Abstract
Haemonchus contortus appears to be the most economically important helminth parasite for small ruminant production in many regions of the world. The two sheep breeds native to the Canary Islands display distinctly different resistant phenotypes under both natural and experimental infections. Canaria Hair Breed (CHB) tends to have significantly lower worm burden and delayed and reduced egg production than the susceptible Canaria Sheep (CS). To understand molecular mechanisms underlying host resistance, we compared the abomasal mucosal transcriptome of the two breeds in response to Haemonchus infection using RNAseq technology. The transcript abundance of 711 and 50 genes were significantly impacted by infection in CHB and CS, respectively (false discovery rate <0.05) while 27 of these genes were significantly affected in both breeds. Likewise, 477 and 16 Gene Ontology (GO) terms were significantly enriched in CHB and CS, respectively (P < 1.0 × 10−4). A broad range of mechanisms have evolved in resistant CHB to provide protection against the parasite. Our findings suggest that readily inducible acute inflammatory responses, complement activation, accelerated cell proliferation and subsequent tissue repair, and immunity directed against parasite fecundity all contributed to the development of host resistance to parasitic infection in the resistant breed.
The intestinal worm Haemonchus contortus is arguably the most economically important helminth parasite for small ruminant production in many regions of the world. As a voracious blood feeder residing in the mucosal layer of the abomasum, H. contortus causes anaemia and hyper-gastrinaemia and alters abomasal secretion. H. contortus infection results in reduced growth, compromised reproduction, and elevated mortality, due to its ubiquitous distribution and severe pathogenicity. Consequently, H. contortus parasitism represents the primary constraint to profitable production of sheep and goats worldwide.
Over the past years, the rapid emergence of drug-resistant H. contortus strains and increasing demands by consumers for inexpensive organic meat and milk products with less drug residues have spurred research on the development of anthelmintic-independent parasite control strategies, such as vaccines1 and novel biologics, nutrient supplements and bioactive compounds, and selective breeding. Among them, selectively breeding sheep and goats with abilities to better resist parasitic infections appears to be a solution to sustainable small ruminant production.
Differences in resistance and susceptibility to parasitic infections between sheep breeds have been long documented2. Over the decades, comparative studies have identified at least 19 sheep breeds displaying varying degrees of resistance to parasitic infections3. For example, St. Croix lambs shed significantly fewer eggs and harbor 99% fewer worms in the abomasum than the age-matched Dorset lambs during both natural and experimental infections4. Locally-adapted breeds such as Santa Ines sheep of Brazil have significantly reduced worm burdens and fewer nodular lesions under natural infections than Suffolk and Ile de France lambs on the same pasture5. In Europe, resistance against H. contortus is better developed in Merinoland sheep than in Rhon sheep6. Red Maasai sheep have been shown to be more resistant to Haemonchus infection than the South African Dorper breed during natural exposure to parasites in Kenya7. Moreover, resistance to parasite infection has a significant genetic component. The contribution of the host genome and genetics has been estimated. For example, additive genetic variation accounts for approximately 30% of the overall variation for parasitic infection8. The resistance traits are often polygenic in nature and not influenced by genes with major effects9. Nevertheless, estimates of heritability for parasite indicator traits in small ruminants are phenotype-dependent, ranging from 0.11 to 0.40 for transformed fecal egg counts (EPG) and 0.19 to 0.26 for packed cell volume (PCV) in German Rhon sheep10. In addition, the host age plays a role. A good example is that in Scottish Blackface lambs at the end of the first grazing season, the heritability of adult worm length is very strong at 0.628. While many efforts have been made to identify genetic variants associated with parasite resistance and tolerance in sheep breeds11,12,13, molecular mechanisms and biological pathways underlying host resistance to parasitic infections in sheep remain largely unknown.
Due to unique geographical characteristics of the Canary Islands, indigenous sheep breeds have been exploited by local farmers for centuries. Among them, the Canaria Hair Breed (CHB) and Canaria sheep (CS) are predominately raised for the production of meat and milk, respectively. Previous studies demonstrate that CHB constantly displays better resistance phenotypes to H. contortus infection than CS, including significantly lower levels of fecal egg counts, fewer adult worm counts, lower number of eggs in utero and female worm stunting14. Further studies15 identified significant negative correlations between two effector cells, eosinophils and γδ/WC1+ T cells, and parasite fecundity in CHB, suggesting that inter-breed difference in regulating immune responses affects Haemonchus infection. In this study, we conducted a RNA-seq based comparative transcriptome analysis in the two indigenous breeds and attempted to understand the molecular basis underlying host resistance.
Results
Parasitology
The total worms recovered from the infected groups of CHB and CS were 1,109.75 (±1,547.73, SD) and 3,280.50 (±2,398.03), respectively. The difference is statistically significant (P < 0.05, Fig. 1). Neither Haemonchus worms nor fecal eggs were recovered from the uninfected group of either breed, as expected. EPG values detected from infected CS sheep were 262.50 ± 287.54 (mean ± SD) while no fecal eggs were detectable in the infected group of CHB sheep at 20 days post infection (dpi). No parasite eggs in either group prior to the experimental challenge were observed.
Figure 1. Differences in worm counts between resistant and susceptible sheep breeds under experimental Haemonchus contortus infection.
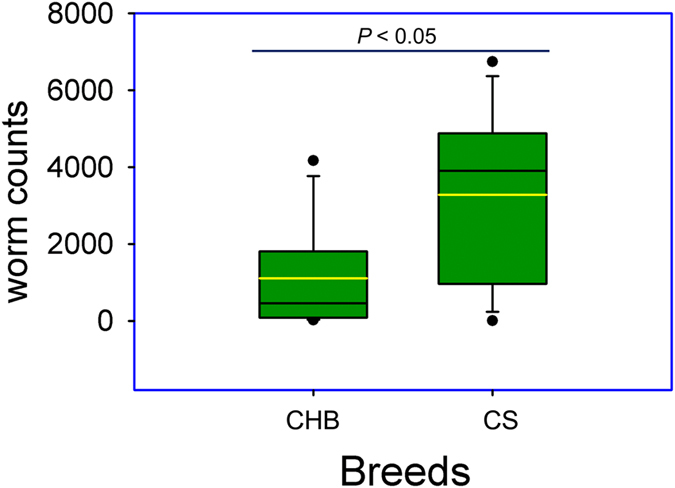
Boxes denote the inter-quartile range between the 1st and 3rd quartiles (25 and 75%, respectively). Black line: mean; Yellow line: median. CHB: Canaria Hair Breed (resistant). CS: Canaria sheep (susceptible).
Haemonchus infection induced distinctly different transcriptome patterns in the abomasal mucosa of CHB and CS breeds
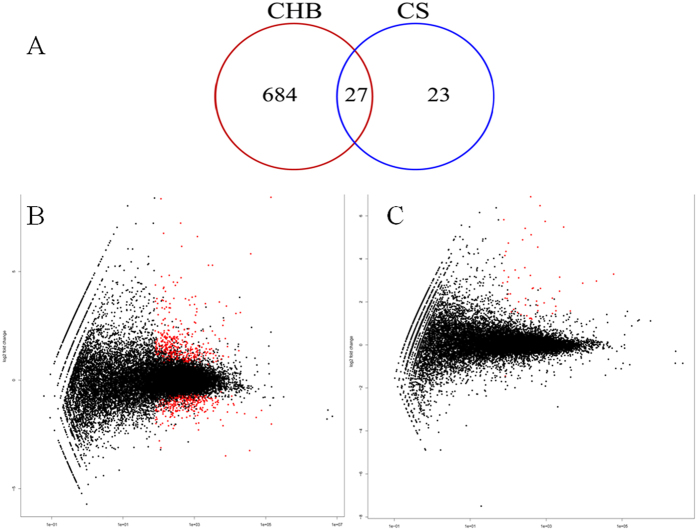
In this study, approximately 79.91% of raw reads (±7.08%; SD) were uniquely mapped to the ovine genome. Compared to their respective uninfected controls, the numbers of genes significantly impacted by infection in CHB and CS breeds at a stringent cutoff value (false discovery rate or FDR < 0.05), were 711 and 49, respectively (Fig. 2). The abundance of 27 genes was significantly changed by infection in both breeds (Table 1). Among them, 25 genes, such as arachidonate 15-lipoxygenase (ALOX15), collagen, type VI, α5 (COL6A5), and serglycin (SRGN), were significantly upregulated while the expression of transthyretin (TTC) was repressed by infection. Intriguingly, the transcript abundance of cadherin 26 (CDH26) was significantly induced by infection in both breeds (adjusted P value or FDR < 1.63 × 10−10); and is strongly correlated with worm counts only in CS (Fig. 3). However, infection had a bidirectional impact on the transcript abundance of a uncharacterized gene containing a unknown microRNA (ENSOARG00000023771), which was significantly upregulated in CHB but downregulated in CS. The genes significantly impacted by infection only in CS included mast cell proteinase-3, γ-glutamyltransferase 5 (GGT5), CD163 as well as those involved in smooth muscle contraction, such as tropomyosin (TPM2), myosin, light chain 9, regulatory (MYL9), and calponin 1, basic, smooth muscle (CNN1).
Figure 2.
(A) Venn Diagram showing the number of genes with significant differences in transcript abundance induced by infection in two sheep breeds compared to their respective uninfected controls at a false discovery rate (FDR) cutoff <0.05. CHB: Canaria Hair Breed (resistant). CS: Canaria Sheep (susceptible). (B,C) Scatter plot of log2 ratio (fold change) vs mean. The red color indicates genes detected as differentially expressed between the infected group and uninfected controls at a false discovery rate (FDR) < 0.05 in resistant Canaria Hair Breed (B) and susceptible Canaria Sheep (C).
Table 1. Genes significantly impacted by Haemonchus contortus infection in both CHB and CS breeds.
| Gene_ID | Symbol |
Fold change |
FDR |
||
|---|---|---|---|---|---|
| CHB | CS | CHS | CS | ||
| ENSOARG00000000338 | ABCA2 | 2.86 | 2.40 | 2.28% | 1.29% |
| ENSOARG00000008480 | ALOX15 | 10.02 | 11.45 | 2.06% | 0.00% |
| ENSOARG00000015249 | CDH26 | 150.19 | 54.19 | 0.00% | 0.00% |
| ENSOARG00000018133 | CFTR | 4.33 | 2.38 | 1.21% | 3.67% |
| ENSOARG00000014842 | COL6A5 | 20.79 | 6.28 | 1.67% | 0.04% |
| ENSOARG00000007787 | FCER1A | 20.69 | 18.23 | 0.00% | 0.00% |
| ENSOARG00000019163 | HBBB | 11.47 | 43.75 | 1.63% | 0.00% |
| ENSOARG00000008994 | IGHE | 22.73 | 35.02 | 0.00% | 0.00% |
| ENSOARG00000013111 | IL1RL1 | 9.73 | 5.26 | 0.00% | 0.00% |
| ENSOARG00000016842 | MCTP1 | 3.89 | 2.95 | 1.26% | 1.03% |
| ENSOARG00000002234 | SLC2A3 | 4.39 | 3.06 | 0.00% | 0.03% |
| ENSOARG00000005322 | SRGN | 4.25 | 3.60 | 0.45% | 0.00% |
| ENSOARG00000012855 | ST3GAL4 | 2.44 | 3.69 | 0.84% | 0.94% |
| ENSOARG00000009990 | SYNM | 2.16 | 6.18 | 2.99% | 0.09% |
| ENSOARG00000005941 | TNC | 3.00 | 4.01 | 2.80% | 3.19% |
| ENSOARG00000014689 | TPSAB1 | 6.96 | 9.43 | 0.00% | 0.00% |
| ENSOARG00000006342 | TTR | 0.28 | 0.37 | 0.00% | 3.17% |
| ENSOARG00000000857 | 8.41 | 7.84 | 0.00% | 1.29% | |
| ENSOARG00000002036 | 38.84 | 44.81 | 0.00% | 0.00% | |
| ENSOARG00000002629 | 13.34 | 23.51 | 0.00% | 0.00% | |
| ENSOARG00000002942 | 7.16 | 7.83 | 0.00% | 0.00% | |
| ENSOARG00000002964 | 12.61 | 29.65 | 0.01% | 0.00% | |
| ENSOARG00000006087 | 13.49 | 26.69 | 0.00% | 0.00% | |
| ENSOARG00000013005 | 3.00 | 3.53 | 4.13% | 0.94% | |
| ENSOARG00000013263 | 71.33 | 91.67 | 0.00% | 0.01% | |
| ENSOARG00000017398 | 5.73 | 7.34 | 2.15% | 0.65% | |
| ENSOARG00000023771 | 0.46 | 3.00 | 0.19% | 1.55% | |
Figure 3. Nonlinear regression between worm counts and normalized transcript abundance per million mapped reads of the gene cadherin 26 (CDH26) in susceptible Canaria Sheep (CS).
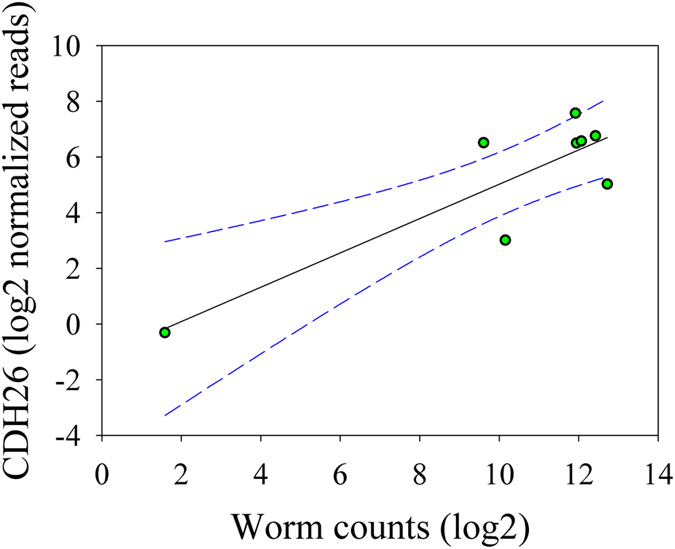
Dotted lines: 95% confidence interval.
Among the genes significantly impacted by infection in CHB sheep, several cytokine receptors and chemokines were strongly upregulated. Notable, the transcript of IL17 receptor beta (IL17RB) was 14.4 fold higher in infected animals than uninfected controls in CHB. IL2 receptor beta (IL2B) was also upregulated. Similarly, chemokine CXC ligand 12 (CXCL12) and chemokine (CXC motif) receptor 6 (CXCR6) were upregulated by infection in CHB. Among the well-known Th2 cytokines, the expression of IL6, IL10 and IL13 was upregulated by infection in both breeds. Moreover, while the extent of upregulation of IL6 by infection remained similar in both breeds (~6.8 fold), overexpression of both IL10 and IL13 mRNA molecules was more profound in the resistant breed (CHB) than in CS. On the other hand, the IL5 mRNA was upregulated by infection in CS but barely detectable in CHB at the sequencing depth in this study. The IL4 expression followed the similar trend: it was upregulated approximately 9 fold by infection in CS but was barely detectable in CHB. However, the IL9 mRNA level remained unchanged by infection in both breeds.
Several genes involved in arachidonic acids metabolism, including eicosanoids metabolism, were significantly impacted by infection, such as arachidonate 5-lipoxygenase (ALOX5) and its activating protein (ALOX5P), prostaglandin reductase 1 (PTGR1), prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (PTGS1, COX1), and thromboxane A synthase 1 (TBXAS1), were all strongly upregulated by infection in CHB. In addition, at least 11 genes implicated in complement activation were significantly impacted by infection in CHB, such as complement factor properdin (CFP, 2.8 fold), complement component 7 (C7, 4.2 fold), and complement factor I (CFI, 12.1 fold). Other known genes involved in protective immunity to helminth infection strongly upregulated by infection in CHB included amphiregulin (AREG, 2.2 fold), granzyme genes A and B (GZMA and GZMB, 6.8 and 12.9 fold, respectively).
41 of the 711 genes significantly impacted by infection in CHB are related to extracellular matrix (ECM, Table 2). Of them, fibronectin 1 (FN1) was strongly upregulated. At least ten collagen genes were significantly upregulated, such as those from Type I, Type III, Type V, Type VI, and Type XII (Table 2). For example, the expression of collagen, type VI, alpha 5 (COL6A5) and collagen, type XII, alpha 1 (COL12A1) was increased 20.8 and 2.4 fold, respectively in CHB, compared to the uninfected controls. Likewise, matrix metallopeptidase 1 (MMP1), MMP2, and MMP14 were significantly up-regulated while the transcript of MMP11 was repressed by infection. Furthermore, several cell adhesion molecules, including integrins, lectins, and cadhesion, were strongly upregulated by infection in CHB, such as conglutinin-like (COLEC8, 451.7 fold), integrin, α11 (ITGA11, 3.4 fold), and lectin, galactoside-binding, soluble, 15 (LGALS15, 340.1 fold).
Table 2. 41 extracellular matrix (ECM) related genes significantly affected by Haemonchus contortus infection in the abomasal mucosa of the Canaria Hair Breed sheep (CHB).
| GeneID | Symbol | Fold change | P value | FDR |
|---|---|---|---|---|
| ENSOARG00000013782 | ALB | 0.20 | 0.0000 | 0.10% |
| ENSOARG00000008507 | ALPL | 10.59 | 0.0000 | 0.00% |
| ENSOARG00000005139 | APLP1 | 0.54 | 0.0001 | 0.49% |
| ENSOARG00000018738 | BMP2 | 1.87 | 0.0004 | 1.11% |
| ENSOARG00000012877 | CFP | 2.78 | 0.0000 | 0.19% |
| ENSOARG00000004871 | COL1A1 | 2.18 | 0.0005 | 1.24% |
| ENSOARG00000001508 | COL1A2 | 1.87 | 0.0029 | 4.42% |
| ENSOARG00000016476 | COL3A1 | 2.09 | 0.0000 | 0.21% |
| ENSOARG00000002129 | COL5A1 | 2.39 | 0.0002 | 0.55% |
| ENSOARG00000016440 | COL5A2 | 1.93 | 0.0001 | 0.28% |
| ENSOARG00000012810 | COL6A1 | 3.72 | 0.0000 | 0.00% |
| ENSOARG00000012880 | COL6A2 | 3.25 | 0.0000 | 0.17% |
| ENSOARG00000019080 | COL6A3 | 2.65 | 0.0003 | 0.83% |
| ENSOARG00000014842 | COL6A5 | 20.79 | 0.0008 | 1.67% |
| ENSOARG00000006410 | COL12A1 | 2.37 | 0.0005 | 1.26% |
| ENSOARG00000009670 | CPXM2 | 2.10 | 0.0007 | 1.63% |
| ENSOARG00000017328 | F3 | 3.03 | 0.0000 | 0.06% |
| ENSOARG00000019404 | FBLN1 | 1.53 | 0.0022 | 3.54% |
| ENSOARG00000017189 | FBN2 | 0.41 | 0.0000 | 0.01% |
| ENSOARG00000016733 | FGA | 0.38 | 0.0009 | 1.93% |
| ENSOARG00000019329 | FN1 | 4.77 | 0.0000 | 0.07% |
| ENSOARG00000018483 | ITGA11 | 3.37 | 0.0015 | 2.75% |
| ENSOARG00000016642 | ITGB7 | 1.93 | 0.0032 | 4.70% |
| ENSOARG00000010344 | LTBP1 | 2.34 | 0.0000 | 0.16% |
| ENSOARG00000005315 | MMP1 | 11.03 | 0.0001 | 0.47% |
| ENSOARG00000013161 | MMP11 | 0.41 | 0.0000 | 0.21% |
| ENSOARG00000019414 | MMP14 | 1.85 | 0.0001 | 0.25% |
| ENSOARG00000018035 | MMP2 | 1.99 | 0.0000 | 0.12% |
| ENSOARG00000008537 | NAV2 | 0.57 | 0.0022 | 3.52% |
| ENSOARG00000010519 | OLFML2B | 2.26 | 0.0004 | 1.03% |
| ENSOARG00000006153 | PDGFA | 0.51 | 0.0009 | 1.85% |
| ENSOARG00000005685 | PLOD2 | 3.06 | 0.0011 | 2.15% |
| ENSOARG00000010041 | POSTN | 6.73 | 0.0000 | 0.00% |
| ENSOARG00000005275 | PXDN | 1.94 | 0.0010 | 1.98% |
| ENSOARG00000004813 | SDC2 | 1.95 | 0.0006 | 1.39% |
| ENSOARG00000005209 | SDC4 | 0.57 | 0.0009 | 1.93% |
| ENSOARG00000006391 | SERPINB5 | 3.99 | 0.0000 | 0.00% |
| ENSOARG00000020413 | SERPINE2 | 1.78 | 0.0003 | 0.80% |
| ENSOARG00000015081 | TGFBI | 3.12 | 0.0000 | 0.13% |
| ENSOARG00000005941 | TNC | 3.00 | 0.0016 | 2.80% |
| ENSOARG00000008334 | VEGFA | 0.44 | 0.0000 | 0.05% |
Of note, approximately 15% of the genes significantly impacted by infection are cell-cycle related. The expression of these cell cycle related genes was predominantly enhanced by Haemonchus infection in CHB. As Table 3 shows, at least 92 genes were significantly upregulated by infection, such as cyclin A2 (CCNA2), cyclin B3 (CCNB3), various centromere proteins (CENPL, CENPN, CENPT, and CENPW) and kinesin family (KIF) members, and at least 5 minichromosome maintenance complex (MCM) components (MCM3, MCM4, MCM5, MCM6, and MCM10). Nevertheless, the infection was also able to repress cell cycle related genes, such as cyclin G1 (CCNG1), regulator of cell cycle (RGCC), and synaptonemal complex protein 3 (SYCP3). Moreover, at least five transcription factors, such as the oncogene MYB, SMAD family members 6 and 9 (SMAD6 and SMAD9), and histone decetylase 5 (HDAC5), were significantly affected by infection in CHB.
Table 3. 100 cell-cycle related genes significantly affected by Haemonchus contortus infection in the resistant breed (CHB).
| Gene ID | Locus (chr:start:end) | Gene symbol | Fold (I/C) | Pvalue | FDR |
|---|---|---|---|---|---|
| ENSOARG00000004361 | 4:60803768:60856028 | ANLN | 5.01 | 5.05E-05 | 0.0022 |
| ENSOARG00000019052 | 11:27395740:27400878 | AURKB | 2.94 | 7.80E-05 | 0.0031 |
| ENSOARG00000012875 | 18:20942195:21007732 | BLM | 2.46 | 8.65E-04 | 0.0185 |
| ENSOARG00000018738 | 13:48462231:48472599 | BMP2 | 1.87 | 4.29E-04 | 0.0111 |
| ENSOARG00000004835 | 11:42540868:42607539 | BRCA1 | 3.09 | 6.51E-04 | 0.0152 |
| ENSOARG00000020126 | 7:32810618:32861855 | BUB1B | 3.38 | 9.57E-05 | 0.0037 |
| ENSOARG00000020247 | 7:33203355:33258031 | CASC5 | 3.52 | 2.94E-03 | 0.0442 |
| ENSOARG00000014176 | 6:3657736:3663110 | CCNA2 | 3.95 | 2.00E-07 | 0.0000 |
| ENSOARG00000009908 | X:51894389:51938870 | CCNB3 | 4.54 | 1.46E-03 | 0.0269 |
| ENSOARG00000010352 | 3:209738381:209761224 | CCND2 | 2.44 | 1.88E-04 | 0.0060 |
| ENSOARG00000004318 | 25:16420008:16437119 | CDC2 | 3.96 | 3.23E-04 | 0.0089 |
| ENSOARG00000020542 | 1:17815061:17818632 | CDC20 | 3.07 | 5.86E-04 | 0.0140 |
| ENSOARG00000001274 | 13:50726485:50734786 | CDC25B | 1.82 | 3.24E-03 | 0.0476 |
| ENSOARG00000014063 | 11:40114667:40125179 | CDC6 | 4.24 | 2.32E-08 | 0.0000 |
| ENSOARG00000009851 | 2:39869389:39909559 | CDCA2 | 2.74 | 1.11E-03 | 0.0218 |
| ENSOARG00000005652 | 3:207550908:207552937 | CDCA3 | 2.65 | 2.88E-05 | 0.0015 |
| ENSOARG00000019830 | 1:12275289:12289262 | CDCA8 | 3.70 | 7.07E-05 | 0.0029 |
| ENSOARG00000011059 | 6:21798087:21861426 | CENPE | 3.68 | 1.30E-05 | 0.0008 |
| ENSOARG00000012529 | 12:53214185:53224480 | CENPL | 2.72 | 6.91E-04 | 0.0158 |
| ENSOARG00000008206 | 14:7100459:7127206 | CENPN | 3.59 | 6.95E-05 | 0.0029 |
| ENSOARG00000003186 | 14:34675201:34680944 | CENPT | 2.71 | 1.54E-03 | 0.0276 |
| ENSOARG00000007744 | 8:12047574:12055213 | CENPW | 3.21 | 1.43E-04 | 0.0049 |
| ENSOARG00000003158 | 22:14531509:14550251 | CEP55 | 4.11 | 2.07E-06 | 0.0002 |
| ENSOARG00000009704 | 5:17254276:17278480 | CHAF1A | 2.25 | 1.82E-04 | 0.0059 |
| ENSOARG00000009289 | 10:21841538:21861932 | CKAP2 | 3.27 | 2.54E-03 | 0.0403 |
| ENSOARG00000007721 | 2:23987476:23992086 | CKS2 | 3.46 | 1.08E-06 | 0.0001 |
| ENSOARG00000019542 | 1:10450581:10480905 | CLSPN | 2.00 | 1.47E-03 | 0.0269 |
| ENSOARG00000021089 | 7:64546752:64587073 | DLGAP5 | 3.93 | 6.21E-04 | 0.0147 |
| ENSOARG00000017620 | 1:186409:193575 | DTYMK | 1.92 | 2.03E-03 | 0.0339 |
| ENSOARG00000007334 | 2:242210461:242229702 | E2F2 | 3.47 | 4.09E-05 | 0.0019 |
| ENSOARG00000008807 | 21:25034831:25052009 | E2F8 | 4.39 | 3.03E-08 | 0.0000 |
| ENSOARG00000020622 | 3:199025742:199151626 | EPS8 | 2.36 | 6.02E-05 | 0.0025 |
| ENSOARG00000005908 | X:61406551:61410223 | ERCC6L | 4.54 | 7.42E-07 | 0.0001 |
| ENSOARG00000014845 | 2:100985329:101014711 | ESCO2 | 3.40 | 5.21E-05 | 0.0022 |
| ENSOARG00000000644 | 13:67321805:67342479 | FAM83D | 3.04 | 9.58E-05 | 0.0037 |
| ENSOARG00000005211 | 19:16642527:16690458 | FANCD2 | 2.84 | 2.96E-05 | 0.0015 |
| ENSOARG00000003968 | 8:76466657:76471151 | FBXO5 | 3.65 | 7.37E-05 | 0.0030 |
| ENSOARG00000015633 | 21:39647404:39648523 | FEN1 | 2.10 | 9.02E-04 | 0.0191 |
| ENSOARG00000011054 | 3:210883196:210893395 | FOXM1 | 3.42 | 6.81E-04 | 0.0158 |
| ENSOARG00000007717 | 13:52921512:52934511 | GINS1 | 3.70 | 8.22E-06 | 0.0006 |
| ENSOARG00000004495 | 22:15498816:15537211 | HELLS | 3.82 | 1.95E-09 | 0.0000 |
| ENSOARG00000019189 | 1:6959524:6970726 | HJURP | 3.11 | 1.52E-04 | 0.0051 |
| ENSOARG00000020743 | 7:42727941:42735567 | KIAA0101 | 4.06 | 1.23E-06 | 0.0001 |
| ENSOARG00000004780 | 19:16405186:16463300 | KIF15 | 3.29 | 2.62E-03 | 0.0413 |
| ENSOARG00000015211 | 15:56874125:56943420 | KIF18A | 2.92 | 1.68E-03 | 0.0295 |
| ENSOARG00000015873 | 5:46994289:47001867 | KIF20A | 3.56 | 6.85E-05 | 0.0028 |
| ENSOARG00000005591 | 24:26450684:26467845 | KIF22 | 2.77 | 7.56E-05 | 0.0030 |
| ENSOARG00000018647 | 7:16078138:16122235 | KIF23 | 2.92 | 1.63E-03 | 0.0288 |
| ENSOARG00000001102 | 1:19196065:19216520 | KIF2C | 3.45 | 2.73E-04 | 0.0079 |
| ENSOARG00000009637 | 20:7678567:7686289 | KIFC1 | 2.48 | 5.81E-04 | 0.0139 |
| ENSOARG00000020216 | 7:33016321:33025738 | KNSTRN | 2.15 | 1.66E-03 | 0.0292 |
| ENSOARG00000009349 | 17:52444861:52510658 | KNTC1 | 2.66 | 1.04E-03 | 0.0208 |
| ENSOARG00000015347 | 11:48337064:48345443 | KPNA2 | 2.30 | 1.21E-04 | 0.0044 |
| ENSOARG00000015665 | 6:5610960:5622911 | MAD2L1 | 2.76 | 1.99E-04 | 0.0062 |
| ENSOARG00000005416 | 13:26912745:26937031 | MCM10 | 2.02 | 3.35E-03 | 0.0487 |
| ENSOARG00000014143 | 20:24477536:24494074 | MCM3 | 2.06 | 3.46E-05 | 0.0017 |
| ENSOARG00000012797 | 9:32400919:32411149 | MCM4 | 2.57 | 1.32E-06 | 0.0001 |
| ENSOARG00000018527 | 3:178690575:178707822 | MCM5 | 2.32 | 1.72E-05 | 0.0010 |
| ENSOARG00000010614 | 2:173834090:173868287 | MCM6 | 2.25 | 3.91E-05 | 0.0019 |
| ENSOARG00000011541 | 2:51686232:51755300 | MELK | 3.87 | 1.90E-06 | 0.0002 |
| ENSOARG00000009575 | 18:53870504:53915925 | MIS18BP1 | 3.74 | 3.46E-03 | 0.0499 |
| ENSOARG00000014562 | 22:46439178:46468099 | MKI67 | 3.15 | 6.36E-04 | 0.0149 |
| ENSOARG00000014901 | 8:60237288:60271515 | MYB | 3.06 | 2.71E-05 | 0.0014 |
| ENSOARG00000003547 | 13:71800636:71832436 | MYBL2 | 3.10 | 1.78E-07 | 0.0000 |
| ENSOARG00000004016 | 6:37256547:37333851 | NCAPG | 3.36 | 5.28E-06 | 0.0004 |
| ENSOARG00000007995 | 4:118730937:118799615 | NCAPG2 | 2.64 | 6.03E-05 | 0.0025 |
| ENSOARG00000009604 | 23:37208584:37244969 | NDC80 | 3.71 | 4.15E-06 | 0.0003 |
| ENSOARG00000011466 | 12:69915815:69927483 | NEK2 | 3.45 | 4.25E-05 | 0.0019 |
| ENSOARG00000011189 | 1:113098396:113135220 | NUF2 | 2.47 | 9.75E-04 | 0.0199 |
| ENSOARG00000005282 | 1:26531082:26561083 | ORC1 | 2.82 | 4.59E-05 | 0.0021 |
| ENSOARG00000014858 | 2:101015056:101043656 | PBK | 4.32 | 1.36E-05 | 0.0008 |
| ENSOARG00000017133 | 13:46615170:46619285 | PCNA | 2.05 | 1.17E-04 | 0.0043 |
| ENSOARG00000010890 | 12:52045755:52079590 | PDPN | 2.08 | 1.43E-03 | 0.0264 |
| ENSOARG00000015691 | 17:29540624:29557094 | PLK4 | 1.97 | 2.02E-03 | 0.0338 |
| ENSOARG00000020607 | 7:39763421:39793166 | POLE2 | 2.83 | 3.61E-04 | 0.0096 |
| ENSOARG00000012267 | 18:20787183:20802103 | PRC1 | 3.88 | 9.65E-04 | 0.0198 |
| ENSOARG00000020254 | 7:33275919:33306843 | RAD51 | 3.12 | 7.61E-05 | 0.0031 |
| ENSOARG00000010707 | 24:15349788:15350439 | RAN | 1.79 | 1.72E-03 | 0.0300 |
| ENSOARG00000017221 | 13:65583237:65642844 | RBL1 | 2.12 | 2.02E-03 | 0.0338 |
| ENSOARG00000017883 | 13:58047850:58061508 | RBM38 | 2.35 | 3.13E-03 | 0.0464 |
| ENSOARG00000015333 | 3:19185137:19191043 | RRM2 | 5.12 | 5.48E-09 | 0.0000 |
| ENSOARG00000017493 | 6:91484955:91532036 | SEPT11 | 2.18 | 2.04E-04 | 0.0063 |
| ENSOARG00000018302 | 2:239211366:239212113 | SFN | 2.06 | 2.68E-03 | 0.0420 |
| ENSOARG00000007399 | 2:18870299:18918858 | SMC2 | 2.60 | 1.89E-03 | 0.0324 |
| ENSOARG00000001001 | 11:19659072:19677569 | SPAG5 | 2.45 | 1.79E-04 | 0.0058 |
| ENSOARG00000017725 | 5:13248115:13254108 | SPC24 | 2.98 | 3.22E-04 | 0.0089 |
| ENSOARG00000002888 | 16:1518544:1544794 | SPDL1 | 1.95 | 2.92E-03 | 0.0442 |
| ENSOARG00000011578 | 18:20124947:20166662 | TICRR | 2.83 | 1.61E-03 | 0.0286 |
| ENSOARG00000001419 | 13:60597535:60651659 | TPX2 | 2.52 | 6.32E-04 | 0.0149 |
| ENSOARG00000016302 | 7:3740229:3789354 | TRIM36 | 10.24 | 1.19E-14 | 0.0000 |
| ENSOARG00000007151 | 8:6885831:6925302 | TTK | 3.59 | 9.22E-06 | 0.0006 |
| ENSOARG00000018884 | 3:136887716:136929972 | TUBA4A | 2.02 | 1.14E-04 | 0.0042 |
| ENSOARG00000006520 | 13:74125916:74129104 | UBE2C | 3.13 | 1.25E-05 | 0.0008 |
| ENSOARG00000008530 | 5:16805815:16842579 | UHRF1 | 3.99 | 2.60E-07 | 0.0000 |
| ENSOARG00000004351 | 8:81449297:81450503 | 1.84 | 8.66E-04 | 0.0185 | |
| ENSOARG00000006991 | 19:59822300:59844027 | 2.08 | 2.93E-04 | 0.0084 | |
| ENSOARG00000005764 | 2:240064407:240068660 | 2.37 | 2.89E-05 | 0.0015 | |
| ENSOARG00000006571 | 21:17310180:17310609 | 2.66 | 3.78E-06 | 0.0003 | |
| ENSOARG00000005759 | 16:10486354:10496192 | 3.81 | 1.78E-04 | 0.0058 | |
| ENSOARG00000000647 | 22:13731293:13771253 | 3.92 | 4.05E-04 | 0.0106 |
Fold is expressed as infected/uninfected controls. FDR: false discovery rate.
Intriguingly, four genes known to regulate abomasal acid secretion and gastric function16 were downregulated by Haemonchus infection in CHB, including ATPase, H+/K+ exchanging, alpha polypeptide (ATP4A), progastricsin (pepsinogen C, PGC), appetite-regulating hormone precursor (GHRL), and forkhead box A2 (FOXA1). However, the transcript abundance of these four genes remained unchanged by infection in CS.
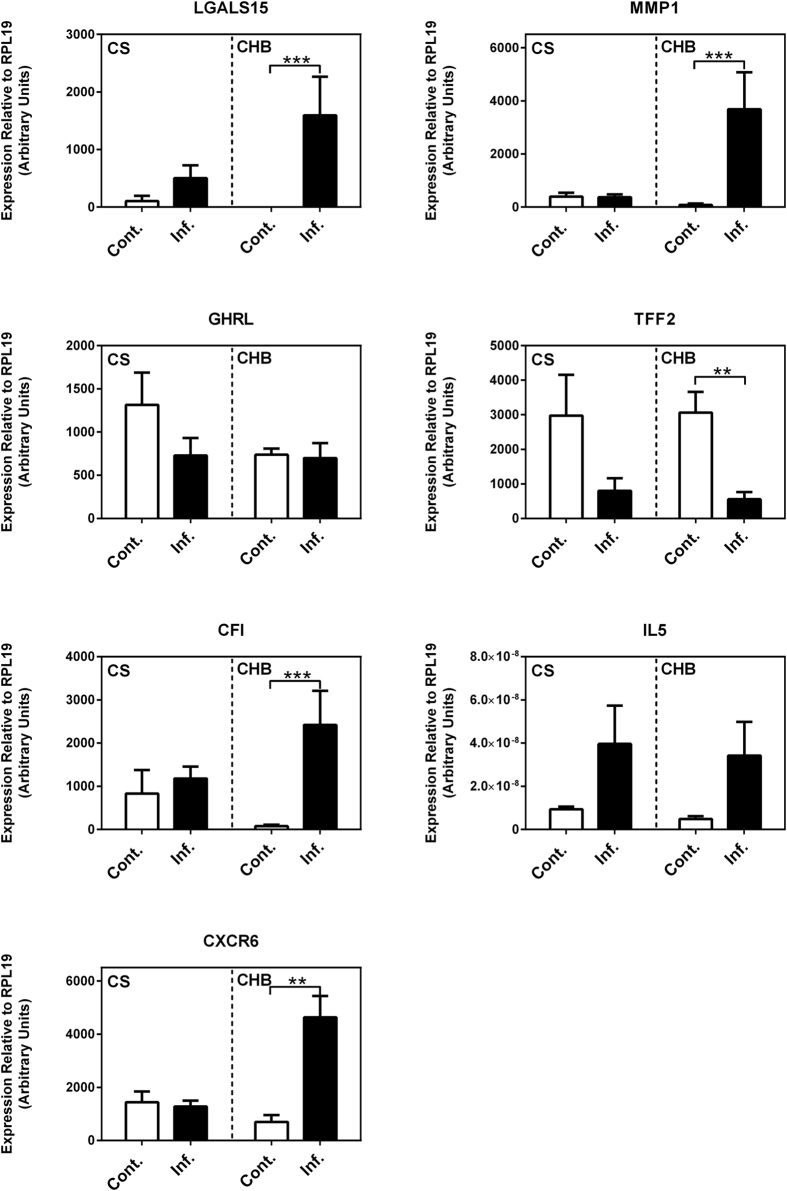
The RNAseq results of selected genes were validated by real-time RT-PCR (Fig. 4). For example, the expression of CFI, CXCR6, LGALS15, and MMP1 was significantly upregulated while TFF2 mRNA level was significantly repressed by infection only in the resistant breed (CHB), in a good agreement with the RNAseq analysis. A strong correlation in log2 transformed fold values between the two platforms, qPCR and RNAseq, was evident (a correlation coefficient R = 0.946; Fig. 5).
Figure 4. Real-Time RT-PCR analysis (qPCR) of selected genes.
Relative expression levels calculated from standard curves were normalized to the endogenous control gene RPL19. Numbers represent mean values plus standard error. Cont.: uninfected controls; Inf.: 20 days post infection by Haemonchus contortus. CS: Canaria Sheep; CHB: Canaria Hair Breed. CFI: Complement factor I; CXCR6: Chemokine (C-X-C motif) receptor 6; GHRL: Ghrelin/obestatin prepropeptide; LGALS15: Galectin 15; IL5: Interleukin 5. MMP1: Matrix metallopeptidase 1; TFF2: Trefoil factor 2. **P < 0.001; ***P < 0.0001.
Figure 5. Linear regression analysis of fold changes calculated from qPCR and RNAseq analysis.
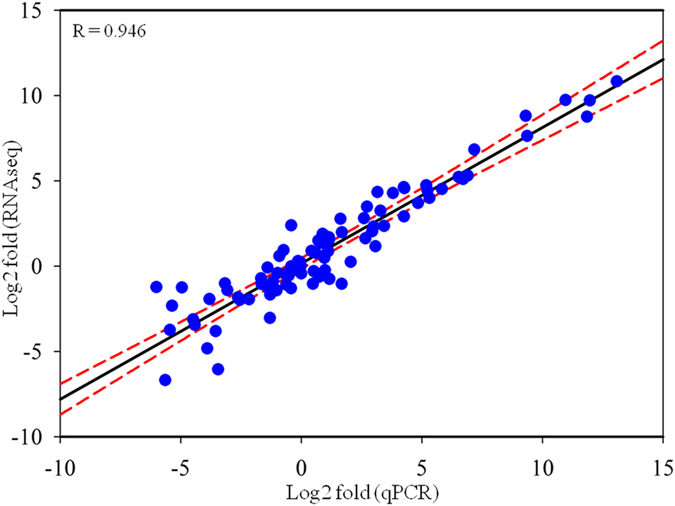
Blue dots represent log2 transformed fold change values of a single gene in an infected sample obtained from qPCR (X-axis) and RNAseq analysis (Y-axis). Dashed lines: 99% Confidence Interval. R: correlation coefficient.
Gene Ontology (GO) implicated in host resistance
Among 477 and 16 GO terms significantly enriched in CHB and CS at a P value cutoff 1.0 × 10−4, respectively, five were significantly enriched in both breeds (Table 4). Select GO terms that may be implicated in the development of host resistance to Haemonchus infection are listed in Table 5. Several GO related to complement activation (both classical and alternative pathways) and its regulation were significantly enriched only in CHB. Numerous cell cycle related GO were significantly enriched as well (Fig. 6). GO related to secretory granule and gastric acid secretion were also enriched, suggesting that the ability to regulate secretory and gastric function of the host may be involved in the development of host resistance. Furthermore, the regulation of inflammation at the site of infection (mucosa), including arachidonic acid metabolism, cyclooxygenase pathway, and positive regulation of MAPK cascade, as well as leukocyte migration were also implicated in host resistance. On the other hand, four of the 11 GO unique to CS were related to muscle contraction.
Table 4. Gene Ontology (GO) biological processes (BP) significantly enriched in both resistant (CHB) and susceptible (CS) breeds.
| GO_ID | Description |
ZScore |
Pvalue |
||
|---|---|---|---|---|---|
| Resistant | Susceptible | Resistant | Susceptible | ||
| GO:0002250 | adaptive immune response | 4.65 | 5.09 | 4.59E-05 | 7.37E-04 |
| GO:0046456 | eicosanoid biosynthetic process | 6.92 | 6.84 | 3.81E-07 | 6.59E-04 |
| GO:0006691 | leukotriene metabolic process | 7.22 | 9.67 | 9.37E-07 | 9.52E-05 |
| GO:1901570 | fatty acid derivative biosynthetic process | 6.92 | 6.84 | 3.81E-07 | 6.59E-04 |
| GO:0006636 | unsaturated fatty acid biosynthetic process | 7.02 | 6.43 | 2.12E-07 | 9.16E-04 |
Table 5. Selected Gene Ontology (GO) terms significantly impacted by Haemonchus contortus infection in the resistant breed (CHB).
| GO_id | Ontology | Description | Observed/Total | Z Score | P Value |
|---|---|---|---|---|---|
| GO:0019369 | BP | arachidonic acid metabolic process | 10/36 | 4.95 | 1.13E-04 |
| GO:0002673 | BP | regulation of acute inflammatory response | 15/38 | 7.93 | 1.16E-08 |
| GO:0002675 | BP | positive regulation of acute inflammatory response | 8/13 | 7.78 | 4.69E-07 |
| GO:0050727 | BP | regulation of inflammatory response | 24/135 | 5.01 | 1.53E-05 |
| GO:0050900 | BP | leukocyte migration | 30/183 | 5.11 | 7.54E-06 |
| GO:0043410 | BP | positive regulation of MAPK cascade | 30/197 | 4.65 | 3.28E-05 |
| GO:0006956 | BP | complement activation | 14/33 | 8.06 | 1.18E-08 |
| GO:0030449 | BP | regulation of complement activation | 11/23 | 7.75 | 9.91E-08 |
| GO:0006957 | BP | complement activation, alternative pathway | 7/10 | 7.87 | 7.26E-07 |
| GO:0006958 | BP | complement activation, classical pathway | 11/25 | 7.33 | 2.87E-07 |
| GO:0031714 | MF | C5a anaphylatoxin chemotactic receptor binding | 5/5 | 8.21 | 1.55E-06 |
| GO:0031715 | MF | C5L2 anaphylatoxin chemotactic receptor binding | 4/4 | 7.35 | 2.25E-05 |
| GO:0007049 | BP | cell cycle | 119/1137 | 5.00 | 1.53E-06 |
| GO:0022402 | BP | cell cycle process | 97/865 | 5.21 | 8.16E-07 |
| GO:0008283 | BP | cell proliferation | 111/1013 | 5.34 | 3.84E-07 |
| GO:0051301 | BP | cell division | 77/504 | 7.59 | 1.83E-11 |
| GO:0051302 | BP | regulation of cell division | 23/140 | 4.47 | 8.26E-05 |
| GO:0051321 | BP | meiotic cell cycle | 15/87 | 3.82 | 8.23E-04 |
| GO:0031577 | BP | spindle checkpoint | 11/40 | 5.15 | 5.72E-05 |
| GO:0007088 | BP | regulation of mitosis | 15/80 | 4.20 | 3.26E-04 |
| GO:0042555 | CC | MCM complex | 5/11 | 5.05 | 5.03E-04 |
| GO:0044818 | BP | mitotic G2/M transition checkpoint | 6/11 | 6.24 | 3.63E-05 |
| GO:0007186 | BP | G-protein coupled receptor signaling pathway | 33/207 | 5.18 | 4.97E-06 |
| GO:0043627 | BP | response to estrogen | 16/83 | 4.46 | 1.48E-04 |
| GO:0001676 | BP | long-chain fatty acid metabolic process | 11/53 | 3.99 | 8.27E-04 |
| GO:0033500 | BP | carbohydrate homeostasis | 15/81 | 4.14 | 3.75E-04 |
| GO:0006865 | BP | amino acid transport | 14/71 | 4.27 | 2.97E-04 |
| GO:0030141 | CC | secretory granule | 28/172 | 4.89 | 1.72E-05 |
| GO:0051048 | BP | negative regulation of secretion | 12/57 | 4.22 | 4.23E-04 |
| GO:0071229 | BP | cellular response to acid | 19/96 | 5.00 | 2.49E-05 |
| GO:0001696 | BP | gastric acid secretion | 6/7 | 8.23 | 7.00E-07 |
BP = Biological processes. MF = Molecular functions. CC = Cellular components.
Figure 6. Gene Ontology (GO) lineage relations.
The Cellular Component Ontology term GO:0000940 (condensed chromosome outer kinetochore) significantly enriched in resistant Canaria Hair Breed (CHB, adjusted P value < 2.30 × 10−4).
Discussion
Parasite resistance refers to the ability of the host to avert infection, resulting in reduced worm burden17. Numerous factors affect this trait. Among them, host genetics play a predominant role in controlling the development of resistance while host sex, age, and prior exposure are also important18. Differences in parasite resistance and susceptibility existing in various sheep breeds have been long recognized3. Moreover, inter- and intra-host variations in resistance are evident in certain sheep populations18. Identifying genetics components controlling inter-, and intra-breed differences in parasite resistance has both pragmatic and theoretical implications. Towards this end, numerous efforts have been made over the decades to unravel genes and/or genetic variants responsible for resistance, partially driven by strong desires to breed farm animals with strong resistance traits. Traditional QTL analysis and Genome-wide Association Studies (GWAS) have led to reports of dozens of QTL or markers on almost every ovine chromosome that are associated with various resistance phenotypes, such as fecal egg counts, packed cell volume, and parasite-specific antibody titers9,11,13. Nevertheless, the development of parasite resistance relies upon the precise control of expression of the host genome. Understanding these regulatory elements will be crucial towards unraveling their functional relevance. As a result, while much progress has been made to identify genes associated with nematode resistance in sheep during the past few years19,20, an in-depth comparison and characterization of transcriptome responses of various breeds and populations, especially those local indigenous breeds harboring varying degrees of parasite resistance and susceptibility, is urgently needed.
The two indigenous breeds of sheep native to the Canary Islands, CHB and CS, display unique and distinct differences in parasite resistance and susceptibility. When co-grazing together on the same pasture under natural infections, differences in fecal trichostrongylid egg counts between CHB and CS are consistently observed14. Under experimental infections with H. contortus, CHB has a significantly lower, by approximately 50%, worm burden than CS, a undeniable trait of parasite resistance14,15, which is confirmed in this study. Moreover, worms recovered in CHB tend to have significantly shorter body length than those in CS. A significantly lower EPG value is consistently observed in the feces of CHB sheep than those of CS animals during experimental infection. For example, at 27dpi, the mean EPG in CS is 5 fold higher than in CHB14. CHB sheep not only shed significantly fewer parasite eggs but also tend to have a delayed egg production, indicating an anti-fecundity effect of the immune response in this breed. The results from this study show that at 20 dpi, no parasite eggs were recovered in the feces of infected CHB animals while EPG in the feces of infected CS sheep reached 262.50 (±287.54, SD). This observation is in agreement with the previous findings14. Haemonchus contortus infection generally elicits a potent Th2 immune response in small ruminants. A strong upregulation of several well-known Th2 cytokines by infection in CHB were observed in this study. Previous studies in the Canary Island breeds suggest that divergence in immune response mechanisms exist between CHB and CS. Among various immune cells, abomasal eosinophil numbers are 2 fold higher in CHB than in CS, suggesting that CHB sheep may have developed abilities for enhanced recruitment of eosinophils to the site of infection (abomasal mucosa). Furthermore, CHB sheep have evolved mechanisms attacking the adult stage of the Haemonchus parasite, especially its reproduction, as evidenced by the fact that fecundity is negatively correlated with eosinophils and γδ T cells in the abomasal mucosa15. However, the precise molecular mechanisms of the parasite resistance in CHB breed remain largely unclear.
In this study, we identified a total of 477 and 16 Gene Ontology (GO) terms that are significantly enriched in the transcriptome of resistant and susceptible sheep breeds in responses to Haemonchus infection, respectively. Among them, only five enriched GO were shared by both breeds. These GO, including leukotriene metabolic process, eicosanoid biosynthesis process, adaptive immune response, and unsaturated fatty acid biosynthesis, likely represents the basic mechanisms of host immune responses to helminth infection in sheep. Indeed, local inflammatory responses are known to be involved in the development of host resistance21. The enriched GO unique to the susceptible CS breed were predominantly muscle contraction-related. In cattle, our previous results suggest that smooth muscle hypercontractility induced by primary infection of the intestinal worm Cooperia oncophora represents an important aspect of host responses22, as in several other host-parasite systems23. In rodent models, helminth infection results in an increase in thickness of jejunal smooth muscle layers. Other studies also support the idea that enhanced muscle contractility appears to be associated with more rapid worm expulsion and stronger host immune responses24. In addition, granzyme-mediated apoptotic signaling pathway (GO:0008626) may play an important role in protecting the host from H. contortus infection in the susceptible CS breed.
Complement activation as one of the earliest events in host immune responses to helminth infection plays an important role in the development of host resistance25. At least 11 complement related genes, such as CFI and C7, were significantly impacted by infection in the resistant CHB breed compared to uninfected controls while none of these genes were affected by infection in the susceptible breed. As a result, both classical and alternative complement pathways appeared to be activated in the resistant breed. Furthermore, two GO molecular functions related to C5a (GO:0031714) and C5L2 anaphylatoxin chemotactic receptor binding (GO:0031715) were significantly enriched in the resistant breed. It is conceivable that these peptides play a critical role in subsequent recruitment of effector cells, such eosinophils and mast cells, to the site of infection.
Intriguingly, approximately 15% of the 711 genes whose transcript abundance were significantly altered by infection in the resistant breed were cell cycle related. The vast majority of these genes were significantly upregulated (Table 3). These genes included several cyclins, minichromosome maintenance complex components, and various kinesin family members (Table 3). In addition, a large class of genes significantly impacted in the resistant breed was ECM related (Table 2). ECM related genes are required during the classical stages of wound repair, inflammation, new tissue formation, and remodeling26. Previous studies identified essential roles of Th2 cytokines in limiting tissue damage during helminth infection in rodent models, especially the involvement of IL17 in the early stage of tissue repair via its role in neutrophil recruitment27. In this study, the transcript abundance of IL17RB was increased approximately14 fold by Haemonchus infection only in the resistant breed. Of note, upregulation of Th2 cytokines IL10 and IL13 by infection was more profound in CHB than in CS. Together, our findings suggest that the accelerated tissue repair ability, likely mediated by Th2 cytokines, has evolved in the resistant CHB breed.
Recently, a significant SNP marker on ovine chromosome (OAR) 6 was reported to affect one of key resistant phenotypes in sheep, EPG9. This maker explains approximately 4% of the variance observed for EPG. It is suggested that there may exist up to 3 QTL within the 5 Mb region of this locus (73.1–78.3 Mb), in addition to a fourth QTL at 55.9–62.6 Mb on OAV6. Several earlier reports also indicate the presence of the QTL lined to EPG in various sheep breeds28,29. Among 21 differentially expressed genes located on OAV6 identified in our study in CHB breeds, at least 5 genes are located within 15 Mb of this marker. The expression of four genes were significantly induced whereas one, albumin, was repressed by infection. Of note, mast cell stem cell factor (SCF) receptor KIT gene (chr6:70189728:70234612) is the closest to the SNP marker. Two major receptors, KIT and the high affinity receptor for IgE, are responsible for regulating various mast cell functions, including chemotaxis, proliferation, apoptosis, and cytokine releases30. The critical roles of mast cells in host immune responses to helminth infection have long been recognized31. Neutralization of KIT and its ligand, SCF, using monoclonal antibodies completely abrogates the mast cell hyperplasia generated by T. spiralis infection in mice, resulting in drastically delayed worm expulsion and a reduced mucosal eosinophilia32. This finding suggests that KIT plays an important in host-parasite interaction. In the past few years, increasing evidence suggests that the epidermal growth factor like molecule, amphiregulin (AREG), plays critical roles in regulating immunity and inflammation as well as in enhancing host resistance to helminth parasites33,34. In rodent models, T. suis infection increases AREG expression, in parallel with the expression of Th2 cytokines IL4 and IL1333. Furthermore, worm clearance is significantly delayed at 14 dpi in AREG deficient mice, which correlates with reduced proliferation of colonic epithelial cells. Recent studies show that AREG is critical for efficient regulatory T cell function35 and may play an important role in orchestrating immunity, inflammation, and tissue repair34. In this study, AREG transcript abundance was significantly enhanced by infection only in the resistant breed, suggesting that this gene may play an important role in the development of host resistance. It would be intriguing to identify SNPs in both coding and promoter regions of the genes located within or closer to the QTL related to parasite resistance on OAV6, including AREG, and correlate the observed genetic variations with various resistant phenotypes. Moreover, dissecting mechanisms of transcriptional regulation of AREG and understanding how it promotes epithelial cell proliferation and regulates host immunity in the gastrointestinal tract warrant further investigation.
In conclusions, the two sheep breeds native to the Canaria Island displayed a distinct difference in several Haemonchus contortus resistant phenotypes under both natural and experimental infections. CHB tends to have significantly reduced worm burden, delayed egg production, and decreased fecal egg yield (counts) than the susceptible Canaria Sheep. A broad range of mechanisms have evolved in resistant CHB to provide protection against H. contortus. Readily inducible acute inflammation responses, complement activation, accelerated cell proliferation and subsequent tissue repair, and innate and acquired immunity directly against worm fecundity are likely to contribute to the development of host resistance to gastrointestinal nematode infection in the CHB breed.
Methods
Animals and parasitology
Male lambs of CHB (11 animals) and CS breeds (12 animals) were obtained from local farms in the Gran Canaria Island (Spain), weaned, and kept in pens at the Faculty of Veterinary Science, University of Las Palmas de Gran Canaria until they were approximately one year old. The animals were fed with a commercial pelleted sheep ration ad libitum and had free access to water throughout the experimental period. The animals were drenched upon arrival with levamisole (Cyber, Fort Dodge, Spain) at the recommended dose (1 ml/10 kg bodyweight) and remained free of parasites (as determined by fecal egg counts) until experimental parasite inoculation. Seven CHB and eight CS animals were inoculated intraruminally with 20,000 H. contortus infective L3 larvae. Four age-matched animals of each breed remained uninfected and served as controls. The experimental infection was allowed to progress for 20 dpi. The time point chosen for this study was based on the results from a previous report that the difference in resistance phenotypes, especially mean EPG values, is most profound between the two breeds14. Furthermore, no fecal parasite eggs become detectable at this time point in CHB. At 20 dpi, both infected and control animals were sacrificed. EPG values were monitored twice during the experiment, prior to the experimental inoculation and immediately prior to necropsy using the MacMaster technique. Adult worms as well as immature larvae from both contents and the tissue of the abomasum were isolated and counted. The fundic abomasum tissue was then sampled and snap frozen in liquid nitrogen prior to storage at −80 °C until total RNA was extracted. The Haemonchus strain used in this trial was initially donated by Drs. Knox and Bartley (Moredun Research Institute, Edinburgh, Scotland) and passaged through successive inoculations in sheep at the premises of the Faculty of Veterinary Science, University of Las Palmas de Gran Canaria (Spain). During the experiment, all animal protocols were approved by the Animal Care and Use Committee of University of Las Palmas per the Institutional Animal Care and Use Committee (IACUC) guidelines. All experimental procedures were carried out in accordance with the approved protocols.
RNA extraction and sequencing using RNA-seq
Total RNA from fundic abomasal samples of both CHS and CS sheep breeds was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) followed by DNase digestion and Qiagen RNeasy column purification (Qiagen, Valencia, CA, USA), as previously described22,36. The RNA integrity was verified using an Agilent BioAnalyzer 2100 (Agilent, Palo Alto, CA, USA). High-quality RNA (RNA integrity number or RIN > 7.5) was processed using an Illumina TruSeq RNA sample prep kit following the manufacturer’s instructions (Illumina, San Diego, CA, USA). Pooled RNAseq libraries were sequenced at 2 × 101 bp/sequence read using an Illumina HiSeq 2000 sequencer, as described previously37. Approximately 56 million paired-end sequence reads per sample (mean ± SD = 55,945,621 ± 41,305,493.24; N = 23) were generated. The metadata and raw sequences files related to this project were deposited in the NCBI Sequence Read Archive (Accession #SRP059627).
Data analysis and bioinformatics
Raw sequence reads were first checked using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). The effect of trimming of low-quality nucleotides on genome alignment was examined using STAR algorithm38. Raw sequence reads (FASTQ files) of 23 samples were mapped against the ovine reference genome Oar_v3.1 using STAR (v2.3.1t) with default parameters. The uniquely mapped read were used to count against the Ensembl annotation Oar_v3.1 for calculating the number of reads per gene. The counts of all samples were tabulated. This table was then inputted to DESeq39 for normalization and identification of differentially expressed genes between infection and control groups of both CHS and CS using the standard workflow as described38. To correct for multiple hypothesis testing, the Benjamini-Hochberg procedure40 was used with an FDR cutoff of 0.05. Gene Ontology (GO) analysis over differentially expressed genes was performed using Fisher’s exact test.
Real-Time RT-PCR (qPCR) validation
In order to validate the results obtained in the RNAseq analysis, the expression of 7 genes (see Supplementary table for their primer sequences) was determined by qPCR as previously described36. Ovine ribosomal protein L19 gene (RPL19), whose expression remained stable among the experimental samples, was used as an endogenous reference gene for all reactions. cDNA was synthesized from high quality total RNA (RIN > 7.5) using Superscript II reverse transcriptase (Invitrogen, Carlsbad, Carlsbad, CA) according to the manufacturer’s instructions. All qPCR reactions were carried out in 96-well plates in a 7500 Real-Time PCR System and analysed with a 7500 Software v2.0.6 (Applied Biosystems, NY). Samples were run in duplicate in a total volume of 25 μl containing the following: 5 μl of cDNA (100 ng), 1 μl of primer mix (forward and reverse, 10 nM each), 0.1 μl of ROX, 12.5 μl of SYBR GreenER qPCR SuperMix (Invitrogen) and 6.4 μl of dd water. The amplification reactions were subjected to a holding stage of 50 °C for 2 min, followed by an initial denaturation at 95 °C for 10 min. The reactions were then followed by 40 cycles of 95 °C for 30 sec, 60 °C for 30 sec and 72 °C for 32 sec. Melting curves were obtained from 60 °C to 95 °C. Relative gene expression values were determined using a standard curve method. Briefly, eight 10-fold serial dilutions of a pool of cDNA samples were used to generate standard curves for each gene to calculate relative gene expression levels. These results were then normalized to RPL19 gene expression levels for each sample.
Additional Information
How to cite this article: Guo, Z. et al. Possible mechanisms of host resistance to Haemonchus contortus infection in sheep breeds native to the Canary Islands. Sci. Rep. 6, 26200; doi: 10.1038/srep26200 (2016).
Supplementary Material
Acknowledgments
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). The USDA is an equal opportunity provider and employer. This trial was partially supported by Spanish National grant (AGL2009/09985) and Fondo Social Europeo (FSE). ZG was supported by funding from TEES-AgriLife Center for Bioinformatics and Genomic Systems Engineering (CBGSE) at Texas A&M University. PY was supported by startup funding from the ECE department and Texas A&M Engineering Experiment Station/Dwight Look College of Engineering at Texas A&M University. JNH was sponsored by Agencia Canaria de Investigación, Innovación y Sociedad de la Información and Fondo Social Europeo (FSE). TNM and DF received funding from the Scottish Government. YCM is supported by European Union’s Horizon 2020 research and innovation programme under grant agreement No. 635408.
Footnotes
Author Contributions Conceived and designed the experiment: J.F.G. and R.W.L. Performed the experiment: J.N.H., T.N.M., T.M., Y.C.-M., D.F., R.W.L. and J.F.G. Analyzed the data: Z.G., P.Y. and R.W.L. Wrote the manuscript: R.W.L. All authors reviewed and approved the manuscript.
References
- Besier B., Lyon J., Michael D., Newlands G. & Smith D. Towards a commercial vaccine against Haemonchus contortus - a field trial in Western Australia. Proc. Australian Sheep Vet Conf 2012, 14–18 (2012). [Google Scholar]
- Sreter T., Kassai T. & Takacs E. The heritability and specificity of responsiveness to infection with Haemonchus contortus in sheep. Int J Parasitol 24, 871–6 (1994). [DOI] [PubMed] [Google Scholar]
- Alba-Hurtado F. & Munoz-Guzman M. A. Immune responses associated with resistance to haemonchosis in sheep. Biomed Res Int 2013, 162158, doi: 10.1155/2013/162158. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble H. R. & Zajac A. M. Resistance of St. Croix lambs to Haemonchus contortus in experimentally and naturally acquired infections. Vet Parasitol 41, 211–25 (1992). [DOI] [PubMed] [Google Scholar]
- Amarante A. F., Bricarello P. A., Rocha R. A. & Gennari S. M. Resistance of Santa Ines, Suffolk and Ile de France sheep to naturally acquired gastrointestinal nematode infections. Vet Parasitol 120, 91–106 (2004). [DOI] [PubMed] [Google Scholar]
- Gauly M., Kraus M., Vervelde L., van Leeuwen M. A. & Erhardt G. Estimating genetic differences in natural resistance in Rhon and Merinoland sheep following experimental Haemonchus contortus infection. Vet Parasitol 106, 55–67 (2002). [DOI] [PubMed] [Google Scholar]
- Mugambi J. M. et al. Resistance of four sheep breeds to natural and subsequent artificial Haemonchus contortus infection. Vet Parasitol 69, 265–73 (1997). [DOI] [PubMed] [Google Scholar]
- Stear M. J., Boag B., Cattadori I. & Murphy L. Genetic variation in resistance to mixed, predominantly Teladorsagia circumcincta nematode infections of sheep: from heritabilities to gene identification. Parasite Immunol 31, 274–82 (2009). [DOI] [PubMed] [Google Scholar]
- Benavides M. V. et al. Identification of novel loci associated with gastrointestinal parasite resistance in a Red Maasai x Dorper backcross population. PLoS One 10, e0122797, doi: 10.1371/journal.pone.0122797 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauly M. & Erhardt G. Genetic resistance to gastrointestinal nematode parasites in Rhon sheep following natural infection. Vet Parasitol 102, 253–9 (2001). [DOI] [PubMed] [Google Scholar]
- Periasamy K. et al. Candidate gene approach for parasite resistance in sheep–variation in immune pathway genes and association with fecal egg count. PLoS One 9, e88337, doi: 10.1371/journal.pone.0088337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M. V. et al. Identification of quantitative trait loci affecting resistance to gastrointestinal parasites in a double backcross population of Red Maasai and Dorper sheep. Anim Genet 43, 63–71 (2012). [DOI] [PubMed] [Google Scholar]
- Marshall K. et al. Quantitative trait loci for resistance to Haemonchus contortus artificial challenge in Red Maasai and Dorper sheep of East Africa. Anim Genet 44, 285–95 (2013). [DOI] [PubMed] [Google Scholar]
- Gonzalez J. F. et al. Comparative experimental Haemonchus contortus infection of two sheep breeds native to the Canary Islands. Vet Parasitol 153, 374–8 (2008). [DOI] [PubMed] [Google Scholar]
- Gonzalez J. F. et al. Fecundity in adult Haemonchus contortus parasites is correlated with abomasal tissue eosinophils and gammadelta T cells in resistant Canaria Hair Breed sheep. Vet Parasitol 178, 286–92 (2011). [DOI] [PubMed] [Google Scholar]
- Ordonez G. R. et al. Loss of genes implicated in gastric function during platypus evolution. Genome Biol 9, R81, doi: 10.1186/gb-2008-9-5-r81 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A. D. et al. Natural selection on individual variation in tolerance of gastrointestinal nematode infection. PLoS Biol 12, e1001917, doi: 10.1371/journal.pbio.1001917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A. D. Causes and consequences of intra- and inter-host heterogeneity in defence against nematodes. Parasite Immuno1 2013, doi: 10.1111/pim.12054 (2013). [DOI] [PubMed] [Google Scholar]
- Diez-Tascon C. et al. Microarray analysis of selection lines from outbred populations to identify genes involved with nematode parasite resistance in sheep. Physiol Genomics 21, 59–69 (2005). [DOI] [PubMed] [Google Scholar]
- Kadarmideen H. N., Watson-Haigh N. S. & Andronicos N. M. Systems biology of ovine intestinal parasite resistance: disease gene modules and biomarkers. Mol Biosyst 7, 235–46 (2011). [DOI] [PubMed] [Google Scholar]
- Li R. W., Sonstegard T. S., Van Tassell C. P. & Gasbarre L. C. Local inflammation as a possible mechanism of resistance to gastrointestinal nematodes in Angus heifers. Vet Parasitol 145, 100–7 (2007). [DOI] [PubMed] [Google Scholar]
- Li R. W. & Schroeder S. G. Cytoskeleton remodeling and alterations in smooth muscle contractility in the bovine jejunum during nematode infection. Funct Integr Genomics 12, 35–44 (2012). [DOI] [PubMed] [Google Scholar]
- Antignano F., Mullaly S. C., Burrows K. & Zaph C. Trichuris muris infection: a model of type 2 immunity and inflammation in the gut. J Vis Exp 2011, doi: 10.3791/2774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallance B. A., Blennerhassett P. A. & Collins S. M. Increased intestinal muscle contractility and worm expulsion in nematode-infected mice. Am J Physiol 272, G321–7 (1997). [DOI] [PubMed] [Google Scholar]
- Li R. W., Hou Y., Li C. & Gasbarre L. C. Localized complement activation in the development of protective immunity against Ostertagia ostertagi infections in cattle. Vet Parasitol 174, 247–56 (2010). [DOI] [PubMed] [Google Scholar]
- Gurtner G. C., Werner S., Barrandon Y. & Longaker M. T. Wound repair and regeneration. Nature 453, 314–21 (2008). [DOI] [PubMed] [Google Scholar]
- Chen F. et al. An essential role for TH2-type responses in limiting acute tissue damage during experimental helminth infection. Nat Med 18, 260–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraldi D. et al. Quantitative trait loci (QTL) mapping of resistance to strongyles and coccidia in the free-living Soay sheep (Ovis aries). Int J Parasitol 37, 121–9 (2007). [DOI] [PubMed] [Google Scholar]
- Gutierrez-Gil B. et al. Quantitative trait loci for resistance to trichostrongylid infection in Spanish Churra sheep. Genet Sel Evol 41, 46, doi: 10.1186/1297-9686-41-46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley S. P. et al. Attenuated type II TGF-beta receptor signalling in human malignant oral keratinocytes induces a less differentiated and more aggressive phenotype that is associated with metastatic dissemination. Int J Cancer 110, 170–6 (2004). [DOI] [PubMed] [Google Scholar]
- Miller H. R. Mucosal mast cells and the allergic response against nematode parasites. Vet Immunol Immunopathol 54, 331–6 (1996). [DOI] [PubMed] [Google Scholar]
- Donaldson L. E., Schmitt E., Huntley J. F., Newlands G. F. & Grencis R. K. A critical role for stem cell factor and c-kit in host protective immunity to an intestinal helminth. Int Immunol 8, 559–67 (1996). [DOI] [PubMed] [Google Scholar]
- Zaiss D. M. et al. Amphiregulin, a TH2 cytokine enhancing resistance to nematodes. Science 314, 1746 (2006). [DOI] [PubMed] [Google Scholar]
- Zaiss D. M., Gause W. C., Osborne L. C. & Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity 42, 216–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss D. M. et al. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity 38, 275–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. W. & Gasbarre L. C. A temporal shift in regulatory networks and pathways in the bovine small intestine during Cooperia oncophora infection. Int J Parasitol 39, 813–24 (2009). [DOI] [PubMed] [Google Scholar]
- Li R. W., Rinaldi M. & Capuco A. V. Characterization of the abomasal transcriptome for mechanisms of resistance to gastrointestinal nematodes in cattle. Vet Res 42, 114, doi: 10.1186/1297-9716-42-114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol 11, R106, doi: 10.1186/gb-2010-11-10-r106. (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 57, 289–300 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.