Abstract
Tritrichomonas species flagellates (IMC strain) were isolated from the biliary tract of an individual who had developed cholecystitis as a complication of acquired agammaglobulinemia. Sequence analysis of Tritrichomonas sp. (IMC clone 2 (cl2)) was performed for several genetic regions including the ITS1-5.8S rDNA-ITS2 region, the cysteine protease (CP)-1, CP-2 and CP-4 to CP-9 genes, and the cytosolic malate dehydrogenase 1 gene. In addition to comparison of the variable-length DNA repeats in the isolate clone with those in T. foetus (Inui cl2) and the T. mobilensis (U.S.A.: M776 cl2) reference strains, this analysis showed that the Tritrichomonas sp. (IMC cl2) was T. foetus (cattle/swine genotype). Injection of T. foetus (IMC cl2) directly into the livers of CBA mice resulted in liver abscess formation on Day 7. Moreover, inoculation via orogastric intubation caused infection in the cecum on Day 5 in CBA mice co-infected with Entamoeba histolytica (HM-1: IMSS cl6). T. foetus (IMC cl2) was able to grow in YI-S medium for over 20 days, even at 5°C. These results indicate that the T. foetus isolate is able to survive in the feces and edible organ meat of the definitive host for a prolonged period of time, and it is possible that the parasite could infect humans.
Keywords: experimental infection, human infection, molecular identification, morphological differentiation, Tritrichomonas foetus
Tritrichomonas foetus (syn. Tritrichomonas suis) has been shown to exist as both cattle/swine and feline genotypes. In heifers, T. foetus causes infertility and early embryonic death via sexually transmitted infection of the reproductive tract [22]. In pigs, T. foetus infects the nasal cavity, stomach and intestine and causes diarrhea in piglets [5, 21]. Feline intestinal tritrichomoniasis caused by T. foetus is usually associated with large bowel diarrhea [2, 9, 20].
To date, only three cases of human tritrichomoniasis caused by T. foetus have been reported, and all three appear to have been rare opportunistic infections in immunocompromised or immunosuppressed individuals [7, 13, 23]. It is suspected that two of those cases were caused by the T. foetus cattle/swine genotype based on the sequence of the ITS1-5.8S rRNA-ITS2 region, which enables differentiation of the T. foetus cattle/swine genotype from the feline genotype and Tritrichomonas mobilensis isolated from a squirrel monkey [3]. The ITS1-5.8S rRNA-ITS2 regions in the cattle/swine and feline T. foetus genotypes differ by a single nucleotide change at position 9 in ITS2 (cattle/swine genotype: cytosine and feline genotype: thymine), and by a single nucleotide change at position 47 in ITS2 in T. foetus and T. mobilensis (T. foetus: thymine and T. mobilensis: cytosine) [5, 8, 16]. The symptoms and location of lesions also differ in human cases; T. foetus-like tritrichomonads have been detected in cerebrospinal fluid (meningoencephalitis) [13], bronchoalveolar lavage specimens (pneumonia) [7] and peritoneal fluid (peritonitis) [23]. To date, no human cases of tritrichomoniasis caused by T. mobilensis have been reported.
In Japan, cattle tritrichomoniasis is a notifiable infectious disease under the domestic animal infectious diseases control law. Based on notification records, no cases of cattle tritrichomoniasis caused by T. foetus occurred during the 51-year period from 1963 to 2014 [1]. In contrast, it is not mandatory to report swine tritrichomoniasis. Although the true prevalence of swine tritrichomoniasis is not known, an epidemiological study in Japan showed that 36 of 64 samples (56.3%) tested positive for T. foetus [5].
In the present study, we conducted a morphological and molecular investigation of a Tritrichomonas sp. human isolate. Moreover, T. foetus infection, which is highly prevalent in Japanese pigs, occurs via the orogastric route. Therefore, we determined whether the isolate is capable of similarly infecting the intestine via the oral route in mice. In addition, we investigated the viability of this parasite in the external environment.
MATERIALS AND METHODS
Tritrichomonas spp. strains: 1) Tritrichomonas sp. human isolate, Tritrichomonas sp. (IMC strain [IMC]). Tritrichomonas flagellates were isolated from the bile fluid of a patient (a 39-year-old Japanese male) with acquired agammaglobulinemia complicated by cholecystitis and were cultured aseptically in YI-S medium [4] containing 10% heat-inactivated adult bovine serum and an antibiotic mixture (potassium penicillin G [1,000 U/ml], streptomycin sulfate [1 mg/ml] and polymyxin B sulfate [130 U/ml]). 2) Tritrichomonas foetus. A reference strain of T. foetus (Inui) [14] was provided by the Bioresource Project of the Institute of Tropical Medicine, Nagasaki University (Nagasaki, Japan). 3) Tritrichomonas mobilensis. A reference strain of T. mobilensis (U.S.A.: M776 [M776]) was obtained from the American Type Culture Collection (No. 50116; Manassas, VA, U.S.A.). All three isolates were cultured at 35.5°C with 4 ml of YI-S medium in screw-capped culture tubes (100 × 13 mm in diameter).
Single-cell cloned cultures: Single-cell clones were established from axenic cultures of Tritrichomonas sp. (IMC), T. foetus (Inui) and T. mobilensis (M776) according to a previously described serial dilution cell cloning protocol for 96-well plates [18], with some modifications. We modified the step for collecting single-celled organisms as follows. First, 2–3 µl drops of a dilute culture suspension in YI-S medium, containing a small but adequate number of organisms, were placed into several wells. Then, the wells were observed under an inverted microscope, and drops containing a single organism were collected by adding 200 µl of fresh medium, mixing gently and aspirating the medium containing the single organism. This medium was then transferred to a culture tube containing fresh YI-S medium and cultured at 35.5°C. Six clones from Tritrichomonas sp. (IMC), three clones from T. foetus (Inui) and five clones from T. mobilensis (M776) were successfully grown. The identities of the second clones (designated as clone 2) from each of these three strains were confirmed, and these were then used for characterization.
Cultivation at various temperatures: For each of the three clones (Tritrichomonas sp. IMC cl2, T. foetus Inui cl2 and T. mobilensis M776 cl2), 2 × 106 organisms were inoculated into two tubes containing 4 ml of YI-S medium each and cultured at 5, 18, 25 or 35.5°C. The number of organisms was counted using a Fuchs-Rosenthal counting chamber every 2–3 days to determine the growth kinetics of the clones at various temperatures. This experiment was performed in order to investigate the viability of each Tritrichomonas sp. strain at various temperatures representative of the external environment.
Scanning electron microscopy (SEM): The Tritrichomonas sp. (IMC cl2) and T. mobilensis (M776 cl2) were fixed with 2.5% glutaraldehyde/0.1 M cacodylate buffer (pH 7.2) for 1 hr in silicon-coated plastic microtubes (2 ml). The fixed cells were washed with 0.1 M cacodylate buffer, pelleted by centrifugation at 275 × g for 3 min and post-fixed with 1% OsO4/0.1 M cacodylate buffer for 1 hr. The post-fixed organisms were attached to a filter with poly-L-lysine-coated pores measuring 0.6 µm in diameter (SEMpore; JEOL DATUM Ltd., Tokyo, Japan) according to the manufacturer’s protocol. The filters were then washed three times with distilled water. Next, the attached cells of the two Tritrichomonas spp. clones were frozen for 40 sec in liquid nitrogen. Each frozen sample was immediately mounted on the specimen stage of the SEM (JSM5600LV; JEOL Ltd.), and the specimen chamber was evacuated to 60 Pa, followed by slow sublimation for 60 min. Then, each of the dry, frozen samples on the SEMpore was sputter-coated with Pt-Pd and observed at an accelerating voltage of 5 kV.
Molecular analysis: DNA was isolated from 50-µl culture samples containing approximately 1 × 106 organisms in 10 mM phosphate-buffered saline (pH 7.4) using a QIAamp DNA Mini kit (Qiagen, Venlo, the Netherlands). The DNA samples from Tritrichomonas sp. (IMC cl1–cl6), T. foetus (Inui cl2) and T. mobilensis (M776 cl2) were used as templates for PCR targeting of the ribosomal RNA internal transcribed spacer region (ITS1-5.8S rRNA-ITS2 region), eight cysteine protease (CP) genes (CP-1, CP-2 and CP-4 to CP-9) and the cytosolic malate dehydrogenase 1 (MDH1) gene. The ITS1-5.8S rRNA-ITS2 region was amplified with a Tricho-F forward primer and a Tricho-R reverse primer [11]. The primers used to amplify CP-1, CP-2 and CP-4 to CP-9, and MDH1 have been previously described by Slapeta et al. [19]. Amplification was performed in 50-µl reaction volumes containing 1× PCR buffer, 250 µM of each dNTP, 0.5 µM of each primer, 1.25 U Takara Ex Taq (Takara Bio Inc., Otsu, Japan) and 2 µl of the DNA samples. The samples were denatured at 94°C for 5 min and then subjected to 35 cycles of 94°C for 30 sec, 55°C for 30 sec (60°C for 30 sec for the ITS region) and 72°C for 1 min, with a final extension at 72°C for 7 min. The PCR products were directly sequenced using an ABI Prism BigDye Terminator v3.1 Cycle Sequencing Ready Reaction Kit and an ABI PRISM 3500 Genetic Analyzer (Applied Biosystems Japan Ltd., Tokyo, Japan). Variable-length DNA repeats were amplified using primers TR7 and TR8 [17, 21]. The PCR was performed by denaturing the DNA at 94°C for 5 min followed by 35 cycles at 94°C for 1 min, 46°C for 1 min and 72°C for 1 min, with a final extension at 72°C for 7 min. The PCR products were separated by electrophoresis on 2% (w/v) agarose gels, and species-specific fingerprints were observed. Fragments of approximately 520 bp in length were isolated from the gels and sequenced.
Experimental infection: Three Tritrichomonas spp.—Tritrichomonas sp. (IMC cl2), T. foetus (Inui cl2) and T. mobilensis (M776 cl2)—were cultured in YI-S medium for 2 days at 35.5°C, and the number of organisms was adjusted to 4 × 108 parasites/ml in YI-S medium. Each suspension (2 × 107 parasites/0.05 ml) was inoculated into the left lobe of the liver in two CBA mice (10-week-old males). This location for infection was chosen to reflect the parasitic location of Tritrichomonas sp. (IMC) in the patient. A suspension (4 × 107 parasites/0.1 ml) of Tritrichomonas sp. (IMC cl2) was also inoculated into the stomach of two NOD.CB17-Prkdcscid/J (NODSCID) mice (8-week-old males) and two CBA mice (10-week-old males), using a mouse feeding needle, to investigate its infectivity and proliferation capability in the intestine (cecum).
Infection of mice with induced chronic colitis: CBA mice with persistent Entamoeba histolytica (HM-1:IMSS cl6) infection and erosion of intestinal mucosa were used as a chronic colitis model. The infectivity of Tritrichomonas spp. in this mouse model was examined and compared with that in healthy mice. Two CBA mice (20-week-old males) with persistent E. histolytica (HM-1:IMSS cl6) infection for 30–40 days [10] received either an oral inoculation of Tritrichomonas spp. using a mouse feeding needle or an intravenous inoculation via the caudal vein. All animal experiments were performed according to protocols approved by the Keio University Institutional Animal Care and Use Committee (authorization number: 08036) and the Institutional Guidelines on Animal Experimentation at Keio University.
Paraffin embedding: Liver and cecum tissues from mice infected with Tritrichomonas spp. were fixed with 10% formalin. The cross sections were processed for paraffin embedding using a graded series of ethanol, xylene and paraffin according to conventional methods. Thin cross sections were stained with hematoxylin and eosin (HE) and periodic acid–Schiff (PAS).
RESULTS
Morphological properties of two Tritrichomonas spp. by SEM: The Tritrichomonas sp. (IMC cl2) and T. mobilensis (M776 cl2) strains examined in the present study exhibited genus-specific characteristics, such as three anterior flagella emerging from the periflagellar canal, an undulating membrane reaching to the posterior end of the body, a recurrent flagellum with a distal free end and a visible axostyle tip. However, the undulating membrane of Tritrichomonas sp. (IMC cl2) appeared to be more acute-angled than that of T. mobilensis. Additionally, a stronger contrast was observed between light and shade than in T. mobilensis (M776 cl2) (Fig. 1A, 1C, 1D and 1F). The morphological aspects of the undulating membranes of the two clones of Tritrichomonas sp. (IMC cl2) and T. mobilensis (M776 cl2) did not change (Fig. 1B and 1E), even in the pre-mitotic phase. In terms of shape, T. mobilensis (M776 cl2) had a typical piriform shape, and egg- and heart-shaped morphologies were also observed (data not shown), consistent with the SEM images presented by Midlej et al. [12]. The morphological aspects of T. foetus (Inui cl2) were almost identical to those of Tritrichomonas sp. (IMC cl2) (data not shown).
Fig. 1.
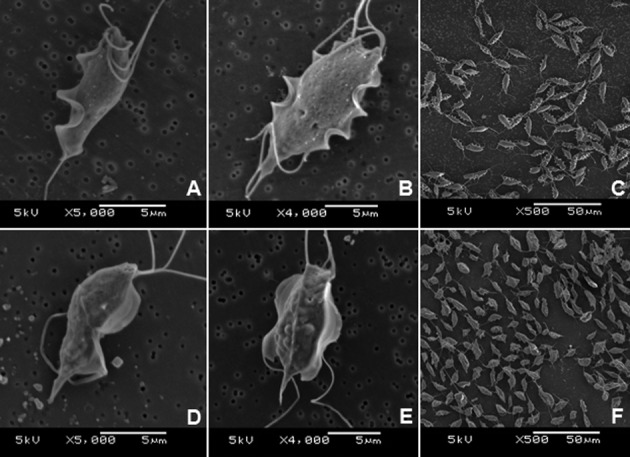
Scanning electron microscopy images of three Tritrichomonas strains: IMC strain clone 2 [IMC cl2] [Tritrichomonas foetus (IMC cl2)] (A, B, C) and Tritrichomonas mobilensis (U.S.A.: M776 cl2) (D, E, F). A and D: organisms in interphase. B and E: organisms in pre-binary division phase. C and F: organisms under low magnification.
Sequence analysis: The sequences of the ITS1-5.8S rDNA-ITS2 regions of the six clones of Tritrichomonas sp. (IMC cl1 to cl6) were all found to be identical to each other and to those of T. foetus (Inui cl2) and the T. foetus cattle/swine genotype reference strains from cattle (AF339836) and pigs (U85966). However, the sequence of the ITS1-5.8S rDNA-ITS2 region of Tritrichomonas sp. (IMC cl2) was found to differ by one nucleotide at position 47 in ITS2 from that of the feline genotype reference strain from a cat (JX960422) [15] and by one nucleotide at position 9 in ITS2 from that of T. mobilensis (M776) (U86612). The sequences of the seven cysteine protease genes (CP-1 and CP-4 to CP-9) and the cytosolic MDH1 gene in the Tritrichomonas sp. (IMC cl2) (CP-1: LC054281, CP-4: LC054283, CP-5: LC054284, CP-6: LC054285, CP-7: LC054286, CP-8: LC054287, CP-9: LC054288 and MDH1: LC054289) and T. foetus (Inui cl2) clones were identical to those of the T. foetus cattle/swine genotype reference strains from cattle (CP-1: JX187023, CP-4: JX187051, CP-5: JX187065, CP-6: JX187081, CP-7: JX187094, CP-8: JX187106, CP-9: JX187118 and MDH1: JX187131) and the T. foetus cattle/swine genotype reference strains from pigs (CP-1: JX648148, CP-4: JX648154, CP-5: JX648157, CP-6: JX648175, CP-7: JX648160, CP-8: JX648163, CP-9: JX648165 and MDH1: JX648169). The number of nucleotide differences in each of the sequences of the seven cysteine protease and MDH1 genes between Tritrichomonas sp. (IMC cl2) (CP-1: LC054281, CP-4: LC054283, CP-5: LC054284, CP-6: LC054285, CP-7: LC054286, CP-8: LC054287, CP-9: LC054288 and MDH1: LC054289) and T. mobilensis (M776) (CP-1: LC054290, CP-4: JX187053, CP-5: JX187070, CP-6: JX187083, CP-7: LC054292, CP-8: LC054293, CP-9: JX187120 and MDH1: JX187133) was 3, 1, 3, 5, 1, 5, 2 and 3, respectively. However, the sequences of CP-2 in the Tritrichomonas sp. clones (IMC cl1–cl6) differed from those of the reference strains of T. foetus (Inui cl2), the T. foetus cattle/swine genotype strain from cattle (JX187036) and the T. foetus cattle/swine genotype strain from pigs (JX648151) by two nucleotides at positions 24 and 50, and the sequences also differed from the T. foetus feline genotype strain from cats (AB973227) and T. mobilensis (M776 cl2) (LC054291) by 22 and 18 nucleotides, respectively (Fig. 2). Moreover, a single nucleotide polymorphism (SNP) at position 50 of CP-2 (highlighted in gray in Fig. 2) resulted in the substitution of leucine [Leu] (codon CTT) in Tritrichomonas sp. (IMC cl2) by proline [Pro] (codon CCT) in the other five strains. PCR-based analysis of the variable-length DNA repeats indicated that the patterns for two Tritrichomonas sp. clones (IMC cl1 and cl2) were identical, and three major PCR products with lengths of approximately 110, 220 and 520 bp were observed (Fig. 3). The pattern for T. foetus (Inui cl2) was similar to that for Tritrichomonas sp. (IMC cl1 and cl2). However, in the PCR pattern from T. mobilensis (M776 cl1 and cl2), four major bands at approximately 110, 220, 520 and 660 bp were observed. The PCR patterns for the Trichomonas vaginalis and Pentatrichomonas hominis clinical isolates clearly differed from those of the two Tritrichomonas strains. Sequence analysis of the 520-bp PCR products from Tritrichomonas sp. (IMC cl2), T. foetus (Inui cl2) and T. mobilensis (M776 cl2) indicated that the sequence of Tritrichomonas sp. (IMC cl2) was identical to that of T. foetus (Inui cl2), but different from that of T. mobilensis (M776 cl2) at four nucleotide positions (Fig. 4). Based on the genetic and morphological characterization, the Tritrichomonas sp. (IMC cl2) human isolate was identified as T. foetus cattle/swine genotype. The sequence data reported in this study were deposited in GenBank.
Fig. 2.
Comparison of the cysteine protease 2 gene sequences Tritrichomonas foetus (IMC cl2) and the five Tritrichomonas spp. reference strains.
Fig. 3.
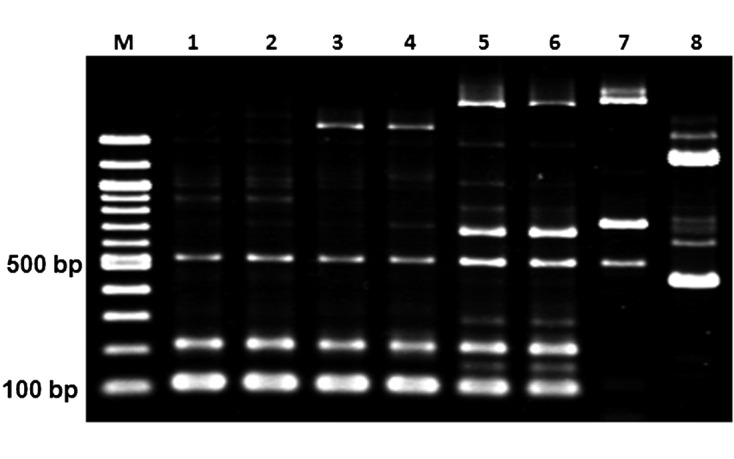
Comparison of variable-length DNA repeats using the TR7/TR8 primers. Lane 1: Tritrichomonas foetus (IMC cl1); Lane 2: T. foetus (IMC cl2); Lane 3: T. foetus (Inui cl1); Lane 4: T. foetus (Inui cl2); Lane 5: Tritrichomonas mobilensis (M776 cl1); Lane 6: T. mobilensis (M776 cl2); Lane 7: Trichomonas vaginalis; Lane 8: Pentatrichomonas hominis.
Fig. 4.
Sequence alignment of the TR7/TR8-amplified variable-length repeats of Tritrichomonas foetus (IMC cl2), Tritrichomonas foetus (Inui cl2) and Tritrichomonas mobilensis (M776 cl2).
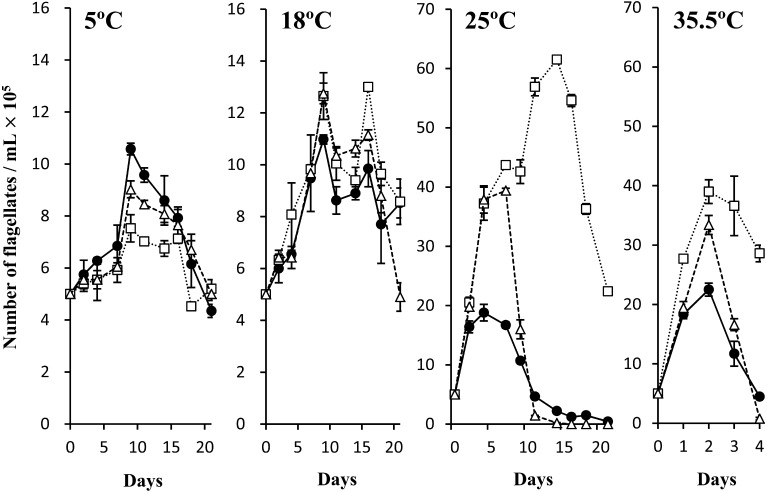
Cultivation at various temperatures: The growth kinetics of the Tritrichomonas sp. (IMC cl2) [Tritrichomonas foetus (IMC cl2)], T. foetus (Inui cl2) and T. mobilensis (M776 cl2) at 5, 18, 25 and 35.5°C are shown in Fig. 5. All three of the examined clones grew at all temperatures, even at 5°C, and survived for more than 20 days. In particular, the number of T. foetus cells (IMC cl2) doubled by Day 9 of cultivation at 5°C. Moreover, proliferation was markedly increased at temperatures above 25°C for all three strains, and the lowest increase was observed for T. foetus (IMC cl2) grown at 25°C and 35.5°C. The highest number of T. mobilensis (M776 cl2) parasites was observed at 25°C and 35.5°C, which was also the highest number among the three clones. This clone also survived the longest.
Fig. 5.
Growth kinetics of Tritrichomonas foetus (IMC cl2), Tritrichomonas foetus (Inui cl2) and Tritrichomonas mobilensis (M776 cl2) at 5°, 18°, 25° and 35.5°C. The mean numbers and standard deviations of duplicate cultures are plotted. T. foetus (IMC cl2): ●–●, T. foetus (Inui cl2): ∆–∆, T. mobilensis (M776 cl2): □–□.
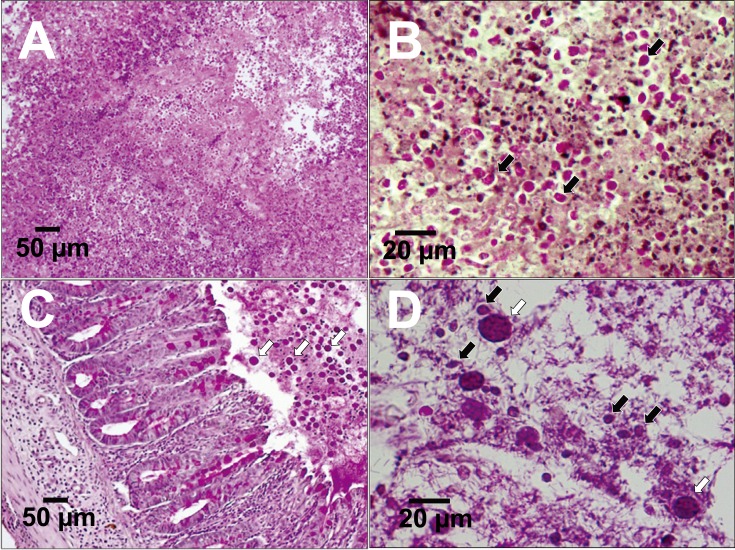
Experimental infection in healthy mice and in mice with induced amoebic colitis: Abscess formation by T. foetus (IMC cl2), T. foetus (Inui cl2) and T. mobilensis (M776 cl2) was confirmed in the livers of CBA mice, and live tritrichomonad organisms were detected in the lesions 7 days after inoculation, as shown in Fig. 6A and 6B. No proliferation of T. foetus (IMC cl2) was observed in the cecum of healthy CBA mice under specific pathogen free (SPF) conditions, even in NODSCID mice, on Day 5 following inoculation via orogastric intubation. No proliferation was observed on Day 5, even after direct injection into the cecum of CBA and NODSCID mice, which was performed as a supporting experiment. However, CBA mice pre-infected with E. histolytica (HM-1: IMSS cl6) with intestinal erosion were successfully infected by T. foetus (IMC cl2). By Day 5, the organism was found to have proliferated in the cecum following inoculation via orogastric intubation, as shown in Fig. 6C and 6D or via intravenous injection into the caudal vein (data not shown). However, the organisms of T. foetus (IMC cl2) could not be found in the other organs including bile duct. PAS staining of thin cross sections of the cecum was observed following T. foetus (IMC cl2) infection by oral or intravenous inoculation; however, tritrichomonad organisms were only detected in the intestinal lumen of the cecum. Moreover, T. mobilensis (M776 cl2) also infected the cecum of two mice co-infected with E. histolytica (HM-1: IMSS cl6), and the infection was similar to that observed following oral inoculation of T. foetus (IMC cl2) (data not shown).
Fig. 6.
Thin sections of liver abscesses from CBA mice infected with Tritrichomonas foetus (IMC cl2) 7 days after inoculation (periodic acid–Schiff (PAS) staining) (A, B). Thin sections of the cecum of CBA mice co-infected with T. foetus (IMC cl2) and Entamoeba histolytica (HM-1:IMSS cl6). Data obtained at 5 days after inoculation via orogastric intubation are shown (PAS staining) (C, D). Black arrows: T. foetus (IMC cl2), white arrows: E. histolytica (HM-1:IMSS cl6).
DISCUSSION
The present Tritrichomonas sp. human isolate was identified as the T. foetus cattle/swine genotype. However, tritrichomoniasis caused by T. foetus in cattle has not been reported in Japan for 51 years [1], suggesting that it is unlikely that this isolate came from cattle.
The patient in the present study had no history of contact with reservoirs or definitive hosts, such as cattle, pigs or cats. Although the source of T. foetus (IMC) in the present case is unknown, T. foetus (IMC, cattle/swine genotype) may be capable of infecting patients orally as well as through direct contact with infected animals via T. foetus-contaminated natural raw foods (e.g., rare liver). Interestingly, of the four patients diagnosed with human tritrichomoniasis, two were Japanese and one was French, and both cultures are known for their consumption of raw meat. However, the patient in this case did not have a certain history of eating undercooked or raw meat, and also in previous cases, we could not find descriptions about those histories [7, 13, 23].
Moreover, the feline genotype in the ITS1-5.8S rDNA-ITS2 region of T. foetus is also found in piglets, indicating that the reverse is true in cats [6, 20]. This suggests that cats may also transmit the T. foetus cattle/swine genotype strain. Moreover, T. foetus (IMC cl2) can survive at low temperatures and shows resistance to low osmotic pressure (75–80 mOsm/l) for over 1 hr (data not shown). To confirm this, we showed that the present T. foetus (IMC cl2, cattle/swine genotype) strain can infect the cecum of CBA mice pre-infected with E. histolytica (HM-1: IMSS cl6) via orogastric intubation. These results indicate that T. foetus (IMC) may also infect the human intestine via the oral route. In addition, our results demonstrate that this parasite may infect the gastrointestinal tract after damage to the mucosa caused by another agent or following co-infection with a collaborative parasite, such as E. histolytica. All four human tritrichomoniasis cases, including the present case, occurred in individuals with underlying immunodeficiency or immunosuppression. These results suggest that, in addition to immunosuppression or immunodeficiency, infection resulting in damage to the intestinal mucosa may also be a risk factor for the onset of tritrichomoniasis in humans.
Although we were unable to determine the specific source or route of infection by T. foetus (IMC, cattle/swine genotype) in the present study, our data demonstrate that tritrichomoniasis is an infectious disease with considerable zoonotic potential, and it may occur in immunosuppressed and immunodeficient individuals with intestinal damage due to pre- or co-infection with another agent.
Acknowledgments
This work was supported in part by a JSPS KAKENHI Grant-in-Aid for Scientific Research ((C) 24590511 and (B) 24406013). We declare no conflicts of interest.
REFERENCES
- 1.Animal Quarantine Service, Ministry of Agriculture, Forestry and Fisheries. 2014. Annual statistics of notifiable animal infectious diseases (1937–2014). Available at: http://www.maff.go.jp/j/syouan/douei/kansi_densen/pdf/h26_todokede_ruinen.pdf
- 2.Bissett S. A., Stone M. L., Malik R., Norris J. M., O’Brien C., Mansfield C. S., Nicholls J. M., Griffin A., Gookin J. L.2009. Observed occurrence of Tritrichomonas foetus and other enteric parasites in Australian cattery and shelter cats. J. Feline Med. Surg. 11: 803–807. doi: 10.1016/j.jfms.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culberson D. E., Pindak F. F., Gardner W. A., Honigberg B. M.1986. Tritrichomonas mobilensis n. sp. (Zoomastigophorea: Trichomonadida) from the Bolivian squirrel monkey Saimiri boliviensis boliviensis. J. Protozool. 33: 301–304. doi: 10.1111/j.1550-7408.1986.tb05611.x [DOI] [PubMed] [Google Scholar]
- 4.Diamond L. S., Clark C. G., Cunnick C. C.1995. YI-S, a casein-free medium for axenic cultivation of Entamoeba histolytica, related Entamoeba, Giardia intestinalis and Trichomonas vaginalis. J. Eukaryot. Microbiol. 42: 277–278. doi: 10.1111/j.1550-7408.1995.tb01579.x [DOI] [PubMed] [Google Scholar]
- 5.Doi J., Abe N., Oku Y.2013. Molecular survey of Tritrichomonas suis (=T. foetus) ‘cat’ and ‘cattle’ genotypes in pigs in Japan. J. Vet. Med. Sci. 75: 475–479. doi: 10.1292/jvms.12-0377 [DOI] [PubMed] [Google Scholar]
- 6.Doi J., Hirota J., Morita A., Fukushima K., Kamijyo H., Ohta H., Yamasaki M., Takahashi T., Katakura K., Oku Y.2012. Intestinal Tritrichomonas suis (=T. foetus) infection in Japanese cats. J. Vet. Med. Sci. 74: 413–417. doi: 10.1292/jvms.11-0171 [DOI] [PubMed] [Google Scholar]
- 7.Duboucher C., Caby S., Dufernez F., Chabé M., Gantois N., Delgado-Viscogliosi P., Billy C., Barré E., Torabi E., Capron M., Pierce R. J., Dei-Cas E., Viscogliosi E.2006. Molecular identification of Tritrichomonas foetus-like organisms as coinfecting agents of human Pneumocystis pneumonia. J. Clin. Microbiol. 44: 1165–1168. doi: 10.1128/JCM.44.3.1165-1168.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey C. F., Schild M., Hemphill A., Stünzi P., Müller N., Gottstein B., Burgener I. A.2009. Intestinal Tritrichomonas foetus infection in cats in Switzerland detected by in vitro cultivation and PCR. Parasitol. Res. 104: 783–788. doi: 10.1007/s00436-008-1255-2 [DOI] [PubMed] [Google Scholar]
- 9.Holliday M., Deni D., Gunn-Moore D. A.2009. Tritrichomonas foetus infection in cats with diarrhoea in a rescue colony in Italy. J. Feline Med. Surg. 11: 131–134. doi: 10.1016/j.jfms.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houpt E. R., Glembocki D. J., Obrig T. G., Moskaluk C. A., Lockhart L. A., Wright R. L., Seaner R. M., Keepers T. R., Wilkins T. D., Petri W. A., Jr2002. The mouse model of amebic colitis reveals mouse strain susceptibility to infection and exacerbation of disease by CD4+ T cells. J. Immunol. 169: 4496–4503. doi: 10.4049/jimmunol.169.8.4496 [DOI] [PubMed] [Google Scholar]
- 11.Jongwutiwes S., Silachamroon U., Putaporntip C.2000. Pentatrichomonas hominis in empyema thoracis. Trans. R. Soc. Trop. Med. Hyg. 94: 185–186. doi: 10.1016/S0035-9203(00)90270-0 [DOI] [PubMed] [Google Scholar]
- 12.Midlej V., Pereira-Neves A., Kist L. W., Bogo M. R., Benchimol M.2011. Ultrastructural features of Tritrichomonas mobilensis and comparison with Tritrichomonas foetus. Vet. Parasitol. 182: 171–180. doi: 10.1016/j.vetpar.2011.05.015 [DOI] [PubMed] [Google Scholar]
- 13.Okamoto S., Wakui M., Kobayashi H., Sato N., Ishida A., Tanabe M., Takeuchi T., Fukushima S., Yamada T., Ikeda Y.1998. Trichomonas foetus meningoencephalitis after allogeneic peripheral blood stem cell transplantation. Bone Marrow Transplant. 21: 89–91. doi: 10.1038/sj.bmt.1701032 [DOI] [PubMed] [Google Scholar]
- 14.Owhashi M., Tomiyoshi F., Hayashi H.1994. Isolation and characterization of a neutrophil chemotactic factor from Tritrichomonas foetus organisms. Immunol. Cell Biol. 72: 249–255. doi: 10.1038/icb.1994.37 [DOI] [PubMed] [Google Scholar]
- 15.Profizi C., Cian A., Meloni D., Hugonnard M., Lambert V., Groud K., Gagnon A. C., Viscogliosi E., Zenner L.2013. Prevalence of Tritrichomonas foetus infections in French catteries. Vet. Parasitol. 196: 50–55. doi: 10.1016/j.vetpar.2013.01.021 [DOI] [PubMed] [Google Scholar]
- 16.Reinmann K., Müller N., Kuhnert P., Campero C. M., Leitsch D., Hess M., Henning K., Fort M., Müller J., Gottstein B., Frey C. F.2012. Tritrichomonas foetus isolates from cats and cattle show minor genetic differences in unrelated loci ITS-2 and EF-1α. Vet. Parasitol. 185: 138–144. doi: 10.1016/j.vetpar.2011.09.032 [DOI] [PubMed] [Google Scholar]
- 17.Riley D. E., Samadpour M., Krieger J. N.1991. Detection of variable DNA repeats in diverse eukaryotic microorganisms by a single set of polymerase chain reaction primers. J. Clin. Microbiol. 29: 2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryan J. A.2008. Cell cloning by serial dilution in 96 well plates protocol. Available at: http://www.level.com.tw/html/ezcatfiles/vipweb20/img/img/34963/3-2Single_cell_cloning_protocol.pdf (Accessed 04 November 2015)
- 19.Slapeta J., Müller N., Stack C. M., Walker G., Lew-Tabor A., Tachezy J., Frey C. F.2012. Comparative analysis of Tritrichomonas foetus (Riedmüller, 1928) cat genotype, T. foetus (Riedmüller, 1928) cattle genotype and Tritrichomonas suis (Davaine, 1875) at 10 DNA loci. Int. J. Parasitol. 42: 1143–1149. doi: 10.1016/j.ijpara.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 20.Stockdale H. D., Givens M. D., Dykstra C. C., Blagburn B. L.2009. Tritrichomonas foetus infections in surveyed pet cats. Vet. Parasitol. 160: 13–17. doi: 10.1016/j.vetpar.2008.10.091 [DOI] [PubMed] [Google Scholar]
- 21.Tachezy J., Tachezy R., Hampl V., Sedinová M., Vanacová S., Vrlík M., Van Ranst M., Flegr J., Kuldaa J.2002. Cattle pathogen tritrichomonas foetus (Riedmüller, 1928) and pig commensal Tritrichomonas suis (Gruby & Delafond, 1843) belong to the same species. J. Eukaryot. Microbiol. 49: 154–163. doi: 10.1111/j.1550-7408.2002.tb00360.x [DOI] [PubMed] [Google Scholar]
- 22.Yule A., Skirrow S. Z., Bonduran R. H.1989. Bovine trichomoniasis. Parasitol. Today (Regul. Ed.) 5: 373–377. doi: 10.1016/0169-4758(89)90298-6 [DOI] [PubMed] [Google Scholar]
- 23.Zalonis C. A., Pillay A., Secor W., Humburg B., Aber R.2011. Rare case of trichomonal peritonitis. Emerg. Infect. Dis. 17: 1312–1313. doi: 10.3201/eid1707.100892 [DOI] [PMC free article] [PubMed] [Google Scholar]