Abstract
Accurate identification and separation of non-classical Bordetella species is very difficult. These species have been implicated in animal infections. B. hinzii, a non-classical Bordetella, has been isolated from mice in experimental facilities recently. We isolated and characterized one non-classical Bordetella isolate from the trachea and lung of an ICR mouse. Isolate BH370 was initially identified as B. hinzii by 16S ribosomal DNA and ompA sequencing. Additionally, isolate BH370 also displayed antimicrobial sensitivity profiles similar to B. hinzii. However, analyses of nrdA sequences determined its identity as Bordetella genogroup 16. The isolation of BH370 from a healthy mouse suggests the possibility of it being a commensal. The nrdA gene was demonstrated to possess greater phylogenetic resolution as compared with 16S ribosomal DNA and ompA for the discrimination of non-classical Bordetella species.
Keywords: Bordetella, infectious disease, laboratory mouse, Malaysia
The genus Bordetella comprises gram-negative, rod-shaped bacteria that can cause respiratory diseases in humans and animals [7]. Classical Bordetella species (B. pertussis, B. parapertussis and B. bronchiseptica) are usually associated with serious human respiratory infections [3, 14]. Differentiation of non-classical Bordetella species that rarely cause human infection is difficult [3]. The most common diagnostic assays include real-time PCR [9] in combination with examination of growth properties, biochemical tests, serotyping and partial 16S rRNA gene sequencing [14]. However, these methods are laborious, and it is still arduous to specifically differentiate non-classical Bordetella species. Recently, researchers have isolated B. hinzii from mice bred in experimental facilities in Japan [5]. The same group also proposed B. hinzii as a possible pathogen in mice [6], potentially interfering with animal research and possibly serving as a source of infection for other mice in the experimental facility. Here, we report the isolation and characterization of a non-classical Bordetella isolate from the trachea and lung of a healthy laboratory mouse.
The Animal Experimental Unit, Faculty of Medicine, University of Malaya, has an established health monitoring program for rodents housed in the facility to ensure the animals are free of specific pathogens that could interfere with scientific experiments. Rodents submitted to the facility for health screening were quarantined in individually ventilated cages (Tecniplast, Via Maggio, Italy). All cages, animal feed, beddings and surgical tools were sterilized before use. The animals were maintained at a room temperature of 19 to 21°C and relative humidity of 54 to 65% under a 12:12 hr light:dark cycle. The current study and the animal handling procedures were approved by the Institutional Animal Care and Use Committee, University of Malaya (Reference no. 2013-11-12/AEU/B/WPF). Healthy mice that were between 6 and 12 weeks old and had no observed signs of illness were randomly chosen and anesthetized with ketamine (80 mg/kg) and xylazine (10 mg/kg) prior to blood collection through cardiac puncture. Blood collected from mice was tested by serology (SMART-SPOT Sentinel Panel Test, Mouse Sera, Biotech Trading Partners, Encinitas, CA, U.S.A.) for the following agents; epizootic diarrhea of infant mice, mouse hepatitis virus, Mycoplasma pulmonis, mouse parvovirus, minute virus of mice, pneumonia virus of mice, respiratory enteric orphan virus, Sendai virus and Theiler’s murine encephalomyelitis virus. Due to the small volume of circulating blood in mice, no euthanasia was performed, as the mice would experience hypovolemia after cardiac puncture, which would result in death. All works were performed in a dedicated procedure room away from breeding facilities. Necropsy and harvesting of selected organs (trachea, lungs, liver and the gastrointestinal tract) were performed inside a biosafety cabinet in the procedure room. The organs were subjected to microbiology culture for Corynebacterium kutscheri, Citrobacter rodentium, Salmonella spp., Helicobacter spp. and Clostridium piliformis. The trachea and lungs of the euthanized rodents were transferred into tubes with ceramic beads, and the specimens were homogenized using a Minilys tissue homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France). A small portion of the homogenate was inoculated onto Columbia agar with 5% sheep blood and incubated at 37°C under ambient conditions for 24 hr. Cultured bacterial isolates were Gram stained, and a lactose fermentation test using phenol red lactose broth was performed. An antimicrobial sensitivity profile was determined using a disc diffusion assay on Mueller-Hinton agar incubated at 37°C for 24 hr. The antibiotic discs (BBL™ Sensi-Disc™, Becton, Dickinson and Co., Franklin Lakes, NJ, U.S.A.) used in the disc diffusion assay were amikacin (30 µg), amoxicillin-clavulanic acid (20/10 µg), ampicillin (10 µg), cefoperazone (75 µg), ceftazidime (30 µg), ciprofloxacin (5 µg), cefuroxime (30 µg), ceftriaxone (30 µg), cefotaxime (30 µg), gentamicin (10 µg), imipenem (10 µg) and meropenem (10 µg). Results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute [1].
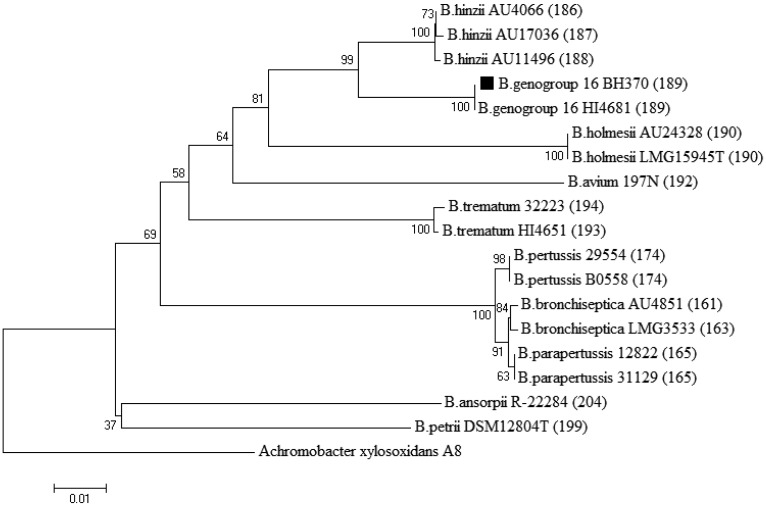
The trachea and lung specimens from an 8-week-old male ICR mouse with no sign of illness grew small, non-lactose fermenting Gram-negative rods (isolate BH370). Serological assays suggested that the mouse was free of the 9 regularly monitored common pathogens. Nucleic acid amplification and sequencing of the 16S ribosomal DNA gene, as described previously [11], were performed to identify the isolate. PCR fragments were purified, sequenced and compared against available sequences in GenBank. BLAST results of the partial 16S ribosomal DNA gene from BH370 (Accession no. LN836016) showed a 100% sequence identity match with B. hinzii strain 3224 (Accession no. AB371725), which had been isolated previously from a sick mouse. However, the isolate was also closely related to other Bordetella species (B. avium, B. holmesii, B. parapertussis and B. bronchiseptica), as their sequences showed as high as a 99% 16S ribosomal DNA sequence identity match. A separate PCR specifically amplifying the ompA gene of B. hinzii [12] to ascertain the identity of the isolate was performed. The resulting PCR fragment from BH370 was sequenced (Accession no. LN836017), and BLAST identified BH370 as B. hinzii with 98% similarity. Sequencing of ompA substantiates the confirmation of non-classical Bordetella species due to its greater sequence variation as compared with the 16S ribosomal DNA gene [12]. Nevertheless, the GenBank public database contained only three B. hinzii ompA entries, which we felt was not quite sufficient to strongly support the identification. The multilocus sequence typing scheme for Bordetella [2] failed to amplify PCR products for DNA sequencing. To resolve the identity ambiguity of isolate BH370, we employed an additional method that has been shown to reliably differentiate species within the genus Bordetella by analyzing the trimmed 765-bp sequences of nrdA [14], which encodes the ribonucleoside-diphosphate reductase alpha chain. At this gene locus, BH370 was distinctively identified as belonging to Bordetella genogroup 16, as shown in Fig. 1, and its nucleotide sequences can be accessed at the PubMLST website (http://pubmlst.org/bordetella) [8]. Resistance to amoxicillin-clavulanic acid, ampicillin, cefuroxime, ceftriaxone and cefotaxime was revealed for BH370 (Table 1).
Fig. 1.
Neighbor-joining phylogenetic tree of Bordetella spp. 765-bp nrdA loci. Isolate BH370 is indicated by a black-filled square symbol before its species name. nrdA locus numbers are indicated in parentheses. Achromobacter xylosoxidans was used as an outgroup. Numbers at nodes indicate bootstrap values (%) for 1,000 replicates. The scale bar indicates nucleotide substitutions per site.
Table 1. Antimicrobial sensitivity results for Bordetella genogroup 16 isolate BH370.
| Antibiotic | Sensitivity testing results by disc diffusion assaya) on isolate BH370 |
|---|---|
| Amikacin | S (19 mm) |
| Amoxicillin/clavulanic acid | R (11 mm) |
| Ampicillin | R (11 mm) |
| Cefoperazone | S (26 mm) |
| Ceftazidime | S (27 mm) |
| Ciprofloxacin | S (22 mm) |
| Cefuroxime | R (12 mm) |
| Ceftriaxone | R (19 mm) |
| Cefotaxime | R (20 mm) |
| Gentamicin | S (18 mm) |
| Imipenem | S (25 mm) |
| Meropenem | S (25 mm) |
a) Zone diameter values are in parentheses. R, resistant; S, sensitive. Results were interpreted according to CLSI breakpoints for Enterobacteriaceae.
The differentiation of non-classical Bordetella species is difficult and labor intensive [14]. Commercial biochemical identification systems, such as API 20NE strips, currently only incorporate two Bordetella species, B. avium and B. bronchiseptica, in their libraries [3]. Even the application of matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry was shown to classify a Bordetella isolate only down to the genus level [7]. Yet confirmation of species is important for the determination of antibiotic treatment and appropriate infection control measures. Since 16S ribosomal DNA and ompA sequences could not provide definite resolution to specifically identify and separate non-classical Bordetella species, we employed a novel genotypic identification scheme based on the nrdA sequences [14]. BH370 was defined as Bordetella genogroup 16 bearing nrdA locus 189. The high sequence similarity of B. hinzii strain 3224 to our isolate suggests the possibility that strain 3224 was misidentified as B. hinzii, since its identification was done only with 16S ribosomal DNA sequencing. Another isolate, which was isolated from a cystic fibrosis mouse, carrying an identical nrdA locus was found in the PubMLST database [8]. The phylogenetic tree (Fig. 1), built using nrdA loci of representative Bordetella isolates clearly showed that Bordetella genogroup 16 and B. hinzii share a very close phylogenetic relationship. However, the reservoir and transmission routes for B. hinzii are currently unknown [7]. B. hinzii has been reported to cause infection in humans [3, 4] and rodents [5, 7], and laboratory mice have also been discovered to harbor B. hinzii in Japan [5, 6]. We suggest that rodents could be the reservoir for Bordetella genogroup 16, even though human infection has not been reported thus far.
Antimicrobial sensitivity assays revealed that isolate BH370 was resistant to amoxicillin/clavulanic acid, ampicillin, cefuroxime, ceftriaxone and cefotaxime, but sensitive to amikacin, cefoperazone, ceftazidime, gentamicin, imipenem and meropenem. Other publications have shown that B. hinzii isolated from clinical cases [3, 4, 13] was resistant or showed intermediate resistance to ampicillin, cefuroxime, ceftriaxone, cefotaxime and ciprofloxacin, which was similar to the antimicrobial sensitivity of BH370, excluding ciprofloxacin. The largely β-lactam resistance profile of isolate BH370 mirrors those observed by others for B. hinzii [3, 4, 13] and B. bronchiseptica [10], reinforcing the close association between Bordetella genogroup 16 and other Bordetella species. In the case of BH370, however, the discovery of this organism in a mouse with the absence of any clinical symptoms could suggest that it exists as a commensal. It may rarely cause noticeable infection in mice, as observed in B. hinzii-positive mice in Japan [5], or it could be an opportunistic pathogen in sick mice, as in the case of B. hinzii strain 3224 [6]. Hence, we suggest that screening for this organism should be considered in routine health monitoring programs for laboratory rodents.
Acknowledgments
This study was supported in part by the University of Malaya Research Officer Grant Scheme (BR009-2014) and High Impact Research grant scheme (E000013-20001). The authors want to thank Mdm. Hamidah Hamid from the Animal Experimental Unit, Faculty of Medicine, University of Malaya, for her assistance in specimen processing.
REFERENCES
- 1.Clinical and Laboratory Standards Institute2012. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Second Informational (Supplement): M100–S21Clinical and Laboratory Standards Institute, Wayne. [Google Scholar]
- 2.Diavatopoulos D. A., Cummings C. A., Schouls L. M., Brinig M. M., Relman D. A., Mooi F. R.2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1: e45. doi: 10.1371/journal.ppat.0010045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fry N. K., Duncan J., Edwards M. T., Tilley R. E., Chitnavis D., Harman R., Hammerton H., Dainton L.2007. A UK clinical isolate of Bordetella hinzii from a patient with myelodysplastic syndrome. J. Med. Microbiol. 56: 1700–1703. doi: 10.1099/jmm.0.47482-0 [DOI] [PubMed] [Google Scholar]
- 4.Funke G., Hess T., von Graevenitz A., Vandamme P.1996. Characteristics of Bordetella hinzii strains isolated from a cystic fibrosis patient over a 3-year period. J. Clin. Microbiol. 34: 966–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashimoto N., Morita H., Yasuda M., Ishida T., Kameda S., Takakura A., Itoh T.2012. Prevalence of Bordetella hinzii in mice in experimental facilities in Japan. Res. Vet. Sci. 93: 624–626. doi: 10.1016/j.rvsc.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 6.Hayashimoto N., Yasuda M., Goto K., Takakura A., Itoh T.2008. Study of a Bordetella hinzii isolate from a laboratory mouse. Comp. Med. 58: 440–446. [PMC free article] [PubMed] [Google Scholar]
- 7.Jiyipong T., Morand S., Jittapalapong S., Raoult D., Rolain J. M.2013. Bordetella hinzii in rodents, Southeast Asia. Emerg. Infect. Dis. 19: 502–503. doi: 10.3201/eid1903.120987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jolley K. A., Maiden M. C.2010. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11: 595. doi: 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koidl C., Bozic M., Burmeister A., Hess M., Marth E., Kessler H. H.2007. Detection and differentiation of Bordetella spp. by real-time PCR. J. Clin. Microbiol. 45: 347–350. doi: 10.1128/JCM.01303-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lartigue M. F., Poirel L., Fortineau N., Nordmann P.2005. Chromosome-borne class A BOR-1 beta-Lactamase of Bordetella bronchiseptica and Bordetella parapertussis. Antimicrob. Agents Chemother. 49: 2565–2567. doi: 10.1128/AAC.49.6.2565-2567.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misbah S., Hassan H., Yusof M. Y., Hanifah Y. A., AbuBakar S.2005. Genomic species identification of Acinetobacter of clinical isolates by 16S rDNA sequencing. Singapore Med. J. 46: 461–464. [PubMed] [Google Scholar]
- 12.Register K. B.2013. Development of a PCR for identification of Bordetella hinzii. Avian Dis. 57: 307–310. doi: 10.1637/10433-102212-ResNote.1 [DOI] [PubMed] [Google Scholar]
- 13.Palacián Ruiz M. P., Vasquez Martinez M. A., Lopez Calleja A. I.2013. Respiratory infection caused by Bordetella hinzii. Arch. Bronconeumol. 49: 409–410. doi: 10.1016/j.arbres.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 14.Spilker T., Leber A. L., Marcon M. J., Newton D. W., Darrah R., Vandamme P., Lipuma J. J.2014. A simplified sequence-based identification scheme for Bordetella reveals several putative novel species. J. Clin. Microbiol. 52: 674–677. doi: 10.1128/JCM.02572-13 [DOI] [PMC free article] [PubMed] [Google Scholar]