflamm. Res. 46: 65–71. [DOI] [PubMed] [Google Scholar]
- 69.Engstrom L., Pinzon-Ortiz M. C., Li Y., Chen S. C., Kinsley D., Nelissen R., Fine J. S., Mihara K., Manfra D. 2009. Characterization of a murine keyhole limpet hemocyanin (KLH)-delayed-type hypersensitivity (DTH) model: role for p38 kinase. Int. Immunopharmacol. 9: 1218–1227. [DOI] [PubMed] [Google Scholar]
- 70.Antonova O., Yossifova L., Staneva R., Stevanovic S., Dolashka P., Toncheva D. 2015. Changes in the gene expression profile of the bladder cancer cell lines after treatment with Helix lucorum and Rapana venosa hemocyanin. J. BUON 20: 180–187. [PubMed] [Google Scholar]
- 71.Gesheva V., Chausheva S., Mihaylova N., Manoylov I., Doumanova L., Idakieva K., Tchorbanov A. 2014. Anti-cancer properties of gastropodan hemocyanins in murine model of colon carcinoma. BMC Immunol. 15: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Michel T., Hentges F., Zimmer J. 2013. Consequences of the crosstalk between monocytes/macrophages and natural killer cells. Front. Immunol. 3: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yokoyama W. M., Kim S., French A. R. 2004. The dynamic life of natural killer cells. Annu. Rev. Immunol. 22: 405–429. [DOI] [PubMed] [Google Scholar]
- 74.Moretta L., Ferlazzo G., Bottino C., Vitale M., Pende D., Mingari M. C., Moretta A. 2006. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol. Rev. 214: 219–228. [DOI] [PubMed] [Google Scholar]
- 75.Sarker M. M., Zhong M. 2014. Keyhole limpet hemocyanin augmented the killing activity, cytokine production and proliferation of NK cells, and inhibited the proliferation of Meth A sarcoma cells in vitro. Indian J. Pharmacol. 46: 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jurincic-Winkler C. D., Gallati H., Alvarez-Mon M., Sippel J., Carballido J., Klippel K. F. 1995. Urinary interleukin-1α levels are increased by intravesical instillation with keyhole limpet hemocyanin in patients with superficial transitional cell carcinoma of the bladder. Eur. Urol. 28: 334–339. [DOI] [PubMed] [Google Scholar]
- 77.Becker M. I., Fuentes A., Del Campo M., Manubens A., Nova E., Oliva H., Faunes F., Valenzuela M. A., Campos-Vallette M., Aliaga A., et al. 2009. Immunodominant role of CCHA subunit of Concholepas hemocyanin is associated with unique biochemical properties. Int. Immunopharmacol. 9: 330–339. [DOI] [PubMed] [Google Scholar]
- 78.Idakieva K., Nikolov P., Chakarska I., Genov N., Shnyrov V. L. 2008. Spectroscopic properties and conformational stability of Concholepas concholepas hemocyanin. J. Fluoresc. 18: 715–725. [DOI] [PubMed] [Google Scholar]
- 79.Thai R., Moine G., Desmadril M., Servent D., Tarride J. L., Ménez A., Léonetti M. 2004. Antigen stability controls antigen presentation. J. Biol. Chem. 279: 50257–50266. [DOI] [PubMed] [Google Scholar]
- 80.Presicce P., Taddeo A., Conti A., Villa M. L., Della Bella S. 2008. Keyhole limpet hemocyanin induces the activation and maturation of human dendritic cells through the involvement of mannose receptor. Mol. Immunol. 45: 1136–1145. [DOI] [PubMed] [Google Scholar]
- 81.Teitz-Tennenbaum S., Li Q., Davis M. A., Chang A. E. 2008. Dendritic cells pulsed with keyhole limpet hemocyanin and cryopreserved maintain anti-tumor activity in a murine melanoma model. Clin. Immunol. 129: 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Burgdorf S., Lukacs-Kornek V., Kurts C. 2006. The mannose receptor mediates uptake of soluble but not of cell-associated antigen for cross-presentation. J. Immunol. 176: 6770–6776. [DOI] [PubMed] [Google Scholar]
- 83.Martinez-Pomares L. 2012. The mannose receptor. J. Leukoc. Biol. 92: 1177–1186. [DOI] [PubMed] [Google Scholar]
- 84.Engering A., Geijtenbeek T. B., van Vliet S. J., Wijers M., van Liempt E., Demaurex N., Lanzavecchia A., Fransen J., Figdor C. G., Piguet V., van Kooyk Y. 2002. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J. Immunol. 168: 2118–2126. [DOI] [PubMed] [Google Scholar]
- 85.McGreal E. P., Rosas M., Brown G. D., Zamze S., Wong S. Y., Gordon S., Martinez-Pomares L., Taylor P. R. 2006. The carbohydrate-recognition domain of Dectin-2 is a C-type lectin with specificity for high mannose. Glycobiology 16: 422–430. [DOI] [PubMed] [Google Scholar]
- 86.Pathak S. K., Basu S., Bhattacharyya A., Pathak S., Kundu M., Basu J. 2005. Mycobacterium tuberculosis lipoarabinomannan-mediated IRAK-M induction negatively regulates Toll-like receptor-dependent interleukin-12 p40 production in macrophages. J. Biol. Chem. 280: 42794–42800. [DOI] [PubMed] [Google Scholar]
- 87.Xaplanteri P., Lagoumintzis G., Dimitracopoulos G., Paliogianni F. 2009. Synergistic regulation of Pseudomonas aeruginosa-induced cytokine production in human monocytes by mannose receptor and TLR2. Eur. J. Immunol. 39: 730–740. [DOI] [PubMed] [Google Scholar]
- 88.McWhorter F. Y., Wang T., Nguyen P., Chung T., Liu W. F. 2013. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. USA 110: 17253–17258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.