SUMMARY
The molecular linkage between neocortical projection neuron subtype and area development, which enables the establishment of functional areas by projection neuron populations appropriate for specific sensory and motor functions, is poorly understood. Here, we report that Ctip1 controls precision of neocortical development by regulating subtype identity in deep-layer projection neurons. Ctip1 is expressed by postmitotic callosal and corticothalamic projection neurons, but is excluded over embryonic development from corticospinal motor neurons, which instead express its close relative, Ctip2. Loss of Ctip1 function results in a striking bias in favor of subcerebral projection neuron development in sensory cortex at the expense of corticothalamic and deep-layer callosal development, while misexpression of Ctip1 in vivo represses subcerebral gene expression and projections. As we report in a paired paper, Ctip1 also controls acquisition of sensory area identity. Therefore, Ctip1 couples subtype and area specification, enabling specific functional areas to organize precise ratios of appropriate output projections.
INTRODUCTION
The remarkable complexity of the neocortex is precisely orchestrated during development, as many millions of neurons are born from progenitors, migrate to birthdate-appropriate layers, adopt subtype-specific gene expression and circuit connectivity, and organize into modality-specific functional areas. Subtype identity is tightly correlated with birthdate, as corticothalamic projection neurons (CThPN) are born around E12.5 in mice, subcerebral projection neurons (SCPN), including corticospinal motor neurons (CSMN), around E13.5, and most callosal projection neurons (CPN) between E14.5-E15.5, with a minority generated and resident with deep-layer subtypes (Woodworth et al., 2012). Nonetheless, functional areas of the cortex vary considerably in their subtype composition; for example, many more SCPN reside in mature motor cortex than in sensory cortex (Greig et al., 2013). This striking reciprocal linkage between subtype and area identity, with functional areas defined in large part by distinctive differences in subtype composition and, therefore, connectivity, has long been appreciated (Brodmann, 1909; Cajal, 1899), but its molecular control remains elusive.
One key transcription factor control over subtype development, Ctip2, is expressed at high levels by SCPN and functions centrally in SCPN terminal differentiation and connectivity (Arlotta et al., 2005). Multiple transcription factors controlling projection neuron specification and postmitotic differentiation operate at least in part by regulating Ctip2 expression. For example, Fezf2 regulates SCPN differentiation partly by promoting expression of Ctip2 (Arlotta et al., 2005; Chen et al., 2008; Molyneaux et al., 2005), and by preventing expression of transcription factors key for specification of alternate subtype fates, including Tbr1 (Bedogni et al., 2010; McKenna et al., 2011) and Satb2 (Alcamo et al., 2008; Britanova et al., 2008). Conversely, repression of Fezf2 and Ctip2 by Sox5 (Kwan et al., 2008; Lai et al., 2008; Shim et al., 2012) and Tbr1 (Bedogni et al., 2010; Han et al., 2011; McKenna et al., 2011) in CThPN, and by Satb2 in CPN (Alcamo et al., 2008; Britanova et al., 2008), is critical for appropriate development of these subtypes. Although these central controls begin to be expressed immediately after neurons exit the cell cycle, subtype identity continues to be refined as neurons mature (Azim et al., 2009), as, for example, initially promiscuous expression of Ctip2 and Satb2 in newly postmitotic layer V neurons resolves to subtype-specific expression in SCPN and CPN by late embryogenesis (Alcamo et al., 2008; Britanova et al., 2008).
A paralogous zinc finger transcription factor, Ctip1 (also known as Bcl11a/Evi9), is also expressed during development. Ctip2 and Ctip1 are closely related, with approximately 60% identity between their nucleotide sequences (Avram et al., 2000; Satterwhite et al., 2001). In the hematopoietic system, Ctip1 controls the development of B cells (Lee et al., 2013; Liu et al., 2006; 2003) and regulates switching between fetal and adult forms of hemoglobin (Bauer et al., 2013; Canver et al., 2015; Sankaran et al., 2008; 2009; Xu et al., 2011). Ctip1 is expressed during development of many areas of the nervous system, including neocortex, hippocampus, striatum, and cerebellum (Arlotta et al., 2008; John et al., 2012; Kuo and Hsueh, 2007; Leid et al., 2004). Although Ctip1 is known to regulate neuronal migration (Wiegreffe et al., 2015), its functions in neocortical subtype development are not known.
Here, we report that Ctip1 controls projection neuron subtype identity and development of proper proportions of subtypes within a given functional area. Ctip1 is expressed by CPN, CThPN, and subplate neurons, but is progressively excluded from Ctip2-expressing CSMN. In the absence of Ctip1, SCPN expand as a population at the expense of CThPN and deep-layer CPN, with higher expression of genes characteristic of SCPN, more neurons projecting to the cerebral peduncle, and a reduction in CThPN and CPN gene expression and projections. In contrast, Ctip1 overexpression in vivo represses Ctip2 expression and reduces the number of neurons projecting subcerebrally, while increasing the number of neurons projecting through the corpus callosum. In accompanying work, we report that Ctip1 controls the acquisition of sensory area identity and area-specific input/output connectivity (Greig et al., 2016), and thus serves to integrate the dual processes of subtype and area development in neocortex.
RESULTS
CTIP1 is expressed by CPN, CThPN, and subplate neurons, but is largely excluded from CSMN
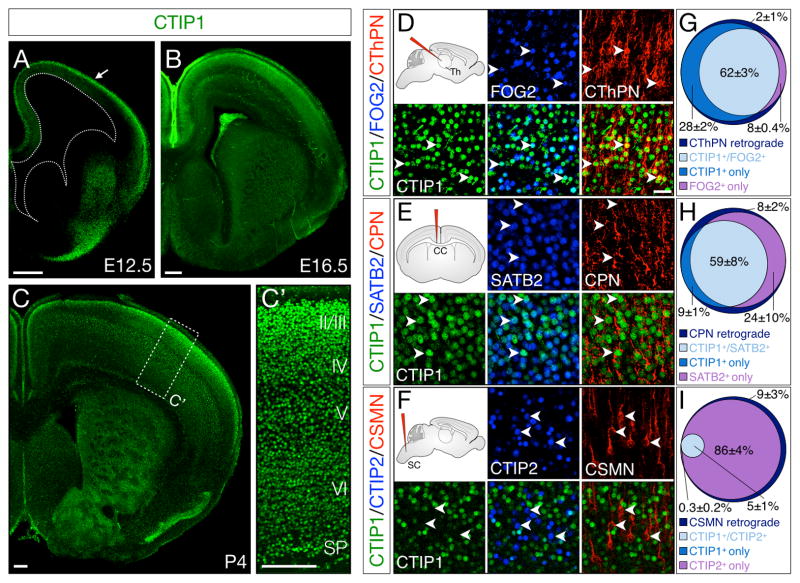
CTIP1 is first detected by immunocytochemistry at approximately E12.5 in young postmitotic glutamatergic projection neurons in the cortical plate (Figure 1A, Figure S1A). Immunostaining is absent from progenitor zones at mid-gestation (Figure S1D–E), although some expression of Ctip1 mRNA can be observed by in situ hybridization at the same age (Figure S1B–C), suggesting that Ctip1 begins to be transcribed as neurons exit the cell cycle, but detectable amounts of protein are not present until postmitotic neurons migrate to the cortical plate. CTIP1 continues to be expressed in postmitotic cortical neurons throughout embryogenesis (Figure 1B), though expression begins to decrease after the first postnatal week. At postnatal day (P) 4, CTIP1 is highly expressed in all six layers of somatosensory cortex (Figure 1C–C′), while expression in motor cortex is largely restricted to subplate, layer V, and the most superficial aspect of layer II/III.
Figure 1. Ctip1 is expressed by postmitotic corticothalamic and callosal projection neurons, but is excluded from corticospinal motor neurons.
(A–C′) CTIP1 immunocytochemistry on coronal brain sections. CTIP1 is expressed by newly-postmitotic projection neurons in the cortical plate beginning around E12.5 (A, arrow) and continuing through E16.5 (B) and P4 (C). At P4, CTIP1 is expressed by neurons in all layers of primary sensory cortex (C′).
(D–I) At P4, CTIP1 is expressed by CThPN and CPN, but not by CSMN. CTIP1 co-localizes with FOG2, as well as with cholera toxin B (CTB) retrograde label after injection into thalamus (D). CPN express both CTIP1 and SATB2, and are retrogradely labeled by injection of CTB into the corpus callosum (E). CSMN are retrogradely labeled by CTB injection into the cervical spinal cord, and express CTIP2, but not CTIP1 (F). Quantification, n=4 (G–I).
CC, corpus callosum; CPN, callosal projection neurons; CSMN, corticospinal motor neurons; CThPN, corticothalamic projection neurons; SC, spinal cord; SP, subplate; Th, thalamus.
Scale bars: 200um (A–C), 40um (D–F). Data are represented as mean ± SEM.
To investigate the subtype specificity of Ctip1 expression, we retrogradely labeled CThPN, CPN, or CSMN with Alexa fluorophore-conjugated cholera toxin B (CTB) at early postnatal ages (P1-P3) and collected labeled brains at P4. In addition, we performed immunocytochemistry for CTIP1 together with FOG2, SATB2, and CTIP2, markers of CThPN, CPN, and SCPN, respectively (Woodworth et al., 2012). We find that CTIP1 is expressed by CPN and CThPN in sensorimotor cortex, but is essentially excluded from CSMN (Figure 1D–I). CTIP1 is expressed by 90% (±0.9%) of retrogradely labeled CThPN in layer VI, and co-localizes with FOG2 in 62% (±2.7%) of CThPN (Figure 1D, G). Similarly, 68% (±9.1%) of retrogradely labeled CPN express CTIP1, and CTIP1 and SATB2 are co-expressed by 59% (±7.9%) of CPN (Figure 1E, H), with no differences between superficial- and deep-layer CPN (61±7.3% superficial-layer, 57±9.0% deep-layer, not significantly different). Very few neurons labeled retrogradely from the spinal cord express CTIP1 (4.9%±1.1%), and there is very little co-expression of CTIP1 and CTIP2 (4.6%±1.4%) in motor cortex (Figure 1F, I).
Having determined that CTIP1 is expressed at high levels by CThPN and CPN, but not by CSMN, we next investigated the temporal course of CTIP1 and CTIP2 expression in embryonic cortex. We find that, at E14.5, nearly all early postmitotic layer V neurons co-express CTIP1 and CTIP2 at high levels, but, by E17.5, CTIP1 and CTIP2 are expressed by two largely distinct populations within layer V (Figure S3A–D). Given the retrograde labeling data above (Figure 1E–I), we reasoned that these populations are likely immature CTIP1-expressing CPN and CTIP2-expressing CSMN.
The early segregation of cortical CTIP1 and CTIP2 expression, together with their functions controlling differentiation of closely-related immune cell types (Liu et al., 2003; Tydell et al., 2007; Wakabayashi et al., 2003), led us to hypothesize that Ctip1 and Ctip2 might interact cross-repressively to control differentiation of deep-layer projection neurons into distinct subtypes. We examined Ctip2 expression in the neocortex of Ctip1−/− mice, and find increased levels of CTIP2 in layers V and VI by qPCR (1.5-fold, p<0.05). Conversely, we find increased CTIP1 expression in layers V and VI of Ctip2−/− neocortex (1.5-fold, p<0.05) (Figure S3E–J). These data indicate that Ctip1 and Ctip2 are genetically cross-repressive, and suggest that these transcription factors might function to sharpen differentiation of distinct neuronal subtypes.
More projection neurons acquire SCPN identity in the absence of Ctip1 function
Since Ctip1−/− mice die almost immediately after birth (Liu et al., 2003), we obtained Ctip1fl/fl mice, which survive to adulthood (Lee et al., 2013; Sankaran et al., 2008), to study Ctip1 function as neocortical projections are established and refined. We deleted Ctip1 specifically from cortical neurons using Emx1-Cre, which is expressed beginning in cortical progenitors (Gorski et al., 2002).
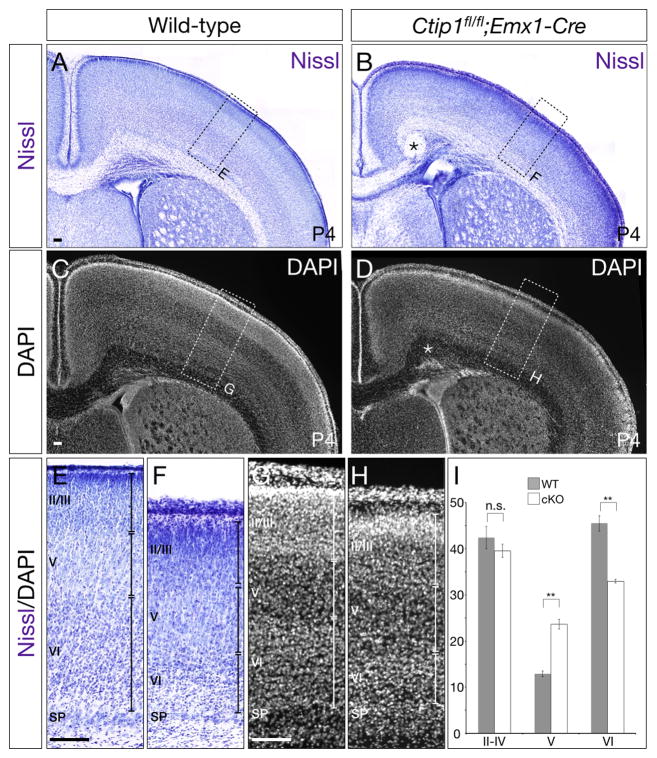
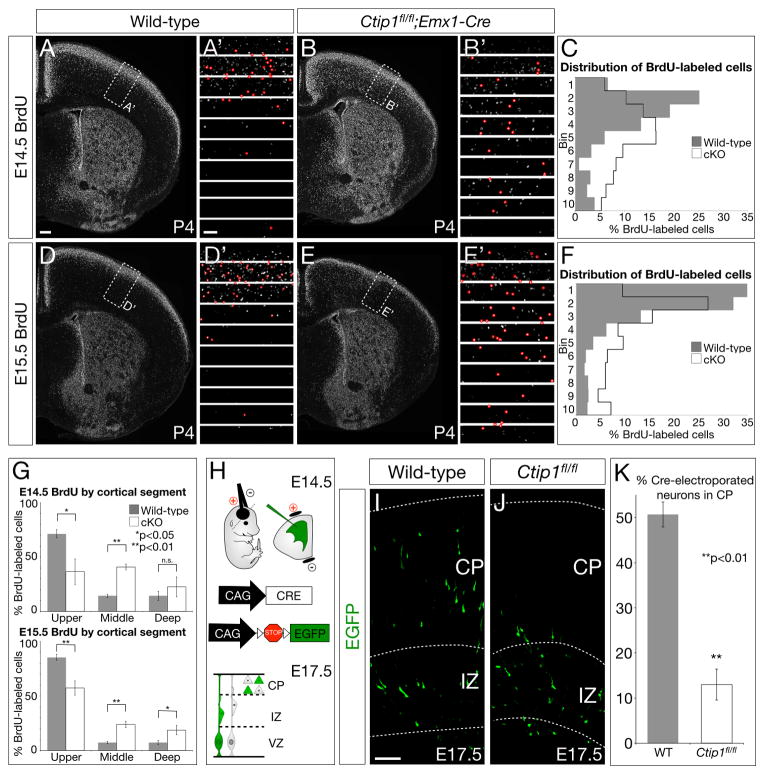
Because Ctip1 is expressed by CThPN and CPN, but not by CSMN, we investigated whether, in the absence of Ctip1 function, more neocortical neurons differentiate into CSMN, and into SCPN more broadly. We compared wild-type and Ctip1fl/fl;Emx1-Cre tissue at P4 by Nissl and DAPI stain, and find that conditional nulls exhibit defective cortical layering, with a lack of clearly distinct boundaries between adjacent layers, but that layer V, which contains SCPN, is expanded relative to other cortical layers (p<0.01, Figure 2A–D, Figure S2). To investigate further, we performed immunocytochemistry and in situ hybridization at P0 for several genes expressed by SCPN, including CTIP2, Fezf2, and Clim1 (Arlotta et al., 2005; Azim et al., 2009; Molyneaux et al., 2005). We find that high-level expression of these genes is expanded radially in Ctip1fl/fl;Emx1-Cre cortex, indicating increased layer V thickness (Figure 3A–F). This expansion is especially striking in somatosensory cortex, where layer V is normally thinner than in motor cortex, and where there are normally fewer SCPN than in motor cortex. In agreement with our Nissl results, this expansion appears to be primarily in the direction of deeper layers, suggesting that layer VIa CThPN are converted to SCPN.
Figure 2. In the absence of neocortical Ctip1, cortical layer V is expanded at the expense of cortical layer VI.
(A–H) Nissl staining and DAPI staining of cortex in wild-type (A, C) and Ctip1fl/fl;Emx1-Cre cortical conditional null (B, D) mice at P4. Layer V is expanded in conditional null cortex (F, H) compared with wild-type (E, G). Asterisk in B and D marks the location of a Probst bundle.
(I) Quantification of layer thickness performed on Nissl-stained tissue, with each layer presented as a percentage of total cortical thickness (n=5).
Scale bars: 100um. Data are represented as mean ± SEM.
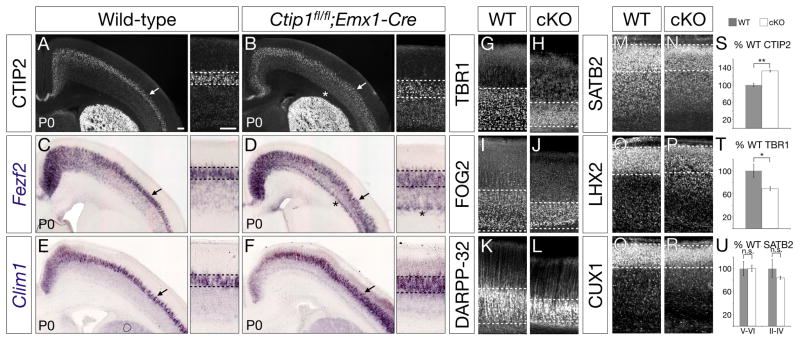
Figure 3. More neurons in Ctip1fl/fl;Emx1-Cre cortex adopt a subcerebral projection neuron identity, and fewer adopt a corticothalamic projection neuron identity.
(A–F) In the absence of Ctip1 function, expression of SCPN marker and control genes CTIP2, Fezf2, and Clim1 increases at P0, especially in somatosensory cortex (arrows in A–F). Dashed lines denote superficial and deep limits of gene expression.
(G–L) In tandem, expression of CThPN marker and control genes TBR1, FOG2, and DARPP-32 is reduced at P0. Dashed lines denote superficial and deep limits of gene expression. Even layer VI neurons that continue to express CThPN identity genes (H, J, L) express aberrantly high levels of CTIP2 and Fezf2 (asterisks in B, D), suggesting mixed CThPN/SCPN identity.
(M–R) CPN marker and control genes SATB2, LHX2, and CUX1 are expressed normally at P0, although boundaries between superficial and deep layers are more difficult to discern. Dashed lines denote superficial and deep limits of gene expression.
(S–U) Quantification of the number of neurons expressing CTIP2 (S), TBR1 (T), and SATB2 (U) in conditional null cortex, presented as a percentage of wild-type neurons, n=3 for each marker.
*, p<0.05; **, p<0.01; n.s., not significant
Scale bars: 100um. Data are represented as mean ± SEM.
Indeed, expression of genes specific to CThPN, including TBR1, FOG2, and DARPP-32 (Molyneaux et al., 2007), is markedly reduced in P0 Ctip1fl/fl;Emx1-Cre cortex, and layer VI is radially thinner (Figure 3G–L). Even within the reduced domain of layer VI, neurons expressing CThPN molecular markers and developmental controls also express inappropriately high levels of CTIP2 and Fezf2 (asterisks in Figure 3B, 3D), suggesting a mixed SCPN/CThPN identity. TBR1 is expressed by 31% fewer neurons in Ctip1fl/fl;Emx1-Cre cortex (p<0.05; Figure 3T), while 32% more neurons express CTIP2 than in wild-type cortex (p<0.01; Figure 3S). These data strongly suggest that, in the absence of Ctip1 function, many neurons that would normally have differentiated into CThPN have instead become SCPN. Expression of CPN genes SATB2, LHX2, and CUX1 (Molyneaux et al., 2009) is approximately normal in deep layers and superficial layers (Figure 3M–R, U). Similarly, in conditional mutants in which Ctip1 is deleted postmitotically (Ctip1fl/fl;Nex1-Cre) (Goebbels et al., 2006), TBR1 expression is reduced relative to wild-type, CTIP2 expression is increased, and SATB2 expression is unchanged in both deep and superficial layers (Figure S3K–N), indicating that Ctip1 functions postmitotically to specify neuronal subtype. These results demonstrate that, in the absence of Ctip1 function, more projection neurons postmitotically adopt an SCPN molecular identity, and fewer adopt a CThPN identity.
More neurons project to subcerebral targets in Ctip1fl/fl;Emx1-Cre mice, and fewer project to the thalamus
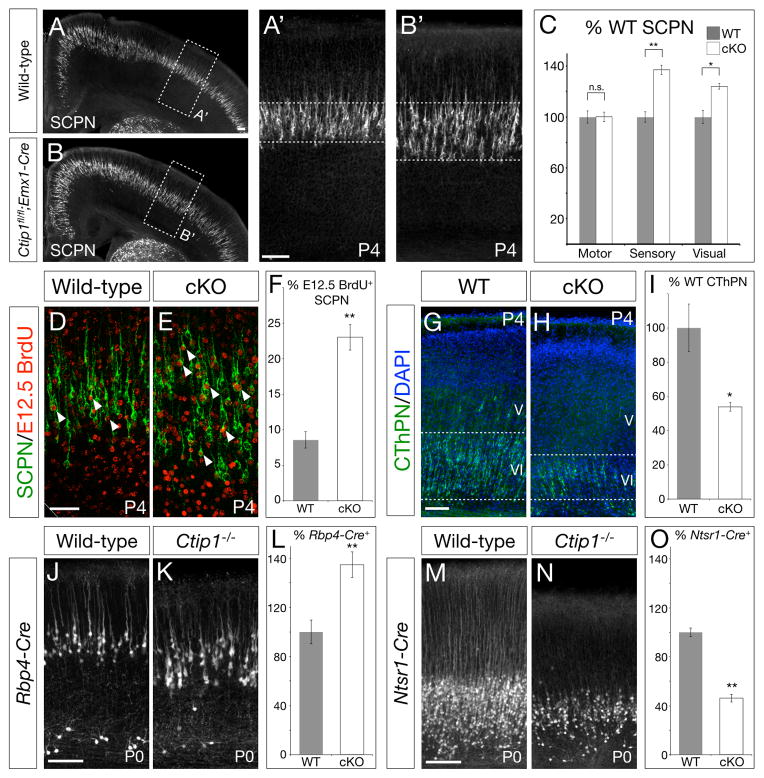
We next investigated whether, beyond dysregulation of characteristic subtype-specific genes, cortical projection targeting is altered in the absence of Ctip1. We retrogradely labeled SCPN from the cerebral peduncle at P1 (labeling all neurons projecting to targets caudal to cerebrum), revealing that significantly more axons project to subcerebral targets in Ctip1fl/fl;Emx1-Cre than in wild-type brains (Figure 4A–C). In particular, the number of SCPN in somatosensory cortex increases 1.4-fold (p<0.01), and the number of SCPN in visual cortex increases 1.2-fold (p<0.05). The number of SCPN in motor cortex, where CTIP1 expression is normally low (Figure 1C), does not change.
Figure 4. More neurons in Ctip1fl/fl;Emx1-Cre somatosensory and visual cortex project toward subcerebral targets, and fewer project toward thalamic targets.
(A–B) More neurons in Ctip1 conditional null somatosensory cortex (B, B′) are retrogradely labeled by injection of cholera toxin B (CTB) into the cerebral peduncle than in wild-type cortex (A, A′).
(C) Quantification of retrogradely-labeled SCPN by area, presented as a percentage of wild-type SCPN in each area, n=3.
(D–F) Additional SCPN in Ctip1fl/fl;Emx1-Cre brains are aberrantly differentiated from E12.5-born neurons. More neurons labeled by injection of BrdU at E12.5 are co-labeled by injection of CTB into the cerebral peduncle in conditional null cortex (E) compared with wild-type cortex (D). Arrowheads mark representative co-labeled neurons. Quantification, n=4 (F).
(G–I) Compared with wild-type (G), fewer CThPN are retrogradely labeled in somatosensory cortex of Ctip1fl/fl;Emx1-Cre mutants (H) following CTB injection into sensory thalamic nuclei. Quantification (I).
(J–O) Ctip1 null cortex contains more mature SCPN and fewer mature CThPN than wild-type. More neurons are marked by SCPN-specific Rbp4-Cre in Ctip1−/− P0 cortex (K) compared with wild-type (J) (n=4). In contrast, fewer neurons are marked by CThPN-specific Ntsr1-Cre in null P0 cortex (N) compared with wild-type (M) (n=3). Quantification (L, O). Recombination by Rbp4-Cre and Ntsr1-Cre is reported by a Rosa26R-tdTomatofl allele.
n.s., not significant; *, p<0.05; **, p<0.01
Scale bars: 100um (A–B, G–H, J–K, M–N), 50um (D–E). Data are represented as mean ± SEM.
In concordance with expression of SCPN-specific genes expanding deeper into layer VI (Figure 3A–F), most additional SCPN in Ctip1fl/fl;Emx1-Cre brains are located in the upper portion of layer VI. By combining BrdU birthdating at E12.5 with retrograde labeling from the cerebral peduncle, we find that 23% of Ctip1 conditional null SCPN have incorporated BrdU at E12.5, when CThPN are usually generated, compared with 9% of wild-type SCPN (Figure 4D–F; p<0.01). In the absence of Ctip1 function, newly born layer VI corticofugal neurons, normally destined to differentiate into CThPN, aberrantly differentiate into SCPN.
Because expression of genes characteristic of CThPN is reduced in Ctip1fl/fl;Emx1-Cre cortex, and because additional neurons projecting through the cerebral peduncle in Ctip1fl/fl;Emx1-Cre cortex are aberrantly differentiated E12.5-born neurons, we investigated CThPN projections in the absence of Ctip1 function. We find that fewer CThPN in Ctip1fl/fl;Emx1-Cre somatosensory cortex are retrogradely labeled by injection of CTB into thalamus than in wild-type cortex (0.5-fold, p<0.05)(Figure 4G–I). Further, we analyzed P0 Ctip1−/− mice carrying a Rosa26R-tdTomato reporter allele (Madisen et al., 2010) and either an Rbp4-Cre or an Ntsr1-Cre allele (Gong et al., 2007), which are specific to SCPN and CThPN, respectively. Fewer CThPN in Ctip1−/− cortex are labeled by Ntsr1-Cre than in wild-type cortex (0.5-fold, p<0.01), and more SCPN are labeled by Rbp4-Cre (1.6-fold, p<0.01) (Figure 4J–O), indicating that fewer neurons differentiate into mature CThPN in the absence of Ctip1. Taken together, these results indicate that Ctip1 controls the development of corticothalamic projection neurons by suppressing their alternative differentiation into subcerebral projection neurons.
CPN pathfinding is disrupted in the absence of Ctip1 function
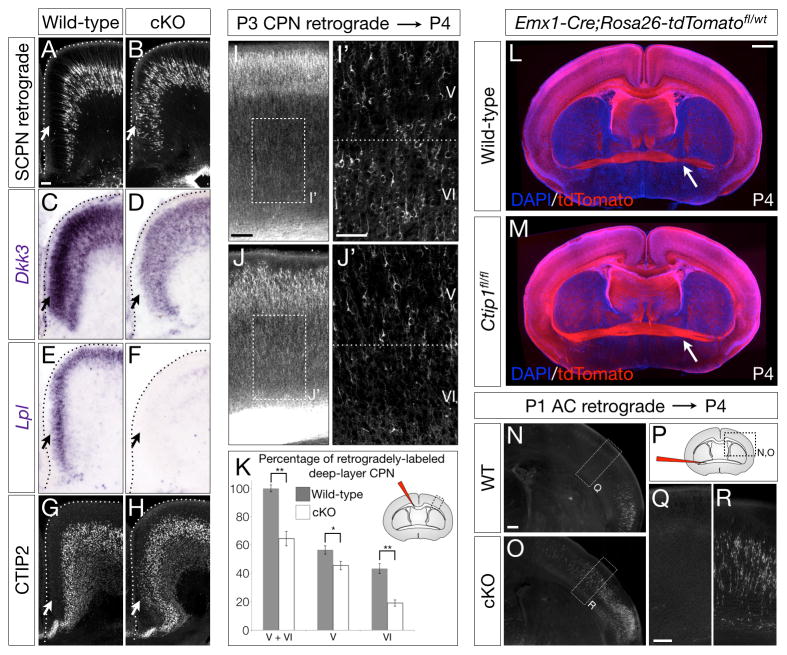
Ctip1fl/fl;Emx1-Cre mice exhibit partial agenesis of the corpus callosum, with prominent Probst bundles containing axons that have failed to cross the midline (asterisk in Figure 2B, D; Richards et al., 2004), and the corpus callosum is thinner in Ctip1fl/fl;Emx1-Cre than in wild-type mice (Figure 2A–D). These data indicate that development of the callosal projection is disrupted in the absence of Ctip1. In addition, many superficial-layer neurons in cingulate cortex, normally the earliest-crossing callosal population (Koester and O’Leary, 1994), are instead retrogradely labeled by injection of CTB from the cerebral peduncle (Figure 5A–B), suggesting that pioneering cingulate cortex CPN in Ctip1fl/fl;Emx1-Cre brains are fate-converted to SCPN. Consistent with this interpretation, superficial-layer neurons in cingulate cortex fail to express genes typical of cingulate cortex CPN, such as Dkk3 and Lpl (Figure 5C–F; (Molyneaux et al., 2009)), and instead aberrantly express CTIP2 (Figure 5G–H).
Figure 5. Cingulate CPN fail to pioneer the callosum in the absence of Ctip1 function, impairing projections of deep-layer CPN.
(A–H) Superficial-layer neurons in Ctip1fl/fl;Emx1-Cre cingulate cortex aberrantly project through the cerebral peduncle (A–B), fail to express cingulate CPN genes Dkk3 (C–D) and Lpl (E–F), and instead express CTIP2 (G–H).
(I–K) Failure of cingulate CPN to pioneer the corpus callosum results in a reduction of retrogradely-labeled deep-layer CPN in Ctip1fl/fl;Emx1-Cre cortex. Fewer CPN are retrogradely labeled in P4 Ctip1fl/fl;Emx1-Cre layers V and VI (J-J′) compared with wild-type (I–I′). Quantification of I–J as a percentage of wild-type deep-layer CPN, n=3 (K). Layer VI CPN are more severely affected than layer V CPN. Schematic of injection site, upper right corner.
(L–R) More neurons in Ctip1 conditional null brains aberrantly project to the contralateral hemisphere via the anterior commissure pathway. The anterior commissure is abnormally enlarged in P4 Ctip1fl/fl;Rosa26R-tdTomatofl/wt;Emx1-Cre brains (arrow in M) compared with Ctip1wt/wt;Rosa26R-tdTomatofl/wt;Emx1-Cre (wild-type) (arrow in L). Retrograde labeling from the contralateral hemisphere at P1 reveals that additional AC-projecting neurons in Ctip1 conditional null brains are aberrantly located in somatosensory cortex, an area where AC-projecting neurons are not normally located (N–O, Q–R). Schematic of injection site (P).
*, p<0.05; **, p<0.01
Scale bars: 100um (A–J), 50um (I′–J′), 500um (L–M), 200um (N–R). Data are represented as mean ± SEM.
Potentially because cingulate CPN fail to pioneer the callosum, fewer deep-layer neurons in somatosensory cortex cross the callosum in Ctip1fl/fl;Emx1-Cre cortex than in wild-type mice (Figure 5I–K; p<0.01). Strikingly, layer VI CPN are more severely affected (56% fewer cross; p<0.01) than layer V CPN (19% fewer cross; p<0.05), suggesting that many later-born CPN are able to follow the few early-born CPN that manage to cross the midline, even in the absence of a fully functional cingulate pioneer population. However, even projections of later-born CPN are cell-autonomously disrupted in the absence of Ctip1, as Cre electroporation into Ctip1fl/fl cortex at E15.5 results in 52% fewer axons crossing the callosum (Figure S5I–K; p<0.01). These data indicate that axon pathfinding is disrupted in CPN across all layers of cortex in the absence of Ctip1.
Although the corpus callosum is thinner in Ctip1fl/fl;Emx1-Cre mice, the anterior commissure is notably enlarged (arrows in Figure 5L–M). Indeed, many neurons in Ctip1fl/fl;Emx1-Cre somatosensory cortex are retrogradely labeled by injection of CTB into the anterior commissure, while retrogradely-labeled neurons are not present in this location in wild-type somatosensory cortex (Figure 5N–R). These data indicate that, in the absence of Ctip1 function, many commissural neurons aberrantly project to the contralateral hemisphere via this abnormal anterior commissure route instead of via the corpus callosum.
Late-born projection neurons migrate abnormally in Ctip1 mutants
Although superficial-layer CPN outside cingulate cortex are not fate-converted to SCPN in Ctip1fl/fl;Emx1-Cre mice (Figure 4A–B), superficial-layer neurons are improperly laminated (Figure 2A–D, Figure 5I–J) and impaired in their projections (Figure S5I–K). To investigate whether Ctip1fl/fl;Emx1-Cre neurons form imprecise layers as a result of abnormal migration, we birthdated cortical neurons in conditional null and wild-type cortex by injecting BrdU into pregnant females at E11.5, E12.5, E13.5, E14.5, and E15.5. We analyzed the laminar position of labeled neurons at P4, after migration is normally complete. We find significant numbers of E14.5- and E15.5-born neurons ectopically located in deep layers in Ctip1fl/fl;Emx1-Cre mutants (Figure 6A–G; p<0.05 for neurons located in bins 5, 6, and 7 at both E14.5 and E15.5), indicating that late-born projection neurons fail to migrate appropriately to superficial layers. These neurons express markers of superficial-layer neurons, such as CUX1, appropriate to their birthdate, but not their laminar position (Figure S5F–H). In contrast, migration of neurons born at E11.5, E12.5, or E13.5 is not significantly affected (Figure S6A–I). These results indicate that many late-born Ctip1fl/fl;Emx1-Cre neurons migrate and become positioned in deep-layer cortex, leading to abnormal lamination of the cortex as a whole.
Figure 6. In the absence of neocortical Ctip1, superficial-layer projection neurons migrate aberrantly and become inappropriately positioned in cortex.
(A–F) Wild-type neurons labeled by BrdU injection at E14.5 (A–A′) or E15.5 (D–D′) are primarily located in superficial layers, while Ctip1fl/fl;Emx1-Cre neurons labeled by BrdU at E14.5 (B–B′) or E15.5 (E–E′) are frequently ectopically located in deep layers. The overall laminar distribution of labeled neurons (marked in red in A′–B′, D′–E′) is strikingly abnormal (C, F). Bin 1 is the most superficial bin, and Bin 10 is the deepest.
(G) Quantification of A–F, with bins 1–4 grouped as “upper” cortical segment, 5–7 as “middle”, and 8–10 as “deep”.
(H–K) Sparse electroporation of Cre at E14.5 causes Ctip1fl/fl neurons to be delayed in the intermediate zone rather than migrate into the cortical plate at E17.5. Schematic of experimental approach (H). Wild-type neurons electroporated with Cre (I) are significantly more likely than electroporated Ctip1fl/fl neurons (J) to have migrated into the cortical plate by E17.5 (quantification, K).
CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; *, p<0.05; **, p<0.01
Scale bars: 200um (A–B, D–E), 50um (A′–B′, C′–D′, I–J). Data are represented as mean ± SEM.
Migration defects in Ctip1fl/fl;Emx1-Cre animals might be due to defective signaling to migrating neurons from post-migratory neurons in the cortical plate, or they might be due to aberrant interpretation of cues by migrating neurons themselves. To distinguish between these possibilities, we electroporated E14.5 wild-type or Ctip1fl/fl embryos with CMV/β-actin promoter-driven Cre and lox-STOP-lox-Egfp constructs, and examined electroporated tissue at E17.5 (Figure 6H). This experiment deletes Ctip1 from only a small fraction of cortical neurons, allowing sparse deletion of Ctip1 in an otherwise wild-type context. Ctip1fl/fl neurons electroporated with Cre at E14.5 are four times less likely than wild-type electroporated neurons to have entered the cortical plate at E17.5 (Figure 6I–K; 13% Ctip1fl/fl vs. 51% wild-type; p<0.01), indicating that migration abnormalities of superficial-layer neurons lacking Ctip1 are cell-autonomous. Further, these defects are specific to neurons migrating to the superficial layers, because migration of Ctip1fl/fl neurons electroporated with Cre at E12.5 and examined at E15.5 is unaffected (not significant; Figure S6J–M).
Ctip1 overexpression represses CTIP2 in layer V neurons, preventing them from extending axons subcerebrally
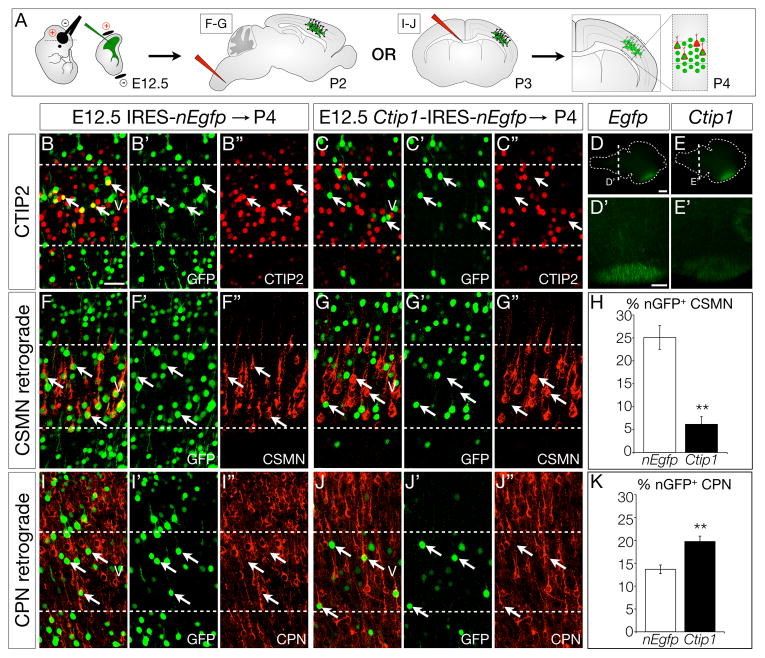
To further investigate potential functions of Ctip1 in repression of CSMN and SCPN specification, we tested whether overexpression of Ctip1 in vivo can repress endogenous expression of Ctip2 by wild-type CSMN. We electroporated CMV/β-actin promoter constructs driving expression of either control IRES-nEgfp (nuclear EGFP) or Ctip1-IRES-nEgfp into the ventricular zone of E12.5 wild-type embryos, and examined CTIP2 expression at P4 by immunocytochemistry (Figure 7B–C). Strikingly, while many control E12.5 Egfp-electroporated layer V neurons are CTIP2-positive (39%), as expected, significantly fewer Ctip1-electroporated layer V neurons are CTIP2-positive (7%; p<0.01). These data demonstrate that high levels of CTIP1 are sufficient to repress expression of Ctip2.
Figure 7. CTIP1 misexpression in vivo represses SCPN identity and projection to the spinal cord.
(A) Schematic of experimental approach. Wild-type embryos were electroporated at E12.5, and retrograde labeling from spinal cord (F–G) or from the contralateral cortical hemisphere (I–J) was performed on electroporated pups at P2 or P3, respectively. Brains were collected at P4 and somatosensory cortex was imaged and analyzed.
(B–C) Many neurons electroporated with control nuclear Egfp (nEgfp) at E12.5 express CTIP2 (B–B″; 39%), while few neurons electroporated with Ctip1-IRES-nEgfp express CTIP2 (C–C″; 7%), n = 3.
(D–K) Misexpression of Ctip1 causes E12.5-born neurons to redirect their axons away from the brainstem and spinal cord, toward targets on the contralateral cortical hemisphere. Neurons electroporated with Ctip1-IRES-Egfp at E12.5 send few axons to the brainstem (wholemount electroporated brains in D–E, coronal brainstem sections in D′–E′). Fewer neurons electroporated at E12.5 with Ctip1-IRES-nEgfp are retrogradely labeled by spinal cord injection at P2 than those electroporated with control nEgfp (F–G; quantification in H, n = 3), while more Ctip1-electroporated neurons than control nEgfp-electroporated neurons are retrogradely labeled by injection into the corpus callosum (I–J; quantification in K, n = 3).
**, p<0.01
Scale bars: 50um (B–C, F–G, I–J), 1mm (D–E), 100um (D′–E′). Data are represented as mean ± SEM.
Conversely, overexpression of Ctip2 or Fezf2 can repress endogenous Ctip1 in layer V neurons. When we electroporate Ctip2-IRES-nEgfp or Fezf2-IRES-nEgfp at E12.5, we find that fewer layer V neurons express Ctip1 at P4 compared with layer V neurons electroporated with control IRES-nEgfp (nEgfp, 64%; Fezf2, 37%, p<0.01; Ctip2, 8%, p<0.01; Figure S7). High-level expression either of Ctip2 or Fezf2 is therefore sufficient to repress expression of Ctip1, although Ctip2 is more efficient than Fezf2.
This cross-repression between Ctip1 and Ctip2/Fezf2, central transcriptional controls over SCPN development, motivated us to investigate whether Ctip1 is also sufficient to prevent layer V neurons from projecting subcerebrally. We electroporated control IRES-nEgfp or Ctip1-IRES-nEgfp constructs into wild-type embryos at E12.5, then retrogradely labeled CSMN or CPN with CTB (Figure 7A). Strikingly, we find a five-fold reduction in the percentage of Ctip1-electroporated neurons projecting to the spinal cord, compared with control nEgfp-electroporated neurons (Figure 7F–H; 25% vs. 6%; p<0.01). Further, we find a significant increase in the number of Ctip1-electroporated layer V neurons projecting across the corpus callosum (Figure 7I–K; 13% nEgfp vs. 20% Ctip1; p<0.05), indicating that the axons of some Ctip1-electroporated neurons are redirected from subcerebral targets and toward contralateral cortical targets. Taken together, these data demonstrate that Ctip1 overexpression alters the balance of deep-layer projection neuron fate specification against SCPN and in favor of CPN, in both gene expression and axonal projections.
DISCUSSION
Although critical molecular controls over projection neuron subtype development have been identified in recent years, it is clear that additional transcriptional regulators remain to be discovered, particularly those that exert fine control over the final distribution, proportions, and balance of subtypes present in specific functionally specialized areas of the cortex. In this work, we identify that the transcription factor Ctip1 directs the proportional allocation of corticothalamic, subcerebral, and callosal projection neurons in deep cortical layers.
Subtype control over deep-layer neurons
Ctip1 controls the balance between specification of distinct, functionally specialized subtypes of deep-layer cortical projection neurons, and, in the absence of Ctip1 function, more SCPN are generated at the expense of CThPN and deep-layer CPN. In Ctip1fl/fl;Emx1-Cre mice, an aberrantly expanded population of neurons with molecular and anatomic characteristics of SCPN occupies an expanded layer V (Figure 3), and more neurons born at E12.5 send axons to the cerebral peduncle instead of to the thalamus, indicating that CThPN located in the upper segment of what would otherwise be layer VI are fate-converted to SCPN (Figure 4).
The population of CThPN located in the most superficial portion of layer VI, near the interface between layer V and layer VI, is particularly vulnerable to switching fate from corticothalamic to subcerebral due to ectopic expression of SCPN genetic controls (Lai et al., 2008; Tomassy et al., 2010). In the absence of Ctip1, all layer VI neurons express higher-than-normal levels of Fezf2 and Ctip2 (asterisks in Figure 3B, 3D); however, only those neurons located in the most superficial portion of layer VI cease to express CThPN controls (Figure 3G–L) and consequently project subcerebrally (Figure 4A–F). The existence of this subpopulation of CThPN, which is generated immediately before SCPN specification begins, and which resides immediately adjacent to layer V, suggests that these closely-related corticofugal neurons require precise transcriptional control by Ctip1 and other genes for correct specification into distinct projection neuron subtypes, and for allocation of correct proportions CThPN and SCPN depending on cortical area.
Intriguingly, Ctip1 controls subtype specification, but is itself non-subtype-specific, as it is expressed at high levels by CPN, CThPN, and subplate neurons. Although some previously-identified controls are expressed by multiple subtypes, they are generally expressed at high levels by one subtype and low levels by another: Ctip2 and Fezf2 by SCPN (high) and CThPN (low); Tbr1 by CThPN (high) and superficial-layer CPN (low); and Sox5 by CThPN (high) and SCPN (low) (Arlotta et al., 2005; Hevner et al., 2001; Lai et al., 2008; McKenna et al., 2011; Molyneaux et al., 2005; Woodworth et al., 2012). Ctip1, in contrast, is specific only in its exclusion from CSMN (Figure 1D–I). This widespread and non-subtype-specific expression is most likely responsible for Ctip1 not being identified as a candidate in microarray-based screens for genes involved in subtype specification (Arlotta et al., 2005; Chen et al., 2005a; 2005b). The data presented here regarding Ctip1 function suggest that further unidentified controls over subtype development might expand or contract populations of neurons by action in developmentally, and likely evolutionarily, related populations.
Abnormalities of commissural neurons
In the absence of Ctip1 function, fewer deep-layer CPN project through the corpus callosum (Figure 5I–K), and, instead, many deep-layer neurons in somatosensory cortex project to the contralateral hemisphere via the anterior commissure (Figure 5L–R). This re-routing might be necessary because the superficial-layer neurons in cingulate cortex that would normally pioneer the callosal projection instead aberrantly project through the pyramidal tract (Figure 5A–H), impairing pathfinding by deep-layer CPN. Alternatively, more AC-projecting neurons might be specified in the absence of Ctip1 function, reflecting a previously unidentified balance between specification of CC-projecting and AC-projecting commissural neurons controlled, at least in part, by Ctip1. We do not exclude the possibility that Ctip1 might regulate expression of axon guidance molecules necessary for pathfinding by CPN. Ctip1 might regulate pathfinding in SCPN and CThPN, as well, particularly in light of aberrant subplate neuron gene expression and axon extension in null mice (Figure S4).
In contrast with these striking abnormalities of deep-layer CPN targeting, Ctip1fl/fl;Emx1-Cre superficial-layer CPN neither express SCPN-specific controls (Figure 3A–F) nor project through the cerebral peduncle (Figure 4A–B), except in cingulate cortex (Figure 5A–H). Although Ctip1 is expressed at high levels by superficial-layer CPN (Figure 1C), these neurons appear to be specified largely normally in the absence of Ctip1 (Figure 3M–R), although many of their axons do not successfully cross the corpus callosum (Figure S5I–K). These data are consistent with the hypothesis that superficial-layer and deep-layer CPN are substantially different from each other molecularly and with regard to their developmental origins, perhaps reflecting distinct evolutionary events of projection neuron diversification (Arlotta et al., 2008; Fame et al., 2011; Molyneaux et al., 2009). Ctip1 is expressed by both superficial-layer and deep-layer CPN, but is necessary for subtype specification only for deep-layer CPN, which appear to bear a closer relationship to concurrently-generated corticofugal projection neurons (Greig et al., 2013; Sohur et al., 2012). Loss of Ctip1 function contrasts starkly with the loss-of-function phenotype observed for another important negative regulator of SCPN development, Satb2, in which large numbers of both superficial-layer and deep-layer neurons aberrantly express SCPN controls such as CTIP2 (Alcamo et al., 2008; Britanova et al., 2008; Srinivasan et al., 2012). This contrast indicates that an increasingly rich set of molecular regulators controls both novel and potentially subtle, but likely functionally critical, aspects of precise projection neuron development.
Ctip1 and Ctip2 interact cross-repressively in developing cortex
Ctip1 and Ctip2, paralogous and highly similar transcription factors, are initially co-expressed in developing cortex (Figure S3A–B), but later are expressed in a complementary fashion, with CTIP1 expressed by CPN and CThPN, and CTIP2 expressed by SCPN (Figure 1D–F, Figure S3C–D). Expression of CTIP2 increases in the absence of Ctip1, and expression of CTIP1 increases in the absence of Ctip2 (Figure S3E–J). Furthermore, overexpression of either Ctip1 or Ctip2 at E12.5 in vivo is sufficient to repress expression of the other (Figure 7B–C, Figure S7A, S7C). From these data, we conclude that Ctip1 and Ctip2 cross-repressively interact to control the development of cortical projection neurons. Ctip1 and Ctip2 exhibit similar patterns of exclusive expression in development of other cell types; Ctip1 expression is sharply downregulated as Ctip2 begins to be expressed in maturing T cells (Tydell et al., 2007), and expression of Ctip1 is significantly increased in P0 Ctip2−/− striatal medium-sized spiny neurons (Arlotta et al., 2008).
Our model of Ctip1 function is at odds with that of Cánovas et al. (Cánovas et al., 2015), who recently proposed that CTIP1 is extensively co-expressed with CTIP2 and is a positive control over SCPN development through repression of TBR1. In direct contrast, we demonstrate that expression of TBR1 and other characteristic CThPN genes is reduced in the absence of Ctip1 (Figure 3G–L, Figure 4M–O), and that expression of CTIP2 is increased (Figure 3A–B), leading to more early-born neurons becoming SCPN (Figure 4A–F). These discrepancies likely result from different loss-of-function approaches, as our investigation uses null and conditional null mouse lines, rather than modest knockdown of Ctip1 using mosaic shRNA electroporation in wild-type animals.
Ctip1 as a candidate human intellectual disability gene
CTIP1 is highly conserved from mouse to human, with only three amino acid substitutions (99.6% identity) between the longest mouse and human CTIP1 isoforms, and Ctip1 and its promoter are located within one of the most ultraconserved non-coding region-rich stretches of the human genome (Sandelin et al., 2004). CTIP1 is expressed in fetal human brain (Satterwhite et al., 2001), although the subtype specificity of its expression in human brain is not known. Intriguingly, Ctip1 is one of three protein-coding genes consistently deleted in 2p15-p16.1 microdeletion syndrome, which is characterized by intellectual disability, microcephaly, and frequently by autistic behavior (Hancarova et al., 2013), and a patient with a de novo Ctip1 microdeletion has been reported with severe speech sound disorder and intellectual delay (Peter et al., 2014). These phenotypes are consistent with functions of Ctip1 in the subtype-specific differentiation and migration of neocortical projection neurons, suggesting that Ctip1 also functions critically in organizing the human neocortex.
Ctip1 couples development of projection neuron subtypes and cortical areas
Although subtype and area development are typically analyzed as independent developmental vectors in the emergence of the mature neocortex, the two are, in fact, highly interdependent, since the proportion and identity of neuron subtypes varies significantly by cortical area (Brodmann, 1909; Cajal, 1899). Molecular programs controlling subtype and area differentiation likely interact at multiple nodes; here, we identify Ctip1 as a central node. Ctip1 directs the proportional allocation of subcerebral and corticothalamic projection neurons in sensory cortex, and, in the absence of Ctip1, sensory cortical areas are transformed, with subtype compositions, gene expression, and output connectivity that is usually present in motor areas (Figure 3; Figure 4; Greig et al., 2016). Strikingly, the expression (Figure 1, Figure S1) and functions (Figure S3) of Ctip1 in postmitotic neurons indicate that this regulation of subtype composition in distinct cortical areas occurs postmitotically. Therefore, the final non-uniform distribution of cortical projection neuron subtypes across cortical areas is based, at least in part, not on differences in subtype production, but in subtype differentiation.
In conclusion, our results indicate that Ctip1 is a central functional control over the precision of neocortical development. Ctip1 directs the development of corticothalamic and callosal projection neurons at multiple stages. First, Ctip1 specifies subtype identity in deep-layer neocortical projection neurons, preventing corticothalamic and callosal projection neurons from acquiring characteristics of subcerebral projection neurons. Second, Ctip1 directs the development of cingulate cortex neurons, allowing these populations to pioneer callosal projections. Finally, Ctip1 enables the proper migration and laminar positioning of superficial-layer callosal projection neurons, permitting these neurons to populate correct laminar locations. Identification of further fine transcriptional controls over the precise differentiation, diversity, positioning, and connectivity of neocortical projection neurons will increasingly illuminate the staggering organizational and functional complexity of the mature neocortex, and provide insight into overt and subtle dysgenesis that can result in human cognitive, sensory, motor, and integrative disorders.
EXPERIMENTAL PROCEDURES
Animals
All mouse studies were approved by the Harvard University IACUC, and were performed in accordance with institutional and federal guidelines. The date of vaginal plug detection was designated E0.5, and the day of birth as P0. Unless noted otherwise, all experiments with Ctip1fl/fl;Emx1-Cre were controlled with Ctip1wt/wt;Emx1-Cre, and Ctip1−/− with Ctip1wt/wt, although no abnormal phenotypes were observed in Ctip1−/wt or Ctip1fl/wt;Emx1-Cre heterozygotes. Sources of mouse lines used are described in Supplemental Information.
Immunocytochemistry and in situ hybridization
Mice were transcardially perfused with 4% paraformaldehyde, and brains were dissected and post-fixed at 4°C overnight. Tissue was sectioned at 50μm on a vibrating microtome (Leica). Non-specific binding was blocked by incubating tissue and antibodies in 8% goat serum/0.3% bovine serum albumin in phosphate-buffered saline. For DAPI staining, tissue was mounted in DAPI-Fluoromount-G (SouthernBiotech). Antibodies and in situ probes used are described in Supplemental Information.
BrdU birthdating
Timed pregnant females were intraperitoneally injected with bromodeoxyuridine (50 mg/kg in PBS) at E11.5, E12.5, E13.5, E14.5, or E15.5. Tissue was collected at P4 and processed for BrdU immunocytochemistry (Magavi and Macklis, 2008).
For analysis of BrdU-labeled SCPN, BrdU labeling was performed as above at E12.5, and littermate pairs were retrogradely labeled from the cerebral peduncle as described below. For acute BrdU administration, timed pregnant females were intraperitoneally injected with BrdU (75 mg/kg) at E14.5, and embryos were collected 30 minutes later.
In utero electroporation
For overexpression experiments, a CMV/β-actin promoter plasmid (derived from CBIG; gift of C. Lois) was used to drive expression of IRES-Egfp (control) or Ctip1 XL-IRES-Egfp (experimental; coding sequence from NM_001242934); or IRES-nEgfp (nuclear EGFP) (control) or Ctip1 XL-IRES-nEgfp, Ctip2-IRES-nEgfp, or Fezf2-IRES-nEgfp (experimental) in CD1 timed pregnant females at E12.5. For some experiments, pups were screened for Egfp expression on a fluorescent dissecting microscope at birth, and pups electroporated in somatosensory cortex were retrogradely labeled (as described below). Brains were collected at P4.
For loss-of-function experiments, two CMV/β-actin promoter plasmids were co-electroporated, driving expression of lox-STOP-lox-Egfp and Cre in Ctip1wt/wt (control) or Ctip1fl/fl (experimental) embryos. Plasmids were electroporated at age indicated in figure (E12.5, E14.5, or E15.5) and brains were collected either 72 hours later (Figure 6, Figure S6) or at P4 (Figure S5). Electroporation conditions were described previously (Molyneaux et al., 2005).
Retrograde labeling
Projection neurons were labeled from their axon termini under ultrasound guidance by pressure injection of Alexa fluorophore-conjugated cholera toxin B (Invitrogen). SCPN were labeled by injection into cerebral peduncle at P1, CThPN were labeled by injection into sensory thalamic nuclei (VPM and VPL) at P1, CSMN were labeled by injection into cervical spinal cord at P2, and CPN were labeled by injection into contralateral corpus callosum at P3. Anterior commissure-projecting neurons were labeled by injection into contralateral anterior commissure at P1. Tissue for all injections was collected at P4, and processed as for immunocytochemistry, above.
Quantification and statistics
Littermate pairs of experimental and control mice were collected at age indicated in figure and processed as for immunocytochemistry, above. Anatomically matched sections from each mouse (generally at the level of the anterior commissure, unless otherwise noted) were selected, and single confocal slices of somatosensory cortex were imaged. Cells positive for indicated marker(s) (retrograde label, protein by immunocytochemistry, BrdU label, GFP by electroporation) were counted within a box of pre-defined size spanning the radial thickness of cortex, with the same size box applied to wild-type and mutant images. Except where specifically noted (Figure 2, Figure S2), data are not corrected for differences in cortical thickness between wild-type and mutant animals (Table S1). Statistical analyses were conducted using unpaired two-tailed t-tests in Microsoft Excel, with a significance threshold of p<0.05.
Supplementary Material
Acknowledgments
We thank L. Pasquina, B. Brandler, P. Davis, and C. Greppi for technical assistance; M. J. Galazo for assistance with retrograde labeling of corticothalamic projection neurons; S. Orkin and J. Xu for generously transferring Ctip1fl/fl mice; K.-A. Nave for the generous gift of Nex1-Cre mice; G. Corfas, P. Arlotta, Z. He, K. Srinivasan, E. Azim, H. Padmanabhan, and V. Sahni for scientific discussions; and members of the Macklis lab for helpful suggestions. This work was supported by grants from the National Institutes of Health (NS045523 and NS075672, with additional infrastructure supported by NS041590 and NS049553), the Harvard Stem Cell Institute, and the Massachusetts Spinal Cord Research Program to J.D.M, and by a grant from the National Institutes of Health (CA031534) to H.O.T. M.B.W. was partially supported by NIH individual predoctoral National Research Service Award NS064730 and the DEARS Foundation. L.C.G. was partially supported by NIH individual predoctoral National Research Service Award NS080343, the Harvard Medical Scientist Training Program, and the DEARS Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
M.B.W. and J.D.M. conceived the project; M.B.W. and L.C.G. designed and performed all experiments; M.B.W., L.C.G., and J.D.M. analyzed and interpreted the data; and M.B.W. and L.C.G. made figures. K.X.L. performed qPCR experiments. G.C.I. and H.O.T. contributed a critical mouse line. M.B.W., L.C.G., and J.D.M. synthesized and integrated the findings, and wrote and revised the paper. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J Biol Chem. 2000;275:10315–10322. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim E, Shnider SJ, Cederquist GY, Sohur US, Macklis JD. Lmo4 and Clim1 progressively delineate cortical projection neuron subtypes during development. Cereb Cortex. 2009;19(Suppl 1):i62–i69. doi: 10.1093/cercor/bhp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RAM, Beyer RP, Bammler TK, Rubenstein JLR, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung AFP, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Brodmann K. Histological cortical localisation in relation to morphology. Brodmann’s Localisation in the Cerebral Cortex, (Brodmann’s Localisation in the Cerebral Cortex: The …) 1909:205–223. trans. 2006. [Google Scholar]
- Cajal SRY. Histology of the nervous system of man and vertebrates. Oxford University Press; USA: 1899. trans. 1995. [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015 doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cánovas J, Berndt FA, Sepúlveda H, Aguilar R, Veloso FA, Montecino M, Oliva C, Maass JC, Sierralta J, Kukuljan M. The Specification of Cortical Subcerebral Projection Neurons Depends on the Direct Repression of TBR1 by CTIP1/BCL11a. J Neurosci. 2015;35:7552–7564. doi: 10.1523/JNEUROSCI.0169-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005a;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proceedings of the National Academy of Sciences. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Šestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 2005b;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame RM, MacDonald JL, Macklis JD. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011;34:41–50. doi: 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, Nave KA. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 2006;44:611–621. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty ML, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JLR, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Greppi C, Macklis JD. Ctip1 controls acquisition of sensory area identity and establishment of sensory input fields in the developing neocortex. Neuron. 2016 doi: 10.1016/j.neuron.2016.03.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MMS, Shin Y, Xu X, Zhu Y, Li M, Šestan N. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancarova M, Simandlova M, Drabova J, Mannik K, Kurg A, Sedlacek Z. A patient with de novo 0.45 Mb deletion of 2p16.1: the role of BCL11A, PAPOLG, REL, and FLJ16341 in the 2p15-p16.1 microdeletion syndrome. Am J Med Genet. 2013;161A:865–870. doi: 10.1002/ajmg.a.35783. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone AA, Goffinet AM, Campagnoni AT, Rubenstein JLR. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- John A, Brylka HH, Wiegreffe CC, Simon RR, Liu P, Jüttner R, Crenshaw EB, Luyten FP, Jenkins NA, Copeland NG, et al. Bcl11a is required for neuronal morphogenesis and sensory circuit formation in dorsal spinal cord development. Development. 2012;139:1831–1841. doi: 10.1242/dev.072850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester SE, O’Leary DDM. Axons of early generated neurons in cingulate cortex pioneer the corpus callosum. J Neurosci. 1994;14:6608–6620. doi: 10.1523/JNEUROSCI.14-11-06608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T-Y, Hsueh Y-P. Expression of zinc finger transcription factor Bcl11A/Evi9/CTIP1 in rat brain. J Neurosci Res. 2007;85:1628–1636. doi: 10.1002/jnr.21300. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MMS, Krsnik Z, Kawasawa YI, Lefebvre V, Šestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proceedings of the National Academy of Sciences. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JRL, Macklis JD. SOX5 Controls the Sequential Generation of Distinct Corticofugal Neuron Subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Lee BS, Dekker JD, Lee BK, Iyer VR, Sleckman BP, Shaffer AL, Ippolito GC, Tucker PW. The BCL11A Transcription Factor Directly Activates RAG Gene Expression and V(D)J Recombination. Mol Cell Biol. 2013;33:1768–1781. doi: 10.1128/MCB.00987-12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dollé P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ippolito GC, Wall JK, Niu T, Probst L, Lee BS, Pulford K, Banham AH, Stockwin L, Shaffer AL, et al. Functional studies of BCL11A: characterization of the conserved BCL11A-XL splice variant and its interaction with BCL6 in nuclear paraspeckles of germinal center B cells. Mol Cancer. 2006;5:18. doi: 10.1186/1476-4598-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magavi SS, Macklis JD. Identification of newborn cells by BrdU labeling and immunocytochemistry in vivo. Methods Mol Biol. 2008;438:335–343. doi: 10.1007/978-1-59745-133-8_25. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JLR, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Peter B, Matsushita M, Oda K, Raskind W. De novo microdeletion of BCL11A is associated with severe speech sound disorder. Am J Med Genet. 2014;164A:2091–2096. doi: 10.1002/ajmg.a.36599. [DOI] [PubMed] [Google Scholar]
- Richards LJ, Plachez C, Ren T. Mechanisms regulating the development of the corpus callosum and its agenesis in mouse and human. Clin Genet. 2004;66:276–289. doi: 10.1111/j.1399-0004.2004.00354.x. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Bailey P, Bruce S, Engström PG, Klos JM, Wasserman WW, Ericson J, Lenhard B. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HKA, Hirschhorn JN, Cantor AB, Orkin SH. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterwhite E, Sonoki T, Willis TG, Harder L, Nowak R, Arriola EL, Liu H, Price HP, Gesk S, Steinemann D, et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–3420. doi: 10.1182/blood.v98.12.3413. [DOI] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Šestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohur US, Padmanabhan HK, Kotchetkov IS, Menezes JRL, Macklis JD. Anatomic and Molecular Development of Corticostriatal Projection Neurons in Mice. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, Leone DP, Bateson RK, Dobreva G, Kohwi Y, Kohwi-Shigematsu T, Grosschedl R, McConnell SK. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proceedings of the National Academy of Sciences. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassy GS, De Leonibus E, Jabaudon D, Lodato S, Alfano C, Mele A, Macklis JD, Studer M. Area-specific temporal control of corticospinal motor neuron differentiation by COUP-TFI. Proceedings of the National Academy of Sciences. 2010;107:3576–3581. doi: 10.1073/pnas.0911792107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tydell CC, David-Fung E-S, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, et al. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- Wiegreffe CC, Simon RR, Peschkes K, Kling C, Strehle M, Cheng J, Srivatsa S, Liu P, Jenkins NA, Copeland NG, et al. Bcl11a (Ctip1) Controls Migration of Cortical Projection Neurons through Regulation of Sema3c. Neuron. 2015;87:311–325. doi: 10.1016/j.neuron.2015.06.023. [DOI] [PubMed] [Google Scholar]
- Woodworth MB, Greig LC, Kriegstein AR, Macklis JD. SnapShot: Cortical Development. Cell. 2012;151:918–918. e1. doi: 10.1016/j.cell.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Peng C, Sankaran VG, Shao Z, Esrick EB, Chong BG, Ippolito GC, Fujiwara Y, Ebert BL, Tucker PW, et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334:993–996. doi: 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.