Abstract
Rat cytochrome P450 (CYP) exhibits inter-strain differences, but their analysis has been scattered across studies under different conditions. To identify these strain differences in CYP more comprehensively, mRNA expression, protein expression and metabolic activity among Wistar (WI), Sprague Dawley (SD), Dark Agouti (DA) and Brown Norway (BN) rats were compared. The mRNA level and enzymatic activity of CYP1A1 were highest in SD rats. The rank order of Cyp3a2 mRNA expression mirrored its protein expression, i.e., DA>BN>SD>WI, and was similar to the CYP3A2-dependent warfarin metabolic activity, i.e., DA>SD>BN>WI. These results suggest that the strain differences in CYP3A2 enzymatic activity are caused by differences in mRNA expression. Cyp2b1 mRNA levels, which were higher in DA rats, did not correlate with its protein expression or enzymatic activity. This suggests that the strain differences in enzymatic activity are not related to Cyp2b1 mRNA expression. In conclusion, WI rats tended to have the lowest CYP1A1, 2B1 and 3A2 mRNA expression, protein expression and enzymatic activity among the strains. In addition, SD rats had the highest CYP1A1 mRNA expression and activity, while DA rats had higher CYP2B1 and CYP3A2 mRNA and protein expression. These inter-strain differences in CYP could influence pharmacokinetic considerations in preclinical toxicological studies.
Keywords: cytochrome P450, metabolism, Rattus norvegicus, regulation, strain difference
Cytochrome P450 (CYP) is one of the key enzymes in drug metabolism. Although the laboratory rat (Rattus norvegicus) has been widely used in the many science fields, including pharmacology and toxicology, it is well known that there are inter-strain differences in CYP in terms of their nucleotide sequence, mRNA and protein expression, as well as their enzymatic activity [23]. For instance, it has been reported that there are strain differences in the mRNA expression and enzymatic activity of CYP between Sprague Dawley (SD) and Wistar (WI) rats [13]. Previous research has also revealed that Dark Agouti (DA) rats have higher CYP3A protein expression and activity than WI rats [16] and that DA rats have higher CYP3A enzymatic activity and mRNA expression than SD rats [12].
Strain differences in CYP have been proposed to be responsible for the strain differences in drug toxicity and drug sensitivity in vivo. There are differences in the hepatotoxicity and nephrotoxicity of acetaminophen between SD rats and Fischer (F344) rats [19]; the toxicity of acetaminophen is attributable to CYP3A metabolic activation [15]. However, in most reports, comparisons were only performed between two strains, and a comprehensive examination of mRNA expression, protein expression and enzymatic activity has not yet been performed.
Therefore, in the current study, we aimed to identify strain differences in CYP by comparing its mRNA expression, protein expression and metabolic activity in vitro in SD, WI, Brown Norway (BN) and DA rats. Analyzing these strains may have ramifications in many fields: WI and SD rats are often used in toxicological studies for pharmaceutical development. DA rats are often used for research in allergic encephalomyelitis and rheumatoid arthritis. The genome of the BN rat has been fully sequenced, so the strain is often used for genetic research. The current study demonstrating the inter-strain differences in CYP could provide further knowledge on pharmacokinetic considerations in preclinical toxicological studies.
MATERIALS AND METHODS
Animals: Male Wistar (Slc:Wistar), Sprague Dawley (Slc:SD), Dark Agouti (DA/Slc) and Brown Norway (BN/SsNSlc) rats (n=4 for each strain) were purchased from Japan SLC, Inc. (Hamamatsu, Japan). They were housed in plastic cages at 22 ± 1°C with a 12 hr light cycle/12 hr dark cycle and given laboratory chow (Labo MR Stock, Nosan Corp., Yokohama, Japan) and tap water ad libitum. When rats were 8 weeks old, liver samples were collected after euthanasia with carbon dioxide. Livers were immediately frozen in liquid nitrogen and stored at −80°C until use. All experiments using animals were performed under the supervision and with the approval of the Institutional Animal Care and Use Committee of Hokkaido University, Japan (approval number: 10-0067, approval day: 7th Dec 2010).
Chemicals: The BCA protein kit was purchased from Thermo Fisher Scientific (Waltham, MA, U.S.A.). ReverTra Ace was obtained from TOYOBO CO., Ltd. (Osaka, Japan). TRI reagent, bovine serum albumin (BSA), diethyl ether and racemic warfarin were from Sigma-Aldrich Inc. (St. Louis, MO, U.S.A.). Warfarin-sodium and dithiothreitol (DTT) were purchased from Wako Pure Chemical Industries (Osaka, Japan). Glucose 6-phosphate (G6P), glucose 6-phosphate dehydrogenase (G6PDH) and β-NADPH were from Oriental Yeast Co., Ltd. (Tokyo, Japan). Warfarin metabolites (4’-, 6-, 7-, 8- and 10-hydroxywarfarin) were obtained from Ultrafine Chemicals (Manchester, U.K.). Anti-rat CYP1A1 (#299124) and 2B1 (#299148) antibodies raised in goats and anti-rat CYP3A2 antibody raised in rabbits (#229223) were purchased from Daiichi Pure Chemicals (Tokyo, Japan). Anti-AHR antibody from rabbits (ab84833-100) was purchased from Abcam (Tokyo, Japan). Anti-goat IgG (sc-2056) and anti-rabbit IgG (sc-2054) antibodies conjugated to horseradish peroxidase were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.).
RNA extraction and cDNA synthesis: Total RNA was extracted from the liver tissue using TRI reagent according to the manufacturer’s instructions. The quality and concentration of extracted RNA was confirmed by agarose gel electrophoresis and via the Nanodrop ND1000 (Thermo Fisher Scientific). First-strand cDNA was generated from 600 ng of total RNA in a final volume of 20 µl with ReverTra Ace.
Quantitative real-time RT-PCR: The reverse-transcribed cDNA was used in quantitative real-time RT-PCR. TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, U.S.A.) were applied to measure the mRNA expression of Cyp1a1 (Rn00487218_m1, NM_012540.2, [4]), Cyp1a2 (Rn00561082_m1, NM_012541.2, [4]), Cyp3a2 (Rn00756461_m1, NM_153312.2, [6]), Ahr (Rn00565750_m1, NM_013149.1, [4]), constitutive androstane receptor (Car) (Hs00231959_m1, NM_005122.3, [6]), pregnane X receptor (Pxr) (Rn00583887_m1, NM_052980.1, [6]) and Gapdh (Rn99999916_m1, NM_017008.3, [4]). The reaction mixture consisted of 4.3 µl of 10x-diluted cDNA, 5 µl of THUNDERBIRD Probe qPCR Mix (TOYOBO), 0.5 µl of TaqMan probe and 0.2 µl of ROX reference dye. The PCR conditions of the real-time PCR (Stepone Plus Real-Time PCR Systems, Applied Biosystems) were as follows; 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec. Analysis was via the comparative Ct method with the Gapdh gene as an internal control. For Cyp2b1 (forward, 5′-GCTCAAGTACCCCCATGGTCG-3′; reverse, 5′-ATCAGTGTATGGCATTTTACTGCGG-3′, [5]), Cyp2c11 (forward, 5′-TGCCCCTTTTTACGAGGCT-3′; reverse, 5′-GGAACAGATGACTCTGAATTCT-3′, [24]) and Gapdh (forward, 5′-ACCACAGTCCATGCCATCAC-3′; reverse, 5′-TCCACCACCCTGTTGCTGTA-3′, [1]), mRNA expression was measured with original constructed primers. The reaction mixture consisted of 1 µl of 10x-diluted cDNA, 2.8 µl of DDW, 5 µl of THUNDERBIRD SYBR qPCR Mix, 0.2 µl of ROX reference dye and 0.5 µl each of the 10 µM forward and reverse primers specific for the target gene. The PCR conditions were as follows: 1 cycle of 95°C for 60 sec, followed by 40 cycles of 95°C for 15 sec and 60°C for 45 sec. Analysis was carried out via the comparative Ct method with the Gapdh gene as an internal control.
Preparation of the microsomal fraction: Liver microsomal fractions were prepared using the method described by Watanabe et al. [28], which was modified slightly from that used by Omura and Sato [20]. In brief, livers were homogenized with three times their volume of sodium phosphate buffer (0.1 M, pH 6.8). The homogenates were centrifuged at 9,000 × g at 4°C for 20 min. The supernatant was decanted to an ultracentrifuge tube and centrifuged at 105,000 × g at 4°C for 70 min. The pellet was homogenized in sodium phosphate buffer on ice and then centrifuged again at 105,000 × g at 4°C for 70 min to wash. The resultant microsomal pellets were resuspended with sodium phosphate buffer. The microsomal fractions were transferred to 1.5 ml tubes and stored at −80°C until use. The protein concentration was determined with the BCA protein assay kit, and the CYP content was determined by the method of Omura and Sato [20].
Western blotting: Protein quantification was performed for CYP1A, CYP2B1, CYP3A2 and AHR proteins by Western blotting. Aliquots of 10–30 µg of microsomal protein (for CYP1A, CYP2B1 and CYP3A2) or cytosolic protein (for AHR) were applied to 8–10% sodium dodecyl sulfate-polyacrylamide gels and separated by electrophoresis using a Mini-Protean II 1-D cell (BioRad, Hercules, CA, U.S.A.). In order to distinguish the bands between CYP1A1 and CYP1A2, the microsomal protein was prepared from WI rats which were orally administered sudan III for the strong induction of CYP1A1 [25]. Clear bands of CYP1A1 and CYP1A2 were observed from the microsomes of sudan III administered WI rats, and these bands were used as positive controls. Western blot analysis was performed as previously reported [14]. Proteins were electrophoretically transferred to nitrocellulose membranes. After membrane blocking with 5% skimmed milk, proteins were identified with specific antibodies and immunostained with diaminobenzidine or ECL Plus Western Blotting Substrate. After scanning the membrane, digital images were analyzed densitometrically using the image analysis program Image J (National Institutes of Health, Bethesda, MD, U.S.A.).
Warfarin metabolism assay: Multiple metabolic routes for warfarin metabolism are mediated by several CYP isoforms that have been described in rats, including 4’-hydroxylation (4’-OH) by CYP2B1 and CYP2C11, 6-hydroxylation (6-OH) by CYP1A1 and CYP2C11, 7-hydroxylation (7-OH) by CYP2C6, 8-hydroxylation (8-OH) by CYP1A1 and 10-hydroxylation (10-OH) by CYP3A2 [3, 7, 8]. Warfarin metabolic activity was measured with liver microsomes using a method described previously [28] with slight modifications. The incubation time for the reaction was 16 min in the present study.
Ethoxyresorufin-O-deethylase (EROD) activity: EROD activity was determined by a fluorescence intensity assay using a previously reported method [2] with slight modifications. In brief, the fluorescence of resorufin was detected at an excitation wavelength of 550 nm and an emission wavelength of 590 nm. Reactions were performed in a 1 ml volume containing 10 mM G6P, 10 mM MgCl2, 0.2 mg/ml microsomal protein and 20 µM ethoxyresorufin in 100 mM sodium phosphate buffer at pH 6.8. The reaction tubes were preincubated in a dark room for 5 min at 37°C, and the reaction was started by adding 20 µl of a mixture of 50 mM NADPH and 200 U G6PDH. The reaction was stopped with 4 ml of ice-cold methanol.
NADPH-P450 reductase (CPR) activity: CPR activity was measured according to the method of Omura and Takesue [21] with slight modifications. Cytochrome c reduction by CPR was monitored at 37°C. The incubation mixture contained 100 mM sodium phosphate buffer (pH 7.4), 20 µM cytochrome c, 50 µM NADPH and 25 µg of microsomal protein. The amount of reduced cytochrome c was calculated by subtracting the data obtained from the blank reaction without microsomes using an extinction coefficient of 21/mM/cm at 550 nm.
Statistical analysis: Statistical analyses were performed with the Tukey-Kramer test using JMP version 7.0 (SAS Institute Inc.; Raleigh, NC, U.S.A.). Statistical significance was considered at P<0.05. Results are shown as average ± standard error (SE).
RESULTS
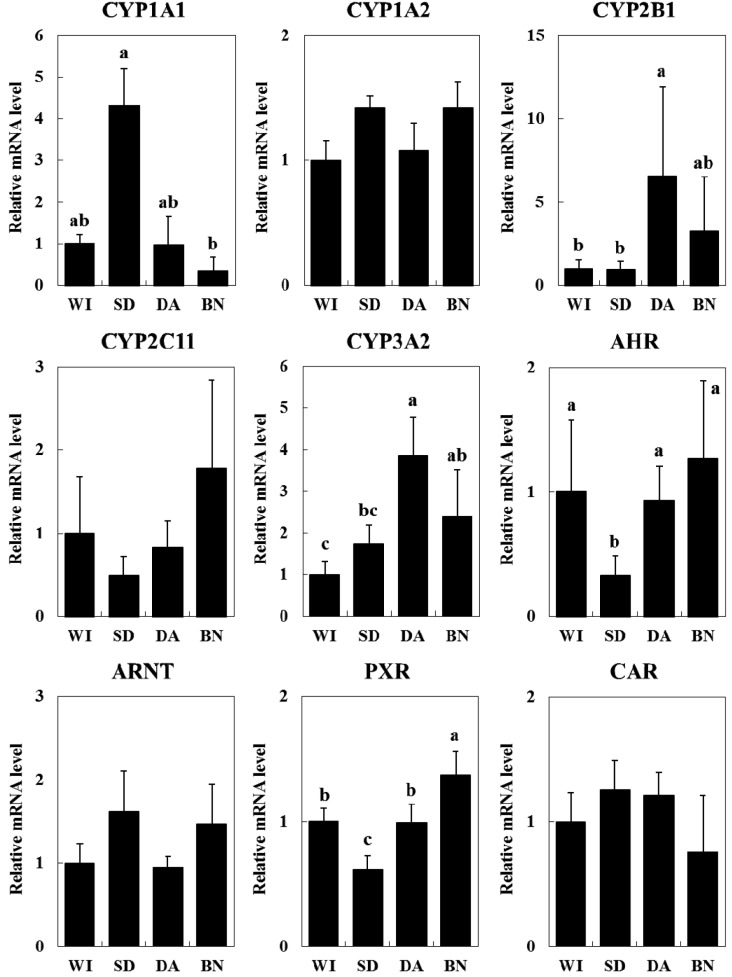
mRNA expression levels of CYP isoforms and their transcriptional factors: Hepatic mRNA expression levels of Cyp1a1, Cyp1a2, Cyp2b1, Cyp2c11, Cyp3a2, Ahr, Arnt, Car and Pxr were measured by real-time RT-PCR (Fig. 1). Cyp1a2, Cyp2c11, Arnt and Car mRNA levels were not significantly different among the four rat strains. On the other hand, the Cyp1a1 mRNA level was 10-fold higher in SD rats than in BN rats. Cyp2b1 and Cyp3a2 mRNA levels were significantly higher in DA rats than in WI and SD rats. Ahr and Pxr mRNA levels were significantly lower in SD rats than in the other strains.
Fig. 1.
Hepatic mRNA expression levels of CYP1A1, 1A2, 2B1, 2C11, 3A2, AHR, ARNT, PXR and CAR in four rat strains. Data are expressed as the ratio of the target mRNA to GAPDH mRNA and normalized to WI. The values are shown as the mean ± SE (n=4). Different characters (a, b) indicate statistically significant differences (Tukey-Kramer test, P<0.05).
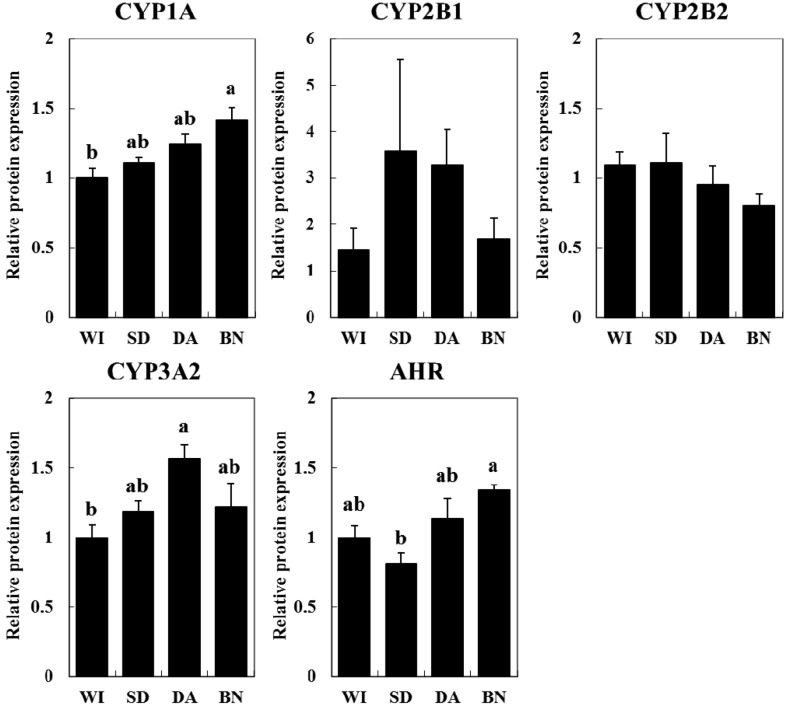
Basal protein expression levels of hepatic CYP and AHR: No significant difference was observed in the CYP content of liver microsomes among the rat strains (data not shown). The basal protein expression levels of hepatic CYP isoforms, i.e., CYP1A, CYP2B1, CYP2B2, CYP3A and AHR (a key transcription factor for CYP1A1), were evaluated (Fig. 2). The expression of CYP1A protein, which consists of CYP1A1 and CYP1A2, was significantly higher in BN rats than in WI rats. CYP3A2 protein expression was higher in DA rats than in WI rats. AHR protein expression was lower in SD rats than in BN rats. Although the expressions of CYP2B1 and CYP2B2 proteins were not statistically different among the strains, CYP2B1 expression did tend to be higher in SD and DA rats than in WI and BN rats.
Fig. 2.
Protein expression levels of CYP1A, 2B1, 2B2, 3A2 and AHR in liver microsomes (CYP1A1, CYP2B1, CYP2B2 and CYP3A2) and the cytosol (AHR) in four rat strains. The values are shown as the mean ± SE (n=4) normalized to WI. Different characters (a, b) indicate statistically significant differences (Tukey-Kramer test, P<0.05).
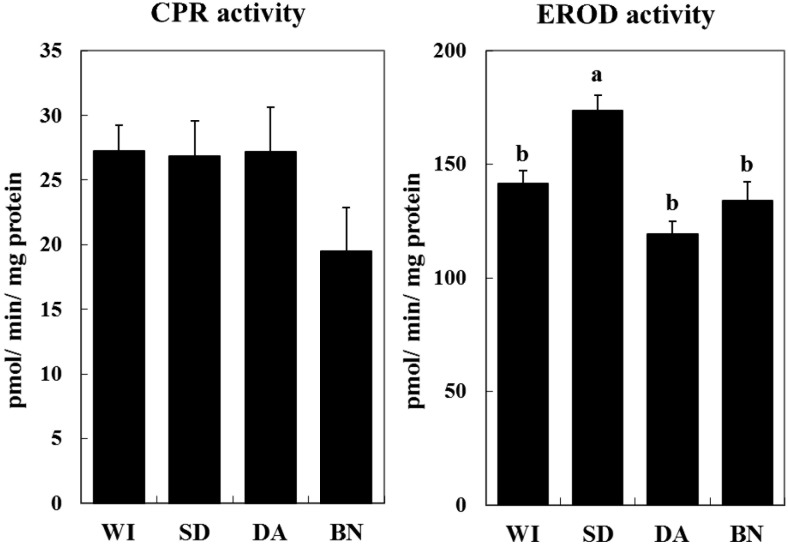
CPR and EROD activity and warfarin metabolism: CPR activity tended to be lower in BN rats, but was not significantly different among the strains (Fig. 3). EROD activity, mainly a reflection of CYP1A1 enzymatic activity, was significantly higher in SD rats than in the other strains (Fig. 3). For warfarin metabolism assay, 4’-OH, 6-OH and 8-OH were the dominant metabolites in all of the rat strains (Table 1). Total warfarin hydroxylation activity was lower in WI rats than in SD and DA rats (Table 1).
Fig. 3.
Hepatic microsomal P450 reductase and EROD activity in four rat strains. NADPH-P450 reductase-dependent cytochrome c reductase (CPR) activity was measured. Reactions were carried out for 1 min at 37°C, and samples were measured using a spectrophotometer. For EROD activity, reactions were carried out for 5 min at 37°C, and the fluorescence was detected at an excitation wavelength of 550 nm and an emission wavelength of 590 nm. The values are shown as the mean ± SE (n=4). Different characters (a, b) indicate statistically significant differences (Tukey-Kramer test, P<0.05).
Table 1. Hepatic microsomal warfarin metabolism activity (pmol/min/mg protein) in four rat strains.
| 4’-OH | 6-OH | 7-OH | 8-OH | 10-OH | total | |
|---|---|---|---|---|---|---|
| WI | 90.0 ± 5.6 b) | 99.7 ± 7.4 b) | 78.1 ± 2.4 b) | 109.9 ± 7.5 b) | 25.5 ± 0.7 b) | 403.2 ± 23.5 b) |
| SD | 132.0 ± 5.8 a) | 124.5 ± 4.4 a,b) | 93.7 ± 1.6 a) | 153.3 ± 3.6 a) | 31.3 ± 0.6 a) | 534.8 ± 16.0 a) |
| DA | 136.9 ± 7.0 a) | 145.6 ± 3.5 a) | 76.1 ± 1.1 b) | 155.0 ± 9.7 a) | 35.9 ± 0.9 a) | 549.5 ± 22.3 a) |
| BN | 123.0 ± 9.2 a) | 116.8 ± 9.1 a,b) | 90.7 ± 5.2 a,b) | 133.8 ± 11.8 a,b) | 30.4 ± 2.0 a,b) | 494.7 ± 37.4 a,b) |
Different characters (a, b) indicate statistically significant differences among the four strains (Tukey-Kramer test, P<0.05).
DISCUSSION
In this study, the mRNA expression, protein expression and enzymatic activity of CYP were determined in WI, SD, DA and BN rats for an integrated analysis and comparison of strain differences in the protein’s activities. On the one hand, mRNA expression showed great differences between the strains. On the other hand, warfarin metabolic activity, a useful method to measure the activity of several CYP isoforms simultaneously, showed comparatively smaller strain differences.
The rank order of protein expression and mRNA expression of CYP3A2 was identical between strains: i.e., DA>BN>SD>WI. CYP3A2 protein expression and its enzymatic activity, which was measured as 10-OH warfarin hydroxylation, showed a similar pattern, and they correlated positively (r=0.62, data not shown) in each rat from the four strains. Expression of mRNA and protein, and enzymatic activity were consistently correlated for CYP3A2. These results provide insight into the strain differences in CYP3A2 activity: i.e., those differences in activity are due to differences in CYP3A2 protein expression, which are in turn caused by differences in Cyp3a2 mRNA expression.
In contrast to our CYP3A2 findings, the rank order of the protein and mRNA expression of CYP1A1 and CYP2B1 varied among the four strains. A lack of correlation between mRNA and protein expression is not uncommon, as protein expression levels largely depend on post-transcriptional and post-translational regulation. Micro-RNAs (miRNAs), which are short, noncoding RNAs that govern gene expression, are one important factor that can explain the difference between mRNA and protein expression levels. For example, strain differences in miRNA expression have been reported in the hippocampus of mice [22]. It has also been reported that Cyp1b1 is regulated by miRNA in humans [26]. From these reports, the difference between the mRNA and protein expression levels observed in the current study suggests possible existence of strain differences in miRNA, which is able to regulate CYP expression. Moreover, post-translational regulation by factors, such as miRNA, is likely to differ among CYP isoforms, since the difference between mRNA and protein expression was only observed in CYP1A and CYP2B, but not CYP3A.
Our study showed that both Cyp3a2 mRNA expression and total warfarin hydroxylation by CYP1A1, CYP2B1, CYP2C6, CYP2C11 and CYP3A2 in WI rats were the lowest among the four strains. Our results agree with the previous report that CYP3A protein expression and its enzymatic activity (C-8 hydroxylation of caffeine) were lower in WI rats than in DA rats [16]. Relatedly, the other report suggested that WI rats (Wistar Lewis) were more sensitive than SD rats and F344 rats to oral administration of clofibrate, a hyperlipidemia drug that is metabolized primarily by CYP3A [27]. Taken together, these observations suggest that the WI rat strain is possibly sensitive to some drugs which are metabolized by CYP.
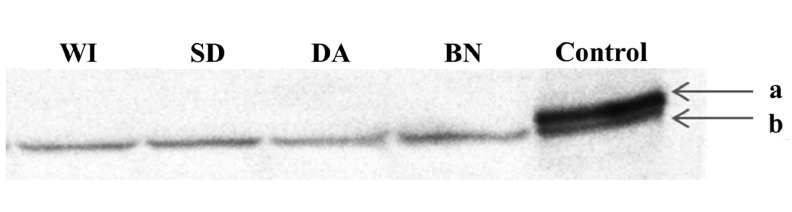
The current study showed that Cyp1a1 mRNA expression and EROD activity were highest in SD rats relative to the other strains examined. According to the results of mRNA in Cyp1a1, the CYP1A1 protein expression in SD rats might be higher than the other strains. However, the protein expression of CYP1A was not the highest in SD rats. We assume that this is because the CYP1A protein mainly consists of CYP1A2. Results of Western blotting (Fig. 4) suggest that the CYP1A1 protein expression was markedly smaller than CYP1A2, and this made it difficult to detect the CYP1A1 protein expression in the liver under physiological conditions. EROD activity, which is indication of both CYP1A1 and CYP1A2 activities, could also be affected by CYP1A2 strongly, because the protein expression level is much more abundant in CYP1A2 than CYP1A1 under physiological conditions. The CYP1A1 enzymatic activity in SD rats might be much higher than the result observed in EROD activity assays. In order to compare CYP1A1 activity among the strains, further studies using the recombinant CYP1A1 from the different rat strains are necessary. CYP1A1 is reported to be the key enzyme involved in the metabolic activation of polycyclic aromatic hydrocarbons [18]. Therefore, SD rats may be more susceptible to chemicals that are metabolically activated by CYP1A1 under physiological conditions.
Fig. 4.
Western blotting of CYP1A. WI; Wistar rat, SD; Sprague Dawley rat, DA; Dark Agouti rat, BN; Brown Norway rat, Control; Wistar rat administered Sudan III, a; CYP1A1, b; CYP1A2 [25].
Our results show that SD rats had high CYP1A1 mRNA expression and enzymatic activity. This is actually inconsistent with previous reports: a study by Kawase et al. [11], Cyp1a1 mRNA expression in SD rats was approximately 10-fold lower than in DA rats, while in a study by Kishida et al. [13], CYP1A1 mRNA expression and enzymatic activity in SD rats were lower than in Wistar Hannover rats. This conflicting result suggests that there may even be intra-strain differences in SD rats. SD rats (Slc:SD) used in the present study were purchased from Japan SLC, Inc., whereas Jcl:SD rats obtained from CLEA Japan, Inc. (Tokyo, Japan) and Crl;CD (SD) rats from Charles River Laboratories Japan, Inc. (Yokohama, Japan) were used in Kawase’s study and Kishida’s study, respectively. Both Slc:SD and Jcl:SD rats were originally obtained from SD rats in Charles River Laboratories in 1968 and 1964, respectively, which is before the enactment of International Genetic Standard in Charles River Laboratories. The intra-strain differences in SD rats are arising among these laboratories over forty years.
DA rats are reported to be poor metabolizers of human CYP2D6 substrates [10], mainly due to a mutation in the promoter region of the Cyp2d2 gene that results in decreased Cyp2d2 mRNA expression [23]. In addition, our study demonstrated that the mRNA expression of Cyp2b1 and Cyp3a2 and their activities were higher in DA rats than in the other strains. Taken together our findings (Cyp2b1 and Cyp3a2) with the reported data of Cyp2d2, it is suggested that DA rats have a unique expression pattern of CYPs, resulting in unique metabolic pattern among the rat strains.
In this study, a unique inversion phenomenon was detected. The mRNA and protein expression levels of AHR tended to be lower in SD rats, although the expression of Cyp1a1 mRNA, which is critically induced via an AHR-mediated pathway, tended to be higher in SD rats. Previous research using Ahr-knockout mice showed that AHR is an essential factor for the induction of CYP1A1 in the liver and for its homeostatic expression [9, 17]. The mechanism of CYP1A1 expression, which has not been elucidated fully, may be different in SD rats compared to the other strains.
In conclusion, our study reveals that hepatic mRNA expression, protein expression and enzymatic activity of CYP1A1, 2B1 and 3A2 in WI rats tend to be lower in comparison to SD, DA and BN rats. On the other hand, in SD rats, the hepatic mRNA expression and enzymatic activity of CYP1A1 is higher than in the other three strains. These strain differences may influence the pharmacokinetics in preclinical toxicological drug studies.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan awarded to M. Ishizuka (No. 24405004 and No. 24248056) and Y. Ikenaka (No. 26304043, 15H0282505, 15K1221305), and the foundation of JSPS Core to Core Program (AA Science Platforms) and Bilateral Joint Research Project (PG36150002 and PG36150003). We also acknowledge the financial support by The Nihon Seimei Foundation. We are grateful to Mr. Takahiro Ichise (Laboratory of Toxicology, Graduate School of Veterinary Medicine, Hokkaido University) for technical support.
REFERENCES
- 1.Chen M. L., Chen C. H.2005. Microarray analysis of differentially expressed genes in rat frontal cortex under chronic risperidone treatment. Neuropsychopharmacology 30: 268–277. doi: 10.1038/sj.npp.1300612 [DOI] [PubMed] [Google Scholar]
- 2.Crespi C. L., Miller V. P., Penman B. W.1997. Microtiter plate assays for inhibition of human, drug-metabolizing cytochromes P450. Anal. Biochem. 248: 188–190. doi: 10.1006/abio.1997.2145 [DOI] [PubMed] [Google Scholar]
- 3.Daly A. K., King B. P.2003. Pharmacogenetics of oral anticoagulants. Pharmacogenetics 13: 247–252. doi: 10.1097/00008571-200305000-00002 [DOI] [PubMed] [Google Scholar]
- 4.Darwish W. S., Ikenaka Y., Nakayama S., Ishizuka M.2014. The effect of copper on the mRNA expression profile of xenobiotic-metabolizing enzymes in cultured rat H4-II-E cells. Biol. Trace Elem. Res. 158: 243–248. doi: 10.1007/s12011-014-9915-9 [DOI] [PubMed] [Google Scholar]
- 5.Darwish W. S., Ikenaka Y., Ohno M., Eldaly E. A., Ishizuka M.2010. Carotenoids as regulators for inter-species difference in cytochrome P450 1A expression and activity in ungulates and rats. Food Chem. Toxicol. 48: 3201–3208. doi: 10.1016/j.fct.2010.08.022 [DOI] [PubMed] [Google Scholar]
- 6.Fery Y., Mueller S. O., Schrenk D.2010. Development of stably transfected human and rat hepatoma cell lines for the species-specific assessment of xenobiotic response enhancer module (XREM)-dependent induction of drug metabolism. Toxicology 277: 11–19. doi: 10.1016/j.tox.2010.08.008 [DOI] [PubMed] [Google Scholar]
- 7.Guengerich F. P., Dannan G. A., Wright S. T., Martin M. V., Kaminsky L. S.1982. Purification and characterization of liver microsomal cytochromes p-450: electrophoretic, spectral, catalytic, and immunochemical properties and inducibility of eight isozymes isolated from rats treated with phenobarbital or β-naphthoflavone. Biochemistry 21: 6019–6030. doi: 10.1021/bi00266a045 [DOI] [PubMed] [Google Scholar]
- 8.Ishizuka M., Okajima F., Tanikawa T., Min H., Tanaka K. D., Sakamoto K. Q., Fujita S.2007. Elevated warfarin metabolism in warfarin-resistant roof rats (Rattus rattus) in Tokyo. Drug Metab. Dispos. 35: 62–66. doi: 10.1124/dmd.106.011775 [DOI] [PubMed] [Google Scholar]
- 9.Jiang W., Welty S. E., Couroucli X. I., Barrios R., Kondraganti S. R., Muthiah K., Yu L., Avery S. E., Moorthy B.2004. Disruption of the Ah receptor gene alters the susceptibility of mice to oxygen-mediated regulation of pulmonary and hepatic cytochromes P4501A expression and exacerbates hyperoxic lung injury. J. Pharmacol. Exp. Ther. 310: 512–519. doi: 10.1124/jpet.103.059766 [DOI] [PubMed] [Google Scholar]
- 10.Kahn G. C., Rubenfield M., Davies D. S., Murray S., Boobis A. R.1985. Sex and strain differences in hepatic debrisoquine 4-hydroxylase activity of the rat. Drug Metab. Dispos. 13: 510–516. [PubMed] [Google Scholar]
- 11.Kawase A., Fujii A., Negoro M., Akai R., Ishikubo M., Komura H., Iwaki M.2008. Differences in cytochrome P450 and nuclear receptor mRNA levels in liver and small intestines between SD and DA rats. Drug Metab. Pharmacokinet. 23: 196–206. doi: 10.2133/dmpk.23.196 [DOI] [PubMed] [Google Scholar]
- 12.Kawase A., Otori T., Fujii A., Yamada A., Komura H., Iwaki M.2011. Strain Differences in the Induction of Cytochrome P450 3A1/3A2 and Nuclear Receptors in the Liver by Phenobarbital and Dexamethasone in Sprague-Dawley Rats and Dark Agouti Rats. J. Health Sci. 57: 414–419. doi: 10.1248/jhs.57.414 [DOI] [Google Scholar]
- 13.Kishida T., Muto S., Hayashi M., Tsutsui M., Tanaka S., Murakami M., Kuroda J.2008. Strain differences in hepatic cytochrome P450 1A and 3A expression between Sprague-Dawley and Wistar rats. J. Toxicol. Sci. 33: 447–457. doi: 10.2131/jts.33.447 [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U. K.1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. doi: 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 15.Li J., Kaneko T., Wang Y., Qin L. Q., Wang P. Y., Sato A.2002. Troglitazone enhances the hepatotoxicity of acetaminophen by inducing CYP3A in rats. Toxicology 176: 91–100. doi: 10.1016/S0300-483X(02)00143-9 [DOI] [PubMed] [Google Scholar]
- 16.Morita K., Maeda Y., Masuda M., Kazusaka A., Imaoka S., Funae Y., Fujita S.1998. Strain differences in CYP3A-mediated C-8 hydroxylation (1,3,7-trimethyluric acid formation) of caffeine in Wistar and Dark Agouti rats. Rapid metabolism of caffeine in debrisoquine poor metabolizer model rats. Biochem. Pharmacol. 55: 1405–1411. doi: 10.1016/S0006-2952(97)00654-0 [DOI] [PubMed] [Google Scholar]
- 17.Mukai M., Tischkau S. A.2007. Effects of tryptophan photoproducts in the circadian timing system: searching for a physiological role for aryl hydrocarbon receptor. Toxicol. Sci. 95: 172–181. doi: 10.1093/toxsci/kfl126 [DOI] [PubMed] [Google Scholar]
- 18.Nebert D. W.1991. Role of genetics and drug metabolism in human cancer risk. Mutat. Res. 247: 267–281. doi: 10.1016/0027-5107(91)90022-G [DOI] [PubMed] [Google Scholar]
- 19.Newton J. F., Yoshimoto M., Bernstein J., Rush G. F., Hook J. B.1983. Acetaminophen nephrotoxicity in the rat. I. Strain differences in nephrotoxicity and metabolism. Toxicol. Appl. Pharmacol. 69: 291–306. doi: 10.1016/0041-008X(83)90311-3 [DOI] [PubMed] [Google Scholar]
- 20.Omura T., Sato R.1964. The carbon monoxide binding pigment of liver microsomes. J. Biol. Chem. 239: 2370–2378. [PubMed] [Google Scholar]
- 21.Omura T., Takesue S.1970. A new method for simultaneous purification of cytochrome b5 and NADPH-cytochrome c reductase from rat liver microsomes. J. Biochem. 67: 249–257. [DOI] [PubMed] [Google Scholar]
- 22.Parsons M. J., Grimm C. H., Paya-Cano J. L., Sugden K., Nietfeld W., Lehrach H., Schalkwyk L. C.2008. Using hippocampal microRNA expression differences between mouse inbred strains to characterise miRNA function. Mamm. Genome 19: 552–560. doi: 10.1007/s00335-008-9116-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakai N., Sakamoto K. Q., Fujita S., Ishizuka M.2009. The importance of heterogeneous nuclear ribonucleoprotein K on cytochrome P450 2D2 gene regulation: its binding is reduced in Dark Agouti rats. Drug Metab. Dispos. 37: 1703–1710. doi: 10.1124/dmd.109.027284 [DOI] [PubMed] [Google Scholar]
- 24.Shaban Z., Soliman M., El-Shazly S., El-Bohi K., Abdelazeez A., Kehelo K., Kim H. S., Muzandu K., Ishizuka M., Kazusaka A., Fujita S.2005. AhR and PPARalpha: antagonistic effects on CYP2B and CYP3A, and additive inhibitory effects on CYP2C11. Xenobiotica 35: 51–68. doi: 10.1080/00498250400021804 [DOI] [PubMed] [Google Scholar]
- 25.Takiguchi M., Darwish W. S., Ikenaka Y., Ohno M., Ishizuka M.2010. Metabolic activation of heterocyclic amines and expression of CYP1A1 in the tongue. Toxicol. Sci. 116: 79–91. doi: 10.1093/toxsci/kfq087 [DOI] [PubMed] [Google Scholar]
- 26.Tsuchiya Y., Nakajima M., Takagi S., Taniya T., Yokoi T.2006. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 66: 9090–9098. doi: 10.1158/0008-5472.CAN-06-1403 [DOI] [PubMed] [Google Scholar]
- 27.Yamoto T., Ohashi Y., Miyakoshi N., Matsunuma N., Manabe S., Makita T.1996. Strain difference in the susceptibility to clofibrate in male rats. Pathophysiology 3: 65–72. doi: 10.1016/0928-4680(95)00056-9 [DOI] [Google Scholar]
- 28.Watanabe K. P., Saengtienchai A., Tanaka K. D., Ikenaka Y., Ishizuka M.2010. Comparison of warfarin sensitivity between rat and bird species. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 152: 114–119. doi: 10.1016/j.cbpc.2010.03.006 [DOI] [PubMed] [Google Scholar]