Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is an epidemic etiology in pigs of all ages causing reproductive failure and respiratory manifestation. PRRSV has been circulating in Chinese pig farms for almost 20 years. The aim of the present study was to fully understand the extent of the genetic diversity and molecular characteristics of PRRSVs in Central China. A strain of PRRSV isolated from a recent outbreak farm in Hunan province in Central China, designated HUN-2014, was sequenced and analyzed with 39 other PRRSVs from 1998 to 2014 in Central China. Comparative results of genomic sequences revealed that all 40 PRRSVs belonged to the North American genotype (NA genotype) and shared 88.8–99.0% homology. Phylogenetic analysis showed three subgenotypes, namely conventional PRRSV (C-PRRSV), specially mutant PRRSV (S-PRRSV) and highly pathogenic PRRSV (HP-PRRSV), in all 40 PRRSVs. Moreover, comparative analysis of amino acid (AA) sequences of NSP2, GP3, GP5 and ORF5a revealed the main evolution trend of PRRSVs in Central China from 1998 to 2014, which was from C-PRRSV to HP-PRRSV, accompanied by different evolving directions to S-PRRSV. In conclusion, both the major evolutionary trend and special features of genetic variation should be emphasized as theoretical basis for development of new vaccines and control strategies for PRRS.
Keywords: analysis of mutation, Central China, epidemic, molecular characteristics, porcine reproductive and respiratory syndrome virus (PRRSV)
Porcine reproductive and respiratory syndrome virus (PRRSV) causes reproductive failure in sows and respiratory dyspnea in piglets and is responsible for tremendous financial losses worldwide [29]. The whole genome length of PRRSV is 14.9–15.5 kb, and it consists of at least 10 open reading frames (ORFs): ORF1a and ORF1b, encoding nonstructural proteins (NSPs), and ORF2–7, encoding structural proteins (SPs) [8]. PRRSV falls into two genotypes, the European genotype (EU genotype) and North American genotype (NA genotype), which share almost 60% homology at the nucleotide level, and the NA genotype was reported to be more virulent than the EU genotype in previous studies [6, 7, 15].
It has been two decades since the first Chinese PRRSV was isolated in 1995 [29], and enormous damage has been caused by PRRSV in that time, especially the outbreak of highly pathogenic PRRSV (HP-PRRSV) in 2007–2008 [23, 31] and reemergence in 2009–2010 [10, 19]. PRRSV is still one of the pandemic diseases in the Chinese pig-breeding industry [25]. In this study, we sequenced the complete genome sequence of the new PRRSV HUN-2014 strain, which was isolated in Hunan province, Central China, in February 2014, and then performed a comparative analysis with 39 other genome sequences of PRRSVs of Central China from 1998 to 2014, to investigate the evolutionary diversity of PRRSV in Central China and to better understand the epidemic characteristics of this virus.
MATERIALS AND METHODS
Ethics statement: All animal experiments were approved by the Animal Care and Use Committee of the China Institute of Veterinary Drug Control (IVDC); we followed the guidelines of the IVDC Animal Care and Use Committee in handling the experimental animals during this study.
Clinical samples: Lungs and lymph nodes were collected from suspected pigs in Hunan province, Central China, in February 2014. All of these pigs displayed typical signs of HP-PRRS, including high fever, labored breathing, pyrexia, lethargy and anorexia. Clinical tissues were homogenized for RNA extraction and virus isolation, and the remaining samples were kept at −80°C until use.
Virus isolation: MARC-145 cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) containing 8% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, U.S.A.), 100 microgram penicillin and 100 units of streptomycin per milliliter of growth medium. Lymph node homogenates were suspended in DMEM (10% v/v) and then subjected to centrifugation. The supernatant was filtered (0.22 µm filter) and then applied to inoculate MARC-145 cells. Then, the isolated viruses were amplified at 37°C with 5% CO2 and monitored daily for cytopathic effects (CPEs). The culture supernatants were harvested when CPEs appeared in 80% of the cells and stored at −80°C as the virus stock until use.
RNA isolation and RT-PCR: Viral RNA was extracted using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) and dissolved in nuclease-free water. Reverse transcription PCR (RT-PCR) was carried out with a PrimeScriptTM One Step RT-PCR Kit (Takara, Dalian, China) according to the manufacturer’s instructions. Four microliter (µl) aliquots of the RNA template were added to 46 µl of the RT-PCR master mix. The cycling conditions were 94°C for 5 min; 30 cycles of denaturation (95°C for 30 sec), annealing (58°C for 30 sec) and extension (72°C for 3 min); and then a final extension at 72°C for 10 min. The primer sets (Table 1) used in RT-PCR were as reported in previous research [14].
Table 1. Primer sets used in this study.
| Oligonucleotides | Sequence | Location |
|---|---|---|
| 1F | ATGACGTATAGGTGTTGGCTCT | 1–22 |
| 1R | CTTACTCTTTCAGGAAGGGTGGT | 1,555–1,577 |
| 2F | AAAACACGCTCTGGTGCGACTAC | 1,357–1,379 |
| 2R | GAGATGGGAAACGAGGCTGAAAAC | 3,793–3,816 |
| 3F | ATGATAGTTCCGCCCGCAGATAC | 3,649–3,671 |
| 3R | GGGTGACGAGACCAGCAATGTTAG | 5,351–5,374 |
| 4F | TGCTTGCTGGTGTTTATGTGACTG | 4,889–4,912 |
| 4R | GCCTCGGACCTTATCAACCTGTA | 6,866–6,888 |
| 5F | GAGACTCACTGACGAGGACTTGGAT | 6,720–6,744 |
| 5R | CAGGCGAGTTCATAAAGAAGATTGG | 8,833–8,857 |
| 6F | TGTGCGAGAAAACTGGCAAACTG | 8,546–8,568 |
| 6R | GCCCTGGTGATAGCAACAAGAGC | 10,596–10,618 |
| 7F | TTCAACCAGATTACAGGGACAAACT | 10,387–10,411 |
| 7R | ATTAGCCATTGCTGAAAATCGTG | 12,592–12,614 |
| 8F | CATTGTCTCGCATTAGTGGTTTG | 12,386–12,408 |
| 8R | CGATAGAGTCTGCCCTTAGTGTC | 14,136–14,158 |
| 9F | CGCTGATTTGCTTTGTCATTAGG | 14,053–14,075 |
| 9R | GCACGGTTCTCGCCAATTATACT | 15,288–15,310 |
Genome cloning and sequencing: The amplified PCR products were subjected to agarose gel electrophoresis and excised from the agarose gel for later purification, which was performed by using a E.Z.N.A. Gel Extraction Kit (OMEGA Bio-tek, Norcross, GA, U.S.A.). The PCR products were cloned into pMD18-T vector according to the manufacturer’s instructions (Takara) and sequenced. For each amplified genomic region, three clones were sequenced in both directions by Life Technologies (Shanghai, China), and the results of sequencing were analyzed.
Genome and amino acid analysis: After sequencing nine fragments, each fragment was spliced together with an overlapping sequence, obtaining the complete genome sequence of HUN-2014. Every ORF and most of the derived amino acids were compared with other isolates. Phylogenetic trees were constructed with MEGA (Version 5.1) using the neighbor-joining method. Bootstrap values were calculated on 1,000 replicates of the alignment. The evolutionary trend of PRRSV in China was analyzed based on nucleotide sequences of HUN-2014 and other known isolates of Central China (Table 2). Multiple sequence alignments were generated with the DNAMAN software (Version 5.1). The nucleotide and amino acid sequence homologies of HUN-2014 with twelve other PRRSVs of Central China were assess further using the BioEdit software (Version 7.0) (Table 3). To explore the genetic variation of PRRSVs of Central China, comparison of NSP2, GP3, GP5 and ORF5a of the HUN-2014 isolate with 39 other strains of Central China were performed.
Table 2. PRRSV strains used in this study.
| No. | Isolate | Identity (%)a) | Province | Year | Access no. | No. | Isolate | Identity (%)a) | Province | Year | Access no. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | S1 | 89.1 | Henan | 1998 | DQ459471 | 21 | 09HEN2 | 97.7 | Henan | 2009 | JF268680 |
| 2 | HN1 | 88.8 | Henan | 2002 | AY457635 | 22 | 09HUN1 | 97.6 | Hunan | 2009 | JF268673 |
| 3 | HuN | 98.4 | Hunan | 2006 | EF517962 | 23 | 09HUN2 | 98.0 | Hunan | 2009 | JF268674 |
| 4 | HN-HW | 98.7 | Hunan | 2006 | FJ797690 | 24 | WUH4 | 99.0 | Hubei | 2011 | JQ326271 |
| 5 | HUN4 | 98.9 | Hunan | 2006 | EF635006 | 25 | HZ-31 | 95.2 | Hubei | 2012 | KC445138 |
| 6 | HUB1 | 98.6 | Hubei | 2006 | EF075945 | 26 | HeNan-A1 | 98.5 | Henan | 2013 | KJ002451 |
| 7 | HUB2 | 98.7 | Hubei | 2006 | EF112446 | 27 | HeNan-A2 | 96.6 | Henan | 2013 | KJ002452 |
| 8 | WUH1 | 98.1 | Hubei | 2006 | EU187484 | 28 | Henan-A3 | 96.9 | Henan | 2013 | KJ019330 |
| 9 | WUH2 | 97.7 | Hubei | 2006 | EU678352 | 29 | Henan-A4 | 97.0 | Henan | 2013 | KJ534539 |
| 10 | Henan-1 | 98.6 | Henan | 2007 | EU200962 | 30 | Henan-A5 | 97.0 | Henan | 2013 | KJ534540 |
| 11 | 07HEN | 98.6 | Henan | 2007 | FJ393457 | 31 | Henan-A6 | 97.2 | Henan | 2013 | KJ534541 |
| 12 | HN2007 | 98.5 | Henan | 2007 | EU880437 | 32 | Henan-A7 | 97.5 | Henan | 2013 | KJ534542 |
| 13 | Em2007 | 93.1 | Hubei | 2007 | EU262603 | 33 | Henan-A8 | 97.5 | Henan | 2013 | KJ534543 |
| 14 | 08HuN | 98.6 | Hunan | 2008 | GU169411 | 34 | Henan-A9 | 96.2 | Henan | 2013 | KJ546412 |
| 15 | WUH3 | 98.6 | Hubei | 2008 | HM853673 | 35 | Henan-A10 | 97.2 | Henan | 2013 | KJ609516 |
| 16 | 09HUB1 | 97.9 | Hubei | 2009 | JF268682 | 36 | Henan-A11 | 96.2 | Henan | 2013 | KJ609517 |
| 17 | 09HUB2 | 97.9 | Hubei | 2009 | JF268683 | 37 | Henan-A12 | 97.2 | Henan | 2014 | KJ819934 |
| 18 | 09HUB5 | 97.9 | Hubei | 2009 | GU168568 | 38 | Henan-A13 | 97.0 | Henan | 2014 | KJ819935 |
| 19 | 09HUB7 | 97.6 | Hubei | 2009 | GU168567 | 39 | Henan-A14 | 96.7 | Henan | 2014 | KJ819936 |
| 20 | 09HEN1 | 97.7 | Henan | 2009 | JF268684 | 40 | HUN-2014 | 100 | Hunan | 2014 | KP330232 |
a) Identity with PRRSV HUN-2014 strain.
Table 3. Detailed comparison of the full-length genomes of HUN-2014 with 12 reference strains of PRRSV of Central China.
| HUN-2014 | HN1 | HUN4 | HUB1 | Em2007 | 09HUB2 | 09HEN2 | HZ-31 | Henan-A4 | Henan-A5 | Henan-A8 | Henan-A10 | Henan-A11 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % identity to HUN-2014 | ||||||||||||
| Nucleotides (length) | ||||||||||||
| 5′ UTR (189) | 88.2 | 99.5 | 99.5 | 94.7 | 98.4 | 98.4 | 98.4 | 98.9 | 98.4 | 98.9 | 98.9 | 99.5 |
| ORF1a (7,422) | 86.5 | 98.8 | 98.4 | 92.7 | 97.6 | 97.4 | 96.5 | 96.8 | 96.2 | 96.8 | 97.9 | 95.7 |
| ORF1b (4,385) | 91.1 | 99.0 | 99.1 | 94.0 | 98.4 | 97.9 | 97.8 | 97.4 | 97.5 | 98.4 | 98.3 | 97.4 |
| ORF2-7 (3,188) | 90.9 | 98.7 | 98.5 | 92.8 | 98.1 | 98.1 | 88.4 | 96.5 | 98.2 | 97.8 | 93.7 | 95.5 |
| 3′ UTR (165) | 93.3 | 98.7 | 97.3 | 91.3 | 98.0 | 97.3 | 96.0 | 96.7 | 96.7 | 96.7 | 97.3 | 98.7 |
| Complete (15,336) | 88.8 | 98.9 | 98.6 | 93.1 | 97.9 | 97.7 | 95.2 | 97.0 | 97.0 | 97.5 | 97.2 | 96.2 |
| Amino acids (length) | ||||||||||||
| NSP1a (166) | 95.8 | 100 | 100 | 97.0 | 100 | 100 | 98.8 | 100 | 100 | 99.4 | 99.4 | 99.4 |
| NSP1b (217) | 82.9 | 98.6 | 97.7 | 87.1 | 96.3 | 97.2 | 92.2 | 94.9 | 96.8 | 98.2 | 97.7 | 97.2 |
| NSP2 (950) | 75.7 | 97.7 | 97.2 | 91.8 | 89.9 | 95.4 | 93.7 | 93.9 | 93.8 | 92.6 | 95.5 | 92.0 |
| NSP3 (446) | 94.4 | 99.8 | 99.3 | 96.4 | 99.6 | 99.1 | 98.7 | 98.2 | 98.9 | 98.0 | 99.6 | 98.7 |
| NSP4 (204) | 94.1 | 100 | 99.0 | 96.1 | 100 | 100 | 99.5 | 99.0 | 98.0 | 99.0 | 100 | 99.0 |
| NSP5 (170) | 92.4 | 98.8 | 98.2 | 93.5 | 98.2 | 98.2 | 94.7 | 97.1 | 96.5 | 97.6 | 98.2 | 97.1 |
| NSP6 (16) | 93.8 | 100 | 100 | 100 | 93.8 | 100 | 100 | 93.8 | 100 | 100 | 100 | 100 |
| NSP7 (259) | 88.8 | 98.8 | 98.8 | 90.7 | 98.8 | 98.8 | 98.1 | 97.7 | 97.7 | 98.1 | 96.5 | 97.7 |
| NSP8 (45) | 97.8 | 100 | 100 | 100 | 100 | 100 | 97.8 | 97.8 | 97.8 | 100 | 100 | 97.8 |
| NSP9 (643) | 97.5 | 99.1 | 99.5 | 97.7 | 98.9 | 98.9 | 98.9 | 98.1 | 98.4 | 98.4 | 98.9 | 98.1 |
| NSP10 (441) | 96.4 | 99.8 | 99.5 | 96.8 | 99.5 | 99.8 | 99.3 | 98.6 | 99.3 | 99.5 | 99.8 | 99.5 |
| NSP11 (223) | 94.6 | 99.1 | 99.6 | 95.5 | 99.1 | 99.6 | 99.1 | 98.2 | 99.1 | 98.2 | 99.1 | 98.7 |
| NSP12 (153) | 94.8 | 100 | 99.3 | 97.4 | 100 | 98.0 | 99.3 | 100 | 100 | 100 | 98.0 | 97.4 |
| GP2 (256) | 91.4 | 97.3 | 97.7 | 92.2 | 97.3 | 96.5 | 86.7 | 94.9 | 97.7 | 97.7 | 93.1 | 93.5 |
| E (73) | 93.2 | 98.6 | 98.6 | 93.2 | 98.6 | 98.6 | 90.4 | 98.6 | 98.6 | 97.3 | 97.3 | 97.3 |
| GP3 (255) | 85.4 | 96.9 | 96.5 | 88.6 | 96.9 | 96.9 | 83.5 | 93.3 | 97.2 | 96.1 | 90.6 | 90.9 |
| GP4 (178) | 88.8 | 97.2 | 96.6 | 94.4 | 96.6 | 96.6 | 87.1 | 95.5 | 96.6 | 95.5 | 96.6 | 96.6 |
| ORF5a (47) | 76.1 | 93.5 | 93.5 | 80.4 | 93.5 | 93.5 | 71.7 | 89.1 | 93.5 | 91.3 | 89.1 | 89.1 |
| GP5 (200) | 86.0 | 97.0 | 97.0 | 91.5 | 96.0 | 96.0 | 83.5 | 93.5 | 95.0 | 96.0 | 90.0 | 94.0 |
| M (174) | 97.7 | 100 | 100 | 96.6 | 100 | 100 | 94.8 | 99.4 | 100 | 99.4 | 98.3 | 100 |
| N (123) | 91.9 | 97.6 | 97.6 | 89.4 | 96.7 | 97.6 | 93.5 | 97.6 | 95.9 | 95.1 | 95.1 | 96.7 |
RESULTS
Analysis of full-length genomic sequences of HUN-2014: The sequence data showed that, excluding the poly (A) tail, the genomic sequence of HUN-2014 was 15,336 nucleotides (nt) in length, consisting of a 189-nt 5′UTR and a 165-nt 3′UTR, and the genome sequence of the HUN-2014 strain has been submitted to GenBank under the accession No. KP330232. HUN-2014 was identified as an HP-PRRSV of the NA genotype that possessed the genetic marker of a 1+29 AA deletion in NSP2.
Analysis on homology of PRRSV in Central China: Comparative analysis of the whole genome sequence and amino acid sequences analysis suggested that all 40 PRRSVs of Central China belonged to the NA genotype and shared 88.8–99.0% homology (Table 2); In addition, we found that more conserved proteins of PRRSV in Central China were NSP1a, NSP4, NSP6, NSP8, NSP12 and M, whereas less conserved proteins were NSP1b, NSP2, GP3, GP5 and ORF5a (Table 3).
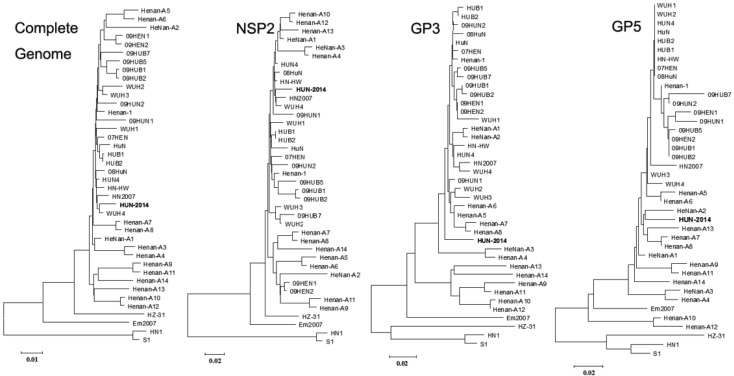
Phylogenetic analysis: Phylogenetic trees were built by applying the nucleotide sequences of the whole genome sequences and amino acid sequences of NSP2, GP3 and GP5 (Fig. 1). Phylogenetic analysis revealed that the 40 PRRSVs of Central China could be categorized into three subgenotypes, namely C-PRRSV, the reference strain of which was HN1, which was isolated between 1996 and 2005 [29]; S-PRRSV, reference strains of which were Em2007 and HZ-31, which possessed specific features of mutation and was considered to be strains of the virus generated by recombination between HP-PRRSV and vaccine for C-PRRSV [11, 18]; and HP-PRRSV, reference strains of which were WUH4 and HUN-2014, which were all isolated after 2006 and contain the consistent gene marker of a 1+29 AA deletion in NPS2 [23, 31].
Fig. 1.
Phylogenetic trees of the 40 PRRSV isolates of Central China based on the complete genomic sequences and amino acids sequences of NSP2, GP3 and GP5. Four unrooted neighbor-joining trees were constructed from the aligned complete genomic sequences and amino acids sequences of NSP2, GP3, GP5 and ORF5a of the 40 PRRSVs of Central China by the distance-based neighbor-joining method using the MEGA software (Version 5.1). Bootstrap values were calculated on 1,000 replicates of the alignment.
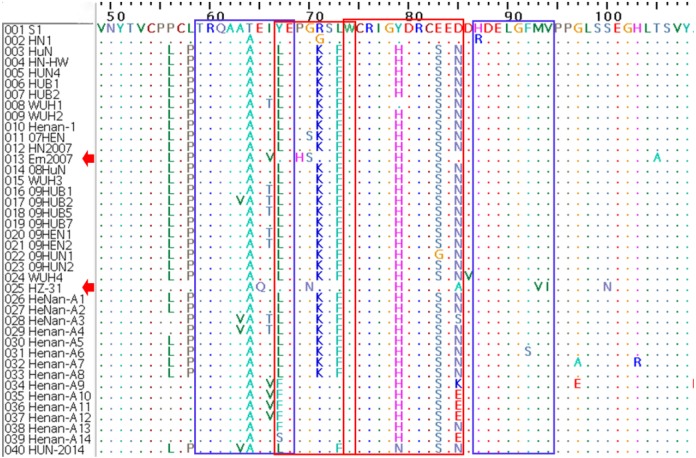
Amino acid analysis of NSP2: NSP2 is the most variable protein in the viral genome of PRRSV and is a multifunctional protein participating in modulation of the host inflammatory response [2], and it is closely related to the replication ability of PRRSV [24]. By comparing the NSP2 amino acid sequences of the 40 PRRSVs of Central China, we found 75.7–97.7% identity between HUN-2014 and the 39 other PRRSVs of Central China (Table 3). Moreover, the genetic marker of a 1+29 AA deletion could be seen in all the 38 PRRSVs isolated since 2006 (denoted with red boxes in Fig. 2), but five of them exhibited special mutations in NSP2, such as a discontinuous 37+2+29 deletion in Em2007; 30+29 deletion in 09HUB1, 09HUB2 and HZ-31; and 49+29 deletion in Henan-A14 (denoted with blue boxes in Fig. 2).
Fig. 2.
Amino acid sequence alignments of the partial NSP2 gene of the 40 PRRSVs of Central China. The discontinuous 1+29 AA deletion regions are indicated by red boxes, and special deletions are indicated by blue boxes.
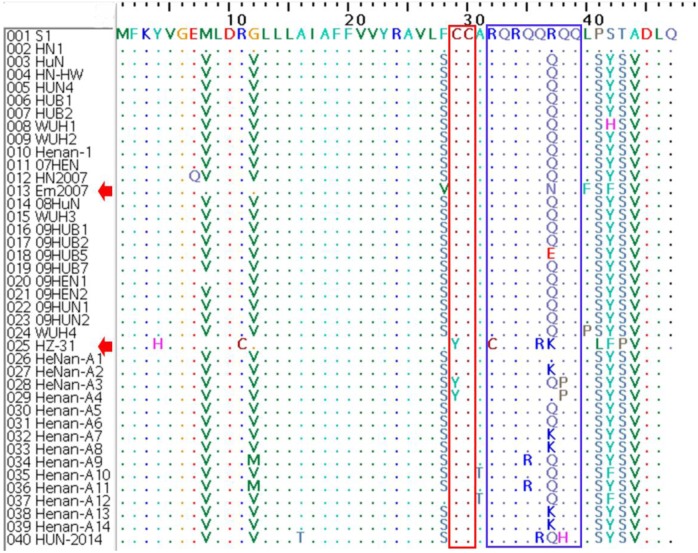
Amino acid analysis of GP3: GP3 has been regarded as a less conserved structural protein PRRSV that interacts with GP2 and GP4, forming multiprotein complexes that are believed to be crucial for the assembly of infectious PRRSV [1]. After comparing the GP3 amino acid sequences of the 40 PRRSVs of Central China, we found 83.5–97.2% identity between HUN-2014 and the 39 other PRRSVs of Central China (Table 3). Previous studies showed that three antigenic epitopes of GP3, 67YEPGRSLW74 (denoted with a red box in Fig. 3), 74WCRIGHDRCGED85 (denoted with a red box in Fig. 3) and 87HDELGFMV94 (denoted with a blue box in Fig. 3), were well conserved among most of the NA-type isolates, whereas the epitope 59TRQAAAEILE68 (denoted with a blue box in Fig. 3) differed in some NA-genotype strains [1, 30]. In this study, most of the HP-PRRSVs were found to have relatively consistent mutations (T64A, Y67L/F, R71K, L73F, Y79H, E83S & D85N/E) in three of the abovementioned epitopes and four sporadic mutations in the relatively conserved epitope (Fig. 3).
Fig. 3.
Amino acid sequence alignments of the partial GP3 gene of the 40 PRRSVs of Central China. The two epitopes, 67YEPGRSLW74 and 74WCRIGHDRCGED85, are denoted by red boxes, and the two epitopes, 87HDELGFMV94 and 59TRQAAAEILE68, are denoted by blue boxes.
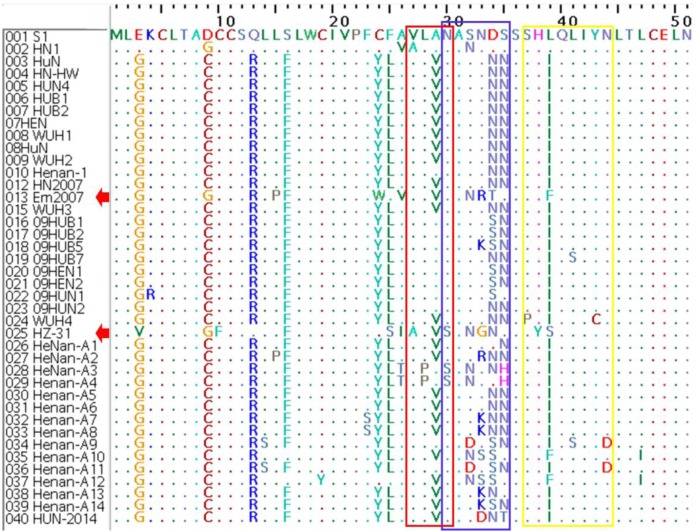
Amino acid analysis of GP5: GP5 is the most heterogeneous structural protein of PRRSV [5], and it could induce IFN-β production in host cells and played a significant role during viral attachment and internalization [4, 13]. After comparing the GP5 amino acid sequences of the 40 PRRSVs of Central China, 83.5–97.0% identity was found between HUN-2014 and the 39 other PRRSVs of Central China (Table 3). A previous study recognized that GP5 and ORF5a coevolved through a fine balance of purifying codon usage to maintain a conserved RQ-rich motif in the ORF5a protein while eliciting a variable N-linked glycosylation motif (30NASNDS35) (denoted with a blue box in Fig. 4) in the alternative GP5 reading frame [16]. In this work, four relatively identical mutations (A29V, D34N/S, S35N and L39I) were seen from C-PRRSV to most of the HP-PRRSVs in the decoy epitope (denoted with a red box in Fig. 4), the abovementioned N-linked glycosylation motif and primary neutralizing epitope (PNE) (denoted with a yellow box in Fig. 4), yet particular mutations observed in S-PRRSV as Em2007 and HZ-31.
Fig. 4.
Amino acid sequence alignments of the partial GP5 gene of the 40 PRRSVs of Central China. The decoy epitope (27V/ALVN30) is denoted with a red box, the primary neutralizing epitope (PNE) (37SHL/IQLIYNL45) is denoted with a yellow box, and the variable N-linked glycosylation motif (30NASNDS35) is denoted with a blue box.
Amino acid analysis of ORF5a: ORF5a is the newest identified structural protein of PRRSV (identified in 2011) and is encoded by an alternative ORF5a that is present in all Arteriviruses [3, 9]. The homology of ORF5a in the 40 PRRSVs of Central China is 71.7–93.5%. Previous reports suggested that ORF5a was essential for virus viability and infectivity [17, 20], whereas another study reported that the ORF5a antibody response is neither neutralizing nor protective against PRRSV [22]. The ORF5a protein possessed two cysteines at positions 29 and 30 that are highly conserved among NA genotype PRRSVs, and a previous study revealed that replacement of cysteine with glycine at position 30 caused the ORF5a protein to interact non-covalently with itself, which may account for the lethal phenotype of mutants carrying substitution of cysteine to glycine at position 30 [21]. No mutation existed at position 30 in this study (denoted with a red box in Fig. 5); additionally, the mutation R37Q/K was observed in the abovementioned RQ-rich motif [16] in most PRRSVs (denoted with a blue box in Fig. 5).
Fig. 5.
Amino acid sequence alignments of the partial ORF5a gene of the 40 PRRSVs of Central China. The region of highly conserved cysteines is denoted with a red box, and the RQ-rich motif is denoted with a blue box.
DISCUSSION
Central China is composed of the three provinces, Henan, Hubei and Hunan, that account for almost 23% of the national pig-breeding industry [26], so work related to surveillance and prevention of major porcine epidemic diseases in Central China is essential for the whole pig industry, especially for pandemic disease like PRRS [12, 29]. PRRSV is a vital pathogen that has caused titanic losses for the global swine industry since its first occurrence in the 1980s. The first Chinese PRRSV strain was isolated in 1995, namely C-PRRSV, and the reference strains, HN1 and S1, were isolated between 1996 and 2005 [12]. The first outbreak of HP-PRRSV occurred in 2006, and it resulted in higher morbidity and mortality than C-PRRSV; this HP-PRRSV possessed the genetic marker of a discontinuous 1+29 AA deletion in NSP2, although the 1+29 AA deletion has subsequently been verified as not related to the high virulence of HP-PRRSV [28]. HP-PRRSV reemerged in 2009 with the same pathogenicity but limited mutations [27, 31], and PRRS has frequently been reported since then [8, 9], which reminds us that the work of monitoring and taking precautions against PRRSV should not be slackened.
On the basis of the whole genome sequences of PRRSV HUN-2014 and the 39 other PRRSVs of Central China from 1998 to 2014, we performed a phylogenetic analysis and a comparative analysis with regard to NSP2, GP3, GP5 and ORF5a. The results demonstrated that these 40 PRRSVs of Central China could be divided into three branches, namely C-PRRSV, S-PRRSV and HP-PRRSV. Since C-PRRSV possessed significantly lower virulence than HP-PRRSV, we tried to seek the characteristics of variation between these 40 PRRSVs, and three particular features were discovered based on Fig. 2 to Fig. 5: (1) abundant concurrent mutations evolved from C-PRRSV to HP-PRRSV, (2) there were diverse mutations at one position from C-PRRSV to S-PRRSV/HP-PRRSV, and (3) there were extraordinary mutations in S-PRRSV but HP-PRRSV. Given this, we speculated that C-PRRSV evolved sophisticated mechanisms to subvert the host defense system by encoding proteins that target key components of the immune signaling pathways in S-PRRSV/HP-PRRSV.
Furthermore, there seems to be numerous concurrent mutations in some particular areas between C-PRRSV and HP-PRRSV, such as the region of 179–782 AA in NSP2, which is associated with the virulence of PRRSV [2]; three conserved epitopes in GP3 that are related to the antigenicity of PRRSV [1, 30]; the decoy epitope and PNE in GP5; and the RQ-rich motif in ORF5a [16], and some of these mutations may be correlated with the virulence or other ability of PRRSV.
To sum up, the genetic diversity of PRRSV should be recognized as a serious issue, and the major variation in PRRSVs should be emphasized, and taken into account along with the crucial changes in phenotype and applied to formulating of preventive for PRRSV.
Acknowledgments
This work was supported by the National Scientific Supporting Program (No. 2007BAD86B01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Chen J. Z., Wang Q., Bai Y., Wang B., Zhao H. Y., Peng J. M., An T. Q., Tian Z. J., Tong G. Z.2014. Identification of two dominant linear epitopes on the GP3 protein of highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV). Res. Vet. Sci. 97: 238–243. doi: 10.1016/j.rvsc.2014.07.011 [DOI] [PubMed] [Google Scholar]
- 2.Fang Y., Fang L., Wang Y., Lei Y., Luo R., Wang D., Chen H., Xiao S.2012. Porcine reproductive and respiratory syndrome virus nonstructural protein 2 contributes to NF-κB activation. Virol. J. 9: 83. doi: 10.1186/1743-422X-9-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Firth A. E., Zevenhoven-Dobbe J. C., Wills N. M., Go Y. Y., Balasuriya U. B., Atkins J. F., Snijder E. J., Posthuma C. C.2011. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J. Gen. Virol. 92: 1097–1106. doi: 10.1099/vir.0.029264-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao J., Ji P., Zhang M., Wang X., Li N., Wang C., Xiao S., Mu Y., Zhao Q., Du T., Sun Y., Hiscox J. A., Zhang G., Zhou E. M.2014. GP5 expression in Marc-145 cells inhibits porcine reproductive and respiratory syndrome virus infection by inducing beta interferon activity. Vet. Microbiol. 174: 409–418. doi: 10.1016/j.vetmic.2014.09.030 [DOI] [PubMed] [Google Scholar]
- 5.Han J., Wang Y., Faaberg K. S.2006. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 122: 175–182. doi: 10.1016/j.virusres.2006.06.003 [DOI] [PubMed] [Google Scholar]
- 6.Han K., Seo H. W., Oh Y., Kang I., Park C., Chae C.2013. Comparison of the virulence of European and North American genotypes of porcine reproductive and respiratory syndrome virus in experimentally infected pigs. Vet. J. 195: 313–318. doi: 10.1016/j.tvjl.2012.06.035 [DOI] [PubMed] [Google Scholar]
- 7.Han K., Seo H. W., Park C., Kang I., Youn S. K., Lee S. Y., Kim S. H., Chae C.2014. Comparative virulence of reproductive diseases caused by type 1 (European-like) and type 2 (North American-like) porcine reproductive and respiratory syndrome virus in experimentally infected pregnant gilts. J. Comp. Pathol. 150: 297–305. doi: 10.1016/j.jcpa.2013.11.205 [DOI] [PubMed] [Google Scholar]
- 8.Huang C., Zhang Q., Feng W. H.2015. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res. 202: 101–111. doi: 10.1016/j.virusres.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson C. R., Griggs T. F., Gnanandarajah J., Murtaugh M. P.2011. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J. Gen. Virol. 92: 1107–1116. doi: 10.1099/vir.0.030213-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leng C. L., Tian Z. J., Zhang W. C., Zhang H. L., Zhai H. Y., An T. Q., Peng J. M., Ye C., Sun L., Wang Q., Sun Y., Li L., Zhao H. Y., Chang D., Cai X. H., Zhang G. H., Tong G. Z.2014. Characterization of two newly emerged isolates of porcine reproductive and respiratory syndrome virus from Northeast China in 2013. Vet. Microbiol. 171: 41–52. doi: 10.1016/j.vetmic.2014.03.005 [DOI] [PubMed] [Google Scholar]
- 11.Li B., Fang L., Xu Z., Liu S., Gao J., Jiang Y., Chen H., Xiao S.2009. Recombination in vaccine and circulating strains of porcine reproductive and respiratory syndrome viruses. Emerg. Infect. Dis. 15: 2032–2035. doi: 10.3201/eid1512.090390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B., Fang L., Liu S., Zhao F., Jiang Y., He K., Chen H., Xiao S.2010. The genomic diversity of Chinese porcine reproductive and respiratory syndrome virus isolates from 1996 to 2009. Vet. Microbiol. 146: 226–237. doi: 10.1016/j.vetmic.2010.05.011 [DOI] [PubMed] [Google Scholar]
- 13.Li J., Tao S., Orlando R., Murtaugh M. P.2015. N-glycosylation profiling of porcine reproductive and respiratory syndrome virus envelope glycoprotein 5. Virology 478: 86–98. doi: 10.1016/j.virol.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo J., Liu Y., Ning Y., Chen J.2010. Sequencing and analysis of complete genome of porcine reproductive and respiratory syndrome virus GD strain and BJ strain. Acta Agriculturae Boreali-Occidentalis Sinica 19: 17–21(In Chinese). [Google Scholar]
- 15.Martínez-Lobo F. J., Díez-Fuertes F., Segalés J., García-Artiga C., Simarro I., Castro J. M., Prieto C.2011. Comparative pathogenicity of type 1 and type 2 isolates of porcine reproductive and respiratory syndrome virus (PRRSV) in a young pig infection model. Vet. Microbiol. 154: 58–68. doi: 10.1016/j.vetmic.2011.06.025 [DOI] [PubMed] [Google Scholar]
- 16.Robinson S. R., Abrahante J. E., Johnson C. R., Murtaugh M. P.2013. Purifying selection in porcine reproductive and respiratory syndrome virus ORF5a protein influences variation in envelope glycoprotein 5 glycosylation. Infect. Genet. Evol. 20: 362–368. doi: 10.1016/j.meegid.2013.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson S. R., Figueiredo M. C., Abrahante J. E., Murtaugh M. P.2013. Immune response to ORF5a protein immunization is not protective against porcine reproductive and respiratory syndrome virus infection. Vet. Microbiol. 164: 281–285. doi: 10.1016/j.vetmic.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen J., Yan X., Dong J., Jiang Y., Fang L., Xiao S., Chen H.2013. Complete genome sequence of a novel deletion porcine reproductive and respiratory syndrome virus strain. Genome Announc. 1: e00486–e13. doi: 10.1128/genomeA.00486-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi Y., Hu Z., Xiong Z., Zhou Y., Jin X., Gu C., Hu X., Cheng G., Song N., Zhang W.2013. Analysis of molecular variation of porcine reproductive and respiratory syndrome virus in Central China from 2006 to 2012. Arch. Virol. 158: 717–721. doi: 10.1007/s00705-012-1542-1 [DOI] [PubMed] [Google Scholar]
- 20.Sun L., Li Y., Liu R., Wang X., Gao F., Lin T., Huang T., Yao H., Tong G., Fan H., Wei Z., Yuan S.2013. Porcine reproductive and respiratory syndrome virus ORF5a protein is essential for virus viability. Virus Res. 171: 178–185. doi: 10.1016/j.virusres.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 21.Sun L., Zhou Y., Liu R., Li Y., Gao F., Wang X., Fan H., Yuan S., Wei Z., Tong G.2015. Cysteine residues of the porcine reproductive and respiratory syndrome virus ORF5a protein are not essential for virus viability. Virus Res. 197: 17–25. doi: 10.1016/j.virusres.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 22.Tian D., Wei Z., Zevenhoven-Dobbe J. C., Liu R., Tong G., Snijder E. J., Yuan S.2012. Arterivirus minor envelope proteins are a major determinant of viral tropism in cell culture. J. Virol. 86: 3701–3712. doi: 10.1128/JVI.06836-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian K., Yu X., Zhao T., Feng Y., Cao Z., Wang C., Hu Y., Chen X., Hu D., Tian X., Liu D., Zhang S., Deng X., Ding Y., Yang L., Zhang Y., Xiao H., Qiao M., Wang B., Hou L., Wang X., Yang X., Kang L., Sun M., Jin P., Wang S., Kitamura Y., Yan J., Gao G. F.2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2: e526. doi: 10.1371/journal.pone.0000526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F. X., Song N., Chen L. Z., Cheng S. P., Wu H., Wen Y. J.2013. Non-structural protein 2 of the porcine reproductive and respiratory syndrome (PRRS) virus: a crucial protein in viral pathogenesis, immunity and diagnosis. Res. Vet. Sci. 95: 1–7. doi: 10.1016/j.rvsc.2013.03.015 [DOI] [PubMed] [Google Scholar]
- 25.Wang X., He K., Zhang W., Zhou Z., Mao A., Yu Z.2014. Genetic diversity and phylogenetic analysis of porcine reproductive and respiratory syndrome virus isolates in East China. Infect. Genet. Evol. 24: 193–201. doi: 10.1016/j.meegid.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 26.Wu X., Chen L., Pan H. M.2013. Dynamic trend of thirty years on pig-breeding in China. Chin. J. Ani. Sci. 16: 7–10(in Chinese). [Google Scholar]
- 27.Yu X., Chen N., Wang L., Wu J., Zhou Z., Ni J., Li X., Zhai X., Shi J., Tian K.2012. New genomic characteristics of highly pathogenic porcine reproductive and respiratory syndrome viruses do not lead to significant changes in pathogenicity. Vet. Microbiol. 158: 291–299. doi: 10.1016/j.vetmic.2012.02.036 [DOI] [PubMed] [Google Scholar]
- 28.Zhou L., Zhang J., Zeng J., Yin S., Li Y., Zheng L., Guo X., Ge X., Yang H.2009. The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J. Virol. 83: 5156–5167. doi: 10.1128/JVI.02678-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L., Yang H.2010. Porcine reproductive and respiratory syndrome in China. Virus Res. 154: 31–37. doi: 10.1016/j.virusres.2010.07.016 [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y. J., An T. Q., He Y. X., Liu J. X., Qiu H. J., Wang Y. F., Tong G.2006. Antigenic structure analysis of glycosylated protein 3 of porcine reproductive and respiratory syndrome virus. Virus Res. 118: 98–104. doi: 10.1016/j.virusres.2005.11.019 [DOI] [PubMed] [Google Scholar]
- 31.Zhou Z., Ni J., Cao Z., Han X., Xia Y., Zi Z., Ning K., Liu Q., Cai L., Qiu P., Deng X., Hu D., Zhang Q., Fan Y., Wu J., Wang L., Zhang M., Yu X., Zhai X., Tian K.2011. The epidemic status and genetic diversity of 14 highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) isolates from China in 2009. Vet. Microbiol. 150: 257–269. doi: 10.1016/j.vetmic.2011.02.013 [DOI] [PubMed] [Google Scholar]